Abstract
Recently, we collected many large-scale datasets for alcohol dependence and EtOH response in five organisms and deposited them in our EtOH-related gene resource database (ERGR, http://bioinfo.mc.vanderbilt.edu/ERGR/). Based on multidimensional evidence among these datasets, we prioritized 57 EtOH-related candidate genes. To explore their biological roles, and the molecular mechanisms of EtOH response and alcohol dependence, we examined the features of these genes by the Gene Ontology (GO) term-enrichment test and network/pathway analysis. Our analysis revealed that these candidate genes were highly enriched in alcohol dependence/alcoholism and highly expressed in brain or liver tissues. All the significantly enriched GO terms were related to neurotransmitter systems or EtOH metabolic processes. Using the Ingenuity Pathway Analysis system, we found that these genes were involved in networks of neurological disease, cardiovascular disease, inflammatory response, and small molecular metabolism. Many key genes in signaling pathways were in the central position of these networks. Furthermore, our protein–protein interaction (PPI) network analysis suggested some novel candidate genes which also had evidence in the ERGR database. This study demonstrated that our candidate gene selection is effective and our network/pathway analysis is useful for uncovering the molecular mechanisms of EtOH response and alcohol dependence. This approach can be applied to study the features of candidate genes of other complex traits/phenotypes.
Introduction
Alcohol dependence (alcoholism) is a chronic and complex disorder with environmental and genetic factors. Twin and adoption studies suggest that genetic factors play more than 50% of the roles in the development of alcohol dependence [1]. Alcohol dependence is likely to be a polygenic, non-Mendelian disorder, and each gene has a small effect. These genes affect a range of genetic endophenotypes that subsequently affect the risk for heavier drinking and alcohol-related life problems [2]. Over the last decade, alcohol dependence in humans and EtOH response in animal models have been extensively studied. Many experimental strategies, including linkage scan, association study, quantitative trait loci (QTL), and microarray expression, have been applied in the studies of alcohol dependence and EtOH response. These studies have identified many EtOH-related chromosome regions and candidate genes in both humans and model organisms [3][4]. Most of these candidate genes are related to neurotransmitter systems, including the dopamine, the GABAergic, the glutamatergic, the opioid, the cholinergic, and the serotonergic systems. Further, they are also related to alcohol-metabolizing enzymes including the alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) system [1][5].
We have seen a rapid generation of large-scale EtOH-related data during the past decade. Recently, we collected these EtOH-related data from five species (human, mouse, rat, fly, and worm) in multiple experimental platforms (e.g., linkage, association, microarray, QTL, and literature search). Then, we annotated these collected EtOH-related candidate genes and constructed the ‘Ethanol-Related Gene Resource’ (ERGR) database [6]. In the ERGR database, we implemented the candidate-gene selection tool, which allows the user to select the EtOH-related candidate genes based on the evidences in multiple datasets and/or multiple species. In this follow-up study, we used the gene selection tool to select the EtOH-related candidate genes. We hypothesized that these candidate genes might play important roles in alcohol dependence and EtOH response at the systems biology level. Therefore, to uncover the molecular mechanisms of alcohol dependence and EtOH response, we performed gene-function enrichment tests and pathway/network analysis for those candidate genes.
Results and Discussion
Candidate-Gene List
In the gene selection page of the current ERGR database [7], we found that there were 42 genes that appeared in at least 5 datasets in all the 5 species, 17 genes appeared in at least 4 human datasets, 10 genes in at least 4 mouse datasets, and 2 genes in at least 3 rat datasets. After mapping all the nonhuman genes to human orthologs and merging them with human genes, we obtained 57 non-redundant human EtOH-related candidate genes. These candidate genes included not only some well-studied EtOH-related genes (e.g., ADH, ALDH, GABA receptor genes, and NPY), but also some less frequently studied genes (e.g., CPE, GFAP, CRYAB, GAD1, and NTRK2). According to the gene–disease association test using the ‘Database for Annotation, Visualization and Integrated Discovery’ (DAVID) bioinformatics web server [8], we found that our candidate genes were highly enriched in alcohol dependence and several brain disorders (Table 1). This finding suggests that our selected gene list is a promising EtOH-related candidate gene list and our candidate-gene selection method is effective.
