Abstract
Nerve growth factor (NGF) signaling begins at the nerve terminal, where it binds and activates membrane receptors and subsequently carries the cell-survival signal to the cell body through the axon. A recent study revealed that the majority of endosomes contain a single NGF molecule, which makes single molecule imaging an essential tool for NGF studies. Despite being an increasingly popular technique, single molecule imaging in live cells is often limited by background fluorescence. Here, we employed a microfluidic culture platform to achieve background reduction for single molecule imaging in live neurons. Microfluidic devices guide the growth of neurons and allow separately-controlled microenvironment for cell bodies or axon termini. Designs of microfluidic devices were optimized and a three-compartment device successfully achieved direct observation of axonal transport of single NGF when quantum dot labeled NGF (Qdot-NGF) was applied only to the distal-axon compartment while imaging was carried out exclusively in the cell-body compartment. Qdot-NGF was shown to move exclusively toward the cell body with a characteristic stop-and-go pattern of movements. Measurements at various temperatures show that the rate of NGF retrograde transport decreased exponentially over the range of 36–14°C. A 10°C decrease in temperature resulted in a threefold decrease in the rate of NGF retrograde transport. Our successful measurements of NGF transport suggest that the microfluidic device can serve as a unique platform for single molecule imaging of molecular processes in neurons.
Introduction
Neurons are highly polarized cells with long axons extending out 1000 times the length of their cell bodies 1, 2. Such long axons preclude effective diffusion of soma-produced proteins for cell growth and maintenance given there are minimal or no ribosomes in axons to synthesize proteins. As a result, neurons rely on active axonal transport for all the materials needed at synaptic terminals, including neurotransmitter receptors, synaptic proteins, ion channels, lipids, and mitochondria 3. In addition, neurotrophic signals (e.g. NGF) derived from the target cells at the distal axon sites, which are essential for the survival and maintenance of neurons, are carried retrogradely by axonal transport to the cell body 4–6. Disruption of axonal transport often precedes the death of the cell body and is linked to many neurodegenerative diseases 7. Therefore, it is of immense interest to understand axonal transport in great detail.
NGF is produced and released by target tissues to activate membrane receptors on the distal axons of innervating neurons. Following the binding of NGF to its TrkA receptor at the axon terminal, the ligand-receptor complex is endocytosed and trafficked to specialized endosomes 4, 8. These endosomes carrying the NGF/TrkA complex together with an array of other signaling molecules are retrogradely transported to the cell body via a microtubule-based transport mechanism 9,10. Our recent report 11 of tracking NGF transport in axons revealed that the majority of endosomes contain single NGF dimer molecule and their movements are intrinsically heterogeneous. Large variations in transport patterns among NGF-containing endosomes call for quantitative characterization of individual behavior, obscured by ensemble averaging. Our initial success of single-molecule observation of fluorescently labeled endosome relied on the use of compartmentalized Campenot chambers, that allow localized application of NGF to axonal termini 12, 13, to spatially filter out the strong background arising from the fluorescence molecules diffusing in solution or binding at the surface of axons. While the Campenot chamber effectively separates distal axons from the cell body, it is technically challenging to combine with single molecule imaging due to a number of reasons. First, the vacuum grease barrier gives rise to huge fluorescence background, preventing any single molecule detection in its vicinity. Second, the soft grease barrier is vulnerable to any mechanical disturbances when transporting the culture from the CO2 incubator to the microscope stage. Third, it takes about two weeks for axons to penetrate through two grease barriers into the distal-axon compartment, by which time the axons are highly bundled in the cell-body compartment and difficult to image individually.
Microfluidics is becoming an increasingly useful tool for molecular and cell biology owing to its ability to manipulate small scale of liquid and precisely control the chemical environment. Previous studies have demonstrated various capabilities of microfluidic devices for biological applications such as on-chip protein/DNA purification 14, 15, crystallization 16, 17, PCR 18, intracellular enzyme reaction and cell manipulation 19, 20. Mammalian cells, such as human KB cancer cells, Jurkat T-cells, HL-60 cells, and cardiac muscle cells, have been cultured and studied inside microfluidic devices 21. Model organism such as Caenorhabditis elegans have also been studied using microfluidics 22. In 2003, Taylor et al 23 reported culturing primary cortical neurons inside microfluidic chambers fabricated with poly(dimethylsiloxane) (PDMS). In their initial 23 and following studies 24–26, Taylor and coworkers have demonstrated that microfluidic chambers composed of two compartments interconnected by microchannels can be used to separate axon termini from their cell bodies and to selectively apply chemicals to axons or cell bodies. Besides being optically transparent in the visible range, PDMS also forms water-tight seal to the glass substrate, which tolerates normal handling process during live-cell imaging 27–29.
In this paper, we used a three-compartment microfluidic chamber for DRG neuronal culture, with which we successfully overcame the abovementioned challenges for NGF tracking using Campenot chamber. Endosomes containing quantum dot labeled NGF were observed to move retrogradely along the axon toward the cell body with a characteristic stop-and-go moving pattern. Their individual transport behaviors vary widely concerning the moving speed and the pausing duration, even for endosomes in the same axon. Statistical analysis of the average transport rate over 250 endosomes showed that NGF transport speed slowed down exponentially over the temperature range of 36–14 °C. Within this temperature range, the Q10 is 3.0 and an Arrhenius plot of the natural logarithm of velocity versus the reciprocal of absolute temperature yielded an apparent activation energy of 22.6 kcal mol−1. Our results demonstrate that microfluidic culture platform offers unique advantages in studying axonal transport at single molecular level.
