Abstract
NMR chemical shifts provide important local structural information for proteins and are key in recently described protein structure generation protocols. We describe a new chemical shift prediction program, SPARTA+, which is based on artificial neural networking. The neural network is trained on a large carefully pruned database, containing 580 proteins for which high-resolution X-ray structures and nearly complete backbone and 13Cβ chemical shifts are available. The neural network is trained to establish quantitative relations between chemical shifts and protein structures, including backbone and side-chain conformation, H-bonding, electric fields and ring-current effects. The trained neural network yields rapid chemical shift prediction for backbone and 13Cβ atoms, with standard deviations of 2.45, 1.09, 0.94, 1.14, 0.25 and 0.49 ppm for δ15N, δ13C′, δ13Cα, δ13Cβ, δ1Hα and δ1HN, respectively, between the SPARTA+ predicted and experimental shifts for a set of eleven validation proteins. These results represent a modest but consistent improvement (2–10%) over the best programs available to date, and appear to be approaching the limit at which empirical approaches can predict chemical shifts.
Keywords: Chemical shift prediction, backbone, protein structure, SPARTA, electric field, hydrogen bonding, torsion angles, SHIFTX, structure database
Introduction
NMR chemical shifts have long been recognized as important sources of protein structural information (Saito 1986; Spera and Bax 1991; Wishart et al. 1991; Iwadate et al. 1999; Wishart and Case 2001). During protein structure calculations, chemical shift derived backbone φ/ψ torsion angles (Luginbühl et al. 1995; Cornilescu et al. 1999; Shen et al. 2009) are often used as empirical restraints, complementing the more traditional restraints derived from NOEs, J couplings and RDCs. More recently, several approaches for generating protein structures have been developed which rely on backbone chemical shifts as the only source of experimental input information (Cavalli et al. 2007; Shen et al. 2008; Wishart et al. 2008). The success of these methods hinges on the accuracy at which chemical shifts can be related to protein structure. Although chemical shifts can be computed for known structures by de novo computational methods (Dedios et al. 1993; Xu and Case 2001; Vila et al. 2008; Vila et al. 2009), database-derived empirically optimized methods yield lower root-mean-square (rms) differences between observed and predicted values. Recent programs of this latter class include ShiftX (Neal et al. 2003), SPARTA (Shen and Bax 2007), and Camshift (Kohlhoff et al. 2009), and these are the chemical shift prediction methods used in chemical shift based structure prediction efforts.
The ShiftX program actually derives predicted 1H, 13C, and 15N chemical shifts from atomic coordinates using a hybrid approach which employs a pre-calculated, database-derived chemical shift hypersurface in combination with classical or semi-classical equations for ring current, electric field, hydrogen bonding and solvent effects. SPARTA is an empirical method which searches a database of assigned proteins of known structure for triplets of residues that most closely match structural and sequence characteristics of any triplet of residues in the query protein. Camshift is a recently introduced program which predicts chemical shifts using an empirically derived complex polynomial function to correlate interatomic distances with chemical shifts. A neural network based method, known as PROSHIFT (Meiler 2003), also makes good chemical shift predictions, albeit at an accuracy slightly below those of the more recent programs.
In this work we introduce the program SPARTA+, also based on the artificial neural network protocol, to predict chemical shifts for backbone and 13Cβ atoms in proteins. Compared to PROSHIFT, SPARTA+ uses an approximately two-fold larger protein database, recently developed for the program TALOS+, which establishes the inverse correlation, i.e., predicts backbone torsion angles from experimental chemical shifts (Shen et al. 2009). As described below, the input parameters for the neural network training procedure differ from those of PROSHIFT, and are more similar to those used by the program SPARTA, hence the naming of the new program.
SPARTA+ employs a well-trained neural network algorithm to make rapid chemical shift prediction on the basis of known structure. Validation on proteins not included in the training set shows modestly improved agreement between the experimental chemical shifts and the SPARTA+ predicted chemical shifts, over chemical shifts predicted by the original SPARTA, Camshift, and ShiftX methods.
Methods
Preparation of the NMR database
This work utilizes the protein structure and chemical shift database, originally used to develop the TALOS program (Cornilescu et al. 1999), and subsequently expanded to 200 proteins for the SPARTA and TALOS+ programs (Shen and Bax 2007; Shen et al. 2009), and most recently expanded further to 580 proteins for developing an empirical relation between chemical shifts and the cis or trans conformation of Xxx-Pro peptide bonds by the program Promega (Shen and Bax 2010). Nearly complete backbone NMR chemical shifts (δ15N, δ13C′, δ13Cα,δ13Cβ, δ1Hα and δ1HN) for these proteins are taken from the BMRB (Doreleijers et al. 1999; Doreleijers et al. 2005), with atomic coordinates taken from the corresponding high-resolution X-ray structures in the PDB (Berman et al. 2000). Residues containing two or less assigned chemical shifts were removed from the database. To minimize the influence of chemical shift outliers, chemical shift values that deviate by more than five standard deviations from the SPARTA-predicted values were also removed from the database. Details regarding the preparation of the database, including calibration of reference frequencies, correction of 2H isotope effects on δ13Cα and δ13Cβ, identification of H-bonds, etc., have been described previously (Shen and Bax 2007; Shen et al. 2009).
Neural network architecture and training
A single-level feed-forward multilayer artificial neural network (ANN) is used in this work to identify the dependence of 15N, 13C′, 13Cα, 13Cβ, 1Hα and 1HN chemical shifts on the local structural and dynamic information as well as amino acid type, and those of its immediate neighbors.