Table 1.
Disease Association of EtOH-Related Candidate Genes
| Disease | No. of genes | p-Value | B–H Testa |
|---|---|---|---|
| Alcohol dependence | 12 | 8.1 × 10−14 | 5.5 × 10−10 |
| Alcohol abuse | 12 | 7.3 × 10−12 | 2.5 × 10−8 |
| Alcoholism | 10 | 2.3 × 10−8 | 5.2 × 10−5 |
| Schizophrenia | 19 | 3.3 × 10−7 | 5.6 × 10−4 |
| Attention deficit disorder conduct disorder oppositional defiant disorder | 9 | 8.0 × 10−7 | 1.1 × 10−3 |
| Heroin abuse | 6 | 2.0 × 10−6 | 2.3 × 10−3 |
Benjamini–Hochberg multiple testing correction on the p-values.
Gene Ontology Term Enrichment and Expression
Examination of Gene Ontology (GO) annotations in a set of genes by comparing with other genes has been proved useful and widely applied in gene function analysis. To exclude some GO terms that are too general to be considered, such as ‘molecular function’ or ‘cell’, we restricted GO term levels 4 and 5 to our candidate genes on the DAVID bioinformatics website. By using 0.01 as a cutoff for the Benjamini–Hochberg multiple testing correction p-value, we found eight significant enriched GO terms from the three ontologies biological process, molecular function, and cellular component (Table 2). Interestingly, six of these GO terms are related to neurotransmitter systems and two of them are related to alcohol metabolism. They all matched the pathophysiological background of alcoholism [5]. The enriched GO annotations provide positive evidence that our candidate genes are highly related to alcohol.
Table 2.
Enriched Gene Ontology (GO) Terms in EtOH-Related Candidate Genes
| GO Term | Annotationa | No. of genes | p-Value | B–H Testb |
|---|---|---|---|---|
| GO: 0007268 | B: Synaptic transmission | 15 | 2.7 × 10−13 | 7.0 × 10−10 |
| GO: 0019226 | B: Transmission of nerve impulse | 16 | 1.0 × 10−12 | 1.9 × 10−9 |
| GO: 0050877 | B: Neurological system process | 18 | 2.4 × 10−7 | 3.2 × 10−4 |
| GO: 0001505 | B: Regulation of neurotransmitter levels | 7 | 1.9 × 10−7 | 2.4 × 10−4 |
| GO: 0006067 | B: EtOH Metabolic process | 4 | 9.4 × 10−7 | 2.4 × 10−4 |
| GO: 0016917 | M: GABA Receptor activity | 5 | 2.6 × 10−6 | 1.3 × 10−3 |
| GO: 0004022 | M: Alcohol dehydrogenase activity | 4 | 2.6 × 10−6 | 2.5 × 10−3 |
| GO: 0045211 | C: Postsynaptic membrane | 7 | 2.9 × 10−6 | 1.7 × 10−3 |
GO-Organizing principles: B, biological process; M, molecular function; C, cellular component.
Benjamini–Hochberg multiple testing correction on the p-values.
To examine the tissue expression profile of our candidate genes, we used BioGPS [9] to check the expressed tissue distribution. Among the 57 candidate genes, 54 of them were found in BioGPS. We observed that approximately two-thirds of these genes were highly expressed in brain or liver tissues. Moreover, among these brain or liver highly expressed genes, approximately half of them were specifically expressed in brain or liver tissues; thus, it further revealed that these candidate genes that are highly expressed in brain or liver tissues tended to have low expression or no detectable expression in non-brain or non-liver tissues. This finding is important for understanding the gene expression profile of EtOH response and alcohol dependence.
