Abstract
The DNA triple helix consists of a third strand of nucleic acid lying in the major groove of an intact DNA duplex. The most stable triplexes form on polypurine:polypyrimidine sequences, and pyrimidine interruptions in the purine strand are destabilizing. Sequence stringency is imparted by specific Hoogsteen hydrogen bonds between third strand bases and the purine bases in the duplex. Appropriate base and sugar modifications of triple helix-forming oligonucleotides (TFOs) confer chromosome targeting activity in living cells. However, broad utilization of TFOs as gene targeting reagents in mammalian cells has been limited by the requirement for homopurine target sequences. Although there have been a number of base analogues described that appear to be promising as candidates for triplex target expansion, none has been examined in a biological system. We have employed a postsynthetic strategy to prepare a collection of TFOs with base analogues at a defined position. Following assessment of affinity for a triplex target with a single C:G inversion, TFOs with a second generation of analogues were synthesized. One of these, TFO-5a, with 2′-OMeguanidinylethyl-5-methylcytosine at the position corresponding to the C:G interruption in the target sequence, was further modified to confer bioactivity. The activity of this TFO, linked to psoralen, was measured in a mammalian cell line that was engineered by directed sequence conversion to carry a triplex target with a single C:G interruption. TFO-5a was active against this target and inactive against the corresponding target with an uninterrupted polypurine:polypyrimidine sequence.
Triple helix-forming oligonucleotides (TFOs)1 have been the subject of research for many years because of their potential as gene targeting reagents for application in living cells (1–4). Triplex formation can occur when a third strand of nucleic acid, either DNA or RNA, lies in the major groove of an intact duplex. The most stable triplexes form on polypurine:polypyrimidine elements with two hydrogen bonds established between the third strand bases and the duplex purines. Third strands may be composed of pyrimidines or purines depending on the nature of the target. Thus, when the duplex sequence is weighted toward A:T pairs, with relatively few G:C pairs, a pyrimidine motif third strand is indicated. Alternatively, with G:C rich duplexes, a purine motif third strand is preferred. Pyrimidine motif third strands form A:T·T and G:C·C+ triplets, while A:T·A and G:C·G triplets are formed by purine motif third strands.
Although triplexes can be readily generated in vitro under the appropriate buffer conditions, formation in vivo is much more challenging. While charge repulsion between the third strand phosphates and those of the duplex can be suppressed by divalent cations such as Mg2+ in vitro, unbound Mg2+ is much less available in cells (5). Conformational restrictions on both the third strand and duplex in a triplex contribute to the much slower rate of triplex formation as compared to the rate of annealing of single strands. A physiological pH is a problem for pyrimidine motif third strands, as the second hydrogen bond formed by cystosine requires protonation with a pKa of 4.5.
These restrictions have been overcome by the incorporation of base and sugar modifications into the third strands. 5-Methylcytosine (5MeC) was found many years ago to ameliorate the pH restriction on pyrimidine motif TFOs (6). The demonstration that RNA third strands formed more stable triplexes than their deoxy counterparts (7) prompted an examination of the influence of sugar analogues on these structures. 2′-O-Methylribose (2′-OMe) in TFOs was shown to further stabilize triplexes (8). Structural studies showed that TFOs with uniform 2′-OMe substitution were preorganized into a conformation very similar to that assumed in a triplex, while requiring minimal conformational adjustments on the part of the duplex (9). Another RNA analogue sugar, 2′-O-aminoethylribose (2′-AE), which is protonated at physiological pH, was shown to greatly enhance the rate of triplex formation and stabilize triplexes once they formed (10–12).
The biochemical and biophysical properties of TFOs with these modifications encouraged examination of their biological activity. We showed that psoralen-linked TFOs with 2′-OMe ribose substitutions and a cluster of 2′-AE residues could introduce mutations at the target site in living cells (13–17). TFO efficacy was influenced by the biology of the cell, with maximal activity during the S phase, probably reflecting the time of greatest access to the targets in a chromatin context (18). The chromosomal targets addressed in these studies were polypurine:polypyrimidine sequences. These results indicated that the combination of 2′-OMe ribose and clustered 2′-AE substitutions conferred bioactivity on pyrimidine motif TFOs against canonical polypurine:polypyrimidine targets.
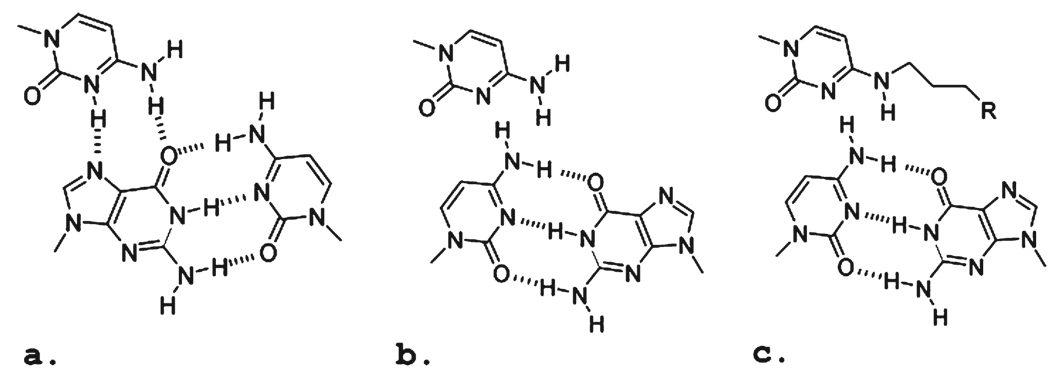
Although the combination of these sugar modifications proved to be important for the biological activity of TFOs against conventional targets, they did not address a major impediment to the broader application of TFOs: the inimical influence of pyrimidines in the homopurine strand of the target sequence. The change of a purine:pyrimidine pair to a pyrimidine:purine pair in the duplex places the purine outside of the hydrogen bonding range of the third strand base (Figure 1). The stability of triplexes formed on these sequences by oligonucleotides with standard nucleoside constituents is generally poor, and inadequate for bioactivity. This problem is well-known, and there have been many attempts to design base analogues that would overcome this limitation (reviewed in refs (19–21)). For structural reasons, the C:G (Figure 1a,b) inversion is more approachable than the T:A inversion, and there has been greater progress with this target.
FIGURE 1.
Schematic of base triplets formed in either an uninterrupted polypurine:polypyrimidine sequence or one with an “inverted” interruption in the sequence: (a) canonical G:C·C+, (b) inverted C:G·C, and (c) inverted C:G·Cmod. The R group is designed to afford additional hydrogen bonding between the base analogue and the purine base of the inverted base in the duplex target sequence.
Several strategies have been employed in this effort. Some have taken advantage of the opportunity to form a single hydrogen bond with the 4-amino of the cytosine of the inverted C:G base pair. TFOs containing the 5-methyl-2-pyrimidone (22) and methylated 3H-pyrrolo[2,3-d]pyrimidine-2(7H)-one (23, 24) analogues form more stable triplexes on duplexes containing a C:G inversion than those with natural bases at the site. Binding has been further enhanced by linkage of the analogue to a bridged 2′-O,4′-C-methylene ribose (25) or a 2′-aminoethoxyribose (26). These modifications are well-known to enhance the stability of triplexes formed by pyrimidine motif third strands (11, 27, 28).
Another approach reflects the attempt to form hydrogen bonds between base analogues in the third strand and the guanine in the inverted base pair. Attention has been given to cytosine derivatives in which extended side chains present hydrogen bond donors to the O6 and N7 acceptors of guanine (Figure 1c). For example, the TFOs with an N4-(3-aminopropyl)cytosine or the N-acetylated derivative were able to form stable triplexes on targets with C:G inversions (29). In an effort to restrict the flexibility of the aminopropyl side chain, an N4-(6-amino-2-pyridinyl)-cytosine derivative was prepared (30). Although a TFO carrying this analogue formed stable triplexes on a C:G inversion target, it also exhibited strong affinity for a target with an A:T base pair at the same site. Consequently, the gain in affinity afforded by the pyridinyl group was offset by the loss of specificity.
The C:G inversion has been addressed with platforms other than cytosine, such as imidazole. Although these analogues showed promise in model studies, they have not been examined in oligonucleotides (31). Encouraging results with heterocyclic based C-glycosides capable of binding C:G inversions were reported by Gold and colleagues (32). A novel candidate based on a bicyclo sugar aminopyrazole appears to have promise for the C:G inversion in antiparallel triplexes (33), although there are nearest neighbor sequence effects.
The activity of analogues with side chains containing aromatic rings has also been examined. Typically, these overcome the triplex destabilization of C:G inversions via intercalation rather than specific hydrogen bond formation (20, 34), and this results in a loss of specificity.
While the focus of this discussion is on analogues designed to overcome the C:G inversion, it should be noted that there has been an effort to address the T:A inversion as well (24, 35, 36).
The analogues discussed in this brief overview have been characterized by biochemical and biophysical analyses, and some appear to have promise. However, despite more than a decade since the demonstration of activity of a TFO against a canonical genomic target in living cells (37), none of the analogues has been introduced into a TFO and then tested for activity in a biological assay.
In this report, we describe the synthesis of N4-alkyl-5-methylcytosine derivatives that improved binding of a pyrimidine motif TFO to a target sequence containing a single C:G inversion. TFOs with several analogues were characterized, and one, containing N4-(2-guanidoethyl)-5-methylcytosine at the inversion site, was active in a gene targeting assay in living mammalian cells.