Results
DRG culture in microfluidic devices
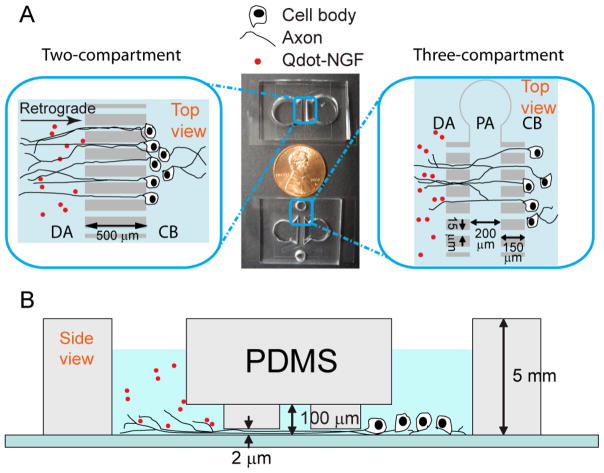
DRG neurons and their axons do not adhere to the substrate as tightly as other types of neurons, such as hippocampal or cortical neurons, making them more susceptible to fluid flow in their microenvironments. The previous reported design (23, 24, 25) for culturing cortical and hippocampal neurons, in which neuron cell bodies and axon termini were cultured in microchannels, often elicited extensive axon-bundling for DRG neurons along the wall perpendicular to the microchannels (supplement figure). We suspect that bundling of axons along the wall is due to the fluid flow along the microchannel, in which small temperature fluctuation or normal handling can exert transient flow with considerable speed. Therefore, to minimize the fluid flow, we modified previous designs so that cell bodies and axon termini were cultured in open wells. Culture wells containing either cell bodies or axon termini were interconnected by one or two columns of microchannels (Figure 1). The height of the microchannels (~2 μm) is designed to be much smaller than the size of the cell body so that neurons cannot cross from the cell-body compartment to the adjacent compartment during the plating process. After embryonic DRG neurons were plated to the right-most compartment, cells extend out long axons (~1 μm diameter) that are smaller than the microchannels and grow across into the distal-axon compartment.
FIGURE 1.
Design of microfluidic devices for DRG neural culture. A) Top view of the two- and three-compartment microfluidic devices. Neurons are initially plated in the cell-body (CB) compartment and their axons grow across the channels into the distal-axon (DA) compartment after a few days. In three-compartment design, a proximal-axon (PA) compartment can be independently accessed through two side holes. Compartmentalized neuronal cultures make it possible to study cell responses to various stimuli in a controlled manner (see main text) B) The side view of the three-compartment device. The height of the microchannel is 2 μm, which prevents cell bodies from crossing into the neighboring compartment and helps flatten axons for ease of imaging.
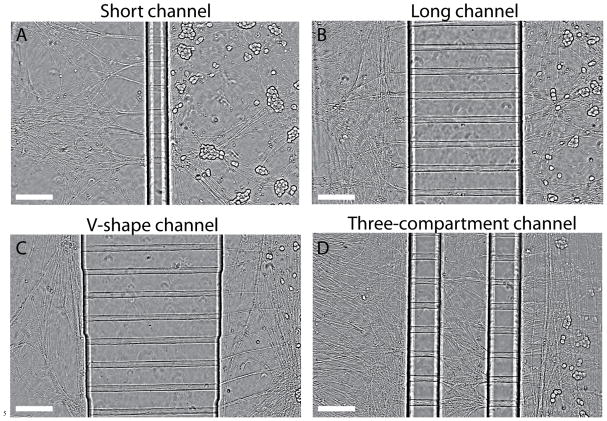
Four different geometries of microfluidic devices that are able to independently control the chemical environment for distal axons and cell bodies were tested for DRG culture. Three of the devices are composed of two fluid compartments separated by: a column of “short” channels 150 μm in length (Figure 2A); a column of “long” channels 500 μm in length (Figure 2B); or a column of varying size (100–1000 μm) channels (Figure 2C). The fourth device is composed of three fluidic compartments, each separated by a column of “short” channels (Figure 2D). The middle compartment of the three-compartment device is about 200 μm wide. The reason for testing different geometries was to optimize the design for both neuronal survival and compatibility with the single molecule imaging. We found that different geometries did not significantly differ in the survival rate or growth of axons through the channels. It is the design of the three-compartment channel, not the absolute length of channel making it best suited for single molecule imaging as discussed in the next section. Figure 2 shows neuronal cultures in four different microfluidic devices 5 days after plating. For all types of microfluidic chambers, axons grow across one or two columns of microchannels in 3–5 days into the distal-axon compartment. Elongation of axons and migration of cell bodies over a period of 2–3 weeks indicate a healthy culture after cell plating. (Figure S1).
FIGURE 2.
Phase contrast images of DRG neurons in four different types of microfluidic devices. A–D) show images of 5-day DRG cultures inside microfluidic devices with short, long, V-shape, and three-compartment channels. Axons grew across one- or two-columns of microchannels into the distal-axon compartment 3 days after plating. At bulk level, fluid isolation between cell-body compartment and distal axon apartment is attained for all four types of devices. However, fluid isolation at single molecule level can only be achieved using the three-compartment microfluidic device shown in (D). Scale bars on each panel represent 200 μm.
