GnT1IP inhibits GlcNAcT-1 to change N-glycosylation patterns on secretory proteins, potentially regulating germ cell adhesion to Sertoli cells during spermatogenesis.
Abstract
Database analyses identified 4933434I20Rik as a glycosyltransferase-like gene expressed mainly in testicular germ cells and regulated during spermatogenesis. Expression of a membrane-bound form of the protein resulted in a marked and specific reduction in N-acetylglucosaminyltransferase I (GlcNAcT-I) activity and complex and hybrid N-glycan synthesis. Thus, the novel activity was termed GlcNAcT-I inhibitory protein (GnT1IP). Membrane-bound GnT1IP localizes to the ER, the ER-Golgi intermediate compartment (ERGIC), and the cis-Golgi. Coexpression of membrane-anchored GnT1IP with GlcNAcT-I causes association of the two proteins, inactivation of GlcNAcT-I, and mislocalization of GlcNAcT-I from the medial-Golgi to earlier compartments. Therefore, GnT1IP is a regulator of GlcNAcT-I and complex and hybrid N-glycan production. Importantly, the formation of high mannose N-glycans resulting from inhibition of GlcNAcT-I by GnT1IP markedly increases the adhesion of CHO cells to TM4 Sertoli cells. Testicular germ cells might use GnT1IP to induce the expression of high mannose N-glycans on glycoproteins, thereby facilitating Sertoli–germ cell attachment at a particular stage of spermatogenesis.
Introduction
Protein N-glycosylation affects glycoprotein structure and function during protein folding, trafficking and clearance, cell–cell interactions, and signal transduction (Dennis et al., 2009; Varki and Lowe, 2009). N-glycans are synthesized by a series of glycosyltransferases and glycosidases along the secretory pathway (Stanley et al., 2009). Golgi resident glycosyltransferases are typically type II transmembrane proteins with a short, N-terminal cytoplasmic tail, a transmembrane domain, and a large, C-terminal catalytic domain (Cantarel et al., 2009). Some glycosyltransferases, including protein O-mannosyltransferases 1 and 2 (Akasaka-Manya et al., 2006) and T-synthase (Ju et al., 2008; Aryal et al., 2010), are regulated by a protein chaperone or complex required for transferase activity. Here we identify a novel inhibitor of a glycosyltransferase activity that is expressed primarily in testicular germ cells.
Glycosyltransferases with similar sequences may catalyze related enzyme reactions, use the same nucleotide-sugar, or have the same substrate specificity. About 90 glycosyltransferase families have been classified in the carbohydrate-active enzymes database (CAZy; Cantarel et al., 2009). However, new glycosyltransferase activities cannot be predicted merely by sequence or homology comparisons. For example, the T-synthase chaperone Cosmc is highly homologous to a β1,3-galactosyltransferase but has no known transferase activity (Ju and Cummings, 2002; Kudo et al., 2002). In addition, glycosyltransferase genes give rise to numerous splice forms that may produce enzymes of different activity or specificity (Thierry-Mieg and Thierry-Mieg, 2006). For example, β4GalT-I has two transcripts with different transcription initiation sites, producing a long form with an extra 13 amino acids at its N terminus that is localized to the cell surface, and a short form localized to the trans-Golgi (Shur et al., 1998; Rodeheffer and Shur, 2002).
To characterize new “orphan” glycosyltransferases, candidates may be expressed in wild-type CHO cells or CHO mutants with altered glycosylation that have a well-characterized glycan complement (Patnaik and Stanley, 2006; North et al., 2010). CHO glycosylation mutants express changes in cell surface glycans that are recognized by cytotoxic plant lectins. Lectin-resistant phenotypes reflect mutations in glycosylation genes, such as glycosyltransferase, glycosidase, or nucleotide-sugar transporter genes (Patnaik and Stanley, 2006; North et al., 2010). For instance, LEC10 CHO mutants have a gain-of-function mutation that induces expression of the Mgat3 gene and Mgat3 (GlcNAcT-III) catalyzes the transfer of the bisecting GlcNAc to complex N-glycans (Campbell and Stanley, 1984; Stanley et al., 2005). The addition of this single residue to N-glycans causes cells to become highly resistant to ricin and hypersensitive to the erythroagglutinin E-PHA (Campbell and Stanley, 1984). Lec1 CHO glycosylation mutants have a loss-of-function mutation in the Mgat1 gene and lack Mgat1 (GlcNAcT-I) activity (Chen and Stanley, 2003). GlcNAcT-I is the medial-Golgi glycosyltransferase that transfers GlcNAc to Man5GlcNAc2 to initiate hybrid and complex N-glycan synthesis (Stanley et al., 2009). Lec1 mutants lack hybrid and complex N-glycans and are consequently resistant to many plant lectins that bind to terminal sugars of N-glycans (Stanley, 1983). By contrast, Lec1 cells are hypersensitive to lectins like Con A that bind high mannose N-glycans. Expression of an orphan glycosyltransferase that alters the lectin resistance phenotype of wild-type CHO cells or of CHO glycosylation mutants allows identification of the new activity.
Using this strategy we characterized a putative mouse glycosyltransferase encoded by the gene 4933434I20Rik with 37% identity to mouse GlcNAcT-IVa and 46% identity to mouse GlcNAcT-IVb (CAZy GT54). Here we show that CHO transfectants expressing membrane-bound forms of this protein have a Lec1 lectin resistance phenotype due to the specific inhibition of GlcNAcT-I activity. The novel activity, termed GlcNAcT-I inhibitory protein (GnT1IP), is expressed mainly in spermatocytes and spermatids of mammalian testis, its transcription and translation are tightly regulated during mouse spermatogenesis, and the changes in glycan complement it induces cause cells to adhere strongly to TM4 Sertoli cells.
Results
A novel inhibitor of complex and hybrid N-glycan synthesis
Database searches identified mouse gene 4933434I20Rik and cDNA NM_026233.2 as encoding sequences similar to the GlcNAcT-IV glycosyltransferase family in CAZy family GT54 (Cantarel et al., 2009). Deduced protein NP_080509.2 is 37% identical to mouse Mgat4A (GlcNAcT-IVa) and 46% identical to mouse Mgat4B (GlcNAcT-IVb) and has an identical sequence to BAB30173.1 except for Asp instead of Asn in NP_080509.2 at the fifth aa. The TMpred program (Hofmann and Stoffel, 1993) predicted that NP_080509.2 contains a transmembrane domain from aa 5–25, whereas SignalP 3.0 (Emanuelsson et al., 2007) predicted a signal peptide from aa 1–26, and a GlcNAcT-IV–like domain from aa 79–373. N-terminal Myc-tagged BAB30173.1 was previously shown to localize to the ER when transiently expressed in HeLa cells (Fink et al., 2006; Sprenger et al., 2008).
The bicistronic vector pIRES2-EGFP was used to stably express N-terminal Myc-tagged NP_080509.2 in wild-type CHO cells. A mixed transfectant population (Fig. 1 A, line 4) contained cells highly resistant to the leukoagglutinin L-PHA, similar to Lec1 mutant cells (Fig. 1 A, line 2). Stable transfectants sorted for high GFP expression were uniformly highly resistant to L-PHA (Fig. 1 A, line 5). Similar results were found for Myc-BAB30173.1 (not depicted). Both proteins contain 253EDD255, a motif that might mediate nucleotide-sugar and metal ion binding in a glycosyltransferase reaction (Breton and Imberty, 1999). However, mutation to either E253A (ADD) or D255A (EDA) did not diminish the L-PHA resistance induced by Myc-NP_080509.2 (not depicted). In addition, when mouse GlcNAcT-I was transiently expressed in Lec1/Myc-NP_080509.2 stable transfectants, it was also inhibited because the introduced GlcNAcT-I failed to reverse the Con A hypersensitivity of Lec1 (not depicted).
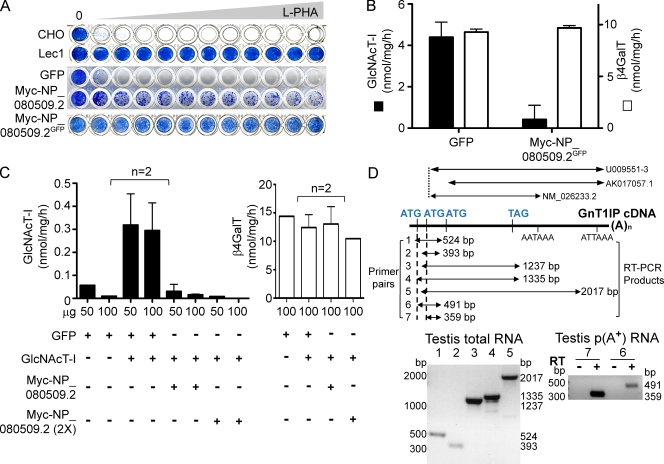
Figure 1.