METHODS
Synthesis, Deprotection, Purification, and Determination of the Mass of TFOs
The oligonucleotides were synthesized on a 0.2 or 1.0 µmol scale on CPG supports (500 Å), or on a deprotection resistant polystyrene support (Oligo-Affinity, Glen Research) using an Expedite 8909 synthesizer. The coupling times for the phosphoramidite monomers of protected 2′-O-methylthymidine and 2′-O-methyl-5-methylcytidine were 360 s. The coupling times for the phosphoramidite monomers of protected 2′-amino-ethylthymidine, 2′-aminoethyl-5-methylcytidine, 2′-O,4′-C-BNA/LNA or ENA monomer, and psoralen were 900 s. The psoralen-containing TFOs were synthesized such that the psoralen coupling step followed the modification of the triazolyl base. The support was treated with an acetonitrile solution of the desired amine (10%) containing DIEA (10%) for 3 h at room temperature. Then the support was washed five times with acetonitrile, after which the psoralen was coupled. The resulting psoralen TFO was deprotected with a 1:1 solution of 28% aqueous NH3 and 40% aqueous methylamine (Aldrich Chemicals) for 90 min at room temperature, immediately evaporated to dryness, and resuspended in HPLC-grade H2O. Support-bound TFOs used in the binding assay were deprotected with 28% aqueous ammonia for 4 h at 55 °C, and then the support was washed five times with water. Purification of psoralen-containing TFOs was conducted by ion-exchange HPLC using a Dionex DNAPac PA-100 column (column sizes of 4.0 mm × 250 mm for analysis and 9.0 mm × 250 mm for purification) on a Shimadzu HPLC system (LC-10ADvp) with a dual wavelength detector (SPD-10AVvp) and an autoinjector (SIL-10ADvp). HPLC conditions were as follows: linear gradient from 0 to 100% buffer B over 60 min, at a rate of 1.0 mL/min [buffer A consisted of 100 mM Tris (pH 7.5) containing 10% CH3CN; buffer B consisted of 1 M NaCl containing 100 mM Tris (pH 7.5) and 10% CH3CN]; UV monitor, 254 and 315 nm (λmax for psoralen). The oligonucleotides were collected, lyophilized, and desalted using NAP 5 columns (GE Healthcare).
The masses of the chemically modified oligonucleotides were determined in the positive ion mode using MALDI-TOF mass spectrometry on a Voyager Applied Biosystems instrument. The matrix used for preparing the MALDI-TOF samples was a mixture of 3-hydroxypicolinic acid (50 mg/mL in 50% CH3CN) and ammonium citrate (50 mg/mL in HPLC-grade water). MALDI-TOF: 5′-Pso-TMeCTTMeCTTTTMeCTMeCtMecttT (lowercase bases were modified with 2′-AE) 6150.46 (calcd) and 6150.57 (found); 5′-Pso-TMeCTTMeCTTTTMeCEtGuaTMeCtMecttT 6235.58 (calcd) and 6233.46 (found).
Binding Assay
The support with covalently bound TFO (5 mg) was suspended in water (200 µL) and divided into four equal portions. The resulting fractions were centrifuged, and the water was removed. Then 3.3 µM hairpin duplex oligonucleotide (5′-TCATTTCTCTYTTTTCTTCTAGATTTTTCTAGAAGAAAAXAGAGAAATGA) in 200 µL of 10 mM HEPES containing 5 mM MgCl2 (pH 7.0) was added to each portion, where X and Y were A and T, T and A, G and C, and C and G, respectively. The suspended support was kept overnight at 60 °C with occasional shaking. The water phase was removed, and after a brief wash, the beads were suspended in 100 µL of 10 mM EDTA in 25 mM NaOH. The suspension was kept at room temperature for 4 h with occasional shaking. Aliquots of 75 µL were taken and diluted to a volume of 150 µL with water, and the UV absorbance measurements were taken.
Guanidinylation Procedure
The support-bound TFO derivatized with alkyldiamine was suspended in 200 µL of dry DMF containing 1 M DIEA and 1 M pyrazole-1-carboxamidine hydrochloride. The suspension was kept at 55 °C overnight. Then the solution was discarded and the support washed four times with DMF and three times with water.
Thermal Denaturation
The thermal melting experiments were conducted using TFOs with a 19-mer Hprt hamster duplex target (5′-GTAGAAGAAAAAAGAGAAA and 3′-CATCTTCTTTTTTCTCTTT) which had a Tm of 53.5 °C. A 1 µM stock solution of target duplex was prepared in a buffer containing 100 mM NaCl, 2 mM MgCl2, and 10 mM sodium cacodylate (pH 7.0). The solution was heated to 80 °C for 15 min, allowed to reach room temperature over 4 h, and then stored in the refrigerator at 4 °C. An aliquot of a TFO solution (1 µM) was added to 1 mL of the stock duplex at room temperature, and the mixture was incubated at room temperature overnight. Thermal melting determinations were conducted using a Cary 3E UV–vis spectrophotometer fitted with a thermostat sample holder and a temperature controller. The triplex solution was heated from 25 to 85 °C at a rate of 0.4 °C/min, and the absorbance at 260 nm was recorded as a function of temperature. The raw data were processed using Sigma plot 5.0 to determine the first-derivative curve, from which the Tm value was obtained. All analyses were performed at least twice with an error of no more than 0.5 °C.
Cells, Transfection, and Assay of Hprt Knockout
Chinese hamster ovary (CHO) AA8 cells were grown in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal calf serum (FBS), penicillin, and streptomycin. Prior to an experiment, the cells were grown in HAT medium (10−4 M hypoxanthine, 5 × 10−5 M aminopterin, and 10−5M thymidine) for 1 week to eliminate Hprt− cells. The cells were synchronized in the G0/G1 phase as described previously (38). Briefly, cells were plated at subconfluent levels, and the next day, the medium was changed to DMEM with 2% FBS and 2%DMSO. After 48 h, the cells were washed and incubated with complete medium containing 100 µM mimosine for 16 h to block them in the early S phase (~90% early S cells) (39). After 16 h, the cells were released from the mimosine block after being fed with a DMEM/10% FBS mixture and further incubated for 4 h. In previous work, we have shown that the Hprt target is most accessible in cells in the mid-S phase (18). Consequently, all bioactivity assays were conducted with mid-S phase cells.
Mid-S phase cells were suspended at a density of 107 cells/mL and mixed with TFO at 5 µM. The cells were electroporated (Amaxa), followed by incubation at room temperature for 3 h, and a 3 min exposure in a Rayonet chamber to UV-A light at 1.8 J/cm2. The cells were passaged for 8 days and then exposed to thioguanine (TG) selection (13). Colonies were fixed, stained, and counted after 10 days. Characterization of the amplification products of the target region from mutant colonies was as described previously (16, 37).
Computational Details
Initial double-stranded (ds) and triple-stranded (tx) DNA coordinates were generated using the Nucleic Acid Builder module of Discovery Studio version 2.1. Consequently, each ds- and tx-DNA was read into CHARMM (40), where the calculations were performed using a CHARMM 27 all-atom additive nucleic acid force field (41, 42). Molecular dynamics (MD) simulations were performed with a combination of CHARMM and NAMD (43).
To construct triplexes with modified bases, the nucleic acid force field was parametrized to include the 2′-OMe moiety of the ribose, the protonated analogue of 5-methylcytosine and N4-(2-guanidoethyl)-5-methylcytosine. Then 32-mer double-stranded (ds) and 32–17-mer triplex (tx) DNA sequences were generated, as summarized in Table 1. The hydrogen atoms were constructed on the basis of the internal coordinates in the CHARMM27 nucleic acid force field. All heavy atoms were harmonically restrained with a force constant of 50 kcal−1 mol Å−2; all force field energy terms were turned on excluding electrostatics, and the ds- and tx-DNAs were energy optimized with 500 steps of steepest descent (SD) (44) followed by 500 steps of conjugate gradient (CD) minimization to relax initial hydrogen atom positions. Next, a pre-equilibrated orthogonal box of explicit TIP3P water molecules was superimposed on the DNA structure. The size of the box was chosen to ensure an at least 20 Å layer of water from the backbone of the DNA and a 10 Å layer of water from both ends of the strands and the edge of the box. Water molecules within 4.1 Å of the DNA were deleted, and sodium counterions were placed by random selection of added water molecules to neutralize the net charge. Prior to the MD calculations, each solute/water/ion system was minimized and equilibrated. For the double-stranded and triplex systems, a rectangular box with approximate dimensions of 130 Å × 71 Å × 71 Å or 128 Å × 71 Å ×72 Å was generated. Periodic boundary conditions (45) were used, and Coulomb interactions were treated with the particle mesh Ewald method (46) with a real space cutoff of 10 Å, a κ value of 0.34, order 6 B-spline interpolation, and a grid spacing of approximately 1.0 Å. Lennard-Jones interactions were truncated from 8 to 10 Å with force switching, and long-range correction (47) was applied to account for the effect of Lennard-Jones interactions beyond truncation. The SHAKE algorithm (48) was applied to all hydrogen atoms with a tolerance limit of 10−8, and the nonbonded pairlist was updated whenever an atom’s relative displacement exceeded 1 Å. All DNA strands were harmonically constrained with a force constant of 50 kcal−1 mol Å−2, to allow the water/ions system to minimize for 2000 steps of steepest descent (SD). Subsequently, the constraints were released to 5 kcal−1 mol Å−2, and the whole solute/water/ion system was minimized for 1000 steps of a conjugate gradient (CG).
Table 1.
Average Strand–Strand Interaction Energies (kilocalories per mole) Calculated over a 5–30 ns MD Simulation for the Five Central Basesa
| strandsb | A:T·T | A:T·5a | G:C·T | G:C·5a | C:GT | C:G·5a |
|---|---|---|---|---|---|---|
| I–II (Watson–Crick) | −81.20 | −61.60 | −76.56 | −56.76 | −97.67 | −94.20 |
| I–III (Hoogsteen) | −78.40 | −55.88 | −65.91 | −53.69 | −56.58 | −67.87 |
| II–III | 3.83 | 4.16 | 0.97 | 4.81 | 1.25 | 3.18 |
| Hoogsteen-Watson–Crick | −74.58 | −51.72 | −64.95 | −48.89 | −55.33 | −64.69 |
| Tm (°C) | 59 | 29 | 39 | 50 | ||
| bioactivity (%) | 100 | <1 | ~10 | ~45 |
All energies are given in kilocalories per mole.
Interaction energies are interstrand base–base contributions calculated for the five central bases.