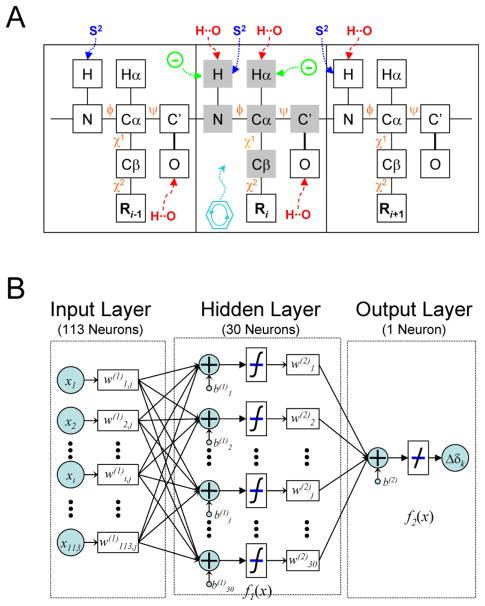
This single-level neural network has an architecture very similar to that of the first level neural network used by TALOS+ (Shen et al. 2009). The input signals to the first layer consist of tri-peptide structural parameter sets derived from the above described protein structural database. For predicting the chemical shifts of any given residue by SPARTA+, the structural input parameters include (1) the backbone and side-chain torsion angles of this residue and its two immediate neighbors, (2) information on interactions such as H-bonding, ring-current effects, and electric field effects, and (3) predicted backbone flexibility (Fig 1A; Table 1, column “Full”). Specifically, each tripeptide is represented by up to 113 nodes, which include for each residue the twenty amino acid type similarity scores, ten numbers representing φ/ψ/χ1/χ2 torsion angles of each tripeptide (the φ value of the first and ψ value of the last residue of the tripeptides are not used), three numbers for the structure-derived predicted N-H order parameter S2 (Zhang and Brüschweiler 2002) of each residue, and twenty numbers representing the H-bonding pattern for the tripeptide (Fig 1B). As was done for the TALOS+ program, amino acid type similarity scores are taken from the 20×20 BLOSUM62 matrix, commonly used for calculating sequence alignment (see http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=sef.figgrp.194). Considering the periodic nature of the torsion angles, each of the φ/ψ/χ1/χ2 torsion angles is represented by its sine and cosine values, thereby avoiding problems associated with the numerical discontinuities that exist when defining torsion angles in the −180° to +180° range (Meiler 2003). For each of the side-chainχ1/χ2 torsion angles, an additional Boolean number [1 or 0] is used to indicate whether a χ1 or χ2 torsion angle is defined for any given residue. For example, [sin(χ), cos(χ), 1] denotes a valid χ1 or χ2 torsion angle; [0, 0, 0] is used for residues lacking χ1 or χ2 torsion angles (χ2,1 torsion angles are used forχ2 of Thr, Val and Ile). The H-bonding input information of each tripeptide is limited to the HN/Hα/O backbone atoms of the center residue, the carbonyl O atom of the first residue, and the HN atom of the last residue. The H-bond information of each atom is denoted by three geometric parameters (Morozov et al. 2004), representing the distance between the donor hydrogen and the acceptor atom (H…A, dHA), the cosine value of the angle at the acceptor atom (B–A…H, Φ), and the angle at the donor hydrogen (A…H–D, Ψ), plus one additional Boolean number [1 or 0] to indicate whether the atom is H-bonded. So, four numbers [dHA,cos(Φ),cos(Ψ),1] are used for each of the potentially H-bonded backbone atoms, and [0,0,0,0] represents the absence of a H-bond.
Figure 1.
(A) Illustration of a protein tripeptide chain together with factors that impact the backbone NMR chemical shifts, considered by the SPARTA+ program. Factors used for prediction of the chemical shifts of the center residue 15N, 13C′, 13Cα, 13Cβ, 1Hα and 1HN (shaded in grey) include the backbone φ/ψ and side-chain χ1/χ2 torsion angles (colored orange), hydrogen bonding (red), electric fields (green), and ring-current effects (aqua), as well as backbone flexibility (blue). (B) Flow chart of the artificial neural network used in this work to study the relation between the protein structural and dynamic parameters (input layer) and NMR chemical shift (output layer).
Table 1.
Statistics of influence of various factors on SPARTA+ chemical shift prediction
| SPARTA | SPARTA+a |
||||||
|---|---|---|---|---|---|---|---|
| Full | Test I | Test II | Test III | Test IV | Test V | ||
| Training Inputb | |||||||
| Residue type | • | • | • | • | • | • | • |
| φ/ψ/χ1/χ2 | •/•/•/× | •/•/•/• | •/•/•/• | •/•/•/• | •/•/•/• | •/•/•/× | •/•/×/× |
| H-bond | × | • | • | • | × | × | × |
| S2 | × | • | • | × | × | × | × |
| Training Target δ−δrcc | |||||||
| −δneighbor | • | × | × | × | × | × | × |
| −δring | • | • | • | • | • | • | • |
| −δEF | × | • | × | × | × | × | × |
| Output Δδpred+δrcd | |||||||
| +δneighbor | • | × | × | × | × | × | × |
| +δring | • | • | • | • | • | • | • |
| +δEF | × | • | × | × | × | × | × |
| +ΔHB | • | × | × | × | × | × | × |
| RMSD(δpred, δobs)e [ppm] | |||||||
| δ15N | 2.56 (2.56) | 2.45 (2.48) | 2.46 (2.48) | 2.47 (2.49) | 2.52 (2.52) | 2.50 (2.51) | 2.62 (2.64) |
| δ1Hα | 0.29 (0.27) | 0.25 (0.25) | 0.27 (0.25) | 0.27 (0.25) | 0.29 (0.29) | 0.29 (0.29) | 0.29 (0.29) |
| δ13C′ | 1.14 (1.13) | 1.09 (1.11) | 1.09 (1.11) | 1.09 (1.11) | 1.13 (1.14) | 1.13 (1.14) | 1.16 (1.16) |
| δ13Cα | 1.04 (1.01) | 0.94 (0.98) | 0.94 (0.98) | 0.97 (0.99) | 0.99 (1.00) | 1.02 (1.03) | 1.02 (1.05) |
| δ13Cβ | 1.16 (1.06) | 1.14 (1.11) | 1.14 (1.11) | 1.14 (1.11) | 1.14 (1.11) | 1.15 (1.12) | 1.16 (1.14) |
| δ1HN | 0.54 (0.51) | 0.49 (0.47) | 0.50 (0.48) | 0.50 (0.48) | 0.58 (0.54) | 0.58 (0.54) | 0.58 (0.54) |
See text (Results and Discussion) for the description of each testing neural network.