Pathways Over-Represented in EtOH Candidate Genes
We used Fisher’s exact test to identify enriched pathways of our candidate genes on all the available KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways [10]. We found ten pathways whose p-values were less than 0.01 after multiple test correction (Table 3). Out of the ten pathways, only the pathway ‘Neuroactive ligand–receptor interaction’ is neuron-related. All the other enriched pathways are related to molecular metabolism. This observation is largely attributed to the five alcohol dehydrogenase (ADH) genes (ADH1A, ADH1B, ADH4, ADH6, and ADH7) in our candidate gene list. Furthermore, we performed a pathway analysis in the Ingenuity Pathway Analysis (IPA) system [11]. We identified the GABA receptor signaling pathway and several other molecular metabolism pathways that have been significantly enriched for our candidate genes. We conducted a similar pathway analysis in the BioCarta system [12], but no significant result was found.
Table 3.
Enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathways in EtOH-Related Candidate Genes
| KEGG ID | Pathway | p-Value |
|---|---|---|
| Hsa00624 | 1- and 2-Methylnaphthalene degradation | 3.6 × 10−7 |
| Hsa00641 | 3-Chloroacrylic acid degradation | 3.6 × 10−7 |
| Hsa00350 | Tyrosine metabolism | 1.4 × 10−5 |
| Hsa00120 | Bile acid biosynthesis | 3.6 × 10−5 |
| Hsa00830 | Retinol metabolism | 1.0 × 10−4 |
| Hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 1.6 × 10−4 |
| Hsa00071 | Fatty acid metabolism | 1.9 × 10−4 |
| Hsa00010 | Glycolysis/gluconeogenesis | 8.2 × 10−4 |
| Hsa00982 | Drug metabolism – cytochrome P450 | 1.5 × 10−3 |
| Hsa04080 | Neuroactive ligand–receptor interaction | 9.9 × 10−3 |
Functional Networks for EtOH Candidate Genes
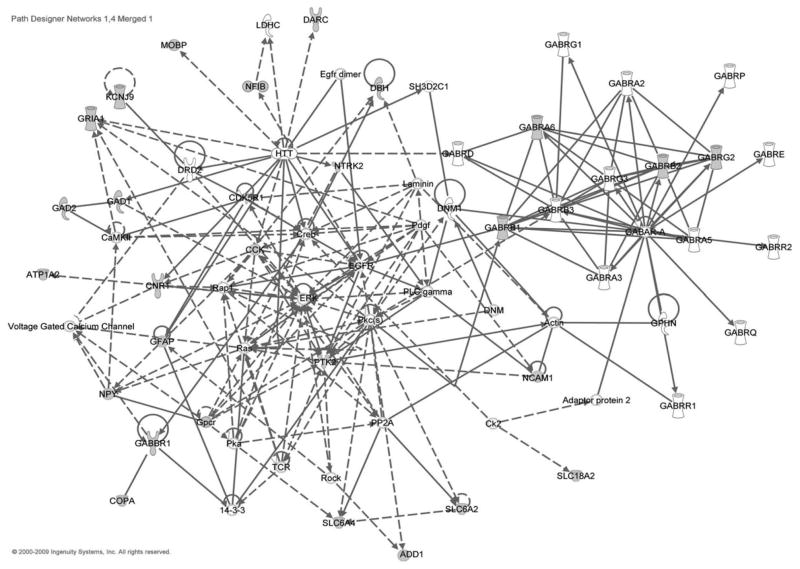
We used the IPA system to examine the potential functional networks of the 57 EtOH-related candidate genes. As a result, we found these genes fell into four significant functional networks (Table 4). Networks 1 and 4 were both related with neurological disease and psychological disorders. We merged these two networks to obtain an integrated network in neurological disease based on our EtOH candidate genes (Fig. 1). We found that most of the EtOH-related candidate genes were linked by genes in signaling pathways. This suggests that many signaling pathways are involved in the processes of alcohol dependence and EtOH response. We observed that proteins encoded by genes PTK2, PKC, ERK, CCK, CREB, RAS, RAP1 (RAP1A and RAP1B), and EGFR had links to ten or more other proteins or molecules and, thus, could serve as the center (hub) of this network. Four of them (PTK2, PKC, ERK, and EGFR) are protein kinases that are involved in important signaling pathways. Among these genes, PTK2 and CCK are EtOH-related candidate genes in our list of 57 genes. Besides, all the central genes have evidence in the ERGR database, except RAP1. Four central genes (PKC, ERK, CREB, and RAS) are in the extracellular signal-regulated kinase (ERK) signaling pathway, which is critical to neuroplasticity and important for us, to understand molecular mechanisms underlying drug addiction [13]. It has been reported that alcohol intake activates PKA signaling, which results in increased phosphorylation of CREB and NPY expression [14][15]. A microarray study also reveals signaling-system abnormalities in postmortem brains of alcohol-dependence subjects [16]. Our results suggest that alcohol triggers many downstream signaling pathways. Furthermore, our results also support the previous findings that ERK and CREB pathways play key roles in alcohol response and alcohol dependence.