MD simulations for all the systems were performed in an isobaric–isothermal ensemble. The equations of motion were integrated with the ’leapfrog’ algorithm (49) and a 2 fs integration time step to propagate the system. The temperature was maintained at 298 K by a Nose′-Hoover heat bath (50, 51) with a thermal piston parameter of 10000 kcal mol−1 ps2. The constant pressure of 1 atm was controlled using the Langevin piston (52) with a mass of 1000 amu. The initial 100 ps of water–ion equilibration was performed by imposing 5 kcal−1 mol Å−2 harmonic constraints on the solute, followed by 500 ps of unconstrained dynamics of the entire system. Finally, 30 ns Langevin dynamics simulations were performed using NAMD version 2.6 with the CHARMM27 force field. The Langevin coupling coefficient of 1 ps−1 with a temperature bath of 298 K was applied to all atoms. The piston oscillation period of 200 fs and the barostat damping time scale of 100 fs were used to maintain a piston pressure of 1 atm.
RESULTS
In previous publications, we identified the oligonucleotide chemistry that supported robust activity against chromosomal targets in living cells (14, 15). The TFOs were designed to bind a polypurine:pyrimidine sequence in the Chinese hamster Hprt gene, adjacent to exon 5. This sequence contains the acceptor site for the splicing of exons 4 and 5 in the primary transcript and terminates in a 5′ TA step that is a preferred site for cross-linking by psoralen. Bioactive psoralen-linked TFOs introduce a psoralen at the target site that can be converted to an interstrand cross-link by photoactivation. Mutagenesis induced by the cross-link inactivates the Hprt gene, yielding cells that survive exposure to selective medium, providing a quantitative measure of TFO bioactivity (see Methods). In earlier work, we have shown that the Hprt deficient clones contain small deletions that include the targeted cross-link site and all or a portion of exon 5 (18, 37).
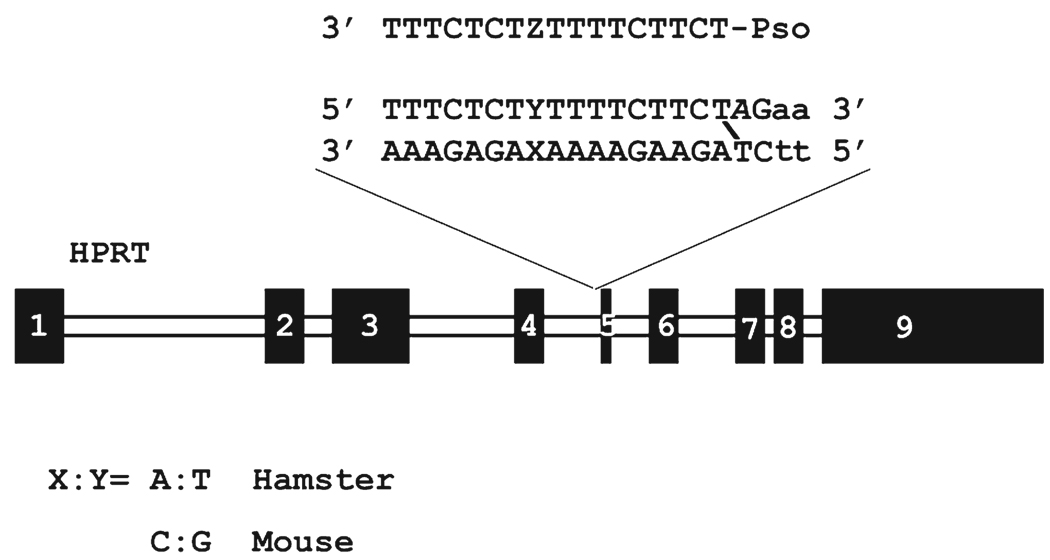
Comparison of the hamster triplex target with the corresponding element from other species revealed that the equivalent murine sequence differed by a single base pair, a C:G inversion in the midst of adjacent T residues (X:Y in Figure 2). Because the Hprt gene is functional in mouse cells, it is apparent that this sequence change does not perturb splicing. Consequently, this target could be employed during the development of the oligonucleotides against targets with a single C:G inversion, with the anticipation that they could be examined in a bioassay based on the knockout of a Hprt gene containing the murine target sequence.
FIGURE 2.
Chinese hamster Hprt gene and psoralen triplex target sequence. The position that varies between mouse and hamster genomes is shown (X:Y) as is the location of base analogues in the third strand (Z). Mutagenesis by a TFO-targeted psoralen cross-link (that links a thymine on each duplex strand) results in small deletions in the target sequence and adjacent exon 5. The frequency of mutations can be determined by quantitation of thioguanine resistant colonies.
Synthesis of Base Analogues
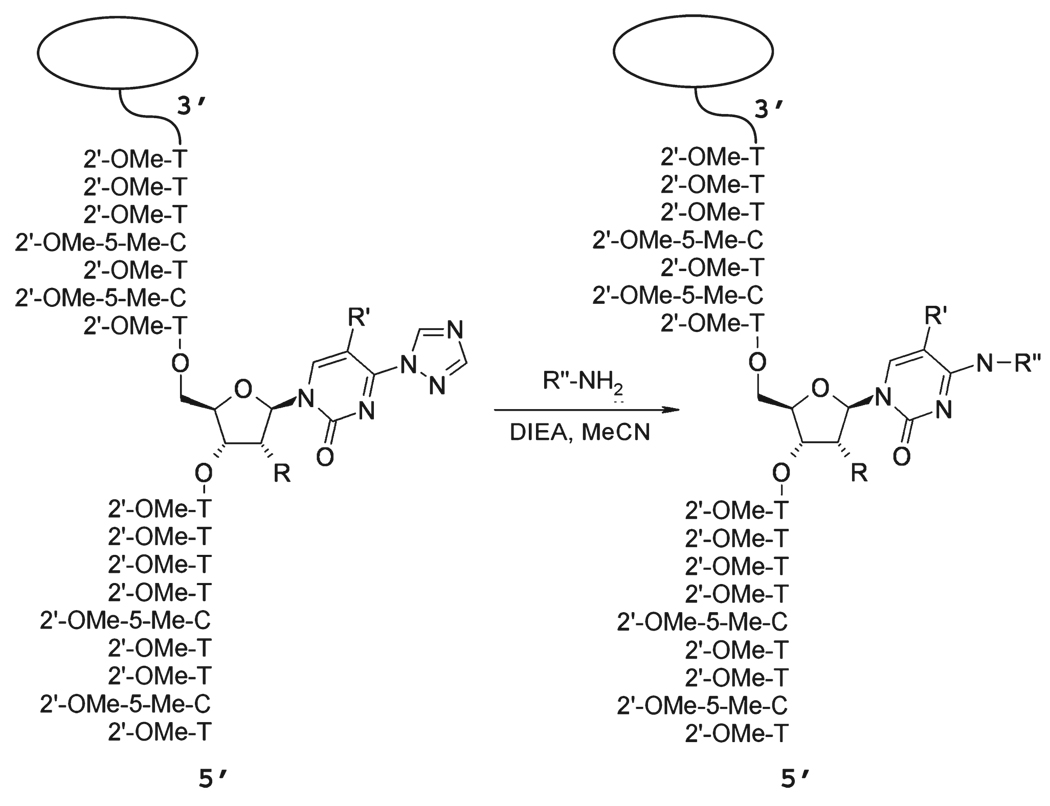
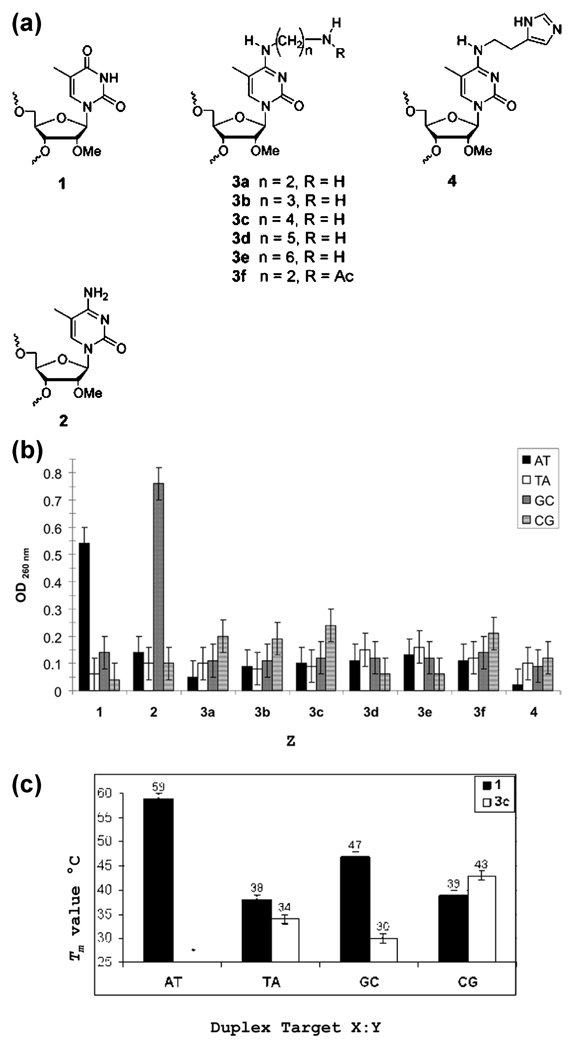
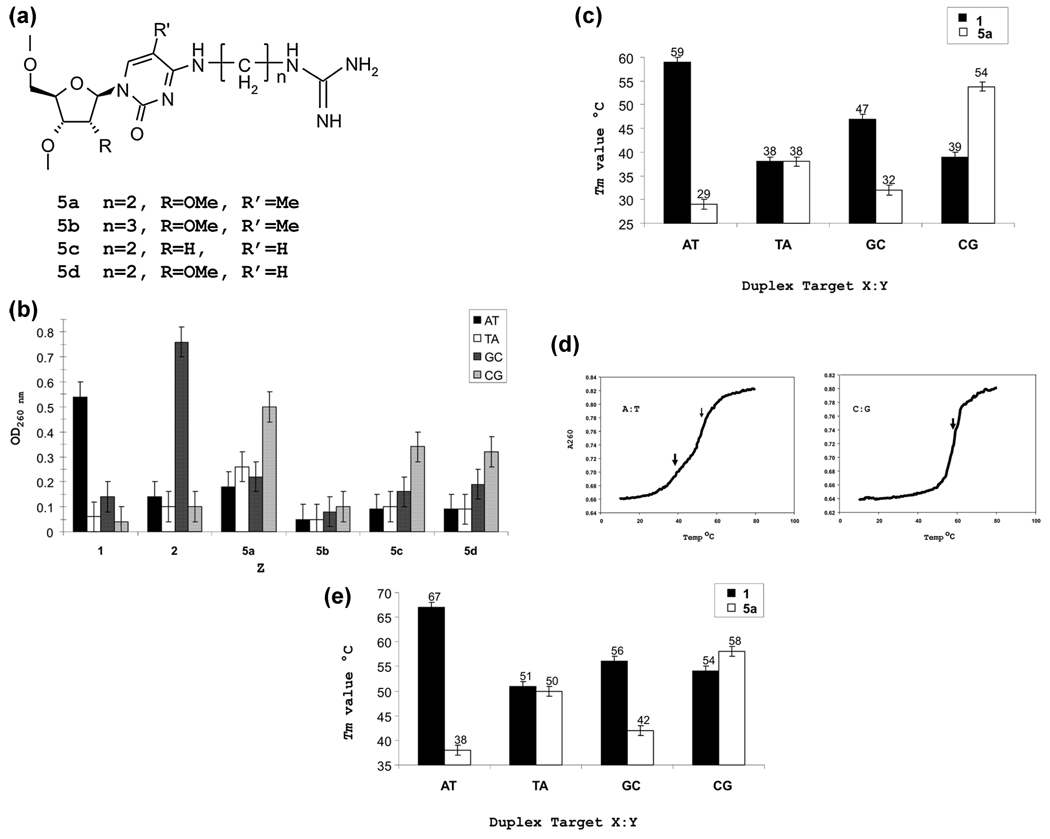
At the outset of this study, we wanted to employ an approach that would allow us to examine multiple candidate analogues without the requirement of synthesizing each as a monomer phosphoramidite prior to incorporation into an oligonucleotide. Accordingly, we chose a strategy of postsynthetic modification of an oligonucleotide containing a triazolyl thymidine (TTri) residue at the Z position in the TFO corresponding to the X:Y position in the duplex. The oligonucleotide was synthesized on a polystyrene support attached by a deprotection resistant linkage (Methods). In our previous work, we found that RNA analogue sugars made an important contribution to TFO bioactivity (14). Consequently, the TTri oligonucleotide was synthesized with 2′-OMe ribose at all positions. Following synthesis and deprotection, the beads with the oligonucleotide attached were removed from the cartridge and loaded in equal amounts into a series of reaction vessels. In the initial synthetic series, triazoyl displacement reactions were performed with alkyl diamines of increasing length (Scheme 1 and Figure 3a). The beads from an individual reaction were washed and divided into four tubes, and each tube was incubated with a hairpin duplex oligonucleotide (Methods) containing the target sequence with one of the four possible base pairs at the X:Y site. The incubation was performed overnight, followed by removal of the aqueous phase (Methods). After being washed, the beads were suspended in elution buffer to remove the bound duplex. The absorbance of the supernatant was then determined. Although this was a very crude measure of binding, it did allow for a rapid preliminary assessment of the influence of chain length on binding to the different targets. Thus, for example, the data suggested that for the C:G target the distance between the N4 atom and the terminal amine should be between three and five atoms. In contrast to the primary amine, histamine, placed within the optimal distance, showed no enhanced binding to the C:G inversion. It was also noteworthy that the C2-N-acetyl analogue was no more effective than the C2-NH2 version (Figure 3b).
Scheme 1.
Postsynthetic Solid Phase Synthesis of Base Analogues
FIGURE 3.
Analysis of base analogues in TFOs containing 2′-OMe ribose. (a) Bases and analogues in TFOs: 1, 2′-OMe thymidine; 2, 2′-OMe 5Me cytosine. (b) Binding assay of analogues shown in panel a. Resin-bound TFOs with base analogues were incubated with hairpin duplexes containing each of four possible base pairs at the X:Y position shown in Figure 2. After being washed, the remaining bound duplex was eluted from the resin (Methods) and the amount measured spectrophotometrically. (c) Thermal stability of 2′-OMe TFOs containing T (against the wild-type target sequence) or analogue 3c in the Z position.