Structural and dynamic factors used as inputs for the database search (SPARTA) or neural network (SPARTA+) training procedure. All factors are for all three residues of a given tripeptide (see Fig 1A). Parameters included and omitted in each input set are marked • and x, respectively.
NMR (secondary) chemical shifts used as the targets (outputs) of the database search (SPARTA) or neural network (SPARTA+) training procedure. The (secondary) NMR chemical shifts were obtained from the difference between the chemical shift δ and the random coil chemical shift δrc, after subtracting the corrections from neighboring residues (δneighbor), the contributions from ring current effects (δring), or electric fields (δEF).
Offsets and corrections, in addition to the random coil chemical shift δrc, applied to SPARTA or SPARTA+ predicted secondary chemical shift (Δδpred), i.e., the final SPARTA/SPARTA+ predicted chemical shifts.
RMS deviation between the predicted and experimental (obs) chemical shifts for eleven proteins which are not present in the SPARTA+ training database. For SPARTA+, the prediction performances for the validation datasets (see Methods) in the training database are provided in parentheses. For the SPARTA predictions, performances listed between brackets are those obtained for the 580-protein training database, but with the protein predicted excluded from this database,
In the hidden layer of the network, where each node receives the weighted sum of the input layer nodes as a signal, 30 such nodes (or hidden neurons) are used. The output of a hidden layer node is obtained through a nodal transformation function (Fig. 1B).
For the purpose of predicting the NMR chemical shifts from protein structural parameters, the secondary chemical shift ΔδX(X = 15N, 13C′, 13Cα, 13Cβ, 1Hα or 1HN) of the center residue of each tri-peptide in the database is used as the target of the first level network, after subtracting the contributions from ring-current effects (δXring) and electric fields effects (δXEF), i.e.,
| (1) |
where δXrc is the random coil chemical shift of nucleus X, δXEF is calculated for 1Hα and 1HN nuclei only, using the Buckingham method (Buckingham 1960) and atom selection criteria analogous to those of the ShiftX program (Neal et al. 2003), δXring is calculated for all six types nuclei using the Haigh-Mallion model (Haigh and Mallion 1979; Case 1995), in the same way as used by the SPARTA program (Shen and Bax 2007). Note that chemical shift corrections from the neighboring residues, as used by the TALOS, SPARTA, and TALOS+ methods, are not included here when calculating the secondary chemical shifts,ΔδX, because the neural network optimally accounts for those effects after training of the network on the database. Each output value has one node with a linear activation function (f2(x) = x; eq 2). The empirical relationship between the NMR secondary chemical shift and the protein structural and sequence data, received by the network (Fig. 1B), is given by
| (2) |
with f1(x) = (1−e−2x)/(1+e−2x), and f2(x) = x. X1×113 is the input data vector consisting of 113 elements; W(1) and b(1) are the weight matrix and bias, respectively, for the connection between the nodes in the input and the hidden layer; W(2) and b(2) are the weight matrix and bias, for the connection between the nodes in the hidden and output layer; Δδ1×1 is the training target or the output vector.
Neural network training
The weight and bias terms were determined by training the artificial neural network on the 580-protein structural database with associated chemical shifts, described above. To prevent over-training, a three-fold training and validation procedure was employed for the neural network model by dividing the input-output training dataset into three separate subsets, followed by separate training of the corresponding neural networks. For each of these three network optimizations, one third of the database was excluded from the training but then used to evaluate the training performance of the neural network on the other two input-output subsets during the training. This subset, referred as the validation dataset, was not used to calculate the weight changes in this network. Training of the network was terminated when the performance of the network on the validation dataset, represented by the mean squared errors between the predicted values and targets, began to degrade. This procedure was repeated three times, each time with a different one third of the database proteins assigned to the validation set.
Neural network testing and validation
In addition to the above three-fold training and validation, a second validation procedure was performed for a set of eleven additional proteins, with also nearly complete chemical shifts, a good quality reference structure, and no homologous protein (≥30% sequence identity) in the 580-protein database. This set of eleven proteins was identified after the original 580-protein database had been assembled and used for training of the ANN.
The final predicted NMR chemical shifts are obtained from:
| (3) |
where ΔδXpred is the ANN-predicted secondary chemical shift (Eq 2) using the weights and biases obtained from the above training steps, after averaging over the outputs from the three separately trained networks.