Expression of NP_080509.2 in CHO cells inhibits GlcNAcT-I. (A) L-PHA resistance of CHO, Lec1 cells, or CHO cells stably expressing GFP or Myc-NP_080509.2 or Myc-NP_080509.2GFP sorted for GFP. Cells were stained with Methylene Blue when control wells with no L-PHA became confluent. The highest L-PHA concentration was 70 µg/ml. (B) GlcNAcT-I and β4GalT activities in lysates from CHO cells stably expressing Myc-NP_080509.2GFP. Error bars = SD; n = 3. (C) GlcNAcT-I and β4GalT activities in lysates of Lec1 cells transiently expressing GFP, GFP and GlcNAcT-I, or Myc-NP_080509.2 and GlcNAcT-I. Myc-NP_080509.2 (2x) received 2x cDNA. Bars represent range (n = 2). (D) Diagram of mouse cDNAs NM_026233.2, AK017057.1, and U009551-3, and the predicted products of RT-PCR. Primer pairs 1, 4, 6 detect only GnT1IP-L; pairs 2, 3, 5, 7 detect both GnT1IP-L and -S. Left panel, RT-PCR of adult testis total RNA with primer pairs 1–5. Right panel, RT-PCR of testis poly(A+) RNA with primer pairs 6 and 7.
In vitro GlcNAcT-I activity was also inhibited in lysates from GFP-sorted CHO/Myc-NP_080509.2 transfectants, but β4GalT activity was not affected (Fig. 1 B). Similarly, when mouse GlcNAcT-I was coexpressed with Myc-NP_080509.2 in Lec1 mutant cells, GlcNAcT-I activity was inhibited, but β4GalT activity was not (Fig. 1 C). However, no inhibition of GlcNAcT-I was observed when lysates from Lec1 cells expressing only GlcNAcT-I or only Myc-NP_080509.2 were mixed (not depicted), indicating that the proteins had to be coexpressed for GlcNAcT-I activity to be inactivated. Thus, the introduction of NP_080509.2 into wild-type CHO cells inhibited GlcNAcT-I but not β4GalT activity, and induced a lectin resistance phenotype characteristic of Lec1 cells that lack GlcNAcT-I. We therefore termed the novel activity GlcNAcT-I inhibitory protein (GnT1IP).
Two GnT1IP transcripts are expressed in mouse testicular germ cells
Previous reports showed that mouse 4933434I20Rik is up-regulated at the transcriptional and translational levels in spermatocytes during spermatogenesis (Iguchi et al., 2006; Chalmel et al., 2007). Northern blot analysis of total RNA from mouse tissues detected GnT1IP transcripts of ∼1.2 kb and ∼2.2 kb (5′ probe), and ∼1.8 kb and ∼2.2 kb (3′ UTR probe) only in testis (unpublished data), as also observed for a 3′ UTR probe in the SymAtlas microarray database (Su et al., 2004). The human homologue of GnT1IP (UniProtKB A6NG13) was also detected only in testis by Northern blot analysis (Hodgson et al., 2006). Rapid amplification of cDNA ends (RACE) from testis RNA revealed that transcription may stop at either of two poly(A+) signals in the 3′ UTR, and that two ATGs might serve as translational start sites for cDNAs NM_026233.2 and U009551-3 (NIA Mouse Gene Index; Fig. 1 D). RT-PCR of total testis RNA using primers from genomic DNA sequence upstream of the NM_026233.2 cDNA detected a 5′ ATG occurring after a CCACG Kozak-like sequence (Fig. 1 D). This transcript, encoding a protein with 44 additional N-terminal amino acids, was termed GnT1IP-L and given the Genbank/EMBL/DDBJ accession no. HM067443 (Fig. S1). GnT1IP encoded by the NM_026233.2 cDNA was termed GnT1IP-S. GnT1IP transcripts were detected in cDNAs generated from adult mouse testis total and poly(A+) RNA (Fig. 1 D). GnT1IP-S homologues exist in rat and human, but GnT1IP-L has been found only in mouse to date. GnT1IP appears to occur only in mammals and aa sequences are well conserved between mammalian species, with mouse and human being 63% identical and 78% similar.
GnT1IP-L is a type II transmembrane protein and GnT1IP-S may be secreted
Constructs of GnT1IP-L and -S with various tags were generated to investigate their intracellular localization (Fig. 2 A). To determine if GnT1IP-L or -S traverses the medial-Golgi, lysates from CHO transfectants were digested with enzymes that remove N-glycans. N-terminally tagged HA-GnT1IP-L (Fig. 2 A, Tag-GnT1IP-L) was sensitive to digestion with both endoglycosidase H (Endo H) and peptide-N-glycosidase F (PNGase F; Fig. 2 B), showing that HA-GnT1IP-L was modified with high mannose or hybrid N-glycans found on glycoproteins localized to the ER and cis- to early medial-Golgi. N-terminally tagged FL-HA-GnT1IP-S (Fig. 2 A, Tag-GnT1IP-S) also carried only high mannose or hybrid N-glycans (Fig. 2 B), supporting a previous finding that Myc-GnT1IP-S localized to the ER and cis- to medial-Golgi in HeLa cells (Fink et al., 2006; Sprenger et al., 2008). However, when GnT1IP-S had a C-terminal tag (Fig. 2 A, GnT1IP-S-Tag) or an internal tag (Fig. 2 A, SP-iTag2-GnT1IP-S), it was partially secreted with complex N-glycans resistant to digestion with Endo H (Fig. S2, A and B). A proportion of GnT1IP-S-HA (Fig. 2 A, GnT1IP-S-Tag) in cell lysate was modified with both complex and high mannose N-glycans (Fig. S2 A), but internally tagged SP-HA-GnT1IP-S (Fig. 2 A, SP-iTag2-GnT1IP-S) in cell lysate had only Endo H–sensitive high mannose N-glycans (Fig. S2 B). By contrast, N-terminally tagged FL-HA-GnT1IP-S (Fig. 2 A, Tag-GnT1IP-S) was not secreted (not depicted). Thus, N-terminal membrane-anchored forms of GnT1IP carry N-glycans reflecting confinement to the ER, cis- and early medial-Golgi compartments.
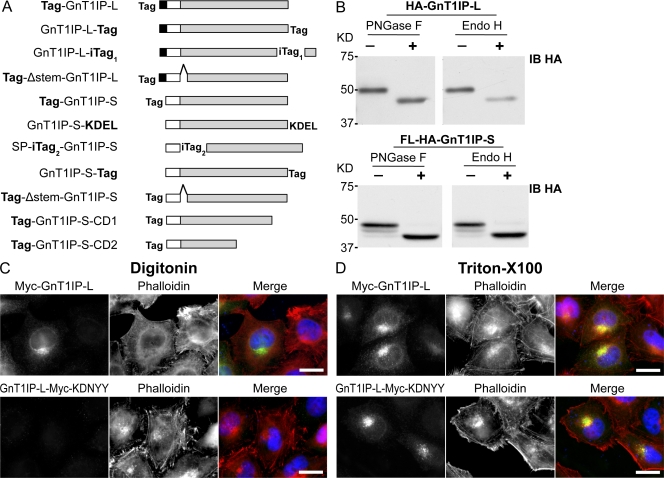
Figure 2.
GnT1IP-L is a type II transmembrane glycoprotein with high mannose N-glycans. (A) GnT1IP constructs showing the N-terminal cytosolic domain of GnT1IP-L (aa 1–48; black box); the predicted transmembrane domain of GnT1IP-L (aa 49–69; white box) or signal peptide (SP) of GnT1IP-S (aa 1–26); the Golgi lumenal domain (aa 70–417 for GnT1IP-L and aa 27–373 for GnT1IP-S; gray box), and the C-terminal deletion mutants Tag-GnT1IP-S-CD1 (39 aa deletion) and -CD2 (122 aa deletion); Tag represents FLAG-HA (FL-HA), HA, or Myc; internal tags were inserted after aa 412 of GnT1IP-L (iTag1) and after aa 26 of GnT1IP-S (iTag2); the 48 aa stem-region deletion (Δ stem) from aa 71–118 in GnT1IP-L and aa 27–74 in GnT1IP-S is shown by a hat. The KDEL sequence was inserted after aa 373 of GnT1IP-S. (B) Lysates from CHO cells expressing HA-GnT1IP-L or FL-HA-GnT1IP-S digested with PNGase F or Endo H (+) or incubated without enzyme (−) and subjected to immunoblotting using anti-HA mAb (IB HA). (C) HeLa cells transiently expressing Myc-GnT1IP-L or GnT1IP-L-Myc-KDNYY were fixed, treated with 5 µg/ml digitonin or (D) 0.2% Triton X-100, immunolabeled for Myc-tagged GnT1IP-L (green) and actin (phalloidin; red), and observed by fluorescence microscopy. Bars, 20 µm.
The C-terminal sequence of GnT1IP is KDNYY, which could be an ER membrane retention signal (Teasdale and Jackson, 1996). To investigate, a Myc tag was inserted before KDNYY (Fig. 2 A, GnT1IP-L-iTag1). GnT1IP-L-Myc-KDNYY was detected primarily in the Golgi in HeLa cells permeabilized using Triton X-100, but was not detected in cells treated with a low concentration of digitonin to permeabilize the plasma membrane and expose cytoplasmic protein domains (Fig. 2, C and D). Antibodies to the C-terminally tagged lumenal domain of CHO GlcNAcT-I-HA in the medial-Golgi and to the lumenal domain of trans-Golgi endogenous β4GalT-I behaved similarly (not depicted). By contrast, and as expected for type II transmembrane glycosyltransferases, N-terminally tagged Myc-GnT1IP-L was detected after treatment with either a low concentration of digitonin or Triton X-100, showing it was accessible to Ab in the cytoplasm (Fig. 2, C and D). Therefore, GnT1IP-L, like GlcNAcT-I and β4GalT-I, is a type II transmembrane protein with an N-terminal cytoplasmic tail. However, neither N- nor C-terminally tagged GnT1IP-L exhibited tight Golgi localization like GlcNAcT-I-HA, and partially colocalized with β4GalT-I (Fig. S2 D).