On the basis of the results of the preliminary binding assay, we synthesized TFO-3c containing all sugar residues as 2′-OMe ribose and the N4-aminobutylcytosine analogue (3c) on a conventional, cleavable, support. The oligonucleotide was purified, and triplexes were formed on duplex targets containing the four possible base pairs at the X:Y position. Triplexes were also formed by an oligonucleotide containing a T at the Z position (TFO-T), as found in the wild-type sequence. The stabilities of the triplexes were determined by analysis of thermal resistance.
The results (Figure 3c and Figures S1 and S2 of the Supporting Information) showed a very striking difference in the stability of the triplexes formed by the TFO-T and TFO-3c on the duplex containing an A:T duplex. While the Tm of the triplex formed by TFO-T was 59 °C, as reported previously (14), we did not observe a stable triplex with TFO-3c on the A:T duplex. In contrast, there were only a few degrees of difference in the Tm value with the T:A duplex, although values for both triplexes were more than 20 °C lower than that of the perfect match TFO-T on the A:T duplex. With the G:C duplex, there was a 17 °C difference, the TFO-T forming a much more stable structure. There was a 5 °C difference between the Tm values of the two triplexes on the C:G duplex, with the TFO-3c species forming the more stable structure. These data indicated that a TFO with the 3c analogue could form a triplex on the C:G duplex that was more stable than on the other duplexes and could serve as a lead compound for additional syntheses.
Synthesis and Characterization of Alkyl Guanido Analogues
The preceding study suggested that extension of a single putative hydrogen bond donor could contribute to the stability of the triplex on the C:G duplex. However, it seemed unlikely that this would be sufficient to be effective in bioassays. In an effort to improve the stability, we decided to add another potential hydrogen bond donor. The guanidine group of arginine is often employed in sequence specific binding proteins as a dual hydrogen bond donor involved in the recognition of guanine (53, 54). Analogues with n = 2 or 3 alkyl diamine side chains (Figure 4a) were synthesized using the postsynthetic strategy on a noncleavable support described above. The TFO was then conjugated, in a second postsynthetic reaction with pyrazole-1-carboxamidine, to yield the guanido derivative. A binding assay clearly demonstrated the superiority of the n = 2 version (Figure 4b). Additional variants, in which triazoyl uracil (rather than triazoyl thymidine) was linked to a deoxy ribose or 2′-OMe-ribose, were also synthesized. The binding assay indicated that the TFO with the 2′-OMe-guanidinylethyl-5-methylcytosine (5a) was the most effective against the C:G duplex target.
FIGURE 4.
Analysis of second generation analogues. (a) Analogue structure. (b) Binding assay as in Figure 3b. (c) Thermal stability of triplexes with 2′-OMe third strands. (d) Thermal stability profile of AE-TFO-5a on the duplex target with A:T or C:G at the X:Y position. The thick arrow denotes the inflection for the triplex, while the thin arrow indicates that for the duplex. There is a single transition in the pattern with the C:G duplex as the triplex is more stable than the underlying duplex. (e) Thermal stability of triplexes with the 2′-AE cluster.
As in the previous experiments, the TFO containing this analogue was synthesized on a conventional support and purified. The stability of triplexes formed by this TFO (TFO-5a) on the duplex targets was determined by thermal stability analysis (Figure 4c and Figure S3 of the Supporting Information). The triplex with the C:G·5MeC-5a triplet had the highest Tm value, while the A:T·5MeC-5a triplex had the lowest. Furthermore, the addition of the guanidine moiety improved the differential between the triplex formed by TFO-5a and that formed by TFO-T.
In our previous work, we found that a cluster of 2′-AE ribose was essential for TFO bioactivity (15). Accordingly, we synthesized a TFO with the 2′-AE cluster with the N4-guanidinylethyl-5-methylcytosine at the Z position (AE-TFO-5a). The oligonucleotide was linked to psoralen so that it could be examined in the Hprt knockout bioassay. Because psoralen is not stable to the conditions of the guanidylation reaction, the derivatization of the triazoyl thymidine was conducted before psoralen coupling (Methods). The resultant oligonucleotide was characterized by thermal stability analysis (Figure 4d,e and Figures S3 and S4 of the Supporting Information). The Tm value of the triplex on the A:T duplex was 38.4 °C, while that of the duplex itself was 52 °C. Consequently, two transitions are apparent (Figure 4d). However, the triplex formed by TFO-5a and the C:G duplex is more stable than the underlying duplex (54.7 °C), and there is a single transition at 58 °C. Single-transition thermal stability profiles are characteristic of TFOs containing a cluster of AE residues and have been discussed previously (14).
The Tm value of the triplex formed by AE-TFO-5a on the C:G duplex was increased relative to that of TFO-5a with only 2′-OMe ribose sugars. However, there was also an increase in the stability of the triplex on the C:G duplex formed by the TFO-T designed for the wild-type sequence. This increase in the Tm value due to the 2′-AE cluster was seen with all the triplexes, although the magnitude of the increase was variable across the targets.
TFO Activity in the Hprt Bioassay
The bioassay is based on the mutagenic activity of a psoralen cross-link targeted by the TFO to a site in the Hprt gene immediately adjacent to exon 5. In most of our studies, we have used Chinese hamster cells because of the ease of manipulation and transfection and the uninterrupted polypurine:polypyrimidine triplex target next to exon 5. As noted above, the corresponding element in the murine genome contains the C:G interruption in the triplex target. Although biological testing of pso-TFOs with base analogues could be performed in murine cells, they do not have the same properties (growth, colony formation, and transfectability) as the CHO cells. Consequently, it would be difficult to compare the results of Hprt knockout by TFO targeting if cells of the two species were employed.