Combining microfluidic cell culturing with single molecule imaging
The fact that most endosomes contain a single NGF dimmer 11 makes single-molecule imaging an essential tool in studying NGF retrograde transport. Reduction of the background fluorescence is crucial in order to achieve single molecule imaging in live cells. Background fluorescence comes from three sources (a) weakly autofluorescent molecules in the cytosol such as NADH and riboflavins; (b) out-of-focus diffusive fluorophores in solution; and (c) non-specific binding of fluorophores to glass or axon surfaces. To solve (a) we conjugate NGF on the surface of quantum dots because their photophysical properties, including brightness and photostability, are far superior to those of organic dyes. The critical reduction of out-of-focus background (source b) was achieved by limiting the penetration depth of the excitation light using a pseudo-TIRF microscope. The pseudo-TIRF set-up (see Figure 4A and the method section for details) resulted in background fluorescence similar to a TIRF microscope while allowing visualization of Qdot-NGF in cell bodies and axons located hundreds of nanometers to a few micrometers away from glass surface, areas generally beyond the illumination depth of TIRF microscopy. In addition to the background fluorescence, challenges for single molecule imaging of NGF transport also come from the signal contamination from those Qdot-NGF molecules that merely bind at the axon surface without being internalized or transported (source c). Surface-binding NGF accounts for more than 80% of total NGF molecules applied to cells 30. Therefore the contamination signal from surface-bound Qdot-NGF often buries the signal of interest, Qdot-NGF molecules that are internalized and transported. The microfluidic chamber design tackles this problem effectively by allowing application of Qdot-NGF only to the distal-axon compartment and imaging exclusively in the cell-body compartment.
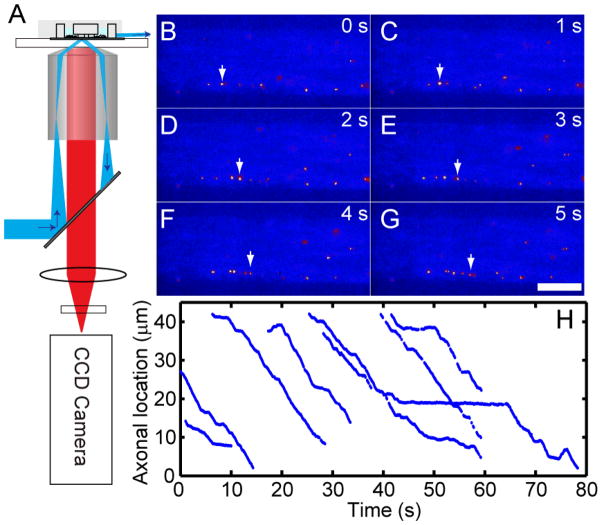
FIGURE 4.
Single molecule imaging of Qdot-NGF axonal transport using pseudo-TIRF microscope and the microfluidic device. A) Illustration of the pseudo-TIRF microscope. Excitation laser light is focused at the back focal plane of the oil-immersion objective. The incident angle at the glass-water interface is controlled to be slightly smaller than the critical angle so that the refracted laser beam could penetrate ~1 μm into the aqueous solution. DRG neurons cultured in a three-compartment microfluidic device were supplied with Qdot-NGFs to the distal-axon compartment. Time-lapse fluorescence images were collected at the microchannels that connect the cell-body compartment and the middle compartment. B–G) illustrate retrograde transport of Qdot-NGF labeled endosomes in axons. Several endosomes containing Qdot-NGFs can be seen within the field of view (bright yellow dots, false color). White arrow in each frame marks the location of the same Qdot-NGF at different time points within the same field of view. Scale bar is 10 μm. H) Representative trajectories of 8 endosomes traveling within the same axon, displaying a typical stop-and-go moving pattern.
The microfluidic device enables fluidic isolation among interconnected compartments except through microchannels, as shown in Figure 1. In order to monitor axonal transport of Qdot-NGF at single molecule level, it is important to avoid any diffusion of free Qdot-NGF into the cell-body compartment through microchannels. For this purpose, the liquid volume in the cell-body chamber is kept slightly larger than that in the distal axon chamber (120 μL vs. 90 μL). The volume difference generates a positive pressure which creates fluidic flow working against diffusion from the distal-axon to the cell-body chamber 23, 31. When measuring the average fluorescence intensity, we detected no leaking into the cell-body compartment 12 h after 1-μM calcein dye was added to the distal-axon compartment, in agreement with previous observations.
However, at single molecule level, diffusive Qdot-NGFs were readily detected in the cell-body compartment within an hour for the two-compartment microfluidic chambers. This single-molecule level fluid mixing between two neighboring compartments is likely due to minuscule fluid flow induced by temperature fluctuations or mechanical disturbances caused by translating the microscope stage to find another field of view. On the other hand, we detect no diffusive Qdot-NGF in the cell-body compartment for the three-compartment microfluidic device 12 h after adding Qdot-NGF to the distal-axon chamber (Figure 3). For the three-compartment geometry, minuscule amount of Qdot-NGF from the distal-axon compartment may enter the middle compartment, where it will be diluted so much that the possibility of Qdot-NGF further reaching cell-body chamber is significantly diminished. The central compartment in the three-compartment microfluidic chamber serves as a buffer space that is an essential feature in order to achieve single molecule imaging of NGF transport in live neurons. Thus, while Qdot-NGF in the distal-axon compartment represents a mixture of surface-bound and internalized Qdot-NGF, the Qdot-NGF observed in the cell-body compartment is exclusively located inside the axon (Figure 3). In addition to its capability to isolate fluid between neighboring compartments, microchannels also limit the curvature of the axons growing in them. As a result, axons are relatively straight and flat inside the microchannels, which is particularly beneficial to single molecule imaging because endosomes remain in the focal plane as they move along the axons.