Only GnT1IP-L and membrane-anchored GnT1IP-S inhibit GlcNAcT-I
The N- and C-terminally tagged forms of GnT1IP-S exhibited different trafficking properties and may therefore have different abilities to inhibit GlcNAcT-I. To investigate, wild-type CHO cells stably expressing different GnT1IP-S constructs were examined for their ability to induce lectin resistance. In contrast to membrane-bound Myc- or FL-HA-GnT1IP-S (Fig. 1 A; Table I), unanchored versions of GnT1IP-S did not induce the Lec1 lectin resistance phenotype in wild-type CHO cells (Table I). In addition, wild-type CHO cells stably expressing GnT1IP-S with a C-terminal KDEL ER retrieval sequence did not become L-PHA resistant (Table I).
Table I.
Lectin resistance of CHO cells stably expressing GnT1IP-L or -S
| Construct transfected | Selection methoda | L-PHA | WGA |
| GnT1IP-L | Hygro | >10Rb | ndc |
| GnT1IP-L | GNA-sorted | >10R | nd |
| Tag-GnT1IP-L | Hygro | >10R | 7R |
| Tag-GnT1IP-L | GNA-sorted | >10R | nd |
| GnT1IP-L-Tag | Hygro | >10R | nd |
| GnT1IP-L-iTag1-KDNYY | Hygro | >10R | nd |
| Tag-Δstem-GnT1IP-L | Hygro | – d | nd |
| GnT1IP-S | G418 | – | – |
| GnT1IP-S-KDEL | Hygro | – | – |
| Tag-GnT1IP-S | G418 or Hygro | >10R | 3R |
| Tag-GnT1IP-S | GFP-sorted | >10R | >8R |
| Tag-GnT1IP-S (E263A or D265A) | G418 | >10R | 3R |
| GnT1IP-S-Tag | Hygro | – | – |
| SP-iTag2-GnT1IP-S | G418 or Hygro | – | – |
| Tag-Δstem-GnT1IP-S | Hygro | – | – |
| Tag-GnT1IP-S-CD1 | G418 or Hygro | – | – |
| Tag-GnT1IP-S-CD2 | G418 or Hygro | – | – |
| CHO untransfected | 5e | 2e | |
| CHO/vector or GFP | G418 or Hygro | – | – |
| Lec1 untransfected | >10R | >8R |
Transfectants were selected for stable expression of constructs in hygromycin (Hygro) or G418 or sorted by flow cytometer for binding of GNA.
Number represents fold-change in lectin resistance from CHO; R, resistant.
nd, not determined.
–, no change in lectin resistance compared to CHO untransfected.
Concentration of lectin in μg/ml that kills 90% CHO cells.
GnT1IP-L differs from GnT1IP-S by 44 N-terminal amino acids, which apparently convert the signal peptide of GnT1IP-S into a type II transmembrane domain in GnT1IP-L. Consistent with this, wild-type CHO cells stably expressing untagged GnT1IP-L or GnT1IP-L-Myc-KDNYY had markedly increased L-PHA resistance, indicating inhibition of GlcNAcT-I (Fig. 3 A). When hygromycin-resistant transfectants expressing Myc-GnT1IP-L were enriched by FACS sorting using the agglutinin Galanthus nivalus (GNA), a plant lectin that binds high mannose N-glycans, the GNA-positive population contained markedly more Myc-GnT1IP-L (Fig. 3 B), and was more resistant to L-PHA (not depicted), similar to cells expressing untagged GnT1IP-L.
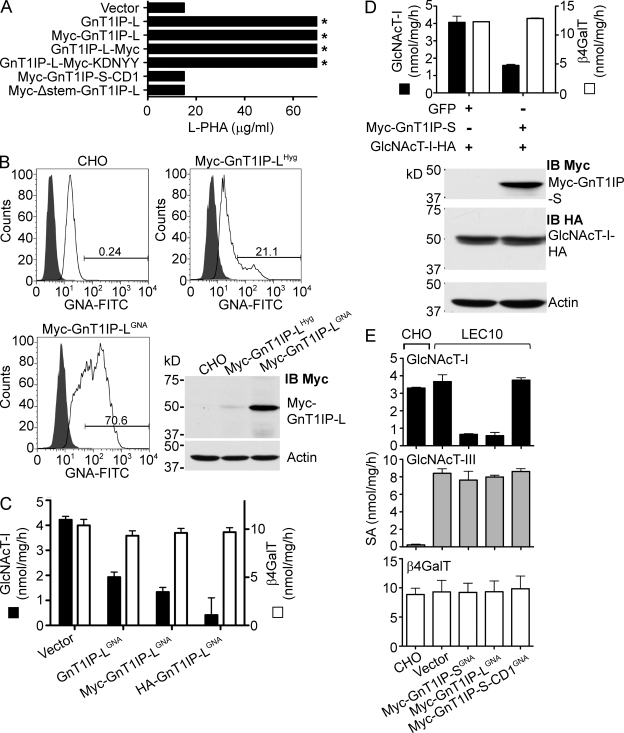
Figure 3.
GnTIP-L inhibition of GlcNAcT-I is specific and deletion mutants do not inhibit. (A) CHO cells stably expressing various GnT1IP-L constructs were tested for L-PHA resistance. Asterisk signifies that ∼50% cells survived at 70 µg/ml L-PHA. (B) CHO cells stably expressing Myc-GnT1IP-L after hygromycin selection (Myc-GnT1IP-LHyg) were sorted for GNA binding (Myc-GnT1IP-LGNA) and analyzed by flow cytometry. Shaded profiles are autofluorescence. Lysates were analyzed by immunoblot (IB Myc), stripped and reprobed with anti-actin mAb. (C) GlcNAcT-I and β4GalT activities in lysates from CHO cells stably expressing vector or GNA-sorted GnT1IP-LGNA or Myc-GnT1IP-LGNA or HA-GnT1IP-LGNA (GNA-sorted twice). Bars represent range (n = 2). (D) Lysates from Lec1 cells stably expressing GlcNAcT-I-HA and transiently expressing GFP or Myc-GnT1IP-S were assayed for GlcNAcT-I and β4GalT activities. Bars represent range of duplicates. Myc-GnT1IP-S and GlcNAcT-I-HA levels were determined by immunoblotting with actin as loading control. (E) GlcNAcT-I, GlcNAcT-III, and β4GalT specific activities (SA) in lysates from CHO and LEC10 cells stably expressing control vector or the GnT1IP construct shown. GnT1IP transfectants were GNA-sorted. Bars represent range (n = 2).
To determine which region(s) of GnT1IP are important for inducing L-PHA resistance, deletion mutants of GnT1IP-S lacking 39 aa (Fig. 2 A, Tag-GnT1IP-S-CD1) or 121 aa (Fig. 2 A, Tag-GnT1IP-S-CD2) from the C terminus, or a stem region deletion of 48 aa (Fig. 2 A, Tag-Δstem-GnT1IP-S or -L), were constructed. None of the deletion mutants induced L-PHA resistance in CHO transfectants (Fig. 3 A; Table I), although all constructs were expressed at similar levels based on immunoblotting, and GnT1IP transcript levels (not depicted). All membrane-bound GnT1IP-L and GnT1IP-S deletion mutants were sensitive to Endo H digestion (not depicted), and thus were localized to the ER and early Golgi compartments.
Consistent with their ability to induce L-PHA resistance, GnT1IP-L and Tag-GnT1IP-L reduced GlcNAcT-I but not β4GalT activity in CHO stable transfectants (Fig. 3 C). When Myc-GnT1IP-S was transiently expressed in Lec1 cells that had been rescued by GlcNAcT-I-HA, GlcNAcT-I activity was reduced but GlcNAcT-I protein levels were not affected (Fig. 3 D). The two GlcNAcT-I-HA forms observed probably reflect O-glycosylation as shown for human GlcNAcT-I (Opat et al., 2000) and as predicted by NetOGlyc 3.1 (Julenius et al., 2005).
To determine if GnT1IP inhibits an unrelated medial-Golgi enzyme, GlcNAcT-III that is endogenously expressed in LEC10 CHO glycosylation mutant cells was examined. Myc-GnT1IP-L and -S induced L-PHA resistance in LEC10 cells and the deletion mutant Myc-GnT1IP-S-CD1 did not (unpublished data), similar to their effects on wild-type CHO cells. LEC10 CHO stable transfectants expressing Myc-GnT1IP-L or -S and sorted for GNA binding had markedly reduced GlcNAcT-I activity, but GlcNAcT-III and β4GalT activities were not affected (Fig. 3 E). The deletion mutant Myc-GnT1IP-S-CD1 had no effect on GlcNAcT-I, GlcNAcT-III, or β4GalT activity (Fig. 3 E). This result was confirmed in wild-type CHO cells stably expressing a mouse cDNA encoding GlcNAcT-III-HA and transiently expressing the different GnT1IP constructs (not depicted). Finally, it was shown that GnT1IP-L and membrane-bound GnT1IP-S induced the same changes in N-glycan complement using MALDI-TOF mass spectrometry of the N-glycans released from glycoproteins of stable CHO cell transfectants (Fig. S3). Cells expressing either GnT1IP-L or Myc-GnT1IP-S produced markedly more of the Man5GlcNAc2 substrate of GlcNAcT-I than wild-type CHO cells, and synthesized very few complex N-glycans, consistent with a similar degree of inhibition of GlcNAcT-I activity.