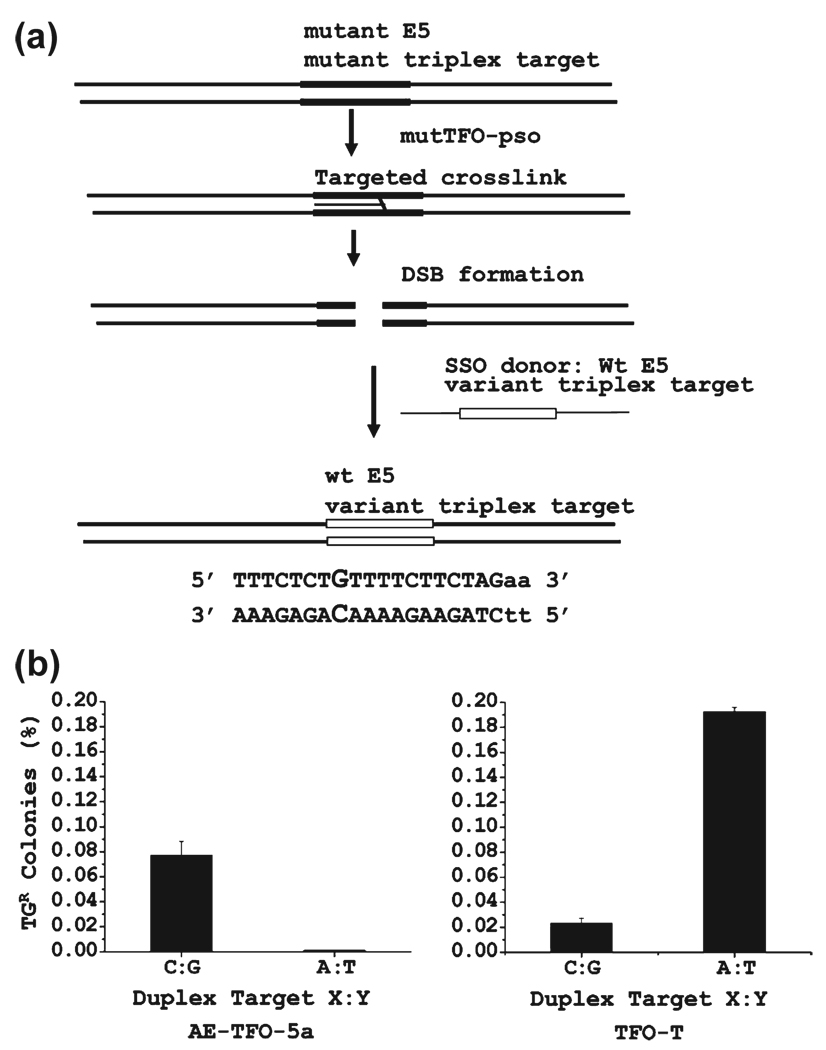
To eliminate this problem, we constructed a variant CHO cell line in which the Hprt target sequence was altered to match the murine sequence. This was done by targeted sequence conversion using a single-strand oligonucleotide donor, as we have described previously (55, 56). The procedure takes advantage of the processing of some fraction of the targeted psoralen cross-links through a double-strand break. These breaks can enter sequence conversion and recombination pathways. We exploited a CHO cell line that we constructed that was deficient in Hprt but had a unique triplex–psoralen target site adjacent to a defective exon 5. The appropriate pso-TFO was cotransfected with a 100-mer single-strand oligonucleotide donor containing the sequence information required to restore exon 5 and replace the hamster triplex target with the murine C:G inversion target (Figure 5a). Following treatment, the cells were incubated in HAT medium to select for cells with the restored Hprt function, and colonies were picked and expanded. Sequence analysis confirmed the expected conversion, and one of the clones was used in the assay of AE-TFO-5a, as were cells with the wild-type hamster sequence.
FIGURE 5.
Bioassay of TFO with the 5a analogue. (a) Construction of the CHO cell line with the murine triplex target site adjacent to exon 5 of Hprt. A cell line with a variant triplex target sequence adjacent to exon 5 was constructed by targeted sequence conversion (55, 56). This cell line is isogenic to the CHO AA8 line, differing only in the conversion of an A:T duplex to a C:G duplex at the X:Y position as shown in Figure 2. (b) Frequency of Hprt deficient colonies after treatment with AE-TFO-T and AE-TFO-5a in cells with the wild-type (A:T) and triplex (C:G) target.
The results of the bioassay indicated that AE-TFO-5a was active against the C:G inversion target in the C:G variant cells and inactive against the A:T target sequence in the wild-type cell line (Figure 5b). In comparison, the TFO against the wild-type sequence (TFO-T) was active against its matched, wild-type, target but exhibited very little activity against the C:G target in the variant cells. The TFO-T had about twice the activity against its matched target as did AE-TFO-5a against the C:G target. Characterization of the target region in the Hprt deficient colonies treated with AE-TFO-5a revealed the same pattern of small deletions found in experiments with bioactive TFOs designed against the wild-type target sequence. These deletions remove the psoralen cross-link site and some portion of exon 5 (13, 18, 37).
Modeling the Triplexes Formed by TFO-5a and TFO-T
The bioassay of AE-TFO-5a demonstrated success against a target with an interruption in the polypurine target sequence. To interpret our thermal stability analyses and bioassay data and guide future effort, we conducted molecular dynamics (MD) simulation studies of the relevant duplex and triplex structures.
Initially, the double-stranded DNAs were constructed, followed by preparation of the modified bases and a TFO strand. The 32-mer polypurine:pyrimidine sequence of interest contained the Chinese hamster Hprt target sequence (Table 1), where X:Y correspond to either an A:T (WT) or a C:G inversion. For comparison, we also modeled the structure formed on a duplex in which the X:Y pair was a G:C duplex. The 17-mer third-strand pyrimidine TFO (T-T-T-C-T-C-T-Z-T-T-T-T-C-T-T-C-T) forms a hydrogen bond to the first strand of the duplex, where Z can be one of the four nucleotides. The bases on the TFO strand were modeled with all sugar residues as 2′-OMe ribose, and all the cytosine bases were methylated at the C5 position. This base and sugar modification pattern corresponds to that of the triplexes examined in the thermal stability determination of Figure 4c. Thus, the results of the MD analysis will be compared to those data.
In the following, all the systems will be cited according to the bases in the midst of the corresponding duplex (X:Y) or triplex (X:Y·Z) structures, where X represents the base in the polypurine strand (first), Y the base in the pyrimidine strand (second), and Z the base in the TFO (third) (as in Figure 2). The systems were simulated using an explicit solvent representation as described in Methods. Production MD simulations were performed for 30 ns, from which the last 25 ns was used in the analyses. Simulations of the duplexes alone showed all the structures to be stable over a 30 ns simulation time and will not be discussed further. With the triplexes, the overall behavior of the simulations was monitored on the basis of the root-mean-square deviation (rmsd) of the backbone atoms (Figure S5 of the Supporting Information). It is evident that during the first few nanoseconds the systems relax, followed by fluctuations about stable values for the remainder of each simulation. This behavior indicates that stable, equilibrated structures were achieved and that the final 25 ns of the trajectories is suitable for more detailed analyses. Interestingly, the A:T·T triplex has the lowest rmsd of the systems studied with an average value of 3.3 (0.5) Å, where the value in parentheses represents the rms fluctuation of the rmsd values. While an indirect measure of stability, the low rmsd compared to those of the other sequences is consistent with this being the most stable triplex as seen in the thermal stability experiments in Figure 4c, which shows the Tm values of the triplexes with 2-OMe ribose at all positions. The other five structures containing the triplets (A:T·5MeC-5a, G:C·T, G:C·5MeC-5a, C:G·T, and C:G·5MeC-5a) demonstrate much larger fluctuations of 4.3 (0.8), 3.9 (0.7), 3.7 (0.5), 4.0 (0.6), and 4.4 (0.7) Å, respectively.
To gain an understanding in molecular detail of the stability of DNA complexes obtained form the thermal melting experiments, the hydrogen bonding patterns were analyzed for all six systems, including Watson–Crick base pairing in the duplexes and hydrogen bonds formed between the TFO strands and their corresponding duplexes. Here, we consider the central 5 bp that encompass the central X:Y·Z sequence, which is the position of the mismatch or inversion. Table S1 of the Supporting Information outlines the heavy atoms involved in the hydrogen bonding interactions for both Watson–Crick and Hoogsteen pairs. The average distances for the corresponding interactions were then calculated and are listed in Tables S2 and S3 of the Supporting Information. Time series of these distances are shown in Figures S6–S11 of the Supporting Information.
Analysis of the Watson–Crick interactions shows the hydrogen bonds to be well maintained in the unmodified systems (A:T·T, G:C·T, and C:G·T) for all five bases shown, including X:Y·Z (Table S2 of the Supporting Information). Notably, for the modified species, Watson–Crick hydrogen bonding is only maintained in C:G·5a while significant disruption of the Watson–Crick interaction is evident in A:T·5a and G:C·5a. These results are consistent with the C:G·5a system having the highest Tm of the modified species and indicate that the guanidinium moiety, in this triplet, is not perturbing the parent duplex.
With regard to the Hoogsteen interactions involving the TFO, these hydrogen bonds are well-maintained in A:T·T, the most stable species (Table S3 of the Supporting Information). Some disruptions of the Hoogsteen interactions occur with G:C·T and to a greater extent with C:G·T, again consistent with the relative stabilities of the species. Upon introduction of 5a into the third strand, significant changes in the Hoogsteen interactions occur. Destabilization of Hoogsteen hydrogen bond interactions occurs in both A:T·5a and G:C·5a, while there is a tendency for the associated hydrogen bonds to become more stable with C:G·5a. These trends are again consistent with the experimental Tm values (Figure 4c). To visualize the molecular interactions leading to the hydrogen bonding patterns outlined in Tables S2 and S3 of the Supporting Information, images of the triplets at the X:Y·Z position for the six simulated systems were extracted from the trajectories (Figure 6). With A:T·T, the ideal nature of both the Watson–Crick and Hoogsteen interactions is evident, leading to the stability of that triplet (Figure 6A). However, when the 5a modification is introduced (Figure 6B), the guanidinium moiety competes with the Watson–Crick interactions, inserting itself between the two Watson–Crick bases. This competition, as well as the lack of other interactions between the TFO and the duplex, would lead to the lower stability of this system. The conformation sampled by the guanidinium moiety in Figure 6B requires minor local unstacking, though the surrounding structure of the TFO is well-maintained. There were no constraints imposed on the guanidinium moiety of the modified base or on the remainder of the system during the MD simulations, thereby allowing the guanidinium moiety to sample all accessible conformations in the vicinity of its neighboring bases. In the course of the 30 ns MD simulations, the guanidinium moiety did not interact with the phosphate backbone or assume an orientation parallel to the helical axis.
