Abstract
With the increasing role of computational tools in the analysis of sequenced genomes, there is an urgent need to maintain high accuracy of functional annotations. Misannotations can be easily generated and propagated through databases by functional transfer based on sequence homology. We developed and optimized an automatic policing method to detect biochemical misannotations using context genomic correlations. The method works by finding genes with unusually weak genomic correlations in their assigned network positions. We demonstrate the accuracy of the method using a cross-validated approach. In addition, we show that the method identifies a significant number of potential misannotations in B. subtilis, including metabolic assignments already shown to be incorrect experimentally. The experimental analysis of the mispredicted genes forming the leucine degradation pathway in B. subtilis demonstrates that computational policing tools can generate important biological hypotheses.
Introduction
As genomic and proteomic databases continue to expand at an accelerating rate, the challenge to accurately annotate gene functions grows in scale and importance. Homology-based methods are now routinely used to annotate protein function in sequenced genomes1–3. Unfortunately, homology methods generate a large number of misannotations due to a relatively high sequence identity (>40%–60%) required for an accurate functional transfer. Sequence-based misannotations can also quickly spread through functional databases based on homology to misannotated genes4–6.
Several ontology-based algorithms have been previously developed to detect potential misannotations. The system Xanthippe7 was used to detect inconsistencies between functional keywords and annotated protein domains. Errors in protein motif (PROSITE patterns) assignments8 were identified by comparing Gene Ontology (GO9) and Swiss-Prot10 annotations. Ambiguous and incomplete EC numbers were identified and shown to result in erroneous functional assignments11.
Context genomic correlations such as chromosomal gene clustering12–14, phylogenetic profiles15, 16, and gene fusion17–19 can provide functional clues even if sequence homology information is remote or absent. We rationalized that the context-based correlations can be used not only to predict gene function, but also to efficiently detect inaccurate annotations. In this paper we develop such a method and demonstrate its ability to identify suspicious functional assignments. In contrast to aforementioned methods, our approach is not based on inconsistencies between several annotations, but rather between annotations and multiple genomic correlations. Therefore, the developed method is able to automatically detect incorrect functional assignments even if only a single annotation is available, or if annotations from several sources are in agreement. Our method can be also used to select the correct assignment among conflicting annotations. In the paper we first demonstrate the power of the method using artificial errors generated in silico, and then apply the algorithm to detect misannotations in the B. subtilis metabolic network.
Results
Strategy of the computational approach
The algorithm presented in this study identifies genes that have either unusually poor genomic correlations with their network neighbors, or alternative network locations with significantly better correlations. The problems of assigning the correct function and identification of misannotations have different objectives and require different algorithms. In many cases, it is possible to reject an existing annotation based on poor genomic correlations, while these correlations are not strong enough, or unique, to accurately predict the correct function.
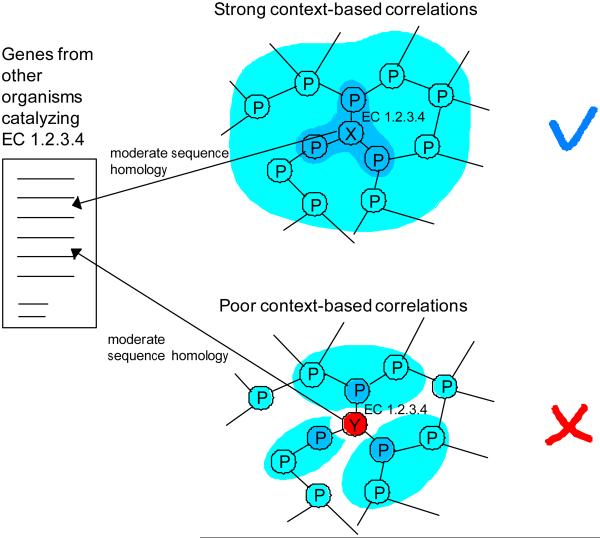
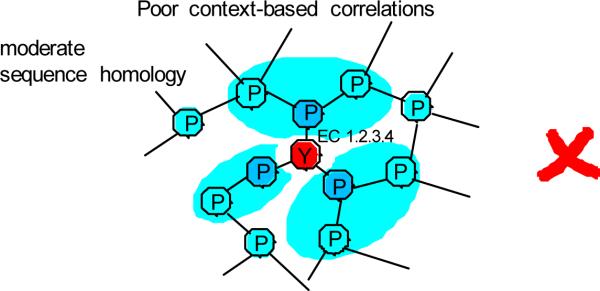
Similar to our previous studies20–22, we represent the metabolic network as a graph with nodes being metabolic genes and edges being connections established by shared metabolites (see Methods). Suppose two genes X and Y in different organisms are annotated to catalyze the metabolic activity specified by the Enzyme Commission (EC) number 1.2.3.4 (Figure 1). The developed approach will suggest that the annotation of the gene X is likely to be correct due to strong context-based correlations with neighboring genes. On the other hand, the gene Y displays poor genomic correlations to its network neighbors, and its annotation is likely to be an error.
Figure 1.