GnT1IP-L is localized to the ER, ERGIC, and cis-Golgi compartments
When N- or C-terminally tagged GnT1IP-L was transiently expressed in HeLa cells, each colocalized with GM130, a cis-Golgi marker (Fig. 4 A; and not depicted). GnT1IP-L also partially colocalized with ERGIC-53 and an ER marker, protein disulfide isomerase (PDI; Fig. 4 A; and not depicted). Membrane-anchored Tag-GnT1IP-S was also primarily localized in the Golgi and ERGIC and partially in the ER, similar to GnT1IP-L (Fig. S4 A). The stem-region deletion mutants of N-terminally tagged GnT1IP-L and -S were also localized to the ER, cis-Golgi, and probably the ERGIC compartment (Fig. 4 B; Fig. S4 B). By contrast, the C-terminal deletion mutants of membrane-bound Myc-GnT1IP-S were primarily localized to the ER (Fig. 4 C; and not depicted).
Figure 4.
GnT1IP-L localizes to the ER, ERGIC, and Golgi. (A–C) HeLa cells transiently expressing different GnT1IP constructs were fixed, immunolabeled for Myc-tagged GnT1IP-L (red) and GM130 or PDI (green), or HA-tagged GnT1IP-L (green) and ERGIC-53 (red), followed by confocal microscopy (A and C) or fluorescence microscopy (B). (D) HeLa cells transiently expressing HA-GnT1IP-L or GlcNAcT-I-HA were treated with CHX for 45 min followed by BFA for 30 min, fixed, immunolabeled for HA-GnT1IP-L or GlcNAcT-I-HA (green) and ERGIC-53 (red), and analyzed by fluorescence microscopy. Bars, 10 µm.
To further investigate GnT1IP-L trafficking, the effects of brefeldin A (BFA) were examined under conditions in which matrix proteins such as GRASP65 and ERGIC-53, normally localized to the cis-Golgi and ERGIC, are redistributed to ER exit sites (Lippincott-Schwartz et al., 1989; Ward et al., 2001). BFA inhibits ADP-ribosylation factor 1 (Arf1) activation on the Golgi membrane by forming a complex with the guanine nucleotide exchange factor GBF1, and thereby causes retrograde transport of Golgi enzymes into the ER (Niu et al., 2005). HeLa cells transiently expressing HA-GnT1IP-L were treated with cycloheximide (CHX) followed by BFA for 30 min after CHX removal. As expected, the medial-Golgi enzymes CHO GlcNAcT-I-HA (Fig. 4 D) and endogenous α-mannosidase II (ManII, not depicted) were relocalized to the ER by BFA treatment. GnT1IP-L was also found in the ER and some GnT1IP-L was observed at ER exit sites, marked by ERGIC-53 (Fig. 4 D).
GnT1IP-L relocates GlcNAcT-I to the cis-Golgi, ERGIC, and ER
When N- or C-terminally tagged GnT1IP-L was transiently coexpressed with GlcNAcT-I-HA in HeLa cells, a major proportion of GlcNAcT-I was mislocalized to the ER (Fig. 5 A; and not depicted). A similar result was obtained when membrane-bound Myc-GnT1IP-S was coexpressed with GlcNAcT-I-HA in HeLa cells (Fig. 5 A). In Lec1 CHO cells, transient expression of GnT1IP-L or membrane-bound GnT1IP-S caused stably expressed GlcNAcT-I-HA to be mislocalized to the ER, and possibly the ERGIC and cis-Golgi compartments (Fig. 5 B). As expected, relocation by GnT1IP was specific for GlcNAcT-I and was not observed when GlcNAcT-I-HA was coexpressed with empty vector or the Golgi GDP-Fuc transporter Slc35c1, or the related multi-transmembrane Golgi protein Slc35c2 (not depicted). Most importantly, all deletion mutants of membrane-bound GnT1IP failed to mislocalize GlcNAcT-I (Fig. 5 C; and not depicted). In GlcNAcT-I-HA–rescued Lec1 cells, Myc-GnT1IP-S-CD1 weakly affected GlcNAcT-I-HA Golgi localization (Fig. 5 B). By contrast, overexpression of Myc-GnT1IP-L or -S did not result in ER localization for endogenous ManII or β4GalT-I (Fig. 5 D; Fig. S4 B). Transient coexpression of either mouse GlcNAcT-III-HA or ManIIx-HA with cDNAs encoding various other proteins, or stable expression of GlcNAcT-III and ManIIx followed by transient expression of other proteins, all resulted in their mislocalization as well as affecting untransfected cells, which prevented us from interpreting specific effects of GnT1IP on their intracellular localization.
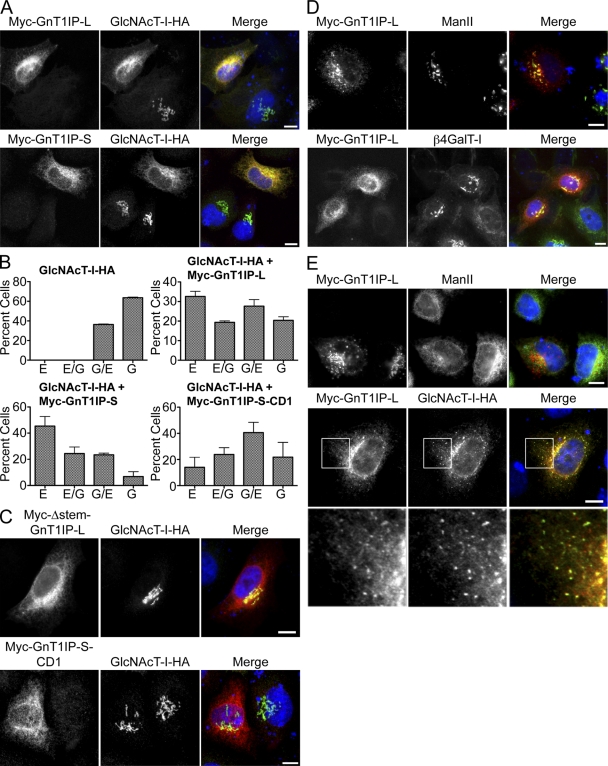
Figure 5.
GnT1IP mislocalizes GlcNAcT-I-HA. (A) Myc-GnT1IP-L and -S were transiently coexpressed with GlcNAcT-I-HA in HeLa cells, fixed, immunolabeled for Myc (red) and for HA (green), and examined by confocal microscopy. Bars 10 µm. (B) Quantitation of effects of transiently expressed Myc-GnT1IP-L, Myc-GnT1Ip-S, or Myc-GnT1IP-S-CD1 on the localization of GlcNAcT-I-HA stably expressed in Lec1 cells. Localization in ∼300 cells per condition was classified into ER (E), more ER than Golgi (E/G), more Golgi than ER (G/E), or Golgi (G), and compared with Lec1 cells stably expressing GlcNAcT-I-HA alone. Error bars = SD; n = 3. (C) Deletion mutants of GnT1IP were coexpressed with GlcNAcT-I-HA in HeLa cells, fixed, immunolabeled for Myc (red) and for HA (green), and examined by immunofluoresence (top) or confocal microscopy (bottom). (D) Myc-GnT1IP-L was transiently expressed in HeLa cells, fixed, immunolabeled for Myc-GnT1IP-L (red) and endogenous ManII or β4GalT-I (green), and observed by fluorescence or confocal microscopy. (E) Myc-GnT1IP-L was either transiently expressed alone or coexpressed with GlcNAcT-I-HA in HeLa cells overnight, CHX was added for 1 h, followed by CHX and BFA for 30 min, cells were fixed, immunolabeled for Myc-GnT1IP-L (red) and GlcNAcT-I-HA or endogenous ManII (green), and observed by fluorescence microscopy. White boxes in the middle panel are enlarged below. Bars, 10 µm.
Because GnT1IP-L caused GlcNAcT-I to go to the ER, BFA treatment was performed to determine if relocalized GlcNAcT-I was also in the cis-Golgi, ERGIC, and at ER exit sites. GlcNAcT-I-HA was coexpressed with Myc-GnT1IP-L in HeLa cells that were treated with BFA in the presence of CHX followed by immunofluorescence microscopy. Cells expressing both GlcNAcT-I-HA and Myc-GnT1IP-L had punctate costaining around the nucleus, typical of ERGIC localization, in addition to reticular costaining of the ER, whereas endogenous ManII localized only to the ER in the presence of Myc-GnT1IP-L and did not colocalize with GnT1IP-L at punctate ER exit sites after BFA treatment (Fig. 5 E).