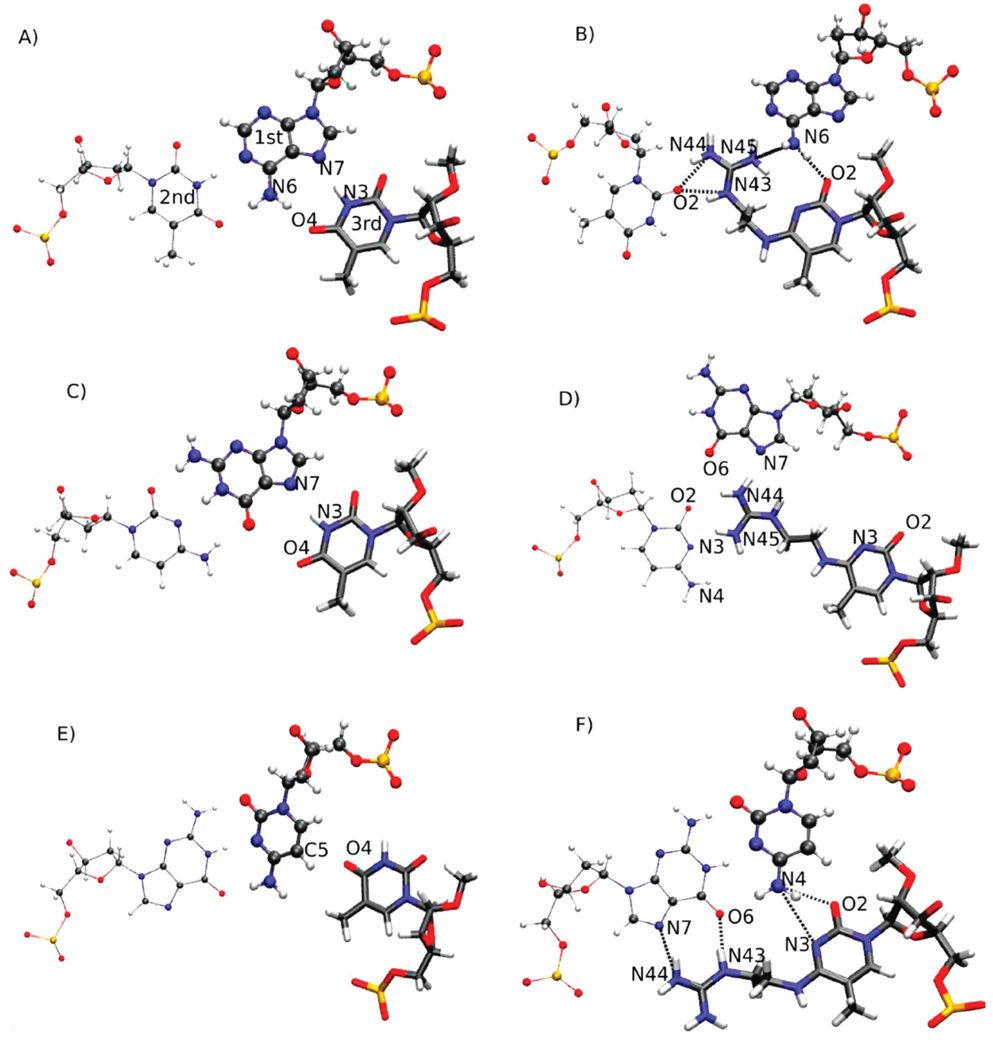
FIGURE 6.
Modeled structures of the central triplets formed by (A) A:T·T, (B) A:T·5MeC-5a, (C) G:C·T, (D) G:C·5MeC-5a, (E) C:G·T, and (F) C:G·5MeC-5a. The structures are snapshots from the 15 ns time step of the MD simulations and are oriented such that the viewer is looking down along the helical axis. In all systems, the base triplets are nearly coplanar. The strand 1 base is shown in standard CPK format, the strand 2 base in thin CPK format, and the strand 3 base in licorice format.
A similar pattern occurs with G:C·T and G:C·5a (Figure 6C,D). In G:C·T, ideal Watson–Crick hydrogen bonding is occurring and the thymine base in the TFO forms strong Hoogsteen hydrogen bonds with the guanine. However, in G:C·5a, the guanidinium again competes for the Watson–Crick interaction, in this case interfering with the approach of the cytosine base to the G in the duplex (Figure 6D). Ultimately, these interactions lead to the guanine in the duplex flipping out of the duplex (Figure S12 of the Supporting Information), further contributing to the destabilization of this species. With C:G·T (Figure 6E), the Watson–Crick interactions are seen to be well-maintained, but the inability of the thymine of the TFO to interact with the cytosine of the duplex via Hoogsteen interactions is readily visualized. This poor interaction leads to the lower Tm value of this species. However, in the C:G·5a triplex (Figure 6F), the guanidinium moiety assumes an orientation that allows it to form well-defined hydrogen bonds with the N7 and O6 atoms of the guanine base in the duplex. These interactions also allow the cytosine of the TFO to form hydrogen bonds with N4 of the cytosine in the duplex, via the N3 and O2 atoms of TFO 5a. It is likely that this collection of hydrogen bonds leads to the stability of this species.
The energetic consequences of the interactions shown in Figure 6 are indicated in Table 1, which lists the interaction energies between five central bases in the three strands. Of the unmodified species, the combination of the Watson–Crick and Hoogsteen interactions is most favorable with A:T·T, followed by G:C·T and then C:G·T, again consistent with the experimental Tm values. With the modified species, the least favorable interactions occur with A:T·5a and G:C·5a in accord with the lower Tm values, while G:C·5a has the most favorable interstrand interaction energies, consistent with its higher Tm value. Thus, while the energies presented in Table 1 are not free energies and, accordingly, cannot be expected to directly predict relative stabilities, they are consistent with the hydrogen bonding analysis and support a model in which modifications that maximize Hoogsteen-type hydrogen bonding, while allowing for the Watson–Crick interactions to be maintained, are predicted to lead to the formation of stable triplexes.
DISCUSSION
In previous work, we identified an effective modification format that conferred activity on TFOs against canonical polypurine:polypyrimidine sequences in living cells. Thus, a major remaining obstacle to the development of TFOs as versatile gene targeting reagents has been the limitation on target sequence options. In the experiments reported here, we have described the synthesis and characterization of a bioactive TFO designed to bind an interrupted target sequence. Several features distinguish our approach from previous efforts to synthesize and characterize effective analogues.
We employed a postsynthetic modification strategy for synthesis of all the analogues examined. This made it possible to synthesize and qualify a number of derivatives without preparing individual analogues as phosphoramidite precursors. A similar approach was described a number of years ago by Huang et al. (29), who employed transamination of a specific cytosine residue in an oligonucleotide. That study took advantage of the resistance of 5-methylcytosine to transamination in the presence of bisulfite, which provided a protective strategy for all cytosines except the one intended for the reaction. However, given the improvement of triplex formation conferred by the 5-methylcytosine, we elected to use the triazoyl-T as the derivatizable base.
We developed a simple support-based binding assay to quickly identify candidates for further analysis. The practical utility of this approach was that oligonucleotides with individual analogues did not have to be purified before examination in the binding assay. Thus, unpromising candidates could be discarded without a major investment of time and expense.
The third strand oligonucleotide contained 2′-OMe ribose at all positions, as a result of our previous experience with bioactive TFOs (57, 58), and the much earlier description of stable triplexes formed by RNA and RNA analogue third strands (7, 59). We have found that the evaluation of TFOs with 2′-OMe ribose provides information that can be used to select candidates for further modification with 2′-AE ribose residues (17). The 2′-AE phosphoramidites are much too costly in time and effort of preparation to be used at all stages of the development process.
After characterization of the TFOs in biochemical and biophysical assays, we examined their activity in a bioassay. To the best of our knowledge, this study is the first to report the activity of a TFO, carrying a code-expanding base analogue, directed toward a chromosomal target in living cells. We took advantage of the quantitative Hprt mutation assay that we have used in previous studies. To establish an isogenic cell line, differing at a single base pair in the triplex target, we exploited our recent development of a strategy for targeted sequence conversion by single-strand oligonucleotide donors (55, 56). This allowed us to construct a matched cell line with the variant target in the identical chromosomal locus as the wild-type cell. We anticipate that this approach can be used to construct additional lines, containing other variant triplex targets, including those with multiple inversions and variations in the bases adjacent to the inversions.
Comparison of the Analogue
The C:G interruption is considered more approachable than the T:A interruption, and efforts to design base analogue “solutions” have received more attention. TFOs containing the cytosine derivatives of the Miller group, N4-propylamino and acetamido, formed triplexes that were more stable on a C:G target than on duplexes with the other bases. These analogues were designed with the expectation that a hydrogen bond would form with one of the Hoogsteen acceptor sites of guanine. In that study, the acetamido derivative was more effective. However, in our experiments, we found little to distinguish them and chose to develop the amino compound (Figure 3b). It is difficult to directly compare the efficacy of the compounds discussed here with those in the prior literature given differences in TFO sequence, and our modification of all ribose units with 2′-OMe or 2′-AE. These modifications make a general contribution to triplex stability. It has been our experience that this can obscure the influence of individual base analogues on stability. Thus, the effect of substitutions that improve the stability of triplexes formed by third strands with deoxy sugars may be less apparent when in RNA analogue third strands (60). The extent to which the sugar modifications improve triplex stability appears to be somewhat variable with TFOs on different targets. Comparison of the stability of the triplexes formed by the 2′-OMe TFOs with those with the 2′-AE residues reveals in some cases a uniform increase in stability; the Tm values of TFOs on the T:A duplex (Figure 4e) were the same (38 °C), and inclusion of the 2′-AE cluster elevated both to the same level (51 and 50 °C). On the other hand, the distinction between the two TFOs on the C:G target, which was 15 °C with the 2′-OMe version, was reduced to 4 °C when the 2′-AE cluster was added.