Illustration of the developed approach. In the figure network nodes represent metabolic genes and edges represent connections established by shared metabolites. Using sequence homology, genes X and Y from different organisms have been assigned to EC 1.2.3.4. Gene X displays strong context-based correlations (darker blue indicating stronger correlations) with neighboring network genes. Consequently, the annotation of X is likely to be correct. In contrast, gene Y does not fit well in the assigned network position and is likely to be misannotated.
To predict potential misannotations we integrated sequence and context correlations using the AdaBoost algorithm with alternating decision trees23, 24. AdaBoost has been successfully applied to several large-scale integration problems in biology, including prediction of gene regulatory response25 and identification of genes responsible for orphan metabolic activities26. The AdaBoost algorithm was trained with a collection of context genomic descriptors: phylogenetic profiles, mRNA co-expression, chromosomal distance between genes, gene clustering across genomes, and protein fusion. For each descriptor, we considered two different scores: the largest pair-wise correlation between the target gene and its direct network neighbors and the average fitness score in the assigned network location calculated as described in Methods.
The average fitness score quantifies the overall context correlations of the target gene with all its network neighbors20, 22. To represent the relative fitness of the existing annotation the AdaBoost score for the best alternative location was also supplied to the algorithm. The highest sequence identity to a Swiss-Prot protein known to catalyze the assigned metabolic activity in another organism was used as the single sequence-based descriptor.
Importantly, the presented approach does not assume a one-to-one relationship between a gene and its function (network location). In cases where a gene is annotated with multiple enzymatic functions, the method calculates, one by one, the likelihoods of each annotation. Only annotations with the likelihood below a certain optimized threshold are marked as potential misannotations. Consequently, multiple annotations are allowed for each gene, as long as they all have good genomic correlations in the assigned network locations.
Method training and optimization
We used the S. cerevisiae metabolic model iLL67227 to train and benchmark the algorithm. The well-curated yeast network allowed us to optimize parameters and evaluate the prediction accuracy using cross-validation. Because the yeast metabolism is relatively well known, we assumed that the vast majority of the network functional assignments are correct, i.e. they represent true positives (TP). To simulate true negative (TN) examples we artificially generated incorrect functional assignments using the three different methods described below. We calculated the ROC curves by sorting the annotations based on their classification scores; annotations with lowest classification scores are more likely to represent true misannotations,
In the first method (TN1), we randomly assigned new metabolic functions to a large fraction (33%) of network genes. The AdaBoost classifier was then trained using TN1 and TP examples (see Methods). The resulting ROC curve for the 50/50 cross-validation is shown in Figure 2a. Due to the random nature of the functional reassignments, the TN1 examples rarely have high sequence identities to the newly assigned functions. Consequently, the algorithm relied primarily on the sequence identity and easily identified the misannotations.
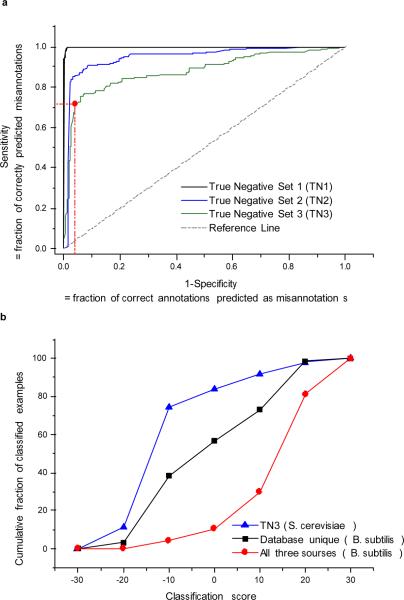
Figure 2.
Performance on identifying misannotations. a) The ROC curves on different types of artificially generated misannotations in the yeast network. The True Negative set 1 (TN1) was generated by randomly assigning incorrect metabolic functions to a fraction of network genes. The TN2 set was generated by reassigning network genes to new metabolic activities only if they had at least 30% sequence identities to newly assigned (incorrect) activities. The TN3 was generated by assigning genes to new activities only if they had similar (within 10%) or higher sequence identities to the reassigned (incorrect) activities. In all cases the remaining (not reassigned) activities were used as true positive examples. For realistic misannotation models, simulated by the sets TN2 and TN3, the method correctly identifies about 70%–90% of misannotations at 5%–15% false positive rate. The red dot in the figure approximately corresponds to 70% true positives and 5% false positives. b) The cumulative distributions of the classification confidence scores for B. subtilis metabolic assignments. The B. subtilis annotations made simultaneously by all analyzed databases (KEGG, MetaCyc and Swiss-Prot) are shown in red, annotations unique to KEGG, MetaCyc, or Swiss-Prot, are shown in black. For comparison we also show the true negative set TN3 from S. cerevisiae in blue. The cumulative distributions demonstrate that the consensus annotations (red) are, on average, more accurate than the ones unique to individual databases (blue, Kolmogorov-Smirnov test P=2*10−19). However, on average, database-specific annotations still score significantly better than true misannotations (KS P=2*10−4).