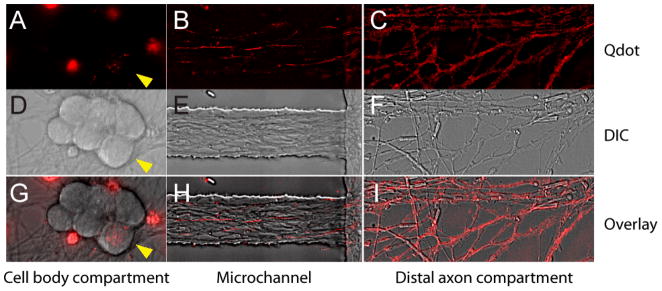
FIGURE 3.
Image of axons containing Qdot-NGF in different compartments of the microfluidic device (three-compartment design). Fluorescent Qdot-NGF (1 nM) was added to the distal axon compartment and incubated for 2hrs prior to imaging. A–C) Fluorescence images of Qdot-NGF in (A) cell-body compartment, (B) microchannel neighboring the cell body compartment and (C) distal-axon compartment. The image in (B) is the projection of 600 time-lapse frames, so that the traces of individual Qdot-NGF depict axons that they traveled along. D–F) DIC images of the same areas as (A–C). G–I) overlaid images of fluorescence and DIC images in each compartment. Note that only one DRG cell body (yellow arrow heads) out of six cell bodies contains Qdot-NGFs. This is because not all neurons were able to extend their axons into the distal axon compartment, which further confirms the liquid isolation between the cell body compartment and the distal axon compartment. The large fluorescence splotch in (A) and (G) arises from the autofluorescent cell debris.
Observing axonal transport of individual NGF-containing endosomes
Using the three-compartment microfluidic chamber, we observed axonal transport of individual NGF-containing endosomes that were internalized at the distal axon termini and retrogradely transported toward the cell bodies (supplementary movie S1). Comparing with Campenot chamber, the fluorescence signal to background ratio is significantly improved (~5 times better) that allows better detection of NGF-containing endosomes and higher spatial localization. In addition, the microfluidic chamber is easier to use, more reliable and compatible with the single molecule imaging setup. Detailed comparisons are provided in the supplementary materials (supplementary figures S3 & S4, movies S2–S5).
Figure 4B–G show time-lapsed fluorescence images of Qdot-NGF moving along axons inside a microfluidic channel. The arrows illustrate the location of the same endosome at different time frames. Photo-blinking of quantum dots indicates that the majority of endosomes contain just a single Qdot-NGF32. Endosomes containing Qdot-NGF were seen moving exclusively toward the cell body with their dynamic processes featuring active movements interrupted by frequent pauses – a characteristic stop-and-go moving pattern (Figure 4H). In most cases, multiple endosomes were moving simultaneously in the same axon (as displayed in Figure 4B–G). We observed as many as 10 endosomes passing a cross section of an axon in 1 min. Individual endosomes vary greatly in their transport rate, pausing duration and frequency. At 36 °C, the average rate of transport is 1.21 μm/s with active moving speed ranging from 0.2 μm/s to 4 μm/s and pausing duration varying from a fraction of a second to tens of seconds. Some fast moving endosomes were seen over-passing the slower moving ones, likely moving on different microtubule tracks 33. During their long journey, a small population (~3–5%) of endosomes appeared to reverse their transport direction toward the axon termini, but only for a short distance before reversing back toward the cell body direction. Real-time tracking of Qdot-NGF in axons allow us to analyze the axonal transport process in great detail.
Temperature dependence of NGF retrograde transport
Retrograde transport of NGF is powered by dynein motor proteins that hydrolyze ATP as energy source 3, 34. It is well-known that ATP hydrolysis is a temperature-dependent chemical process 35, 36, thus temperature is likely to play an important role in NGF transport. Previous experiments measuring the accumulation of radio-labeled proteins on a distal ligature had indicated a strong temperature dependence of axonal transport 35, but the exact relationship is debatable. For example, the rate of anterograde protein transport in the optic and the olfactory nerves of the garfish were observed to decrease linearly as the temperature decreased 37, while an exponential temperature dependence of the rates of anterograde protein transport in cat and rat sciatic nerve were reported 38, 39. With the capability to accurately measure the speed of individual endosomes in microfluidic chambers, we examined the temperature dependence of NGF transport in DRG neurons.
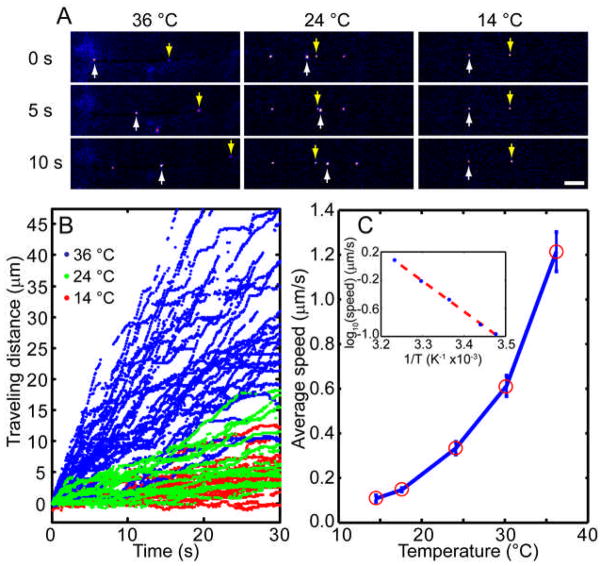
The rate of transport was measured at different temperatures from 14 °C to 36 °C. Fluorescence snapshots at 0, 5, and 10 sec of individual moving endosome at 14 °C, 24 °C, and 36 °C are shown in Figure 5A (also see supplementary movies S2–S4 at those temperatures). Locations of two representative endosomes labeled with Qdot are marked with white and yellow arrows. Speeds of endosomes within the same axons can vary drastically. For instance, center columns (T=24 °C) of Figure 5A shows that one endosome (white arrow) can catch up with and pass another one in the same axon (yellow arrow). Such endosome-to-endosome speed variations can be clearly visualized from Figure 5B, where more than 40 individual trajectories of single endosomes were collected at each specific temperature. Average traveling speeds of endosome transport were then constructed from individual trajectories. Average speed decreases over an order of magnitude from 1.2 μm/s at 36 °C to 0.1 μm/s at 14 °C. To rule out the possibility that cultures degrade at lower temperatures, we measured the axonal transport of the same neural culture at 36 °C, cooling down to 10 °C and warming up again to 36 °C. We found that the transport rate decreased significantly from 36 °C to 10 °C, but recovered completely when returned to 36 °C (supplement figure). These results not only confirmed previous finding that fast axonal transport is markedly impaired at lower temperatures and but also showed that the temperature effect is reversible.