Table 4.
Networks of EtOH-Related Candidate Genes in the IPA System
| ID | Molecules in the networka | Score | No. of focus genesb | Function |
|---|---|---|---|---|
| 1 | 14-3-3, ADD1, ATP1A2, CaMKII, CCK, CDK5R1, CNR1, COPA, Creb, DBH, ERK, ERK1/2, GABBR1, GAD1, GAD2, GFAP, Gpcr, GRIA1, Laminin, NCAM1, NPY, NTRK2, Pdgf, Pka, Pkc(s), PLC gamma, PP2A, PTK2, Rap1, Ras, Rock, SLC6A2, SLC6A4, TCR, voltage gated calcium channel | 38 | 18 | Genetic disorder, neurological disease, psychological disorders |
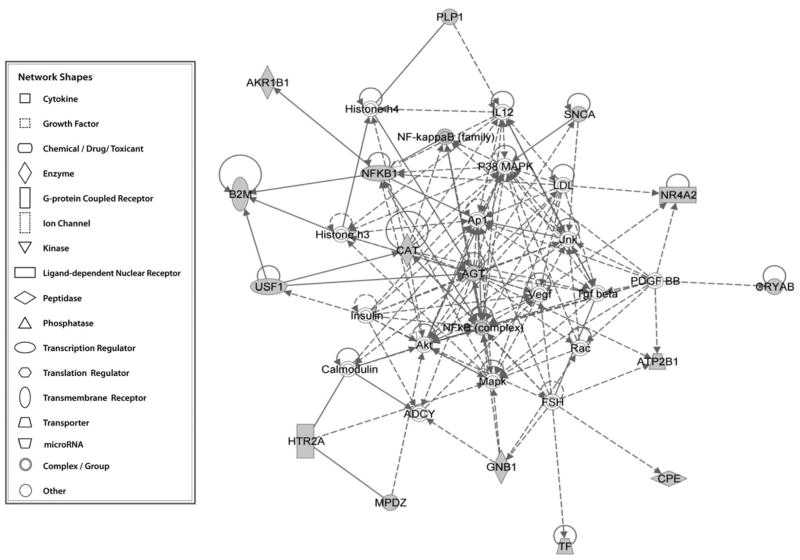
| 2 | ADCY, AGT, AKR1B1, Akt, Ap1, ATP2B1, B2M, Calmodulin, CAT, CPE, CRYAB, FSH, GNB1, Histone h3, Histone h4, HTR2A, IL12, Insulin, Jnk, LDL, Mapk, MPDZ, NF-kappaB (family), NFKB1, NFkB(complex), NR4A2, P38 MAPK, PDGF BB, PLP1, Rac, SNCA, TF, Tgf beta, USF1, Vegf | 32 | 16 | Cardiac inflammation, cardiovascular disease, inflammatory response |
| 3 | ADH4, ADH6, ADH7, ADH1A, ADH1B, ADH1C, ADHFE1, alcohol dehydrogenase, ALDH1A1, APOD, beta-estradiol, C11ORF10, C9ORF 5, CCDC82, CDC123, CHCHD8, cholesterol, CPA2, EPHX1, ERLIN1, GINS3, HNF 4A, IFT122, IGSF8, ISOC1, MRPS23, NUDT11, PMP22, PSMB6, RCADH5, SOX9, TRAF 6, UFC1, VHL, WDR42A | 29 | 14 | Small molecule biochemistry, lipid metabolism, vitamin and mineral metabolism |
| 4 | Actin, adaptor protein 2, Ck2, DARC, DNM, DNM1, DRD2, EGFR, Egfr dimer, GABAR-A, GABRA2, GABRA3, GABRA5, GABRA6, GABRB1, GABRB2, GABRB3, GABRD, GABRE, GABRG1, GABRG2, GABRG3, GABRP, GABRQ, GABRR1, GABRR2, GPHN, HTT, KCNJ9, LDHC, MOBP, NFIB, SH3D2C1, SLC18A2 | 16 | 9 | Neurological disease, developmental disorder, psychological disorders |
The underlined gene symbols are those in our list of 57 EtOH-related candidate genes. Names in upper case denote gene symbols, and those in lower case denote gene’s full names, molecular complexes, or chemical molecules.
Number of EtOH-related candidate genes in the network, corresponds to the number of underlined genes listed in the ‘Molecules in the network’ column.
Fig. 1. Neurological disease network of EtOH-related candidate genes in the IPA system.