In the second method (TN2), to simulate misannotations due to a residual sequence homology to nonnative metabolic activities, genes were only reassigned to incorrect activities for which they had >30% sequence identity . A random choice was made if several reassignments were possible for a gene. The classification algorithm was then independently trained using TN2 examples (Figure 2a). The mean area under the ROC curve for the TN2 set, based on four independent reassignment experiments, was 0.93 (95% CI: 0.90 – 0.95). In spite of the large fraction (40%) of misannotations in the reassigned network, the algorithm identified about 90% of true misannotations with only 20% of correct annotations misclassified as misannotations.
Finally, in the third method (TN3), the genes were reassigned only if they had similar (within 10%) or even higher sequence identities to the newly assigned (incorrect) activities. This test simulated misannotation especially difficult to detect using sequence homology. In total 26% of the network genes were reassigned using the third method. The mean area under the ROC curve for the TN3 examples (Figure 2a), based on four independent reassignment runs, was 0.87 (95% CI: 0.86 – 0.88). The algorithm identified about 80% of misannotations while misclassifying 20% of correct annotations. Because many metabolic assignments in existing databases have been made based primarily on sequence homology, it is likely that the errors simulated using the second and the third methods dominate real world misannotations.
To understand the transferability of our approach to other species we repeated the analysis using the curated E. coli metabolic model iJR90428. The negative examples TN2 and TN3 for the bacterial metabolic model were generated in the same way as for the yeast network. The classifiers optimized for the yeast network were directly applied to the bacterial model without further modification or optimization. The resulting performance for the E. coli network was similar to S. cerevisiae (Supplementary Fig. 1). Consequently, the optimized method is able to detect misannotations in different species. The policing approach should be also quite effective in non-model organisms because the context correlations, with the exception of co-expression, can be calculated directly from genomic sequences; the decrease in sensitivity without expression information was less than 3% (at 25% false positive rate). The accuracy of other context correlations will only improve as more genomes are sequenced.
Potential misannotations in B. subtilis metabolic network
To test our algorithm on a less-studied network we applied it to the model gram-positive bacterium B. subtilis. We investigated the B. subtilis metabolic annotations available in KEGG29 (655 genes), Swiss-Prot10 (528 genes), and MetaCyc30 (369 genes). The different number of annotated genes in these databases is a consequence of different annotation strategies. While some databases strive for maximum coverage, others focus on the most accurate annotations. There are 277 B. subtilis annotations shared by all three databases and additional 122, 10 and 20 unique annotations in KEGG, MetaCyc and Swiss-Prot, correspondingly. We applied the developed algorithm to all B. subtilis metabolic assignments in the three databases using the parameters optimized for the TN3 yeast examples. The cumulative distributions of the AdaBoost classification scores for B. subtilis annotations (Figure 2b) show that the metabolic assignments shared by all databases (red curve) are on average more accurate compared to annotations present exclusively in a single database (black curve, Kolmogorov-Smirnov test P=2*10−19). Nevertheless, the database-unique annotations display, on average, significantly better scores compared to the scores of misannotated genes (TN3 yeast examples, blue curve, P=2*10−4). This demonstrates that it is not possible to detect potential misannotations simply by identifying database-unique functional assignments.
Based on the ROC characteristics (Figure 2a) the most efficient part of the TN3 curve allows to identify 70% of misannotations, while classifying only about 5% of correct assignments as misannotations. Considering the total number of analyzed B. subtilis metabolic assignments (679) and assuming that about 10% of the database assignments are misannotations4, 5, the red point in Figure 2a corresponds to the analysis of 80 genes with the worst classification scores; about half of these genes should represent true misannotations. Indeed, we manually analyzed the list of 80 genes with the worst classification scores and for 34 cases we either found counter-evidence or could not identify any experimental study supporting the annotations (Table 1). Although the potential misannotations usually have weak sequence homology (usually <40% identity) to known enzymes, the classifier is not simply relying on homology to identify misannotations. For about 35% of the annotations with good classification scores sequence identity was also weak (<40%), but these metabolic assignments are supported by good context-based correlations.
Table 1.
Potential misannotations in the B. subtilis metabolic network. Annotation source: K: KEGG, M: MetaCyc, and S: Swiss-Prot. Homology score is the highest proteinprotein sequence identity to another Swiss-Prot protein with the target activity; the corresponding BLAST E-value is also shown. The context genomic correlations are represented as the relative percentile ranks. For example, the “expression profile” rank of 20% indicates that the target gene has better co-expression values in 20% of all other possible network locations compared to the location assigned in the database. Lower percentile ranks indicate better consistencies with genomic context correlations. For the protein fusion, “Y” (“N”) indicates that fusion event(s) between an ortholog of the candidate gene and a network neighbor was detected (not detected). The presence of a significantly better alternative location (“Y”/“N”) was determined by the ALR ratio as described in Supplementary Methods.