Estimated errors for the predicted NMR chemical shifts
The original SPARTA program estimates the chemical shift prediction errors on the basis of an empirical correlation between this error and the spread in chemical shifts among the 20 best matched tripeptides (Shen and Bax 2007). In the present study, an estimate for the chemical shift prediction error, σ, can be obtained by using an empirical Δδ(φ,ψ) error surface (Spera and Bax 1991), which is calculated by:
| (4) |
where the prediction errors between ANN-predicted δ(φk,ψk)pred and experimental δ(φk, ψk)obs chemical shifts are convoluted with a Gaussian function and then summed over all residues (k) of the validation subsets in the training database, followed by normalization.
The SPARTA+ chemical shift prediction, accomplished by the above described ANN procedure, is carried out by a program largely written in C++, which is ten times faster than the original SPARTA method. On a PC with a single 2.4 GHz CPU, the SPARTA+ chemical shift prediction takes ca 2 seconds for a 100-residue protein, the majority of which is actually attributed to loading of the error surfaces.
Results and discussion
Neural network chemical shift prediction
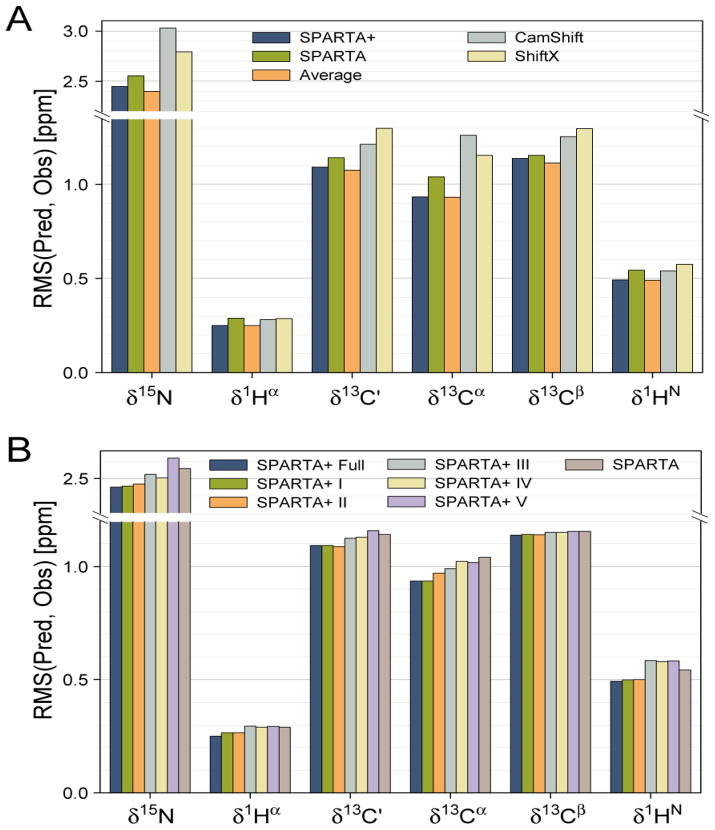
For each type of nucleus (15N, 13C′, 13Cα, 13Cβ, 1Hα and 1HN), three artificial neural networks were trained separately to predict the chemical shift, using a three-fold training and validation procedure. The trained weights and biases obtained for each network are then used to calculate the chemical shifts for each of a protein’s backbone and 13Cβ atoms (except for the N- and C-terminal residues), using Eqs 2 and 3. The low rms difference between the predicted and observed NMR chemical shifts, evaluated over the validation datasets (Table 1), indicates that the networks are well-trained.
To further inspect the chemical shift prediction performance of the trained neural networks, eleven additional proteins were used which were not present in any of the training or validation sets. The chemical shifts predicted for these eleven proteins were obtained by averaging the outputs of the three separately trained neural networks, obtained from the above described three-fold training procedure. The predicted chemical shifts show good agreement with the experimental chemical shifts, with standard deviations of 2.45, 1.09, 0.94, 1.14, 0.25, and 0.49 ppm for δ15N, δ13C′, δ13Cα, δ13Cβ, δ1Hα and δ1HN, respectively, including outliers. The rmsd’s for δ15N, δ13C′ andδ13Cα in this set of eleven proteins are slightly lower than those for the validation datasets used during the network training (Table 1), most likely the result of the three-fold averaging procedure used for this set, which is not applicable for the validation sets (see below). The performance of alternate chemical shift prediction programs was also evaluated on this set of eleven proteins, including SPARTA (Shen and Bax 2007) and webserver versions of ShiftX (Neal et al. 2003), CamShift (Kohlhoff et al. 2009), and PROSHIFT (Meiler 2003).
Comparison of the predicted with experimental chemical shifts (Fig. 2A; Table S1) indicates that SPARTA+ slightly outperforms SPARTA, with rmsd values that are ca 10–15% lower for δ13Cα, δ1HN and δ1Hα, 5% for δ15N and δ13C′, with the smallest improvement (2%) for δ13Cβ. SPARTA+ outperforms the ShiftX and Camshift programs by slightly larger margins (ca 10–20%) for all six nuclei (Fig. 2A), and the alternate ANN-based PROSHIFT program by somewhat larger margins (Table S1). Interestingly, the fractional improvement in chemical shift prediction accuracy is largest for 13Cα, often used as the most significant indicator of protein secondary structure.
Figure 2.