FIGURE 5.
Temperature dependence of NGF endosomal transport. A) Snap short images show transport of Qdot-NGF in DRG axons at 36 °C, 24 °C or 14 °C. The yellow and white arrowheads point to two different Qdot-NGFs that migrated retrogradely along the axon toward the cell body. In general, higher temperature results in much faster endosome transport. Scale bar is 10 μm. B) Trajectories of Qdot-NGF axonal transport show that the moving speed varies greatly from one endosome to another at the same temperature. But the overall transport rate slows down noticeably as the temperature drops from 36 °C to 14 °C. C) The average transport speed is plotted as a function of temperature in the linear scale and in the Arrhenius plot (inset). The measured data clearly favors an exponential relation between the transport rate and the temperature (goodness of fitting r2= 0.9988).
Our measurements favor an exponential rather than a linear relation between the transport rate and the temperature (Figure 5C). The temperature dependence of the rate for NGF transport is also presented in the form of an Arrhenius plot in the insert of Figure 5C. Over this range of temperatures, 14–36 °C, a linear relationship between the natural logarithm of velocity versus the reciprocal of absolute temperature was obtained. The Q10 was approximately 3.0, and the Arrhenius activation energy calculated from the slop was 22.5±0.65 kcal mol−1. This value falls in the range of reported activation energy 15–26 kcal mol−1 for molecular motors, but is somewhat higher than the previously reported activation energy for dynein motors using radio-labeled protein assay (~15 kcal mol−1).
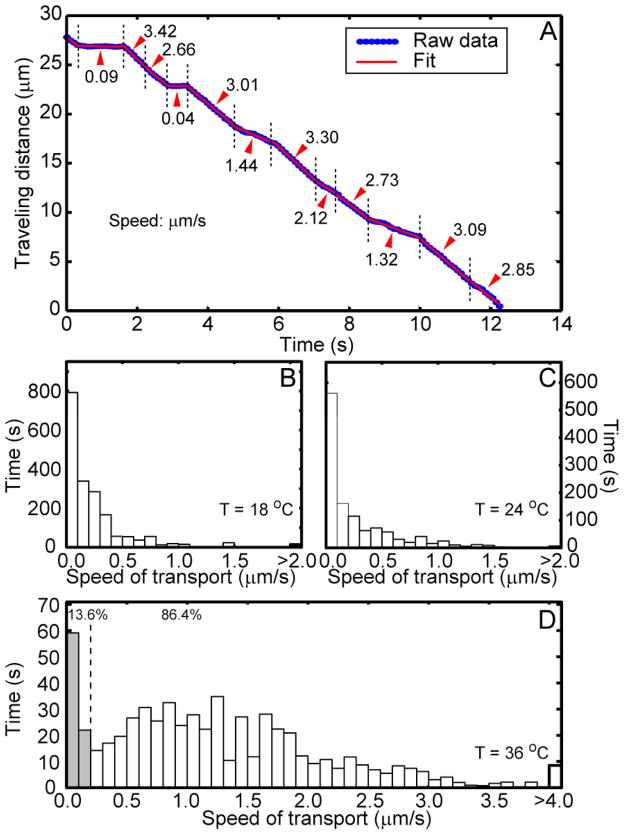
The displacement vs. time trajectories often display abrupt changes in velocity, therefore each trajectory was parsed into a series of connected straight-line segments (Figure 6A). The slope of each segment is the instantaneous velocity at that time. A histogram analysis of accumulated time duration at certain speed of transport was displayed in Figure 6B–D. At 36 °C, the distribution show a clear “pausing” phase (< 0.15 μm/s) and a broadly distributed moving phase peaked around 1.3 μm/s while tailing all the way to 4μm/s. As temperature is lowered to 24 °C and 18 °C, the moving speed profile shifted toward the lower vale with any speed faster than 1.5 μm/s rarely observed. The considerable decrease of NGF transport speed from 36 °C to 14 °C is consistent with the molecular motor process in which ATP turn over is the rate-limiting step 40, considering microtubules are stable over this temperature range. Disintegration of microtubules at low temperatures 41, 42, however, could be responsible for the complete stopping of transport at temperatures below 10 °C (data not shown).
FIGURE 6.
A) A representative time trajectory of Qdot-NGF containing endosome. Notice that speed of the endosomal transport spans over a large range (0.04–3.30 μm/s). An automatic algorithm was used to identify sections with different speeds and their time durations. Histograms of the accumulated traveling time vs. transport speed were plotted in B) 18 °C, C) 24 °C and D) 36 °C. At 36 °C, the percentage of time that endosomes stay “static” (< 0.2 μm/s, gray bars) and “mobile” (> 0.2 μm/s, white bars) has also been highlighted.