Shapes represent different molecule types (details are illustrated in Fig. 2). Filled shapes represent EtOH-related candidate genes. Solid lines denote protein–protein interactions and dashed lines denote regulation relationships.
It is worthy to note that there are 16 genes in our EtOH candidate gene list involved in cardiac inflammation, cardiovascular disease, and inflammatory response (Table 4 and Fig. 2). We observed that genes AGT, AKT, CAT, and MAPK were in the center of the network. Alcohol has both protective and harmful effects on cardiovascular disease, depending on the volume of EtOH consumed [17][18]. Moderate alcohol consumption protects against cardiovascular disease and promotes anti-inflammatory processes, while heavy alcohol consumption is a risk factor for cardiovascular disease and hypertension [18]. Furthermore, several alcohol metabolites including acetaldehyde and fatty acid ethyl esters are specific toxins of myocardial tissue and might cause alcoholic heart disease [19]. Both acute and chronic alcohol consumptions induce various functions of the immune system, including inflammatory cell activation and effects on lung, liver, and cardiovascular diseases [20]. Specifically, the AGT gene, an angiotensinogen, locates at the center of the network (Fig. 2). Several studies have reported that AGT was differentially expressed in alcohol dependence samples. Also, one association study showed that alcohol drinking might be specifically associated with the high-normal blood pressure in M allele carriers of the AGT gene T174M polymorphism [21–23]. These cardiovascular disease and inflammatory response genes may locate in the alcohol-triggered downstream pathways, and they may serve as a feedback to affect alcohol drinking.
Fig. 2. Network of EtOH-related candidate genes in cardiovascular disease and inflammatory response.
Shapes represent different molecule types, as illustrated in the left panel of the figure. Filled shapes represent EtOH-related candidate genes. Solid lines denote protein–protein interactions and dashed lines denote regulation relationships.