Chemical shift prediction performance of various methods, evaluated over a set of eleven proteins not included in the neural network training database. The prediction performance (vertical axis) for the 15N, 13C′, 13Cα, 13Cβ, 1Hα and 1HN chemical shifts is represented by the rms difference between the experimental and the predicted chemical shifts. Colors of the bars indicate the program used for predicting the chemical shifts, as marked in the panel. The orange bar corresponds to the weighted average (70%/30%) of the SPARTA+ and SPARTA predicted chemical shifts. (B) Impact of various structural and dynamic input parameters on SPARTA+ chemical shift predictions. Dark blue columns correspond to using the full set of SPARTA+ input parameters; the adjacent 5 bars correspond to input parameters defined in Table 1. The most right hand bar in each set corresponds to the original SPARTA prediction method.
Although with Pearson’s correlation coefficients in the 0.7–0.8 range the prediction errors of SPARTA and SPARTA+ are correlated (data not shown), there clearly is considerable scatter. Averaging the predictions made by the original SPARTA program with those of SPARTA+, using weight factors of 0.3 and 0.7, respectively, yields a slight further improvement in prediction accuracy for 15N, 13C′, and 13Cβ (Fig. 2A; Table S1).
Impact of structural parameters on prediction accuracy
The SPARTA program uses the φ/ψ/χ1 torsion angles and residue type information of a query tripeptide to predict the chemical shifts for the atoms of its center residue, followed by applying corrections for the ring-current shift and H-bonding (H-bond distance only). Compared with SPARTA, the SPARTA+ procedure considers more H-bond geometric factors for the H-bonded atoms, as well as additional side-chain χ2 torsion angle information, electric field effects, and structure-based prediction of backbone flexibility (see Methods; Table 1).
In order to investigate the impact of the different structural factors on the prediction accuracy of SPARTA+, multiple neural networks with different input of the protein structural/dynamic parameters and output of the (secondary) chemical shifts are evaluated. The network trained with the full set of the listed input parameters (see Methods) is named “Full” (Table 1). Five additional testing networks are implemented too and referred to as “Test I” (lacking the electric field effect contribution relative to “Full”), “Test II” (additionally lacking the predicted backbone order parameter), “Test III” (additionally lacking H-bonding information), “Test IV” (additionally lacking χ2 torsion angles), and finally “Test V” (additionally lacking χ1 torsion angles). All five testing networks have 30 and 1 neurons in their hidden and output layers, respectively; the number of input neurons are 113, 110, 90, 81 and 72, respectively (Table 1; see Methods for details on the number of neurons/nodes used for each individual structural/dynamic parameter). All testing networks are trained in the same three-fold training and validation procedure, and using the same training database, as used for the network “Full”. The accuracy of the chemical shift predictions performed by the trained testing networks is used to evaluate the importance of the various parameters for chemical shift prediction (Fig. 2B).
When only the residue type, backbone φ/ψ and side-chain χ1 torsion angles, and ring-current effects are considered (network “Test IV”), the ANN remains capable of capturing the relation between NMR chemical shifts and protein structure reasonably well for all six types of nuclei (Table 1). Compared with the original SPARTA method, the overall prediction accuracy for the validation datasets is 1–2% worse for 13C′ and 13Cα predictions, 5–7% worse for 13Cβ, 1Hα and 1HN, and about 2% better for the 15N (Table 1). Considering that the H-bond correction applied by SPARTA after its initial database search contributes a ca 5% improvement to its chemical shift prediction performance for 1Hα and 1HN, the accuracy of the chemical shifts predicted by the Test IV network actually is quite close to that of the database search component of the original SPARTA method, with the exception of the ca 5% lower prediction accuracy for 13Cβ. This result applies for both the validation datasets in the training database and for the eleven test proteins which are absent in the training database (Table 1). Moreover, the three-fold training and validation procedure results in three networks that are trained separately with “half-independent” training datasets, making the contribution to chemical shift prediction errors from imperfect training data somewhat uncorrelated. As a result, averaging the chemical shifts predicted by the three separately trained networks then further improves the accuracy of the predicted chemical shifts by 2–4% (Table S2), making it slightly better than that of the SPARTA predicted shifts (except for 1H predictions).
The effects of side-chain conformation on backbone chemical shifts have been well recognized (Dedios et al. 1993; Wang and Jardetzky 2004; Villegas et al. 2007; London et al. 2008; Mulder 2009). As indicated by the results of the Test V network, which lacks χ1 torsion angle input information relative to network Test IV, the accuracy of the predicted chemical shifts decreases by 5% for 15N and by about 1–2% for the other nuclei. When additionally considering the impact of the χ2 torsion angle by comparing the difference in prediction accuracy of networks Test III and Test IV, a small improvement (~3%) of the δ13Cα prediction is observed (Fig. 2B; Table 1), but with the other nuclei virtually unaffected. Further inspection indicates that the observed improvement in δ13Cα prediction is almost entirely accounted for by the aromatic amino acids (Phe, His, Tyr and Trp) and Met (Fig. S2).
When H-bonding parameters are additionally included as input parameters when training the network (Test II), accuracy of the predicted chemical shifts further increases, both for the validation datasets in the training database and the set of eleven test proteins (Fig 2B; Table 1). The improvement in prediction accuracy upon of inclusion of H-bond input parameters is largest for proton chemical shifts (10–13%), but an improvement of 1–3% is also seen for 13C′, 15N, and 13Cα. A small further improvement (2–3%) in chemical shift prediction accuracy of the network is observed for 13Cα chemical shifts when the predicted backbone flexibility, as represented by the structure-predicted S2 order parameter of Zhang and Brüschweiler (2002), is included with the input parameters (network Test I). Finally, the accuracy of the network-predicted 1Hα and 1HN chemical shifts is improved by several percentage points, when the electric field contribution to the 1Hα and 1HN chemical shifts is excluded prior to the network training and added back later to the predicted chemical shifts (as present by the network Full).