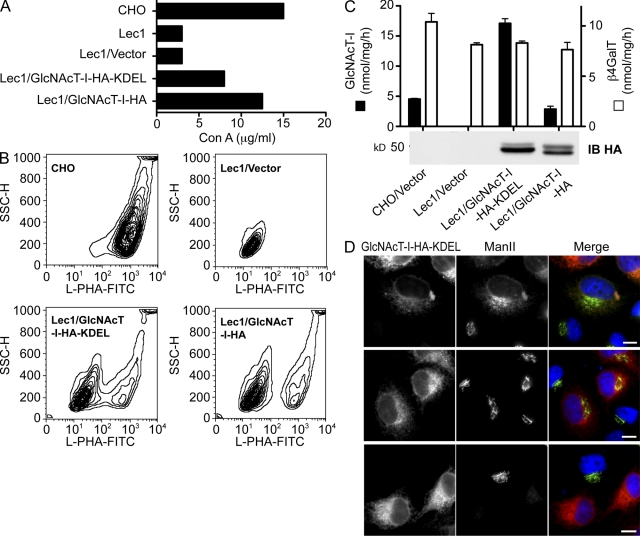
To determine if relocation of GlcNAcT-I to the ER might be sufficient to cause its inactivation, a CHO GlcNAcT-I-HA-KDEL construct was investigated. In Lec1 cells, GlcNAcT-I-HA-KDEL that colocalized in the ER with PDI (not depicted) partially rescued the Lec1 phenotype in a Con A resistance test (Fig. 6 A), and increased L-PHA binding (Fig. 6 B). Surprisingly, and despite its somewhat modest induction of lectin resistance, GlcNAcT-I-HA-KDEL had higher GlcNAcT-I activity than GlcNAcT-I-HA in the in vitro GlcNAcT-I assay of cell lysate, though GlcNAcT-I-HA-KDEL was not more highly expressed than GlcNAcT-I-HA (Fig. 6 C). Transiently expressed GlcNAcT-I-HA-KDEL also displayed higher activity than GlcNAcT-I-HA in Lec1 cell lysates (not depicted). The higher activity of GlcNAcT-I-HA-KDEL may reflect the environment of the ER versus the Golgi. In the Golgi GlcNAcT-I occurs in a kin recognition complex with ManII and GlcNAcT-II (Nilsson et al., 1994; Opat et al., 2000; Hassinen et al., 2010), which may be disrupted in the ER. Although endogenous ManII mislocalized to the ER with GlcNAcT-I-HA-KDEL in some cells (Fig. 6 D, top), in others its Golgi localization was unchanged (Fig. 6 D, middle). Interestingly, cells in which GlcNAcT-I-HA-KDEL was highly expressed lost endogenous ManII completely, presumably due to ER degradation (Fig. 6 D, bottom).
Figure 6.
GlcNAcT-I-KDEL localized mainly in the ER is active. Lec1 cells stably expressing empty vector, GlcNAcT-I-HA-KDEL, or GlcNAcT-I-HA were analyzed for (A) Con A resistance and (B) L-PHA-FITC binding by flow cytometry. (C) GlcNAcT-I and β4GalT activities in transfectant lysates were determined. Bars represent range (n = 2). (D) HeLa cells transiently expressing GlcNAcT-I-HA-KDEL were fixed and immunofluorescently labeled for GlcNAcT-I-HA-KDEL (red) and endogenous ManII (green). Bars, 10 µm.
GnT1IP interacts directly with GlcNAcT-I
Colocalization of GlcNAcT-I with GnT1IP-L in the ER indicated that GnT1IP-L might interact with GlcNAcT-I. To investigate, Myc-GnT1IP-L or -S or GlcNAcT1-Myc were cotransfected into CHO cells with various HA-tagged Golgi enzymes or a control HA-tagged protein, aa 321–600 of the p85 subunit of PI,3 kinase (HA-p85ni). Lysates were incubated with anti-Myc beads and bound proteins (IP Myc) were analyzed by immunoblotting (IB HA and IB Myc) as described in Materials and methods. Protein input blots showed that constructs were equivalently expressed. IP Myc blots determined bound protein levels, as well as equivalence of anti-Myc beads based on the signal from immunoglobulin light chain.
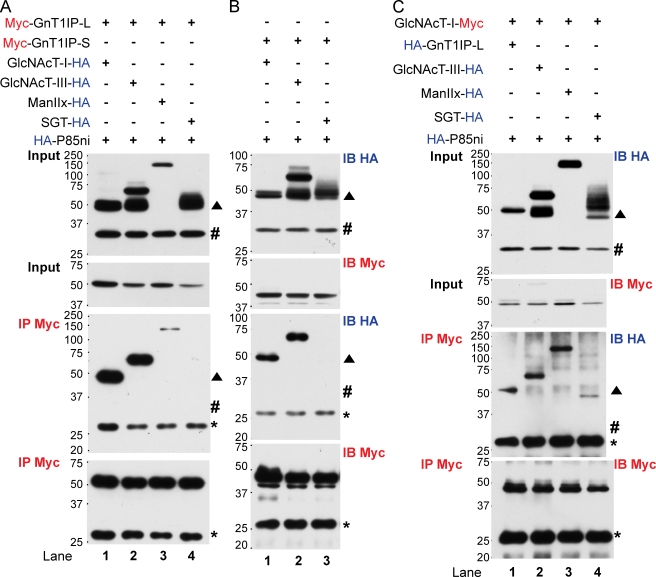
Myc-GnT1IP-L on beads bound GlcNAcT-I-HA but did not bind the control HA-p85ni (Fig. 7 A, lane 1; blot IP Myc/IB HA). Coimmunoprecipitaiton also occurred between GlcNAcT-I-HA and Myc-GnT1IP-S (Fig. 7 B, lane 1). When the tags were reversed, HA-GnT1IP-L was pulled down by GlcNAcT-I-Myc on beads (Fig. 7 C, lane 1). GlcNAcT-I-Myc also pulled down the medial-Golgi resident enzymes GlcNAcT-III-HA and ManIIx-HA (Fig. 7 C, lanes 2 and 3), as anticipated based on previous evidence of kin recognition complexes between GlcNAcT-I and ManII. However, only a minor fraction of the short form of the trans-Golgi resident bovine β4GalT-I (SGT-HA; Fig. 7 C, lane 4) was pulled down. Myc-GnT1IP-L and -S also pulled down the full-length form of medial-Golgi GlcNAcT-III-HA (Fig. 7 A, lane 2; Fig. 7 B, lane 2) and ManIIx-HA was moderately coimmunoprecipitated by Myc-GnT1IP-L (Fig. 7 A, lane 3). However, trans-Golgi SGT-HA was not pulled down by either Myc-GnT1IP-L or -S (Fig. 7 A, lane 4; Fig. 7 B, lane 3). In summary, medial-Golgi enzymes GlcNAcT-I, GlcNAcT-III, and ManIIx coimmunoprecipitated with membrane-bound GnT1IP-L and -S. However, only GlcNAcT-I enzyme activity was inhibited by membrane-bound GnT1IP-L and -S (Fig. 3 E).
Figure 7.
GnT1IP-L interacts with medial-Golgi but not trans-Golgi enzymes. (A) Myc-GnT1IP-L or (B) Myc-GnT1IP-S were coexpressed with GlcNAcT-I-HA, GlcNAcT-III-HA, ManIIx-HA, β4GalT-I lacking 13 N-terminal amino acids (SGT-HA), or aa 321–600 of the p85 subunit of PI3 kinase (HA-p85ni) in CHO cells. Lysates were immunoprecipitated with anti-Myc beads, and 40% of the beads were analyzed by immunoblotting on two blots (IP-Myc with IB-HA or IB-Myc). Input (1/40th lysate) was immunoblotted with IB-HA or IB-Myc. (C) GlcNAcT-I-Myc was coexpressed with HA-GnT1IP-L, GlcNAcT-III-HA, ManIIx-HA, or SGT-HA in CHO cells and lysates were treated as in A. ▲, Fragment of GlcNAcT-III-HA not bound by GnT1IP; #, position or predicted position of HA-p85ni; *, immunoglobulin light chains from anti-Myc beads show equal concentration of beads.
Interestingly, secreted SP-HA-GnT1IP-S also interacted with GlcNAcT-I, the full-length GlcNAcT-III and ManIIx, but not the trans-Golgi sialyltransferase STX (Fig. S5 A). This is consistent with confocal microscopy results in which secreted SP-Myc-GnT1IP-S was partially colocalized with GlcNAcT-I-HA in the Golgi (Fig. S2 C). The C-terminal deletion mutant Myc-GnT1IP-S-CD2 pulled down GlcNAcT-I-HA, GlcNAcT-III-HA, and ManIIx-HA equivalently, but did not bind to SGT-HA (Fig. S5 B). Similarly, the stem-deletion mutant Myc-Δstem-GnT1IP-L pulled down GlcNAcT-I-HA, full-length GlcNAcT-III, and a minor fraction of SGT-HA (Fig. S5 C, lanes 2–4). However, the interaction between Myc-Δstem-GnT1IP-L and GlcNAcT-I-HA was somewhat weaker than between Myc-GnT1IP-L and GlcNAcT-I-HA (Fig. S5 C, lanes 1 and 2). All coimmunoprecipitations were specific because the N-terminal deletion fragment of GlcNAcT-III-HA at ∼50 kD or control HA-p85ni were not pulled down by GlcNAcT-I, GnT1IP-L, or mutant constructs.