Protein–Protein Interaction Network
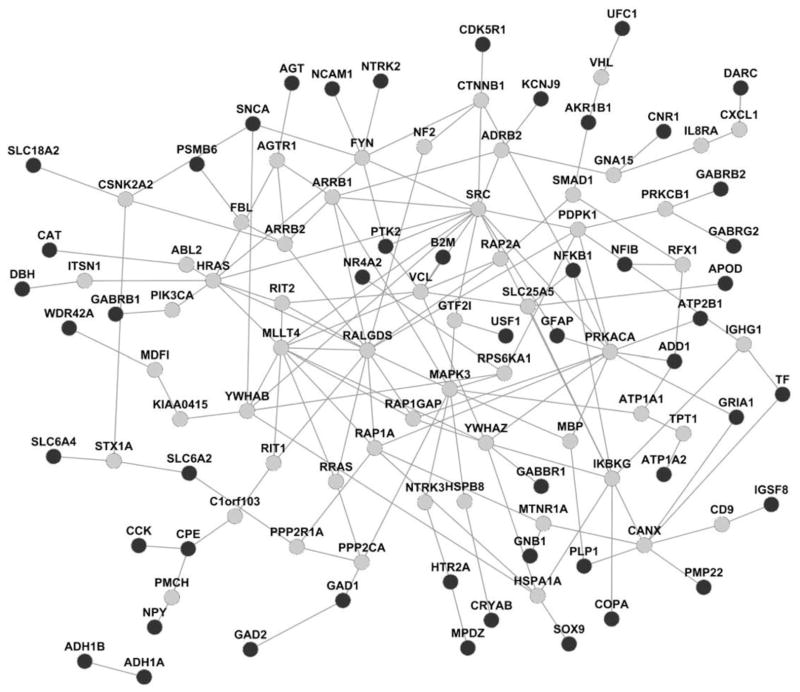
We explored the protein–protein interactions (PPIs) of the 57 candidate genes in the human interactome, and we then extracted their subnetwork using the Steiner minimal tree algorithm (Fig. 3). The extracted subnetwork had 106 nodes and 158 edges (links). Among these nodes, 50 were in our 57 EtOH-related candidate genes and 56 were new genes. Among these new genes, 25 (ca. 45%) had evidence related to alcohol in the ERGR database. In this subnetwork, we observed six nodes (FYN, VCL, VHL, CANX, PMCH, and STX1A) that were linked to more proteins encoded by EtOH-related genes and had EtOH-related evidence in the ERGR database. Specifically, genes CANX and STX1A have EtOH-related evidence in all human, mouse, and rat. Gene FYN is a tyrosine kinase of the SCR family that phosphorylates the NMDA receptors and regulates the sensitivity to EtOH [24]. Gene FYN has been reported to be associated with alcohol dependence in several studies [24–26].
Fig. 3. Protein–protein interaction network of EtOH-related candidate genes.
Black nodes are EtOHrelated candidate genes and gray nodes are other genes.
Conclusions
In this study, we obtained 57 EtOH-related candidate genes from our recently constructed ERGR database, based on the overlap of genetic datasets (i.e., evidence). We hypothesized that these candidate genes play important roles in alcohol dependence and EtOH response at systems and cellular levels. We tested this hypothesis by examining functional and network features including enrichment tests in genetic association disease databases, GO, KEGG, expression, pathways, and networks. The enrichment-test results consistently indicated that these candidate genes are over-represented in alcohol-related disease, alcohol metabolism, and neurotransmitter systems. Examination of the expression pattern indicated that most of these genes were highly expressed or specifically expressed in brain or liver tissues. In our network and pathway analysis, we found that alcohol dependence is involved in many signaling pathways. Our analysis supports the scenario of alcohol consumption being related to cardiovascular disease and inflammatory response. Overall, these results indicated that our candidate gene selection method is effective and that the biological process of alcohol dependence might have been involved in many pathways and diseases.
Experimental Part
EtOH-Related Candidate Genes
In our previous work, we collected more than 50 large-scale datasets for alcohol dependence or EtOH response in 5 species (human, mouse, rat, fly, and worm) and constructed the ERGR database [6]. We developed a candidate gene selection strategy based on dataset overlap in the database. On the gene selection page of the ERGR [7], we listed the potential EtOH-related candidate genes including genes in five or more datasets of all five species, genes in four or more human datasets, genes in four or more mouse datasets, and genes in three or more rat datasets. We first mapped genes in non-human species (e.g., mouse) to human orthologs and then merged all these human genes. This resulted in a total of 57 non-redundant potential EtOH-related candidate genes in human, based on the evidence in multiple species and multiple datasets.