Application of SPARTA+ to CS-Rosetta
Recently introduced procedures to generate protein structures using NMR chemical shifts as the only experimental input data have been quite successful in generating good quality models for small to medium-sized proteins (Cavalli et al. 2007; Shen et al. 2008; Wishart et al. 2008). Here, we evaluate the impact of improved chemical shift prediction on the effectiveness of one such protocol, CS-Rosetta (Shen et al. 2008).
CS-Rosetta utilizes NMR chemical shifts at two distinct steps of its protocol: fragment selection, and selection of its final models. The impact of improved chemical shift prediction on these two stages will be discussed below.
CS-Rosetta relies on the existence of a large database of protein structures from which fragments are selected to function as building blocks for the query protein. Similarity between the experimental chemical shifts of short segments in the query protein and chemical shifts of fragments in the protein database is used to guide the selection of the most suitable fragments. As the procedure requires a large database of high quality structures with known chemical shifts, and the database of experimentally determined NMR structures remains relatively small, CS-Rosetta utilizes a much larger database of X-ray structures, to which chemical shift values are added by prediction methods. A considerable improvement was found when the program SPARTA was used for adding chemical shifts to the protein database compared to predictions obtained using a less advanced program, known as DC, even though the accuracy of chemical shift predictions by SPARTA is only 10–20% better than those obtained by DC (Shen et al. 2008).
Considering that SPARTA+ offers a similar level of improvement over SPARTA, a comparable improvement in fragment quality might be expected when using the database with more accurately predicted chemical shifts, where fragment quality is measured by the backbone coordinate rms difference between the query segment and selected database fragments that most closely match the experimental secondary chemical shifts. However, on average, we find no improvement in fragment quality when using the protein structural database to which chemical shifts have been added by SPARTA+ over the database where these chemical shifts were added by SPARTA (data not shown). A likely reason for the lack of improvement is that the Rosetta structure generation procedure only utilizes the backbone torsion angles (φ/ψ/ω) from the selected fragments, whereas the improved chemical shift prediction above was shown to be dominated by sidechain and hydrogen bonding contributions (Fig. 2B; Table 1).
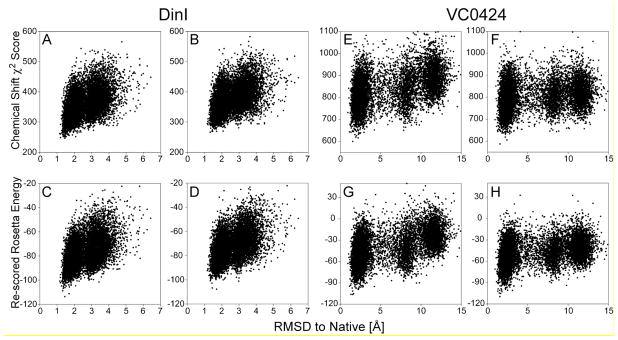
The second stage where accuracy of the chemical shift prediction plays a role during the CS-Rosetta protocol is during selection of the final models, from the very large ensemble of structures generated by its Monte Carlo procedure. Model selection is based on a combination of lowest empirical energy, as scored by the classic Rosetta program (Rohl et al. 2004), combined with a weighted chemical shift error score,χ2, that accounts for the agreement between experimental chemical shifts and values predicted for each model. These latter models are full atom structures, including sidechains, H-bonds, etc, and improved ability to predict the chemical shifts for such structures is therefore expected to somewhat increase the ability to distinguish between accurate and less accurate models. We evaluate the impact of SPARTA+ on model selection for two proteins, DinI and Vc0424, neither of which is included in the SPARTA+ training database. For both proteins, a standard CS-Rosetta procedure (Shen et al. 2008) is performed, using a SPARTA+ assigned protein structural database. For each protein, the 10,000 structures generated by CS-Rosetta are then evaluated by calculating the total χ2 score between the experimental chemical shifts and values predicted either by SPARTA+ or by SPARTA. For both proteins, models with the lowest total chemical shift χ2 value are closer to the experimental reference structure (Fig. 3A,B,E,F) when using SPARTA+ chemical shifts. This small advantage remains when combining the χ2 value with the Rosetta empirical energy function in the standard manner (Shen et al. 2008), again yielding slightly lower backbone rms differences between the models with the lowest total score and the corresponding reference structures (Fig. 3C,D,G,H; Table S2).
Figure 3.
CS-Rosetta model selection using either SPARTA+ or SPARTA chemical shift predictions. For proteins DinI (A-D; PDB entry 1GHH (Ramirez et al. 2000)) and VC0424 (E-H; PDB entry 1NXI (Ramelot et al. 2003)), 10,000 structures each were generated by a standard CS-Rosetta protocol, using a protein structural database with chemical shifts added by SPARTA+. For each CS-Rosetta model, the totalχ2 error function between the experimental chemical shifts and values predicted by SPARTA+ (A,E) or SPARTA (B,F) are plotted against the Cα coordinate rmsd relative to the experimental PDB structure. The re-scored Rosetta energy, calculated by adding the scaled SPARTA+ (C,G) or SPARTA (D,H) chemical shift χ2 score to the raw Rosetta energy, is also plotted and used to select the final models (Table S3).