GnT1IP expression is highly regulated during mouse spermatogenesis
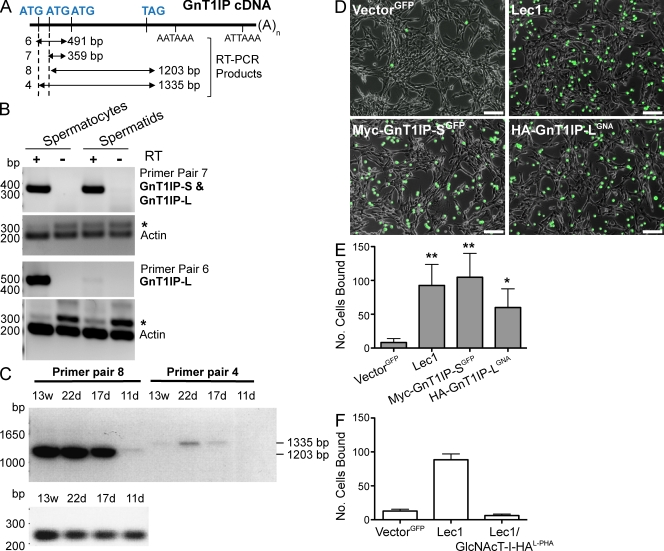
GnT1IP transcripts are present in testicular germ cells but not Sertoli cells (Chalmel et al., 2007). In mouse testis from postnatal day 22 to the adult, GnT1IP transcripts are transported from ribonucleoprotein (RNP) particles that contain translationally inactive mRNA into translationally active polysomes (Iguchi et al., 2006). To investigate GnT1IP-L and -S in germ cells, total RNA from purified mouse spermatocytes and spermatids was subjected to RT-PCR with specific primer sets (Fig. 8 A). GnT1IP-L was expressed well in spermatocytes, but not in spermatids (Fig. 8 B). GnT1IP-S was expressed well in spermatids and probably in spermatocytes (Fig. 8 B). Transcriptional regulation of GnT1IP was examined in testis during the first wave of spermatogenesis at postnatal days 11, 17, and 22 and in post-pubertal adult males at 13 wk. Transcription of GnT1IP-L and -S was up-regulated in 17-d mice and GnT1IP-S maintained a similar expression level from day 17 until the adult (Fig. 8 C). However, GnT1IP-L reached its highest expression level in 22-d testis, and was barely detected in the adult (Fig. 8 C). Primer pair 6, which detected only GnT1IP-L cDNA (Fig. 8 A), also showed that GnT1IP-L was reduced in expression after day 22 (not depicted). In summary, GnT1IP-L and -S are expressed in mouse testicular germ cells, and GnT1IP-L expression is highly regulated during the first wave of spermatogenesis.
Figure 8.
GnT1IP expression is regulated during spermatogenesis and enhances binding to Sertoli cells. (A) Diagram of GnT1IP cDNA and primer pairs for RT-PCR. (B and C) Total RNA from spermatocytes and spermatids partially purified from testes at postnatal day 11 (11 d) to 13 wk (13 w) was subjected to RT-PCR. Primer pair 6 detected only GnT1IP-L; pair 7 detected both GnT1IP-L and -S. Actin primers generated a 280-bp cDNA or 320-bp product from genomic DNA (*). Primer pairs 4 and 8 detected GnT1IP-L and -S coding regions. (D) Equal numbers of CHO cells stably expressing vector GFP, Myc-GnT1IP-S, HA-GnT1IP-L, or Lec1 cells labeled with CFDA-SE were assayed for binding to TM4 Sertoli cells. Images of wells after washing were taken by fluorescence microscopy (FITC channel merged with phase contrast, 10x). Bars, 40 µm. (E) Three pictures taken from fields similar to those shown in D close to the center of each well were processed by NIH ImageJ software, green cells were counted, and triplicates averaged. Error bars = SD; n = 4 experiments. **, P < 0.01; *, P < 0.05 based on the two-tailed Student’s t test. (F) Lec1 cells expressing GlcNAcT-I-HA selected for hygromycin resistance or sorted for L-PHA binding (GlcNAcT-I-HAL-PHA) were tested for binding to TM4 Sertoli cells as in D. Bars represent range (n = 2).
It was previously shown that truncated N-glycans occurring in the ManIIx knockout mouse inhibit the binding of germ cells to Sertoli cells (Akama et al., 2002). To investigate whether high mannose N-glycans induced by the inhibition of GlcNAcT-I affect binding to Sertoli cells, CHO cells stably expressing HA-GnT1IP-LGNA (GNA-sorted) or Myc-GnT1IP-SGFP (GFP-sorted) were tested for binding to mouse TM4 Sertoli cells (Mather, 1980). Both transfectants bound much better to TM4 Sertoli cells than control CHO cells expressing GFP alone (Fig. 8, D and E). Similar results were found for Myc-GnT1IP-S with fixed 15P-1 Sertoli cells (not depicted). Lec1 mutant CHO cells bound to Sertoli cells as well as the GnT1IP transfectants. Importantly, Lec1 cells rescued with GlcNAcT-I-HA, which were sorted for L-PHA binding and the expression of complex N-glycans, lost their ability to adhere strongly to Sertoli cells (Fig. 8 F). Therefore, inhibition of GlcNAcT-I by GnT1IP during spermatogenesis, leading to enhanced expression of high mannose N-glycans on germ cell glycoproteins, might be important for facilitating Sertoli–germ cell interactions during specific stages of spermatogenesis (Fig. 9).
Figure 9.
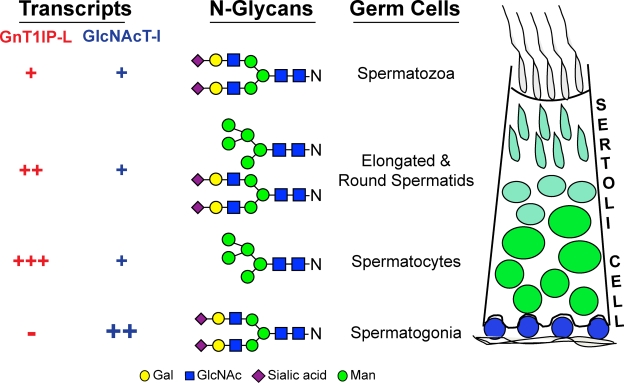
Summary diagram. The relative levels of GnT1IP-L and GlcNAcT-I transcripts in germ cells vary during spermatogenesis as indicated (Chalmel et al., 2007; Fig. 8), and the N-glycans on germ cells vary as shown by changes in lectin binding (Jones et al., 1992). In spermatogonia, negligible levels of the new GlcNAcT-I inhibitor GnT1IP-L described herein, and high GlcNAcT-I leads to the synthesis of complex N-glycans. In spermatocytes, increased expression of GnT1IP-L accompanied by down-regulation of GlcNAcT-I leads to the expression of high mannose N-glycans on germ cell glycoproteins. This change in N-glycans is predicted to enhance the binding of spermatocytes to Sertoli cells (Fig. 8). At later stages of spermatogenesis GnT1IP-L expression levels decrease while GlcNAcT-I levels increase, and spermatids express complex N-glycans and have reduced binding to Sertoli cells.
Discussion
We describe here a novel mechanism of regulated inhibition of complex and hybrid N-glycan synthesis, by a newly identified activity termed GlcNAcT-I inhibitory protein, or GnT1IP. Membrane-bound forms of GnT1IP specifically inhibit GlcNAcT-I activity, leading to the termination of N-glycan synthesis in the medial-Golgi at the intermediate Man5GlcNAc2Asn. This causes cell surface glycoproteins to carry mainly high mannose N-glycans, thereby profoundly altering recognition of the cell by glycan-binding proteins such as galectins (Patnaik et al., 2006). GnT1IP physically interacts with GlcNAcT-I and specifically inhibits GlcNAcT-I activity. Membrane-bound forms of GnT1IP cause GlcNAcT-I to become mislocalized to the cis-Golgi, ERGIC, and ER. However, mislocalization by itself does not cause GlcNAcT-I to be inactivated because mislocalized GlcNAcT-I-KDEL is able to rescue the Lec1 CHO mutant to a good extent, and exhibits robust GlcNAcT-I activity in cell lysates. Cells highly expressing GlcNAcT-I-HA-KDEL in the ER either mislocalized endogenous ManII to the ER or led to its disappearance. Previous reports have shown that GlcNAcT-I occurs in a complex with ManII and GlcNAcT-II (Nilsson et al., 1994, 1996; Opat et al., 2000; Hassinen et al., 2010).
GnT1IP deletion mutants did not inactivate or redirect GlcNAcT-I to the ER, but surprisingly did coprecipitate with GlcNAcT-I (Fig. S5). Secreted GnT1IP-S (SP-Myc-GnT1IP-S) also interacted with GlcNAcT-I, consistent with immunofluorescence microscopy showing GnT1IP-S colocalized in the Golgi with GlcNAcT-I. Soluble GlcNAcT-I also localizes to the Golgi in a complex with medial-Golgi enzymes before being secreted (Opat et al., 2000). We show here that GnT1IP-L associates with the medial-Golgi enzymes ManIIx and GlcNAcT-III, but does not inhibit the activity of GlcNAcT-III. GlcNAcT-I dynamically cycles between the ER and Golgi (Hoe et al., 1995; Opat et al., 2001), a cycle obviously interrupted by membrane-bound GnT1IP. Perhaps membrane-bound GnT1IP disrupts the medial-Golgi complex that contains GlcNAcT-I by interacting with each component, prevents the GlcNAcT-I cycle, and specifically inhibits GlcNAcT-I activity. GnT1IP does not associate with or mislocalize the trans-Golgi or TGN glycosyltransferases β4GalT-1 and STX, and does not inhibit β4GalT or GlcNAcT-III enzyme activity.