Organization of Bioactive TFO-5a
Recognition of C:G inversions by analogues that exploit the single hydrogen bond option with an N4 proton of cytosine has been described: a 5-methyl-2-pyridone by the Imanishi lab (61) and a 5-methyl-2-pyrimidinone by the Leumann group (26). The efficacy of these analogues was greatly enhanced by combination with a sugar analogue that improves triplex stability, a 2′-O,4′-C-methylene-bridged ribose in the former report and a 2′-AE ribose in the latter. The combination of the 2′-AE sugar and the “S” analogue {N-[4-(3-acetamidophenyl)thiazol-2-yl]acetamide}, designed to recognize the T:A inversion, has also been discussed (62). This combinational approach clearly has promise, and it would be of interest to examine the activity of the guanidyl analogue linked to a 2′-AE ribose. However, for our initial study, we considered TFOs with a different organization of 2′-AE substitutions.
This approach was chosen for the following reasons. We have found that TFO bioactivity correlates with the association rate of triplex formation. TFOs with clustered 2′-AE residues (three or four) had the highest kon values, and the greatest bioactivity (15). We have argued that the grouping promoted the nucleation step that initiates triplex formation. Consequently, we felt it was important to maintain the 2′-AE cluster. One approach would have been to prepare the guanidyl derivative coupled to a 2′-AE ribose and embed the mixed analogue in the 2′-AE cluster. This would require that the analogue not only support a stable interaction with the C:G base pair but also contribute to the activity of the cluster in stimulating the nucleation phase of triplex formation. To avoid this, we chose the alternative design, placing the analogue away from the cluster so that it was required to interact effectively only with the C:G pair. We also considered the possibility of combining the 2′-AE cluster with an isolated 2′-AE-5a analogue. However, we have found that isolated 2′-AE residues, in addition to the cluster, actually reduce TFO bioactivity, although they improve affinity in vitro (63). Consequently, we maintained the 2′-AE cluster as previously described and synthesized the guanidyl derivative on a 2′-OMe ribose, in keeping with the other positions outside the cluster. This organization maintained, in AE-TFO-5a, the kinetic properties of our previous AE-TFOs. Thus, we found that the rate of association of AE-TFO-5a against the C:G target duplex, and that of TFO-T against the A:T (wt) target, were the same (not shown).
Molecular Modeling
Molecular modeling, including MD simulations, of six of the experimentally studied triplexes allows for a model of the types of interactions required to obtain stable triplexes with duplex targets containing pyrimidine interruptions in the purine strand. As anticipated, the presence of an inversion, as in C:G·T, disallows the formation of Hoogsteen hydrogen bonding, though it appears that disruption of Watson–Crick interactions in the duplex does not occur. To overcome the loss of stability due to the inversion, a successful base modification must maximize interactions with the hydrogen bonding groups in the major groove of the DNA while still allowing for the Watson–Crick interactions to be maintained. Even in the case of strong interactions of the modified bases with the major groove, perturbation of the Watson–Crick interactions would both disrupt the favorable intermolecular interactions between the strands in the duplex and allow water to penetrate the helix, further destabilizing the complex.
The results of the MD simulations were in accord with the thermal stability data of Figure 4c. They were also in good agreement with the bioactivity data. The interaction energy calculations of Table 1 are consistent with the 10-fold greater bioactivity of AE-TFO-T against the matched A:T target (−74.58 kcal/mol) and the much lower bioactivity against the C:G inversion target (−55.3 kcal/mol). Similarly, the bioactivity of AE-TFO-5a against the C:G inversion target (−64.69 kcal/mol) was greater than that against the A:T target (−51.72 kcal/mol). It should be noted that when considering the “perfect G:C” target the MD simulations predict some level of bioactivity of AE-TFO-T (wt) (−64.95 kcal/mol) but minimal activity of AE-TFO-5a (−48.89 kcal/mol). Experiments are in progress to test these predictions.
These results demonstrate the utility of modeling based on MD simulations that yield a molecular picture of the factors stabilizing triplexes. They suggest that these approaches may be used in a predictive fashion to facilitate the design of TFOs that overcome inversions in triplex target sequences. We anticipate that the strategy presented here, combining base and sugar modification chemistry, molecular modeling, and the bioassay based on isogenic cell lines containing variant triplex targets, will contribute to the expansion of the triplex binding code, a challenge for the past five decades.
Supplementary Material
Footnotes
This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute on Aging (Grant Z01 AG000746-10), and by National Institutes of Health Grant GM051501 (A.D.M.).
Abbreviations: 2′-AE, 2′-O-aminoethylribose; Hprt, hypoxanthine phosphoribosyltransferase; 5MeC-EtGua, 2′-OMe-guanidinylethyl-5-methylcytosine; TG, thioguanine; TTri, triazolylthymidine; TFO, triple helix-forming oligonucleotide.
SUPPORTING INFORMATION AVAILABLE
Primary thermal stability profiles (Figures S1–S4), support for the molecular modeling (Tables S1–S3 and Figures S5–S11), and the distortion of the G:C·5a triplet (Figure S12). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Thuong NT, Helene C. Sequence specific recognition and modification of double helical DNA by oligonucleotides. Angew. Chem., Int. Ed. 1993;32:666–690. [Google Scholar]
- 2.Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidman MM, Glazer PM. The potential for gene repair via triple helix formation. J. Clin. Invest. 2003;112:487–494. doi: 10.1172/JCI19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin JY, Glazer PM. Repair of DNA lesions associated with triplex-forming oligonucleotides. Mol. Carcinog. 2009;48:389–399. doi: 10.1002/mc.20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartwig A. Role of magnesium in genomic stability. Mutat. Res. 2001;475:113–121. doi: 10.1016/s0027-5107(01)00074-4. [DOI] [PubMed] [Google Scholar]
- 6.Lee JS, Woodsworth ML, Latimer LJ, Morgan AR. Poly(pyrimidine) 3 poly(purine) synthetic DNAs containing 5-methylcytosine form stable triplexes at neutral pH. Nucleic Acids Res. 1984;12:6603–6614. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts RW, Crothers DM. Stability and properties of double and triple helices: Dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 8.Escude C, Sun JS, Rougee M, Garestier T, Helene C. Stable triple helices are formed upon binding of RNA oligonucleotides and their 2′-O-methyl derivatives to double-helical DNA. C. R. Acad. Sci. III. 1992;315:521–525. [PubMed] [Google Scholar]
- 9.Asensio JL, Carr R, Brown T, Lane AN. Conformational and thermodynamic properties of parallel intramolecular triple helixes containing a DNA, RNA, or 2′-OMeDNA third strand. J. Am. Chem. Soc. 1999;121:11063–11070. [Google Scholar]
- 10.Blommers MJ, Natt F, Jahnke W, Cuenoud B. Dual recognition of double-stranded DNA by 2′-aminoethoxy-modified oligonucleotides: The solution structure of an intramolecular triplex obtained by NMR spectroscopy. Biochemistry. 1998;37:17714–17725. doi: 10.1021/bi9816352. [DOI] [PubMed] [Google Scholar]
- 11.Cuenoud B, Casset F, Husken D, Natt F, Wolf RM, Altmann KH, Martin P, Moser HE. Dual recognition of double stranded DNA by 2′-aminoethoxy-modified oligonucleotides. Angew. Chem., Int. Ed. 1998;37:1288–1291. doi: 10.1002/(SICI)1521-3773(19980518)37:9<1288::AID-ANIE1288>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 12.Carlomagno T, Blommers MJ, Meiler J, Cuenoud B, Griesinger C. Determination of aliphatic side-chain conformation using cross-correlated relaxation: Application to an extraordinarily stable 2′-aminoethoxy-modified oligonucleotide triplex. J. Am. Chem. Soc. 2001;123:7364–7370. doi: 10.1021/ja002592r. [DOI] [PubMed] [Google Scholar]
- 13.Puri N, Majumdar A, Cuenoud B, Natt F, Martin P, Boyd A, Miller PS, Seidman MM. Targeted gene knockout by 2′-O-aminoethyl modified triplex forming oligonucleotides. J. Biol. Chem. 2001;276:28991–28998. doi: 10.1074/jbc.M103409200. [DOI] [PubMed] [Google Scholar]
- 14.Puri N, Majumdar A, Cuenoud B, Natt F, Martin P, Boyd A, Miller PS, Seidman MM. Minimum Number of 2′-O-(2-Aminoethyl) Residues Required for Gene Knockout Activity by Triple Helix Forming Oligonucleotides. Biochemistry. 2002;41:7716–7724. doi: 10.1021/bi025734y. [DOI] [PubMed] [Google Scholar]
- 15.Puri N, Majumdar A, Cuenoud B, Miller PS, Seidman MM. Importance of Clustered 2′-O-(2-Aminoethyl) Residues for the Gene Targeting Activity of Triple Helix-Forming Oligonucleotides. Biochemistry. 2004;43:1343–1351. doi: 10.1021/bi035808l. [DOI] [PubMed] [Google Scholar]
- 16.Richards S, Liu ST, Majumdar A, Liu JL, Nairn RS, Bernier M, Maher V, Seidman MM. Triplex targeted genomic crosslinks enter separable deletion and base substitution pathways. Nucleic Acids Res. 2005;33:5382–5393. doi: 10.1093/nar/gki851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shahid KA, Majumdar A, Alam R, Liu ST, Kuan JY, Sui X, Cuenoud B, Glazer PM, Miller PS, Seidman MM. Targeted cross-linking of the human β-globin gene in living cells mediated by a triple helix forming oligonucleotide. Biochemistry. 2006;45:1970–1978. doi: 10.1021/bi0520986. [DOI] [PubMed] [Google Scholar]