Discussion
Monitoring the transport of single molecules within DRG neurons requires both successful cell culturing and delicate imaging strategies. The most important factor in culturing DRG neurons is the surface property of the microfluidic device. At micron scale, the surface-to-volume ratio is so large that it results in drastic consequences for the viability of a neuronal culture. Freshly prepared PDMS surface is highly hydrophobic with a water contact angle of 110° 43, which does not support cell attachment. Furthermore, freshly prepared PDMS chips reduce surface hydrophilicity of the glass substrate over time when two surfaces are less than 100 μm apart. This is likely due to leaking and diffusing of un-crosslinked monomer or oligomers from the PDMS to the glass substrate, which not only deteriorates cell attachment to the PLL-coated glass surface but also intoxicates cells. As a result, microfluidic chambers assembled from freshly prepared PDMS chip resulted in poor neuronal survival rates. To address this problem, we first increased PDMS polymerization reaction to a higher degree by baking at 70 °C for 24 h. Then, PDMS chips were soaked in soap water for 12 h followed by ethanol for 12 h to remove the remaining uncrosslinked monomers. Finally, PDMS chips were sterilized by dry-autoclaving at 120 °C for 30 min. The autoclaving process is repeated three times to further increase the polymer crosslinking and to completely expel any remaining diffusive monomers and organic solvent molecules trapped inside the crosslinked PDMS. The baking-soaking-autoclaving surface treatment significantly improved the viability of DRG neurons inside the microfluidic chamber and cell survival rate is comparable to that of mass culture.
Materials and Methods
Chemicals and reagents
PDMS composed of silicone elastomer and curing agent (Sylgard 184 elastomer kit, Dow Corning, Corning, NY) was purchased from Fisher Scientific (Pittsburgh, PA). Poly-L-Lysine (PLL, molecular weight 70,000–150,000), boric acid, sodium tetraborate, methylcellulose and trichloromethyl silane (TCMS) was purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). Phosphate-buffered saline (PBS), Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), and other cell culture reagents were purchased from Mediatech Inc. (Herndon, VA). CO2 independent medium and streptavidin-coated quantum dot (multiple color sampler kit) was purchased from Invitrogen (Carlsbad, CA). Negative photoresist SU8-2 and SU8-50 were purchased from Microchem (Newton, MA).
PDMS microfluidic device fabrication
The microfluidic device for DRG neuron culture was fabricated using soft-lithography 23, 44. The master mold is a silicon wafer with two photoresist patterns of different height (2 μm and 100 μm). A 4-inch silicon wafer was first cleaned in piranha solution (3:1 concentrated sulfuric acid to 30% hydrogen peroxide solution), thoroughly rinsed in DI water and dried at 200 °C for 10 min. The first layer of photoresist SU8-2 was spin coated (Headway spin coater, Headway Research Inc., Garland, TX) onto the silicon wafer substrate at 2000 rpm for 60 s to create ~2.5-μm thick layer. This layer was patterned by exposing to 365-nm UV light through a high-resolution transparency mask. A subsequent development step dissolves the photoresist in the unexposed area. The second photoresist, SU8-50, was spun on top of the first layer for 60 s at 1000 rpm to obtain a thickness of ~100 μm. This second layer was aligned with the first layer and exposed to UV light through another patterned transparency mask. After the second photoresist layer was fully developed, the master mold was annealed at 150 °C for 2 h to facilitate attachment to the silicon substrate. All the procedures to make master wafer were carried out at the Stanford Nanofabrication Facility Center.
PDMS microfluidic chips were fabricated by replica-molding - curing a liquid mixture of degassed silicone elastomer and curing agent (weight ratio 10:1) on the master wafer patterned with SU8 photoresist. Prior to addition of prepolymers, the wafer was treated with TCMS vapor for 1 min to facilitate removal of PDMS from the master mold after curing. After curing at 70 °C for 2 h, the PDMS layer was peeled off from the master mold and cut into chips ~25 × 15 mm2. Two ~7 mm diameter wells were punched into the PDMS chips and the PDMS chips were then fully cured by placing in a 70 °C oven for 24 h. PDMS surfaces were further washed with detergent, water, ethanol, and sterilized by autoclaving. Glass coverslips (24 × 40 mm2, No. 1.5 thickness, VWR, West Chester, PA) were cleaned by sonicating in detergent, DI water, and ethanol (30 min for each step). After cleaning, glass coverslips were coated with 1 mg/ml PLL, washed twice in superQ water, and dried in sterilized hood. PDMS microfluidic devices were finished by pressing the PDMS chips onto the PLL coated coverslips.
Plating and culturing of dorsal root ganglia neurons
Primary embryonic rat dorsal root ganglia neurons were isolated from Sprague-Dawley fetal rats (age E15-E16) as described previously 45. Briefly, dorsal ganglia of rat embryos were dissected in Hank’s buffer solution and enzymatically treated in 0.25% trypsin at 37 °C temperature for 30 min followed by mechanical dissociation by passing through a fire-polished Pasteur pipette for less than 10 times. DMEM containing 10% fetal calf serum was used to stop the trypsinization process and cells were spun down at the bottom of the collecting tube. Dissociated DRG neurons were resuspended, counted and plated at a density of ~15,000 cells per microfluidic chamber. Cultures were maintained in neurobasal medium supplemented with B27, 2 mM L-glutamine/GlutaMAX, 0.5% cellulose, and 50 ng/ml NGF. All cultures were housed in a humidified incubator at 37 °C supplied with 5% CO2 and DRG neurons were selected by applying 4 μM cytosine arabinoside (1-β-D-arabinofuranosylcytosine) for 24 h to cultures 2 days after plating cells. This method produces neuronal cultures that are free of non-neuronal cells. NGF was removed from the culture medium 5 h before addition of Qdot-NGF. NGF was purified from mouse submaxillary glands, biotinylated and coupled to Qdot via biotin-streptavidin interaction according to published protocols 46.