The inhibitory function of GnT1IP contrasts with previous examples of glycosyltransferase complexes, which are required to induce, promote, or preserve enzyme activity (Giraudo et al., 2001; Akasaka-Manya et al., 2006; Seko and Yamashita, 2008; Aryal et al., 2010). Membrane-bound GnT1IP causes dominant inhibition of GlcNAcT-I that could be useful for glycosylation engineering. Thus, transient expression of GnT1IP in cells expressing recombinant therapeutic antibodies could be used for rapid testing of the effect of converting complex to high mannose N-glycans on antibody-dependent cellular cytotoxicity (ADCC; Carter, 2001; Kanda et al., 2007; Mori et al., 2007). High mannose N-glycans have little core fucosylation and induce ADCC much more effectively than IgG with a complex fucosylated N-glycan (Zhou et al., 2008). Although most glycoproteins with high mannose N-glycans are removed from serum by the liver within minutes, antibodies with high mannose N-glycans have a longer lifespan (Millward et al., 2008; Zhou et al., 2008).
Importantly, mouse GnT1IP is expressed almost solely in testes (Chalmel et al., 2007), and the splice form producing transmembrane GnT1IP-L is confined to testicular germ cells, beginning with spermatocytes and being down-regulated after the spermatid stage (Fig. 8). GnT1IP-S is found in spermatids and probably spermatocytes, but is predicted not to inhibit GlcNAcT-I. GnT1IP-L expression is highly regulated during spermatogenesis (Fig. 8) and translation of GnT1IP transcripts is also strictly controlled (Iguchi et al., 2006). We have shown that GnT1IP-induced high mannose N-glycans markedly increase cell adhesion to Sertoli cells (Fig. 8), indicating a potential role for the regulated expression of GnT1IP in the modulation of interactions between Sertoli and germ cells essential to spermatogenesis. Consistent with this, lectin studies suggest complementary expression of high mannose and complex N-glycans in rat testes (Jones et al., 1992). Thus, L-PHA binding to complex N-glycans is decreased during the transition from spermatogonia to spermatocytes, whereas Con A binding to high mannose N-glycans increases. Later in spermatogenesis, complex N-glycans replace high mannose N-glycans to become the major N-glycans in germ cells. These dynamics coincide with the timing and cell specificity of GnT1IP expression in testes, especially GnT1IP-L (Fig. 9). Other evidence of the importance of N-glycans in germ-Sertoli cell interactions comes from the ManIIx knockout mouse that permanently lacks a subset of complex N-glycans, leading to increased expression of high mannose and hybrid N-glycans (Akama et al., 2002, 2006). Males lacking ManIIx exhibit defective adhesion of germ cells to Sertoli cells, release immature germ cells into epididymis, and are sterile (Akama et al., 2002).
Complex N-glycans are essential for mammalian development beyond mid-gestation (Ioffe and Stanley, 1994; Metzler et al., 1994). They carry sugar determinants recognized by selectins and galectins that mediate cell adhesion and migration (Cummings and Esko, 2009; Cummings and Liu, 2009) and regulate growth factor signaling (Zhao et al., 2008; Dennis et al., 2009; Song et al., 2010). Interestingly, mouse GlcNAcT-I is highly expressed in spermatogonia and down-regulated in spermatocytes and spermatids, in a complementary expression pattern with GnT1IP (Chalmel et al., 2007). Thus, reduced expression of GlcNAcT-I coincides with up-regulation of GnT1IP, both leading to increased expression of high mannose N-glycans and enhanced Sertoli–germ cell binding during differentiation from spermatocytes to spermatids (Fig. 9). The question of whether temporary inhibition of GlcNAcT-I during spermatogenesis is required for male fertility awaits the generation of GnT1IP conditional knockout mice, but is of interest for human spermatogenesis. GnT1IP expression is also highly restricted to human testes (Hodgson et al., 2006; Chalmel et al., 2007) and transcript levels of human GnT1IP (Ensembl, gene ID: AC093671.1) are misregulated in some forms of human spermatogenic failure including patients with Sertoli Cell Only syndrome, or men with many early spermatids but no late spermatids, in which a fourfold decrease in GnT1IP transcripts was observed (Spiess et al., 2007).
Materials and methods
RNA isolation and RT-PCR
Testes from adult mice were homogenized in TRIzol (Invitrogen), extracted with chloroform and isopropyl alcohol, and the RNA precipitate was dissolved in RNase-free water and stored at −80°C. Total RNA (1–2 µg) was used to synthesize cDNA primed by oligo-dT (T12-18; Invitrogen) and extended using SuperScript II reverse transcription (Invitrogen). Control reactions contained double-distilled water in place of reverse transcription. After 50 min elongation at 42°C, reactions were stopped at 70°C for 10 min and stored at −20°C. Partially purified mouse spermatocytes or spermatids provided by Paula Cohen (Cornell University, NY) were lysed and converted into cDNA using the Cells-to-cDNA II kit (Applied Biosystems). Primer pairs used for detecting GnT1IP-S and -L transcripts were: pair 1, 5′-GATTGTAAAGTGTGGTCAAGAGGGGC-3′: 5′-GGGTAAAGTTTCCCTGCTGTCCTTA-3′; pair 2, 5′-TGTGCAGCCTGGTGTTGGCAGT-3′: 5′-GGGTAAAGTTTCCCTGCTGTCCTTA-3′; pair 3, 5′-TGTGCAGCCTGGTGTTGGCAGT-3′: 5′-GAGAAAATTTGATATTTGCTGTGCC-3′; pair 4, 5′-ATGTGCCTGGGAGAAAGTGTTGGGGACC-3′: 5′-GAGAAAATTTGATATTTGCTGTGCC-3′; pair 5, 5′-TGTGCAGCCTGGTGTTGGCAGT-3′: 5′-TCGGTCATGCATTCCACATGAGTTTACAGGA-3′; pair 6, 5′-ATGTGCCTGGGAGAAAGTGTTGGGGACC-3′: 5′-GGGTAAAGTTTCCCTGCTGTCCTTA-3′; pair 7, 5′-ATGAAGGCCAAGGACGTTAACTTGC-3′: 5′-GGGTAAAGTTTCCCTGCTGTCCTTA-3′; pair 8, 5′-ATGAAGGCCAAGGACGTTAACTTGC-3′: 5′-GAGAAAATTTGATATTTGCTGTGCC-3′. GlcNAcT-I transcripts were detected with primers 5′- CCAAGCTTCTCCCKGYGGGGGCCAGG-3′: 5′-GGCRGAGCCCAGRARGGAMAGGCAGGWGCT-3′ (Chen and Stanley, 2003) and β-actin with primers 5′-GTGGGCCGCTCTAGGCACCA-3′: 5′-TGGCCTTAGGGTTCAGGGGG-3′.