| Gene name | Annotated function (EC number) | Homology Score | Phylogenetic profile rank (%) | Expression profile rank (%) | Clustering profile rank(%) | Gene distance rank (%) | Protein fusion? | Significantly better alternative location? |
|---|---|---|---|---|---|---|---|---|
| adhB | 1.1.1.284 (K) | 40.7/3E-74 | 90 | 90 | 91 | 83 | N | Y |
| alaT | 2.6.1.17 (K) | 48.5/3E-98 | 58 | 23 | 28 | 72 | N | Y |
| bcsA | 2.3.1.74 (K, S, M) | 29.7/1E-04 | 74 | 79 | 73 | 84 | N | Y |
| bsaA | 1.11.1.9 (K, S, M) | 55/2E-51 | 47 | 57 | 55 | 52 | N | Y |
| Cad | 4.1.1.18 (M) | 22.8/2E-14 | 45 | 80 | 60 | 84 | N | Y |
| dgkA | 2.7.1.107 (K, S, M) | 32.3/2E-11 | 64 | 21 | 55 | 81 | N | N |
| hipO | 3.5.1.32 (K, M) | 35.9/6E-59 | 45 | 64 | 54 | 8 | N | N |
| Pps | 2.7.9.2 (K, M) | 43.5/0.002 | 44 | 38 | 71 | 30 | N | Y |
| xpt | 2.4.2.7 (M) | 29.2/5E-07 | 4 | 1 | 7 | 12 | N | N |
| ybbD | 3.2.1.52 (K) | 34.2/1E-27 | 49 | 1 | 54 | 36 | N | N |
| ycgT | 1.8.1.9 (K) | 29.8/2E-25 | 50 | 21 | 33 | 16 | N | N |
| yhcV | 1.1.1.205 (K) | 37/0.002 | 22 | 68 | 44 | 46 | Y | N |
| yhdR | 2.6.1.1 (K) | 30.1/3E-30 | 2 | 1 | 9 | 25 | N | Y |
| yhfR | 5.4.2.1 (K) | 38.3/1E-12 | 22 | 65 | 22 | 17 | N | N |
| yisP | 2.5.1.32 (K) | 27.8/8E-24 | 87 | 49 | 60 | 73 | N | Y |
| yitC | 3.1.3.71 (K, S) | 38.7/4E-18 | 87 | 42 | 72 | 10 | N | N |
| yjmC | 1.1.1.37 (K) | 39.8/2E-60 | 68 | 30 | 48 | 37 | N | Y |
| yktC | 3.1.3.25 (K,S) | 38.1/2E-28 | 73 | 49 | 49 | 61 | N | N |
| ykuR | 3.5.1.47 (K) | 35.6/3E-43 | 75 | 70 | 50 | 79 | N | Y |
| yngE | 6.4.1.3 (K) | 40.1/8E-92 | 1 | 4 | 2 | 12 | Y | Y |
| yngF | 4.2.1.17 (K, M) | 38.9/5E-39 | 1 | 2 | 2 | 14 | Y | Y |
| yngI | 6.2.1.3 (K) | 31/6E-63 | 1 | 10 | 56 | 31 | Y | Y |
| yoaD | 1.1.1.95 (K) | 33.8/1E-39 | 1 | 1 | 24 | 74 | N | Y |
| yogA | 1.1.1.1 (K) | 29.7/2E-21 | 39 | 81 | 71 | 30 | N | Y |
| yqhT | 3.4.11.9 (K) | 34.9/4E-22 | 50 | 54 | 11 | 78 | N | Y |
| yrhE | 1.2.1.2 (K) | 37.5/1E-129 | 2 | 60 | 58 | 51 | Y | Y |
| ysfC | 1.1.3.15 (K) | 27.3/4E-10 | 55 | 66 | 76 | 34 | N | Y |
| yumB | 1.6.99.3 (K) | 26.6/3E-25 | 1 | 18 | 26 | 18 | N | Y |
| yumC | 1.8.1.9 (K) | 29.3/1E-21 | 81 | 32 | 35 | 52 | N | Y |
| yvcN | 2.3.1.5 (K) | 28.6/6E-13 | 78 | 36 | 77 | 78 | N | N |
| yvcT | 1.1.1.215 (K, S) | 47.3/8E-79 | 53 | 59 | 47 | 49 | N | Y |
| ywrD | 2.3.2.2 (K) | 31.4/9E-55 | 25 | 85 | 43 | 27 | N | N |
For each potential misannotation we show in Table 1 the gene name, annotation source, the highest sequence identity to enzymes responsible for the annotated activity in other species, the relative strength of various context-based correlations, and the existence of good alternative network locations (see Supplementary Methods). In the table the context correlation values are represented by their relative percentile ranks based on the average fitness scores (see Methods). For example, the “expression profile” rank of 10% indicates that the target gene has better co-expression scores in 10% of all possible network locations compared to the location assigned in the database. Overall, the results in Tables 1 suggest that Swiss-Prot and MetaCyc are more conservative in their functional assignments compared to KEGG, which has the largest number of annotations as well as potential misannotations. We want to emphasize that the majority of KEGG-unique annotations displayed good confidence scores, indicating that only a fraction of them is likely to be incorrect.