Pseudotota l-internal-reflection-fluorescence microscopy
An inverted Nikon microscope (TE300) was modified for pseudo-TIRF illumination. The 532-nm laser beam (Melles Griot, Carlsbad, CA) was first expanded to 3 cm and then focused at the back focal plane of the objective lens (Plan Apo 60×, NA 1.45, Olympus, Center Valley, PA). The focal point was moved off-axis so that the light underwent total internal reflection at the glass-water interface. The angle of refraction was then carefully tuned by moving the focusing lens that was mounted on a translation stage. The incident angle was adjusted to be slightly smaller than the critical angle so that the laser beam could penetrate ~1 μm into the aqueous solution (compared to ~200 nm in true TIRFM). Three hours prior to imaging, the distal-axon compartment of the microfluidic chamber was supplied with 1 nM Qdot-NGF. Immediately before imaging, the microfluidic chamber was transferred from the incubator to a customized microscope stage with controlled temperature between 10–40 °C. Then, free Qdot-NGF in solution was washed off and the culture medium was replaced by CO2 independent medium to minimize pH change induced by CO2 withdrawal. Fluorescence emission from Qdot605 was collected by the objective lens, transmitted through the 550DCLP dichroic mirror, filtered with a Qdot605/20 emission filter (Chroma Technology, Rockingham, VT) and focused onto a sensitive EMCCD CCD camera (Cascade 512B, Photometric, Tucson, AZ). Time-lapsed images were collected at a speed of 10 frames/s. A home-made temperature control system was used to regulate the temperature of both the microscope stage and the objective. Actual temperature at the sample was measured using a thermister.
Imaging and data analysis
Endosomes containing single internalized Qdot-NGF molecules being transported through the axons were imaged. On average, the fluorescence signal from a single Qdot-NGF spans an area of 4 × 4 pixels on the digitized image. After background subtraction, the endosomes were identified as the local maxima with a signal-to-noise ratio larger than 3. Spatial distribution of the fluorescence intensity of individual endosomes was fitted to a two-dimensional Gaussian function. The fitted center, corresponding to the position of the endosome, was located with a precision of 10 nm. After measuring the position of endosomes at consecutively timed images, the trajectories were constructed by linking the time-resolved endosome positions. The average speed was determined by measuring the distance the endosome traveled over the observation time. The fluorescence intensity of each endosome was measured by integrating the total fluorescence over the 4 × 4 pixel area centered on the endosome.
Conclusion remarks
The combination of microfluidic culture chamber with single molecule imaging techniques makes a powerful tool for axonal transport studies. By applying fluorescent probes to cell body compartment or the axon termini compartment, one can selectively probe retrograde or anterograde transport process. Ultimate detection sensitivity at single molecular level makes it possible to study low abundance molecules that have important biological functions in neurons.
Supplementary Material
Additional Supporting Information may be found in the online version of this article:
Figure S1: Monitoring the growth of DRG in a microfluidic chamber for two weeks.
Figure S2: Comparing the DRG growth in the two-well design and the previous reported four-well design.
Movie S1: Endosome transporting within microfluidic channels.
Movie S2: Transport of endosome at 36°C.
Movie S3: Transport of endosome at 24°C.
Movie S4: Transport of endosome at 14°C.
Acknowledgments
We thank Drs. Chengbiao Wu, Wei Wang, and Janice S. Valletta for their assistance in carrying out the experiments and helpful discussions. BC wishes to express her gratitude to Prof. William W. Mobley and Prof. Yanmin Yang for their generosity in sharing resources and knowledge that significantly accelerated the research. This work was supported by National Institute of Health (NIH) grant NS057906, Bio-X interdisciplinary initiatives program and Dreyfus new faculty award to BC.