Concluding Remarks
By using the artificial neural network approach, including a more complete consideration of various structural/dynamic parameters in proteins, SPARTA+ is able to predict chemical shifts for backbone and 13Cβ atoms with modestly improved accuracy, compared with other similar chemical shift prediction approaches. The improvement of the accuracy in the SPARTA+ predicted chemical shifts is mostly credited to the additional structural/dynamic factors, i.e., χ2 torsion angle, H-bonding and electric fields, as well as an averaging procedure over the outputs from three separated neural networks. Of all predicted chemical shifts, δ13Cα appears to benefit most from incorporation of the structure-predicted effect of backbone dynamics, used as an input parameter by SPARTA+. Conceivably, further improvements in this regard could be obtained by recording very extended (~1 μs) molecular dynamics trajectories, and averaging predicted chemical shifts over such a trajectory (Li and Brüschweiler 2010). However, from a practical perspective, such a computationally demanding approach is not yet practical.
Two interesting questions remain: Have we reached the limit of how well empirical methods can predict chemical shifts from known structure, and what is the reason for such a limit? Indeed the finding that only small increments in prediction accuracy are obtained when including additional input parameters suggests that we are asymptotically approaching the limit at which empirical approaches can predict chemical shifts. One may wonder whether the accuracy of the coordinates plays a role in prediction accuracy, for example. For the program ShiftX, a correlation between the accuracy of the prediction and the quality of the structure was reported (Neal et al. 2003). However, the SPARTA+ database uses far more stringent criteria for its database, including a crystallographic resolution threshold of 2.4 Å. Comparing the prediction accuracy for the 10 highest resolution structures (all ≤1Å) with those of the lowest resolution structures (all at ~2.4 Å) also shows a modest improvement for the higher resolution structure, although the effect is much smaller than found for ShiftX (Table S4). When evaluating proteins of even lower crystallographic resolution, the SPARTA+ accuracy further deteriorates (Table S4). However, with structures solved at a crystallographic resolution of 1Å representing the most favorable case, and prediction errors remaining rather large, further progress by using a better reference database will not substantially improve results any further.
At a crystallographic resolution of 1Å, atom positions are defined very well, and errors in backbone torsion angles are small compared to the gradient of the chemical shift surface with respect to these angles. However, two important sources of potential error remain. First, many sidechains are highly disordered in solution as judged, for example, by NMR relaxation measurements (Palmer 1997; Kay 1998; Yang et al. 1998; Lee and Wand 2001), an effect not easily accounted for by an empirical approach such as SPARTA+. Second, ab initio calculations indicate chemical shifts to be extremely sensitive to relatively small deviations from ideal geometry and small steric clashes. Even at the highest level of resolution, the atomic coordinate precision is usually insufficient to accurately account for such distortions (Karplus 1996), and empirical characterization by an approach such as SPARTA+ appears beyond reach. Even if we were to add corrections for specific geometry distortions to the SPARTA+ values, predicted by density functional theory (DFT) computations, this would not be of immediate practical use, as the precise magnitude of a local geometric distortion almost invariably remains subject to high experimental uncertainty.
Although the improvement of the chemical shifts prediction performance is modest, chemical shift prediction by SPARTA+, using Eq 2 with its trained weights and biases, is more than an order of magnitude faster than SPARTA. Moreover, the neural network equation (Eq 2) used by SPARTA+ is differentiable with respect to the torsion angles, making it potentially possible to be used (on the fly) by the protein structure calculation and refinement procedures in combination with other, standard input restraints, in a manner similar to that proposed for CamShift (Kohlhoff et al. 2009).
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the NIDDK, NIH, and by the Intramural AIDS-Targeted Antiviral Program of the Office of the Director of the NIH.
Footnotes
Software availability
SPARTA+ and detailed instructions on its use can be downloaded from http://spin.niddk.nih.gov/bax/software/SPARTA+. Source code is available upon request.
References
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham AD. Chemical shifts in the nuclear magnetic resonance spectra of molecules containing polar groups. Canadian Journal of Chemistry-Revue Canadienne De Chimie. 1960;38:300–307. [Google Scholar]
- Case DA. Calibration of ring-current effects in proteins and nucleic acids. J Biomol NMR. 1995;6:341–346. doi: 10.1007/BF00197633. [DOI] [PubMed] [Google Scholar]
- Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci U S A. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Dedios AC, Pearson JG, Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts - an ab initio approach. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- Doreleijers JF, Nederveen AJ, Vranken W, Lin JD, Bonvin A, Kaptein R, Markley JL, Ulrich EL. BioMagResBank databases DOCR and FRED containing converted and filtered sets of experimental NMR restraints and coordinates from over 500 protein PDB structures. J Biomol NMR. 2005;32:1–12. doi: 10.1007/s10858-005-2195-0. [DOI] [PubMed] [Google Scholar]
- Doreleijers JF, Vriend G, Raves ML, Kaptein R. Validation of nuclear magnetic resonance structures of proteins and nucleic acids: Hydrogen geometry and nomenclature. Proteins-Structure Function and Genetics. 1999;37:404–416. doi: 10.1002/(sici)1097-0134(19991115)37:3<404::aid-prot8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Haigh CW, Mallion RB. Ring current theories in nuclear magnetic resonance. Prog Nucl Magn Reson Spectrosc. 1979;13:303–344. [Google Scholar]