Disease Association and GO Term-Enrichment Test
The Genetic Association Database (GAD) [27] is an archive of human genetic association studies of complex diseases and disorders. To evaluate our candidate genes with disease association, we mapped our candidate genes to the GAD database by the DAVID bioinformatics tools [8], which provides a comprehensive set of functional annotation tools for a set of genes of interest [28]. We also examined the Gene Ontology (GO) terms for our candidate genes based on the whole human genes as a background using the ‘Functional Annotation Chart’ tool on the DAVID bioinformatics web server. We selected those GO terms enriched in our candidate gene list by requiring Benjamini–Hochberg multiple testing corrected p-values less than 0.01. Because the top 3 levels of GO terms are too general, we restricted our GO term enrichment search at levels 4 and 5, which is an option suggested on the DAVID web server.
Gene Expression
We checked the expression tissue distribution of these candidate genes on BioGPS [9], which is initially from Gene Atlas [29]. It includes gene expression information in 79 human tissues by a microarray experiment. Among the tissues in the BioGPS, there were 17 brain-related tissues: amygdala, caudatenucleus, cerebellum, cerebellum peduncles, cingulate cortex, fetal brain, globus pallidus, hypothalamus, medulla oblongata, occipital lobe, parietal lobe, pons, prefrontal cortex, subthalamic nucleus, temporal lobe, thalamus, and whole brain.
Pathway Analysis
To check the pathway enrichment of our EtOH-related candidate genes, we performed Fisher’s exact test of our candidate genes in all pathways using pathway information in both KEGG [10] and BioCarta [12]. In detail, we counted the number of candidate genes in a specific pathway and the number of candidate genes not in the pathway by comparing our candidate gene list with the gene list of the pathway. We also counted the number of genes that were in the pathway, but were not in our candidate gene list and the number of genes that were neither in our candidate genes list nor in the specific pathway. These numbers were added in a 2 × 2 contingency table. The statistical significance of the contingency table was calculated by Fisher’s exact test. To make the analysis biologically meaningful, only those pathways with five or more candidate genes were included in our statistical test. Furthermore, we checked the enriched pathways with the DAVID bioinformatics web server [8] and the Ingenuity Pathway Analysis (IPA) tool [11].
Gene Network and Subnetwork Analysis
We used the network modules in the IPA system to identify the network and subnetworks of our candidate gene list. These genes were clustered into several (sub)networks, based on their protein–protein interaction, regulation, and other relationships. To obtain a comprehensive protein–protein interaction network of these candidate genes, we used our previous integrated human protein–protein interaction data from six databases [30]. We mapped these candidate genes into the human interactome and extracted the subnetwork using the Steiner minimal tree algorithm, which was designed to find the smallest tree connecting all the nodes of interest from a whole network [31].
Acknowledgments
We thank the financial support of this project by the National Institute of Health (AA017437, AA017828 and LM009598), the Thomas F. and Kate Miller Jeffress Memorial Trust Fund (J-900), and the NARSAD Young Investigator Award to Z. Z.
References
- 1.Quickfall J, el-Guebaly N. Can J Psychiatry. 2006;51:461. doi: 10.1177/070674370605100708. [DOI] [PubMed] [Google Scholar]
- 2.Mayfield RD, Harris RA, Schuckit MA. Br J Pharmacol. 2008;154:275. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick DM, Foroud T. Alcohol: Clin Exp Res. 2003;27:868. doi: 10.1097/01.ALC.0000065436.24221.63. [DOI] [PubMed] [Google Scholar]
- 4.Schumann G, Spanagel R, Mann K. Alcohol: Clin Exp Res. 2003;27:880. doi: 10.1097/01.ALC.0000065437.18136.86. [DOI] [PubMed] [Google Scholar]
- 5.Kçhnke MD. Biochem Pharmacol. 2008;75:160. doi: 10.1016/j.bcp.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Guo A-Y, Webb BT, Miles MF, Zimmerman MP, Kendler KS, Zhao Z. ‘Ethanol-Related Gene Resource (ERGR)’ homepage. Nucleic Acids Res. 2009;37:D840. doi: 10.1093/nar/gkn816. http://bioinfo.mc.vanderbilt.edu/ERGR/ [DOI] [PMC free article] [PubMed]
- 7.‘ERGR Gene Selection’ homepage. http://bioinfo.mc.vanderbilt.edu/ERGR/topgenes.php.