The B. subtilis gene dgkA is a typical example of a potential misannotation. The gene is annotated in all considered databases as “diacylglycerol kinase” (DagK, EC 2.7.1.107), possibly based on weak sequence homology. However, dgkA has poor context-based correlations with the network neighbors of the EC 2.7.1.107 activity (Table 1). In a recent study31, the authors confirmed that dgkA is not a diacylglycerol kinase but rather an undecaprenol kinase. Another example is the B. subtilis gene ywrD which is annotated in KEGG as an ortholog of the gamma-glutamyltransferase (EC 2.3.2.2). Weak context-based correlations (Table 1) with neighboring network genes suggest that ywrD is unlikely to catalyze the EC 2.3.2.2 function. The gamma-glutamyltransferase activity (EC 2.3.2.2) is required for growth on extracellular glutamyl compounds, such as glutathione (GSH) (1), as the source of sulfur. However, it was demonstrated32 that ywrD- mutant grows well on minimal media with GSH as the sole sulfur source. In addition, His-tag purified ywrD could not hydrolyze GSH. These findings strongly suggest that ywrD is not gamma-glutamyltransferase. Further analysis of each case in Table 1 is presented in Supplementary Table 1.
Leucine degradation pathway in B. subtilis
The developed method can be used to identify suspicious functional assignments for several genes in a pathway. An interesting example is the yngJIHGFE gene cluster in B. subtilis (Figure 3a). The yngJ gene is listed in KEGG as a hypothetical protein, yngI as acyl-CoA synthetase (EC 2.3.1.86) (until recently as long-chain-fatty-acid---CoA ligase, EC 6.2.1.3), yngH as acetyl-CoA carboxylase biotin carboxylase subunit (EC 6.4.1.2/6.3.4.14), yngG as hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4), yngF as enoyl-CoA hydratase (EC 4.2.1.17), and yngE as propionyl-CoA carboxylase beta chain (EC 6.4.1.3). In MetaCyc yngE is listed as similar to propionyl-CoA carboxylase and yngF as enoyl-CoA hydratase (EC 4.2.1.17). In Swiss-Prot, yngJ is listed as probable acyl-CoA dehydrogenase (EC 1.3.99.-), yngH as biotin carboxylase 2 (EC 6.3.4.14/6.4.1.2), yngG as hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4), and yngF as putative enoyl-CoA hydratase/isomerase.
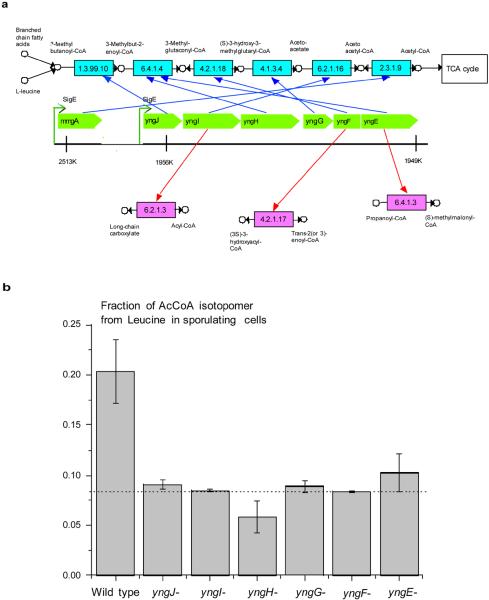
Figure 3.
Function of genes forming the yng cluster in B. subtilis. a) The genomic positions of the yng genes are shown in green. The detected misannotations are indicated in red. The predicted functions, forming the degradation pathway, are shown in blue. The expression of all yng gene is controlled by the σE transcription factor34; the gene mmgA is also under the σE control and is responsible for the last step of the leucine catabolism. b) Fractional 13C labeling of Acetyl-CoA in the wild type sporulating cells and in the sporulating yng mutants. The 13C labeling in the figure indicates the fraction of the Acetyl-CoA isotopomer generated from leucine in sporulating cells only (see Methods). The errors in the figure represent SEM. The background Acetyl-CoA isotopomer labeling is shown by the dash line.
Our algorithm predicted as potential misannotations the assignments of the EC 6.4.1.3 function to yngE, EC 4.2.1.17 to yngF, and EC 6.2.1.3 to yngI. These genes have significantly better genomic correlations in different network locations (functions): yngE in EC 6.4.1.4, yngF in EC 4.2.1.18, and yngI in EC 6.2.1.16. Overall, the yng genes form the consecutive reactions in the leucine (2) degradation pathway33. Based on the corrected functional assignments we can also suggest the likely functions for yngJ (EC 1.3.99.10) and yngH (EC 6.4.1.4 subunit, forming the enzyme complex with yngE). Consequently, the yng cluster forms a complete degradation pathway from 3-methylbutanoyl-CoA (3) to acetoacetyl-CoA (4), which can be further catabolized through the bacterial TCA cycle.
What is the biological role of the leucine degradation pathway in B. subtilis? In early stages of sporulation B. subtilis cells divide into two unequal compartments. The smaller compartment develops into a bacterial spore, and the larger compartment forms the mother cell which protects and nurtures the spore until the spore is fully developed. Interestingly, the yng genes are under transcriptional control of the σE sigma factor and are primarily expressed early in the mother cell during sporulation34, i.e. when extracellular nutrients are limited. The expression of the gene mmgA responsible for the last step of the leucine catabolism, acetoacetyl-CoA to Acetyl-CoA (5) conversion (EC 2.3.1.9, see Figure 3a), is also controlled by the σE factor.