References
- 1.Craig AM, Banker G. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- 2.Witte H, Bradke F. Curr Opin Neurobiol. 2008;18:479–487. doi: 10.1016/j.conb.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Holzbaur EL. Trends Cell Biol. 2004;14:233–240. doi: 10.1016/j.tcb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Beattie EC, Zhou J, Grimes ML, Bunnett NW, Howe CL, Mobley WC. Cold Spring Harb Symp Quant Biol. 1996;61:389–406. [PubMed] [Google Scholar]
- 5.Campenot RB, MacInnis BL. J Neurobiol. 2004;58:217–229. doi: 10.1002/neu.10322. [DOI] [PubMed] [Google Scholar]
- 6.Zweifel LS, Kuruvilla R, Ginty DD. Nat Rev Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
- 7.Coleman M. Nat Rev Neurosci. 2005;6:889–898. doi: 10.1038/nrn1788. [DOI] [PubMed] [Google Scholar]
- 8.Grimes ML, Beattie E, Mobley WC. Proc Natl Acad Sci U S A. 1997;94:9909–9914. doi: 10.1073/pnas.94.18.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsui-Pierchala BA, Ginty DD. J Neurosci. 1999;19:8207–8218. doi: 10.1523/JNEUROSCI.19-19-08207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riccio A, Pierchala BA, Ciarallo CL, Ginty DD. Science. 1997;277:1097–1100. doi: 10.1126/science.277.5329.1097. [DOI] [PubMed] [Google Scholar]
- 11.Cui BX, Wu CB, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S. Proc Natl Acad Sci U S A. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campenot RB. Proc Natl Acad Sci U S A. 1977;74:4516–4519. doi: 10.1073/pnas.74.10.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campenot RB. Brain Res. 1987;465:293–301. doi: 10.1016/0165-3806(87)90250-1. [DOI] [PubMed] [Google Scholar]
- 14.Yao S, Anex DS, Caldwell WB, Arnold DW, Smith KB, Schultz PG. Proc Natl Acad Sci U S A. 1999;96:5372–5377. doi: 10.1073/pnas.96.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Effenhauser CS, Bruin GJM, Paulus A, Ehrat M. Anal Chem. 1997;69:3451–3457. doi: 10.1021/ac9703919. [DOI] [PubMed] [Google Scholar]
- 16.Hansen CL, Skordalakes E, Berger JM, Quake SR. Proc Natl Acad Sci U S A. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen CL, Classen S, Berger JM, Quake SR. J Am Chem Soc. 2006;128:3142–3143. doi: 10.1021/ja0576637. [DOI] [PubMed] [Google Scholar]
- 18.Fang TH, Ramalingam N, Xian-Dui D, Ngin TS, Xianting Z, Lai Kuan AT, Peng Huat EY, Hai-Qing G. Biosens Bioelectron. 2009;24:2131–2136. doi: 10.1016/j.bios.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Rhee SW, Taylor AM, Tu CH, Cribbs DH, Cotman CW, Jeon NL. Lab Chip. 2005;5:102–107. doi: 10.1039/b403091e. [DOI] [PubMed] [Google Scholar]
- 20.McWhirter JL, Noguchi H, Gompper G. Proc Natl Acad Sci U S A. 2009;106:6039–6043. doi: 10.1073/pnas.0811484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millet LJ, Stewart ME, Sweedler JV, Nuzzo RG, Gillette MU. Lab Chip. 2007;7:987–994. doi: 10.1039/b705266a. [DOI] [PubMed] [Google Scholar]
- 22.Chronis N, Zimmer M, Bargmann CI. Nat Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- 23.Taylor AM, Rhee SW, Tu CH, Cribbs DH, Cotman CW, Jeon NL. Langmuir. 2003;19:1551–1556. doi: 10.1021/la026417v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. Nat Protoc. 2006;1:2128–2136. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 25.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor AM, Rhee SW, Jeon NL. Methods Mol Biol. 2006;321:167–177. doi: 10.1385/1-59259-997-4:167. [DOI] [PubMed] [Google Scholar]
- 27.Sia SK, Whitesides GM. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 28.McDonald JC, Whitesides GM. Acc Chem Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 29.Merkel TC, Bondar VI, Nagai K, Freeman BD, Pinnau I. J Polym Sci Part B: Polym Phys. 2000;38:415–434. [Google Scholar]
- 30.Ure DR, Campenot RB. J Neurosci. 1997;17:1282–1290. doi: 10.1523/JNEUROSCI.17-04-01282.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooksey GA, Sip CG, Folch A. Lab Chip. 2009;9:417–426. doi: 10.1039/b806803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S. Proc Natl Acad Sci U S A. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mudrakola HV, Zhang K, Cui B. Structure. 2009;17:1433–1441. doi: 10.1016/j.str.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vale RD, Malik F, Brown D. J Cell Biol. 1992;119:1589–1596. doi: 10.1083/jcb.119.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brimijoin S, Olsen J, Rosenson R. J Physiol. 1979;287:303–314. doi: 10.1113/jphysiol.1979.sp012660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cosens B, Thacker D, Brimijoin S. J Neurobiol. 1976;7:339–354. doi: 10.1002/neu.480070406. [DOI] [PubMed] [Google Scholar]
- 37.Gross GW. Brain Res. 1973;56:359–363. doi: 10.1016/0006-8993(73)90353-3. [DOI] [PubMed] [Google Scholar]
- 38.Ochs S, Smith C. J Neurobiol. 1975;6:85–102. doi: 10.1002/neu.480060112. [DOI] [PubMed] [Google Scholar]
- 39.Bisby MA, Jones DL. Exp Neurol. 1978;61:74–83. doi: 10.1016/0014-4886(78)90182-6. [DOI] [PubMed] [Google Scholar]
- 40.Kawaguchi K, Ishiwata S. Biochem Biophys Res Commun. 2000;272:895–899. doi: 10.1006/bbrc.2000.2856. [DOI] [PubMed] [Google Scholar]
- 41.Tilney LG, Porter KR. J Cell Biol. 1967;34:327–343. doi: 10.1083/jcb.34.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Echandia EL, Piezzi RS. J Cell Biol. 1968;39:491–497. doi: 10.1083/jcb.39.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaudhury MK, Whitesides GM. Langmuir. 1991;7:1013–1102. [Google Scholar]
- 44.Quake SR, Scherer A. Science. 2000;290:1536–1540. doi: 10.1126/science.290.5496.1536. [DOI] [PubMed] [Google Scholar]
- 45.Chan JR, Rodriguez-Waitkus PM, Ng BK, Liang P, Glaser M. Mol Biol Cell. 2000;11:2283–2295. doi: 10.1091/mbc.11.7.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pathak S, Cao E, Davidson MC, Jin S, Silva GA. J Neurosci. 2006;26:1893–1895. doi: 10.1523/JNEUROSCI.3847-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article:
Figure S1: Monitoring the growth of DRG in a microfluidic chamber for two weeks.
Figure S2: Comparing the DRG growth in the two-well design and the previous reported four-well design.
Movie S1: Endosome transporting within microfluidic channels.
Movie S2: Transport of endosome at 36°C.
Movie S3: Transport of endosome at 24°C.
Movie S4: Transport of endosome at 14°C.