- Iwadate M, Asakura T, Williamson MP. C-alpha and C-beta carbon-13 chemical shifts in proteins from an empirical database. J Biomol NMR. 1999;13:199–211. doi: 10.1023/a:1008376710086. [DOI] [PubMed] [Google Scholar]
- Karplus PA. Experimentally observed conformation-dependent geometry and hidden strain in proteins. Protein Science. 1996;5:1406–1420. doi: 10.1002/pro.5560050719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE. Protein dynamics from NMR. Nature Structural Biology. 1998;5:513–517. doi: 10.1038/755. [DOI] [PubMed] [Google Scholar]
- Kohlhoff KJ, Robustelli P, Cavalli A, Salvatella X, Vendruscolo M. Fast and Accurate Predictions of Protein NMR Chemical Shifts from Interatomic Distances. J Am Chem Soc. 2009;131:13894–13895. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- Lee AL, Wand AJ. Microscopic origins of entropy, heat capacity and the glass transition in proteins. Nature. 2001;411:501–504. doi: 10.1038/35078119. [DOI] [PubMed] [Google Scholar]
- Li DW, Brüschweiler R. Certification of Molecular Dynamics Trajectories with NMR Chemical Shifts. Journal of Physical Chemistry Letters. 2010;1:246–248. [Google Scholar]
- London RE, Wingad BD, Mueller GA. Dependence of amino acid side chain C-13 shifts on dihedral angle: Application to conformational analysis. J Am Chem Soc. 2008;130:11097–11105. doi: 10.1021/ja802729t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbühl P, Szyperski T, Wüthrich K. Statistical basis for the use of 13Cα chemical shifts in protein structure determination. J Magn Reson Ser B. 1995;109:229–233. [Google Scholar]
- Meiler J. PROSHIFT: Protein chemical shift prediction using artificial neural networks. J Biomol NMR. 2003;26:25–37. doi: 10.1023/a:1023060720156. [DOI] [PubMed] [Google Scholar]
- Morozov AV, Kortemme T, Tsemekhman K, Baker D. Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations. Proc Natl Acad Sci U S A. 2004;101:6946–6951. doi: 10.1073/pnas.0307578101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder FAA. Leucine Side-Chain Conformation and Dynamics in Proteins from C-13 NMR Chemical Shifts. Chembiochem. 2009;10:1477–1479. doi: 10.1002/cbic.200900086. [DOI] [PubMed] [Google Scholar]
- Neal S, Nip AM, Zhang HY, Wishart DS. Rapid and accurate calculation of protein H-1, C-13 and N-15 chemical shifts. J Biomol NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- Palmer AG. Probing molecular motion by NMR. Curr Opin Struct Biol. 1997;7:732–737. doi: 10.1016/s0959-440x(97)80085-1. [DOI] [PubMed] [Google Scholar]
- Ramelot TA, Ni SS, Goldsmith-Fischman S, Cort JR, Honig B, Kennedy MA. Solution structure of Vibrio cholerae protein VC0424: A variation of the ferredoxin-like fold. Protein Science. 2003;12:1556–1561. doi: 10.1110/ps.03108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez BE, Voloshin ON, Camerini-Otero RD, Bax A. Solution structure of DinI provides insight into its mode of RecA inactivation. Protein Science. 2000;9:2161–2169. doi: 10.1110/ps.9.11.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using rosetta. Meth Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- Saito H. Conformation-dependent C13 chemical shifts - A new means of conformational characterization as obtained by high resolution solid state C13 NMR. Magn Reson Chem. 1986;24:835–852. [Google Scholar]
- Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR. 2010;46:199–204. doi: 10.1007/s10858-009-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu GH, Eletsky A, Wu YB, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spera S, Bax A. Empirical correlation between protein backbone conformation and Ca and Cb 13C nuclear magnetic resonance chemical shifts. Journal of American Chemical Society. 1991;113:5490–5492. [Google Scholar]
- Vila JA, Aramini JM, Rossi P, Kuzin A, Su M, Seetharaman J, Xiao R, Tong L, Montelione GT, Scheraga HA. Quantum chemical C-13(alpha) chemical shift calculations for protein NMR structure determination, refinement, and validation. Proc Natl Acad Sci U S A. 2008;105:14389–14394. doi: 10.1073/pnas.0807105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila JA, Arnautova YA, Martin OA, Scheraga HA. Quantum-mechanics-derived C-13(alpha) chemical shift server (CheShift) for protein structure validation. Proc Natl Acad Sci U S A. 2009;106:16972–16977. doi: 10.1073/pnas.0908833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas ME, Vila JA, Scheraga HA. Effects of side-chain orientation on the C-13 chemical shifts of antiparallel beta-sheet model peptides. J Biomol NMR. 2007;37:137–146. doi: 10.1007/s10858-006-9118-6. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Jardetzky O. Predicting N-15 chemical shifts in proteins using the preceding residue-specific individual shielding surfaces from phi, psi(i-1), and chi(1) torsion angles. J Biomol NMR. 2004;28:327–340. doi: 10.1023/B:JNMR.0000015397.82032.2a. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G. CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36:496–502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Xu XP, Case DA. Automated prediction of N-15, C-13(alpha), C-13(beta) and C-13 ‘ chemical shifts in proteins using a density functional database. J Biomol NMR. 2001;21:321–333. doi: 10.1023/a:1013324104681. [DOI] [PubMed] [Google Scholar]
- Yang DW, Mittermaier A, Mok YK, Kay LE. A study of protein side-chain dynamics from new H-2 auto-correlation and C-13 cross-correlation NMR experiments: Application to the N-terminal SH3 domain from drk. J Mol Biol. 1998;276:939–954. doi: 10.1006/jmbi.1997.1588. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Brüschweiler R. Contact model for the prediction of NMR N-H order parameters in globular proteins. J Am Chem Soc. 2002;124:12654–12655. doi: 10.1021/ja027847a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.