Due to the structure of its TCA cycle, B. subtilis cannot grow on leucine as the sole carbon source35. Nevertheless, the catabolism of the leucine and fatty acids through the TCA cycle can provide additional energy during early sporulation stages. The selection of the energy source becomes logical if one considers the membrane and amino acid composition of B. subtilis. Leucine is one of the most abundant amino acid in logarithmically growing B. subtilis cells36, responsible for about 8–10% of all protein residues (see also Supplementary Fig. 2). In addition, B. subtilis lipids are predominantly (>90%) composed of branched chain fatty acids35, 37; odd-iso fatty acids can be oxidized to 3-methylbutanoyl-CoA. It is likely that during sporulation branched chain fatty acids and amino acids are present in the extracellular media due to the bacterial cannibalism process38, 39, which allows a fraction of B. subtilis cells to kill their non-sporulating siblings and feed on the released nutrients.
To experimentally investigate the role of the yng cluster during sporulating we used 13C labeling experiments. First, we analyzed B. subtilis 168 cells in non-sporulating minimal medium supplemented with [U13C]-L-Leucine (see Methods). Because the degradation pathway leads from leucine to Acetyl-CoA (Figure 3a), we measured the fractional labeling of the Acetyl-CoA m2 mass isotopomer using liquid chromatography-tandem mass spectroscopy (see Methods) and calculated the fraction of Acetyl-CoA originating directly from leucine. No 13C labeling above the natural abundance of the m2 isotopomer (8%) was detected in cell during vegetative growth. This result confirmed that the leucine degradation pathway is not active during favorable environmental conditions34.
Next, we investigated the activity of the leucine pathway during sporulation. It was previously shown that 2.5 hours after the start of sporulation the activity of σE-regulated genes is at the highest34. We inoculated bacterial cells into sporulation medium supplemented with [U13C]-Leucine, and extracted metabolites after 2.5 hours. In sporulating cells the fraction of Acetyl-CoA derived from leucine was about 2.5–3 times higher than background, while all yng mutants displayed essentially background labeling levels (Figure 3b). Consequently, the yng pathway is indeed active during sporulating.
Several genes from the yng cluster have been assigned in KEGG to isoleucine (6) degradation pathway: yngE as an ortholog of EC 6.4.1.3, yngF as an ortholog of EC 4.2.1.17. To investigate the possibility that the yng genes also play a role in the isoleucine to Acetyl-CoA degradation we tested the activity of the isoleucine degradation pathway during sporulation. Similar to the leucine experiments, we measured the labeling of Acetyl-CoA in sporulation conditions supplemented with [U13C]-L-Isoleucine. No labeling above background was detected (Supplementary Fig. 3). Consequently, the yng genes are unlikely to participate in the isoleucine degradation. Although B. subtilis can utilize isoleucine and valine (7) as the sole nitrogen source40, our experiments demonstrate that the isoleucine pathway is either not active during sporulation or its products are not primarily degraded to Acetyl-CoA.
Discussion
The main idea of the presented approach is to employ functional genomic correlations essentially in reverse. Instead of using them to assign protein function41, 42, we utilize the correlations to predict potential misannotations. The developed method, or similar approaches, can be automatically applied to many thousands of metabolic assignments in various functional databases. Based on this analysis the potential misannotations can be marked with corresponding confidence scores. As topologies of protein-protein interaction networks are discovered, similar methods can be also developed and optimized to identify misannotations in the context of molecular interaction networks. Importantly, the developed method was not conceived as a criticism of such valuable resources as Swiss-Prot, KEGG, and MetaCyc. Our results clearly demonstrate that the majority of annotations in these databases are correct. Nevertheless, we think that the method can help the existing resources to improve the annotation quality and reduce the spread of misannotations.
Methods
Metabolic networks construction
The metabolic networks were constructed using known enzymatic reactions for the considered organisms: the iLL672 model27 for S. cerevisiae, iJR904 model28 for E. coli, and B. subtilis metabolic reactions from KEGG43, MetaCyc30 and Swiss-Prot10. Only genes with assigned EC numbers were considered; activities representing non-metabolic reactions, such as EC 2.7.11.1 (non-specific serine/threonine protein kinase) or EC 2.7.7.6 (RNA polymerase) were excluded. Each metabolic network was represented as a graph with nodes as metabolic genes and edges as functional connections established by metabolites shared between enzymes20–22. The shortest path between a pair of nodes was used as the metabolic network distance between the corresponding genes. The 40 most connected co-factors and metabolites were not considered in calculating metabolic distances22(Supplementary Table 2).
Context genomic correlations
We used the following context correlations: phylogenetic profiles15, 16, mRNA co-expression44, 45, chromosomal distance, gene clustering (chromosomal co-localization across a set of genomes)12, 14, fusion of protein domains17, 18. The phylogenetic profile correlations were constructed using BLASTP searches, using E-value cutoff 10−3, against a collection of 70 evolutionary distinct genomes22; pair-wise phylogenetic profiles were calculated using Pearson's correlation coefficient. The co-expression values were calculated using Spearman's rank correlation between expression profiles obtained from the Rosetta Compendium dataset for S. cerevisiae46, Stanford Microarray Database (SMD) for E. coli and the GEO database47 for B. subtilis. The physical distance between genes from target genomes was used as chromosomal distance. To calculate the chromosomal clustering of genes across genomes orthology mapping was established using the KEGG SSDB database29; the chromosomal clustering values were calculated based on a collection of 105 diverse genomes26. A pair of genes was considered fused if at least 70% of each protein could be aligned to non-overlapping regions of a third protein in the NCBI NR database (using BLAST E-value cutoff 10−3). A detailed description of the data sources and the methods used to calculate the context-based correlations are given in our previous publications22, 26.