- 8.‘Database for Annotation, Visualization and Integrated Discovery (DAVID)’ homepage. http://david.abcc.ncifcrf.gov/ [PubMed]
- 9.‘BioGPS’ homepage. http://biogps.gnf.org/
- 10.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. Nucleic Acids Res. 2006;34:D354. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.‘Ingenuity Pathway Analysis’ homepage. http://www.ingenuity.com/
- 12.‘BioCarta’ homepage. http://www.biocarta.com/
- 13.Zhai H, Li Y, Wang X, Lu L. Cell Mol Neurobiol. 2008;28:157. doi: 10.1007/s10571-007-9240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wand G. J Clin Invest. 2005;115:2697. doi: 10.1172/JCI26436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandey SC, Zhang H, Roy A, Xu T. J Clin Invest. 2005;115:2762. doi: 10.1172/JCI24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokolov BP, Jiang L, Trivedi NS, Aston C. J Neurosci Res. 2003;72:756. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- 17.Bau PF, Bau CH, Rosito GA, Manfroi WC, Fuchs FD. Alcohol. 2007;41:479. doi: 10.1016/j.alcohol.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Collins MA, Neafsey EJ, Mukamal KJ, Gray MO, Parks DA, Das DK, Korthuis RJ. Alcohol: Clin Exp Res. 2009;33:206. doi: 10.1111/j.1530-0277.2008.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J, Wold LE. Ther Adv Cardiovasc Dis. 2008;2:497. doi: 10.1177/1753944708095137. [DOI] [PubMed] [Google Scholar]
- 20.Szabo G, Mandrekar P. Alcohol: Clin Exp Res. 2009;33:220. doi: 10.1111/j.1530-0277.2008.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Alcohol: Clin Exp Res. 2000;24:1873. [PubMed] [Google Scholar]
- 22.Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Neuropsychopharmacology. 2006;31:1574. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- 23.Takashima Y, Kokaze A, Matsunaga N, Yoshida M, Sekiguchi K, Sekine Y, Sumiya Y. J Physiol Anthropol Appl Hum Sci. 2003;22:187. doi: 10.2114/jpa.22.187. [DOI] [PubMed] [Google Scholar]
- 24.Pastor IJ, Laso FJ, Inés S, Marcos M, González-Sarmiento R. Eur Psychiatry. 2009;24:191. doi: 10.1016/j.eurpsy.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Schumann G, Rujescu D, Kissling C, Soyka M, Dahmen N, Preuss UW, Wieman S, Depner M, Wellek S, Lascorz J, Bondy B, Giegling I, Anghelescu I, Cowen MS, Poustka A, Spanagel R, Mann K, Henn FA, Szegedi A. Biol Psychiatry. 2003;54:1422. doi: 10.1016/s0006-3223(03)00635-8. [DOI] [PubMed] [Google Scholar]
- 26.Ishiguro H, Saito T, Shibuya H, Toru M, Arinami T. Am J Med Genet, B. 2000;96:716. [PubMed] [Google Scholar]
- 27.‘Genetic Association Database (GAD)’ homepage. http://geneticassociationdb.nih.gov/
- 28.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 29.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. Proc Natl Acad Sci USA. 2004;101:6062. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo AY, Sun J, Riley BP, Thiselton DL, Kendler KS, Zhao Z. Mol Psychiatry. 2009;14:18. doi: 10.1038/mp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein P, Ravi R. J Algorithms. 1995;19:104. [Google Scholar]