- 18.Majumdar A, Puri N, Cuenoud B, Natt F, Martin P, Khorlin A, Dyatkina N, George AJ, Miller PS, Seidman MM. Cell Cycle Modulation of Gene Targeting by a Triple Helix-forming Oligonucleotide. J. Biol. Chem. 2003;278:11072–11077. doi: 10.1074/jbc.M211837200. [DOI] [PubMed] [Google Scholar]
- 19.Gowers DM, Fox KR. Towards mixed sequence recognition by triple helix formation. Nucleic Acids Res. 1999;27:1569–1577. doi: 10.1093/nar/27.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purwanto MG, Weisz K. Non-natural nucleosides for the specific recognition of Watson-Crick base pairs. Curr. Org. Chem. 2003;7:427–446. [Google Scholar]
- 21.Fox KR, Brown T. An extra dimension in nucleic acid sequence recognition. Q. Rev. Biophys. 2005;38:311–320. doi: 10.1017/S0033583506004197. [DOI] [PubMed] [Google Scholar]
- 22.Prevot-Halter I, Leumann CJ. Selective recognition of a C-G base-pair in the parallel DNA triple-helical binding motif. Bioorg. Med. Chem. Lett. 1999;9:2657–2660. doi: 10.1016/s0960-894x(99)00451-5. [DOI] [PubMed] [Google Scholar]
- 23.Ranasinghe RT, Rusling DA, Powers VE, Fox KR, Brown T. Recognition of CG inversions in DNA triple helices by methylated 3H-pyrrolo[2,3-d]pyrimidin-2(7H)-one nucleoside analogues. Chem. Commun. 2005:2555–2557. doi: 10.1039/b502325d. [DOI] [PubMed] [Google Scholar]
- 24.Rusling DA, Powers VE, Ranasinghe RT, Wang Y, Osborne SD, Brown T, Fox KR. Four base recognition by triplex-forming oligonucleotides at physiological pH. Nucleic Acids Res. 2005;33:3025–3032. doi: 10.1093/nar/gki625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okiba S, Hari Y, Sekiguchi M, Imanishi T. A 2′,4′-Bridged Nucleic Acid Containing 2-Pyridone as a Nucleobase: Efficient recognition of a C-C Interruption by Triplex Formation with a Pyrimidine Motif. Angew. Chem., Int. Ed. 2002;40:2079–2081. doi: 10.1002/1521-3773(20010601)40:11<2079::AID-ANIE2079>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 26.Buchini S, Leumann CJ. Stable and selective recognition of three base pairs in the parallel triple-helical DNA binding motif. Angew. Chem., Int. Ed. 2004;43:3925–3928. doi: 10.1002/anie.200460159. [DOI] [PubMed] [Google Scholar]
- 27.Obika S, Uneda T, Sugimoto T, Nanbu D, Minami T, Doi T, Imanishi T. 2′-O,4′-C-Methylene bridged nucleic acid (2′,4′-BNA): synthesis and triplex-forming properties. Bioorg. Med. Chem. 2001;9:1001–1011. doi: 10.1016/s0968-0896(00)00325-4. [DOI] [PubMed] [Google Scholar]
- 28.Orum H, Wengel J. Locked nucleic acids: A promising molecular family for gene-function analysis and antisense drug development. Curr. Opin. Mol. Ther. 2001;3:239–243. [PubMed] [Google Scholar]
- 29.Huang CY, Cushman CD, Miller PS. Triplex formation by an oligonucleotide containing N4-(3-acetamidopropyl)-cytosine. J. Org. Chem. 1993;58:5048–5049. [Google Scholar]
- 30.Huang CY, Bi G, Miller PS. Triplex formation by oligonucleotides containing novel deoxycytidine derivatives. Nucleic Acids Res. 1996;24:2606–2613. doi: 10.1093/nar/24.13.2606. [published erratum, (1997) Nucleic Acids Res. 25, 3750] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purwanto MG, Weisz K. Binding of imidazole-derived nucleosides to a CG base pair. J. Org. Chem. 2004;69:195–197. doi: 10.1021/jo035597q. [DOI] [PubMed] [Google Scholar]
- 32.Li JS, Chen FX, Shikiya R, Marky LA, Gold B. Molecular recognition via triplex formation of mixed purine/pyrimidine DNA sequences using oligoTRIPs. J. Am. Chem. Soc. 2005;127:12657–12665. doi: 10.1021/ja0530218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi Y, Uchida Y, Takaki T, Aoki E, Sasaki S. Recognition of CG interrupting site by W-shaped nucleoside analogs (WNA) having the pyrazole ring in an anti-parallel triplex DNA. Bioorg. Med. Chem. 2009;17:6803–6810. doi: 10.1016/j.bmc.2009.08.040. [DOI] [PubMed] [Google Scholar]
- 34.Griffin LC, Kiessling LL, Beal PA, Gillespie P, Dervan PB. Recognition of all four base pairs of double helical DNA by triple helix formation: Design of nonnatural deoxyribonucleosides for pyrimidine-purine base pair binding. J. Am. Chem. Soc. 1992;114:7976–7982. [Google Scholar]
- 35.Obika S, Inohara H, Hari Y, Imanishi T. Recognition of T·A interruption by 2′,4′-BNAs bearing heteroaromatic nucleobases through parallel motif triplex formation. Bioorg. Med. Chem. 2008;16:2945–2954. doi: 10.1016/j.bmc.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi Y, Nakamura A, Aoki E, Sasaki S. Modification of the aromatic ring of the WNA analogues for expansion of the triplex recognition codes. Nucleic Acids Res. Suppl. 2005:173–174. doi: 10.1093/nass/49.1.173. [DOI] [PubMed] [Google Scholar]
- 37.Majumdar A, Khorlin A, Dyatkina N, Lin FL, Powell J, Liu J, Fei Z, Khripine Y, Watanabe KA, George J, Glazer PM, Seidman MM. Targeted gene knockout mediated by triple helix forming oligonucleotides. Nat. Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 38.Sawai M, Takase K, Teraoka H, Tsukada K. Reversible G1 arrest in the cell cycle of human lymphoid cell lines by dimethyl sulfoxide. Exp. Cell Res. 1990;187:4–10. doi: 10.1016/0014-4827(90)90108-m. [DOI] [PubMed] [Google Scholar]
- 39.Orren DK, Petersen LN, Bohr VA. Persistent DNA damage inhibits S-phase and G2 progression, and results in apoptosis. Mol. Biol. Cell. 1997;8:1129–1142. doi: 10.1091/mbc.8.6.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks BR, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 41.Mackerell AD, Jr, Wiorkiewicz-Kuczera J, Karplus M. An all atom empirical energy function for the simulation of nucleic acids. J. Am. Chem. Soc. 1995;117:11046–11975. [Google Scholar]
- 42.Mackerell AD, Jr, Banavali NK. An all atom empirical energy function for the simulation of nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 2000;21:105–120. [Google Scholar]
- 43.Phillips JC, Braun R. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levitt M, Lifson S. Refinement of protein conformations using a macromolecular energy minimization procedure. J. Mol. Biol. 1969;46:269–279. doi: 10.1016/0022-2836(69)90421-5. [DOI] [PubMed] [Google Scholar]
- 45.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford, U.K: Oxford University Press; 1987. [Google Scholar]
- 46.Darden T, York D. Particle Mesh Ewald: An N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 47.Steinbach PJ, Brooks BR. New spherical cutoff methods for long range forces in macromolecular simulation. J. Comput. Chem. 1994;15:667–683. [Google Scholar]
- 48.Ryckaert JP, Ciccotti G. Numerical integration of Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 49.Hockney RW. In: The potential calculation and some applications. Alder B, Fernbach S, Rotenberg M, editors. San Diego: Academic Press; 1970. pp. 136–211. [Google Scholar]
- 50.Nose S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 51.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1659–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 52.Feller SE, Zhang H. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 53.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. U.S.A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 55.Majumdar A, Muniandy PA, Liu J, Liu JL, Liu ST, Cuenoud B, Seidman MM. Targeted Gene Knock In and Sequence Modulation Mediated by a Psoralen-linked Triplex-forming Oligonucleotide. J. Biol. Chem. 2008;283:11244–11252. doi: 10.1074/jbc.M800607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Majumdar A, Liu J, Thompson LH, Seidman MM. Sequence conversion by single strand oligonucleotide donors via non-homologous end joining in mammalian cells. J. Biol. Chem. 2010;285:23198–23207. doi: 10.1074/jbc.M110.123844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majumdar A, Puri N, McCollum N, Richards S, Cuenoud B, Miller P, Seidman MM. Gene Targeting by Triple Helix-Forming Oligonucleotides. Ann. N.Y. Acad. Sci. 2003;1002:141–153. doi: 10.1196/annals.1281.013. [DOI] [PubMed] [Google Scholar]
- 58.Seidman MM, Puri N, Majumdar A, Cuenoud B, Miller PS, Alam R. The development of bioactive triple helix-forming oligonucleotides. Ann. N.Y. Acad. Sci. 2005;1058:119–127. doi: 10.1196/annals.1359.020. [DOI] [PubMed] [Google Scholar]
- 59.Escude C, Francois JC, Sun JS, Ott G, Sprinzl M, Garestier T, Helene C. Stability of triple helices containing RNA and DNA strands: Experimental and molecular modeling studies. Nucleic Acids Res. 1993;21:5547–5553. doi: 10.1093/nar/21.24.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Miller PS, Seidman MM. Selectivity and affinity of DNA triplex forming oligonucleotides containing the nucleoside analogues 2′-O-methyl-5-(3-amino-1-propynyl)uridine and 2′-O-methyl-5-propynyluridine. Org. Biomol. Chem. 2008;6:4212–4217. doi: 10.1039/b810709b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Obika S, Hari Y, Sekiguchi M, Imanishi T. Stable oligonucleotide-directed triplex formation at target sites with CG interruptions: Strong sequence-specific recognition by 2′,4′-bridged nucleic-acid-containing 2-pyridones under physiological conditions. Chemistry. 2002;8:4796–4802. doi: 10.1002/1521-3765(20021018)8:20<4796::AID-CHEM4796>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Rusling DA, Powers VE, Lack O, Osborne SD, Fox KR, Brown T. Stable recognition of TA interruptions by triplex forming oligonucleotides containing a novel nucleoside. Biochemistry. 2005;44:5884–5892. doi: 10.1021/bi050013v. [DOI] [PubMed] [Google Scholar]
- 63.Alam MR, Majumdar A, Thazhathveetil AK, Liu ST, Liu JL, Puri N, Cuenoud B, Sasaki S, Miller PS, Seidman MM. Extensive sugar modification improves triple helix forming oligonucleotide activity in vitro but reduces activity in vivo. Biochemistry. 2007;46:10222–10233. doi: 10.1021/bi7003153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.