Context-based fitness functions
We calculated the “fitness” of every gene in its assigned network position using the following equation:
| (1) |
where x is the gene to be tested at the target network position, y is a neighboring gene from the ith network Layeri, c(x, y) is a context-based correlation between genes x and y, wi is the weight factor for Layeri, and p is the optimized power factor for the context-based correlation. The summation in Equation 1 is, first, over all genes in a given Layeri around the network position of the tested gene and, second, over all layers up to the layer R (R = 3 in our calculation). |N| is the total number of genes in all considered layers. The parameters for each context-based method were optimized using simulated annealing (SA) algorithm48 so that the log sum of the ranks of the correct functions for all known metabolic genes were minimized.
Sequence homology information
The sequence homology descriptor of protein function was represented as the highest sequence identity to a Swiss-Prot protein (using BLAST E-values cutoff 5*10−2) annotated to carry out the target function excluding genes that are 1) from the query genome or 2) likely annotated based on computational methods, i.e., genes with keywords probable, like, by similarity, hypothetical, or putative in their annotations.
Combining sequence-based and context-based descriptors
All context and sequence homology descriptors were combined using the AdaBoost algorithm with alternating decision trees (ADT)23, 24. For each classification the algorithm also generates a confidence measure (classification score).
The highest sequence identity to a protein known to catalyze the target enzymatic activity in other species was supplied to the classification algorithms as the sequence-based descriptor. The context-based descriptors were supplied to the classification algorithm as the gene-specific ranks, i.e. context correlation ranks of the target gene at the annotated location compared to all other network positions. For each context descriptor, we consider two separate ranks. First, the rank based on the overall fitness of the target gene in the annotated location calculated using Equation 1. Second, the rank based on the largest pairwise correlation of the target gene and its immediate network neighbors. For each target gene, we also supplied the classification algorithm with two additional AdaBoost scores: 1) the total score for the target gene in the annotated location, and 2) the score in best alternative network location.
Cross-validation
The performance of the method was benchmarked using the S. cerevisiae networks using the 50/50 cross validation in which all samples were randomly divided into two sets with approximately equal number of TN and TP cases. Results from the two sets were pooled to estimate the overall performance. We also applied multivariable logistic regression to combine the different descriptors and predict misannotations. Although the AdaBoost algorithm tends to slightly outperform logistic regression, a comparable performance was observed for the two methods (Supplementary Fig. 4). All results reported in the paper are based on the AdaBoost algorithm.
Labeling experiments
B. subtilis 168 mutants (yngE-, yngF-, yngG-, yngH-, yngI-, and yngJ-) were obtained from the Medicago Main Collection. Growth of these strains was tested using the minimal medium M9 supplemented with various carbon sources. The strains were grown on sporulation agar medium (DSM) and incubated overnight at 37°C. On the following day, cells were inoculated into sporulation medium (Schaeffer et al., 1965) supplemented with 5 mM of [U13C]-L-Leucine or [U13C]-LIsoleucine (Cambridge Isotope Laboratories) at the beginning of the growth curve. The cells were harvested 2.5 hours after the onset of the sporulation.
Cellular metabolites were extracted using EtOH:H2O (60:40) and 10mM ammonium acetate solution at 70°C. Cell debris was removed from the extract by centrifugation and the supernatant was completely dried. Samples were injected in a liquid chromatography-tandem mass spectrometer (LC-MS/MS, Agilent) with a C18 column (Waters Atlantis T3 150×2.1×3). The identity of the peaks was establishes by verifying the peak retention time and mass spectrum for each mass isotopomer of Acetyl-CoA. The natural (background) abundance of the m2 isotopomer of Acetyl-CoA (8%) was calculated by the Analyst software (Agilent).
Supplementary Material
Figure 4.
Acknowledgements
We would like to thank Igor Feldman, Andrey Rzhetsky, Michiel de Hoon, Sarah Gilman, Chani Weinreb for comments on the manuscript and valuable discussions. This work was supported in part by the National Institutes of Health GM079759 grant to D.V. and the National Centers for Biomedical Computing (MAGNet) U54CA121852 grant to Columbia University.
References
- 1.Andrade MA, et al. Automated genome sequence analysis and annotation. Bioinformatics. 1999;15:391–412. doi: 10.1093/bioinformatics/15.5.391. [DOI] [PubMed] [Google Scholar]
- 2.Rost B. Enzyme function less conserved than anticipated. J. Mol. Biol. 2002;318:595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- 3.Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 4.Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- 5.Gilks WR, Audit B, De Angelis D, Tsoka S, Ouzounis CA. Modeling the percolation of annotation errors in a database of protein sequences. Bioinformatics. 2002;18:1641–1649. doi: 10.1093/bioinformatics/18.12.1641. [DOI] [PubMed] [Google Scholar]
- 6.Linial M. How incorrect annotations evolve--the case of short ORFs. Trends Biotechnol. 2003;21:298–300. doi: 10.1016/S0167-7799(03)00139-2. [DOI] [PubMed] [Google Scholar]
- 7.Wieser D, Kretschmann E, Apweiler R. Filtering erroneous protein annotation. Bioinformatics. 2004;20(Suppl 1):i342–347. doi: 10.1093/bioinformatics/bth938. [DOI] [PubMed] [Google Scholar]
- 8.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bairoch A, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green ML, Karp PD. Genome annotation errors in pathway databases due to semantic ambiguity in partial EC numbers. Nucleic Acids Res. 2005;33:4035–4039. doi: 10.1093/nar/gki711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends. Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 13.Lee JM, Sonnhammer EL. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13:875–882. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynen MA, Bork P. Measuring genome evolution. Proc. Natl. Acad. Sci. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad Sci. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 18.Marcotte EM, et al. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 19.Yanai I, Derti A, DeLisi C. Genes linked by fusion events are generally of the same functional category: a systematic analysis of 30 microbial genomes. Proc. Natl. Acad. Sci. 2001;98:7940–7945. doi: 10.1073/pnas.141236298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kharchenko P, Vitkup D, Church GM. Filling gaps in a metabolic network using expression information. Bioinformatics. 2004;20:I178–I185. doi: 10.1093/bioinformatics/bth930. [DOI] [PubMed] [Google Scholar]
- 21.Kharchenko P, Church GM, Vitkup D. Expression dynamics of a cellular metabolic network. Molecular Systems Biology. 2005;1 doi: 10.1038/msb4100023. doi:10.1038/msb4100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Vitkup D. Predicting genes for orphan metabolic activities using phylogenetic profiles. Genome Biol. 2006;7:R17. doi: 10.1186/gb-2006-7-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freund Y, Mason L. The Alternating Decision Tree Learning Algorithm. In: Bratko I, Dzeroski S, editors. Proceedings of the Sixteenth International Conference on Machine Learning; San Francisco: Morgan Kaufmann Publishers Inc.; 1999. pp. 124–133. [Google Scholar]
- 24.Freund Y, Schapire RE. A short introduction introduction to Boosting. Journal of Japanese Society for Artificial Intelligence. 1999;14:771–780. [Google Scholar]
- 25.Middendorf M, Kundaje A, Wiggins CH, Freund Y, Leslie C. Predicting genetic regulatory response using classification. Bioinformatics. 2004;20:I232–I240. doi: 10.1093/bioinformatics/bth923. [DOI] [PubMed] [Google Scholar]
- 26.Kharchenko P, Chen L, Freund Y, Vitkup D, Church GM. Identifying metabolic enzymes with multiple types of associated evidence. BMC Bioinformatics. 2006;7:177. doi: 10.1186/1471-2105-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuepfer L, Sauer U, Blank LM. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res. 2005;15:1421–1430. doi: 10.1101/gr.3992505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12. Genome Biol. 2003;R54 doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caspi R, et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D511–516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jerga A, Lu YJ, Schujman GE, de Mendoza D, Rock CO. Identification of a soluble diacylglycerol kinase required for lipoteichoic acid production in Bacillus subtilis. J. Biol. Chem. 2007;282:21738–21745. doi: 10.1074/jbc.M703536200. [DOI] [PubMed] [Google Scholar]
- 32.Minami H, Suzuki H, Kumagai H. Gamma-glutamyltranspeptidase, but not YwrD, is important in utilization of extracellular blutathione as a sulfur source in Bacillus subtilis. J. Bacteriol. 2004;186:1213–1214. doi: 10.1128/JB.186.4.1213-1214.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichenberger P, et al. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 2003;327:945–972. doi: 10.1016/s0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 35.Sonenshein AL, Hoch J, Losick R. Bacillus subtilis and its Closest Relatives. ASM Press; Washington, DC: 2001. [Google Scholar]
- 36.Sauer U, et al. Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Appl. Environ. Microbiol. 1996;62:3687–3696. doi: 10.1128/aem.62.10.3687-3696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Pastor JE, Hobbs E, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 39.Ellermeier CD, Hobbs E, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 40.Debarbouille M, Gardan R, Arnaud M, Rapoport G. Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letovsky S, Kasif S. Predicting protein function from protein/protein interaction data: a probabilistic approach. Bioinformatics. 2003;19(Suppl):197–204. doi: 10.1093/bioinformatics/btg1026. [DOI] [PubMed] [Google Scholar]
- 42.Borenstein E, Shlomi T, Ruppin E, Sharan R. Gene loss rate: a probabilistic measure for the conservation of eukaryotic genes. Nucleic Acids Res. 2007;35:e71–75. doi: 10.1093/nar/gkl792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG database at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 45.Wu LF, et al. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Genet. 2002;31:255–265. doi: 10.1038/ng906. [DOI] [PubMed] [Google Scholar]
- 46.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 47.Barrett, et al. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by simulated annealing. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.