cDNA constructs
GnT1IP-L and related cDNAs were cloned into pcDNA3.1/Hygro (hygromycin) or pIRES2-EGFP/Neo (neomycin or G418) or pcDNA3/Neo vectors using primers 5′-GAGACTGAGCTCCACCATGTGCCTGGGAGAAAGTGTTGGGGACC-3′ and 5′-TCGAGTCGACCTAGTAATAATTATCCTTGAGGTGCTG-3’. HA-GnT1IP-L and Myc-GnT1IP-L were constructed with HA (2YPYDVPDYASL12) and Myc (2EQKLISEEDL11) fused at the N terminus of GnT1IP-L, respectively. Myc was also fused at the C terminus of GnT1IP-L after 417 aa (418EQKLISEEDL427) for GnT1IP-L-Myc or after 412 aa (413EQKLISEEDL422) for GnT1IP-L-Myc-KDNYY (GnT1IP-L-iTag1). For Myc-Δstem-GnT1IP-L, aa 71–118 of GnT1IP-L were deleted. GnT1IP-S and related constructs were cloned into pcDNA3.1/Hygro or pIRES2-EGFP/Neo or pcDNA3/Neo expression vectors using primers 5′-GAGACTGAGCTCCACCATGAAGGCCAAGAACGTTAACTTGCTCTTC-3′ and 5′-CTCTGAGGATCCCTAGTAATAATTATCCTTGAGGTGCTG-3′. GnT1IP-S was fused with Myc (2EQKLISEEDL11) for Myc-GnT1IP-S or fused with FL-HA (2DYKDDDDKGSYPYDVPDYASLRPLE26) for FL-HA-GnT1IP-S. For Myc-Δstem-GnT1IP-S, aa 27–74 of GnT1IP-S were deleted. For Myc- or FL-HA-GnT1IP-S-CD1, the C-terminal 334–373 aa of GnT1IP-S were deleted. For Myc- or FL-HA-GnT1IP-S-CD2, the C-terminal 252–373 aa of GnT1IP-S were deleted. For the E253A or D255A mutations of GnT1IP-S, Myc-GnT1IP-S was used as template in combination with the QuikChange XL site-directed mutagenesis kit (Agilent Technologies) and primers 5′-CCAAGGCCACGTATTATTTACAGCTAGCAGATGATATCGTAGCAA-3’ and 5′-TTGCTACGATATCATCTGCTAGCTGTAAATAATACGTGGCCTTGG-3’ for Myc-GnT1IP-S (ADD), or with primers 5′-CCAAGGCCACGTATTATTTACAGCTAGAAGATGCTATCGTAGCAA-3’ and 5’-TTGCTACGATAGCATCTTCTAGCTGTAAATAATACGTGGCCTTGG-3’ for Myc-GnT1IP-S (EDA). For GnT1IP-S-KDEL, the 374KDEL377 sequence was fused after aa 373 of GnT1IP-S by PCR. An HA (27YPYDVPDYASL37) or Myc (27EQKLISEEDL36) tag was inserted between aa 26 and 27 of GnT1IP-S by PCR for SP-iTag2-GnT1IP-S. For GnT1IP-S-HA, HA (374YPYDVPDYASL384) was fused after aa 373 of GnT1IP-S. For C-terminal HA or Myc-tagged CHO GlcNAcT-I, HA (448YPYDVPDYASL458) or Myc (448EQKLISEEDL457) was fused after aa 447 of GlcNAcT-I (Chen and Stanley, 2003) and cloned into pcDNA3.1/Hygro between HindIII and XbaI for GlcNAcT-I-HA or HindIII and BamHI for GlcNAcT-I-Myc. For GlcNAcT-I-HA-KDEL, the HA tag and 458SEKDEL464 sequences were fused after aa 447 of CHO GlcNAcT-I and cloned into pcDNA3.1/Hygro between HindIII and BamHI. For C-terminal HA-tagged mouse GlcNAcT-III, HA (539YPYDVPDYA547) was fused after aa 538 of GlcNAcT-III (Bhattacharyya et al., 2002) and cloned into pcDNA3.1/Hygro between HindIII and BamHI. For C-terminal HA-tagged mouse ManIIx, ManIIx DNA sequence was amplified from mouse testis cDNA and the HA (1153YPYDVPDYA1161) sequence was fused after aa 1152 of ManIIx (Akama et al., 2006) and cloned into pcDNA3.1/Hygro between HindIII and BamHI. For C-terminal HA-tagged bovine SGT, which lacks N-terminal 13 aa of bovine β4GalT-I, HA (390YPYDVPDYA398) was fused after aa 389 of bovine SGT. Bovine SGT was amplified from a pSVL vector containing a bovine β4GalT-I cDNA, a gift of Joel Shaper (Johns Hopkins Medicine, Baltimore, MD; Russo et al., 1990), and cloned into pcDNA3.1/Hygro between BamHI and XbaI. STX-HA was amplified from pcDNA3 containing a mouse St8sia2 cDNA, a gift from Rita Gerardy-Schahn (Medizinische Hochschule, Hannover, Germany), and cloned in pcDNA3.1/Hygro between KpnI and XbaI. N-terminal HA-tagged human p85ni, consisting of the iSH2 and nSH2 domains of PI3K (aa 320–600) (Wu et al., 2009), was a gift of Jonathan Backer (Albert Einstein College of Medicine, New York, NY).
Cell culture
HeLa cells were from the late Dennis Shields (Albert Einstein College of Medicine, New York, NY). CHO cells were grown and maintained in α-modified Eagle’s medium with 10% FBS (Gemini Bio-Products) in suspension or on plates incubated in 5% CO2 at 37°C. HeLa cells were grown and maintained in DME with 10% FBS on plates in 5% CO2 at 37°C. Sertoli TM4 cells (CRL-1715) and 15P-1 (CRL-2618) Sertoli cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in medium containing a 1:1 mixture of DME and Ham’s F12 medium, 5% horse serum, 2.5% FBS, 2.5 mM l-glutamine, 15 mM Hepes, 0.5 mM sodium pyruvate, and 1.2 g/L sodium bicarbonate.
Transfection methods
Expression constructs were transfected using Fugene 6 (Roche) or polyethylenimine (PEI). For Fugene 6, cells cultured on plates overnight were transfected according to the manufacturer’s instructions with 3 µl Fugene per μg vector. For PEI, 11 mg PEI (PEI87K, a gift of Robert Haltiwanger, Stony Brook University, NY) was dissolved in 10 ml distilled water, sterilized by filtration, and frozen at −80°C. After 2 h, the PEI solution was thawed at 37°C and the cycle was repeated two more times before transfection. PEI and DNA were added to separate tubes containing medium in a ratio of 3 µl:1 µg, then mixed and incubated for 10 min at room temperature. The mixture was added drop-wise to cells on a plate containing culture medium and incubated at 37°C overnight. For immunoprecipitation experiments, 3.4 × 107 CHO cells were plated on a 10-cm plate overnight before PEI transfection. For immunofluorescence microscopy, HeLa cells were plated on poly-l-lysine–coated coverslips on a 6-well plate at 1.5 × 105 cells/well and incubated for 16 h at 37°C in 5% CO2 before Fugene 6 or PEI transfection. Cells were collected from plates by scraper, trypsin, or enzyme-free Cell Dissociation Solution (Millipore) or fixed for immunofluorescence microscopy.
To obtain stable transfectant cell populations, antibiotic selection was started 24–48 h after transfection by adding ∼106 transfectants to selection media containing 1 mg/ml active G418 (Gemini Bio-Products) for 5–7 d or 1.4 mg/ml Hygromycin (EMD) for 1 d before switching to 0.7 mg/ml Hygromycin for 4–6 d. Resistant colonies were pooled and used or sorted by fluorescence-activated cell sorting (FACS) for expression of GFP, or binding of GNA-FITC or L-PHA-FITC before use.
Antibodies and lectins
Anti-HA mouse mAb (HA.11) and anti-Myc mouse mAb (9E10) were from Covance; anti-HA rabbit polyclonal antibody (pAb; Y-11) was from Santa Cruz Biotechnology, Inc.; anti-Xenopus laevis β-actin mAb (AC-15) was from Abcam; anti–human Golgi matrix protein GM130 rabbit pAb and anti–rat Golgi matrix protein GM130 mouse mAb (35/GM130) were from EMD and BD, respectively; goat HRP-conjugated anti–mouse secondary antibody was from Thermo Fisher Scientific; anti–bovine PDI rabbit pAb and anti–rat PDI mouse mAb (1D3) were from Stressgen; Phalloidin conjugated with Alexa 568 (red) was from Invitrogen; anti–human ERGIC-53 mouse mAb (G1/93) was from Enzo Life Sciences, Inc.; anti–human β4GalT-I rabbit polyclonal antibodies (pAb) were a gift from Eric Berger (University of Zurich, Zurich, Switzerland); anti–human ManII rabbit pAb was a gift from Kelly Moremen (University of Georgia, Athens, GA); and Alexa 488 (green) goat anti–rabbit or anti–mouse secondary antibodies and Alexa 568 (red) goat anti–mouse IgG (H+L) secondary antibody were from Invitrogen. Plant lectins Con A, Phaseolus vulgaris leucoagglutinin (L-PHA), wheat germ agglutinin (WGA), Galanthus nivalis lectin GNA-FITC, and L-PHA-FITC were from Vector Laboratories.
Lectin resistance test
To determine lectin resistance, 2,000 CHO cells in medium containing 10% FBS (100 µl) were added to each well of a 96-well plate. Lectins (100 µl) in medium containing 10% FBS were added at increasing concentrations. After 4 d at 37°C in the 5% CO2 incubator, or when control wells were confluent, the medium was removed and the cells were fixed and stained with Methylene Blue in 50% methanol.
Flow cytometry analysis and FACS
For flow cytometry, cells were washed once with 1 ml cold FACS binding buffer (1x HBSS containing 1 mM CaCl2, 1 mM MgCl2, 0.05% or 0.1% sodium azide, and 2% BSA Fraction V [Sigma-Aldrich]). Cells were incubated with 12 µg/ml GNA-FITC lectin in FACS buffer for 30 min on ice, washed with 1 ml FACS binding buffer, and the cell pellet was resuspended in 0.5 ml FACS buffer, without BSA, and subjected to FACSscan (BD) flow cytometer for FACS analysis. Raw data were processed by Flowjo software (Tree Star, Inc.) after 7-Amino-actinomycin D (BD)–positive cells were gated out. For cell sorting, cells were washed with FACS binding buffer without sodium azide and incubated in the same buffer containing 12 µg/ml GNA-FITC or L-PHA-FITC. After 1 h on ice-water the cells were washed once with 1 ml cold FACS buffer, resuspended in 0.5 ml FACS buffer containing penicillin (100 units) and streptomycin (100 µg/ml; Invitrogen) and amphotericin B (2.50 µg/ml; Invitrogen), and subjected to flow cytometry (Dako MoFlo and Dako MoFlo XDP) to sort GNA- or L-PHA-binding cells after gating out 7-Amino-actinomycin D–positive cells. Cells expressing GFP were sorted similarly for high GFP expression.
GlcNAcT-I, GlcNAcT-III, and β4GalT enzyme assays
For enzyme assays, exponentially growing cells were washed three times in saline and lysed in 1.5% Triton X-100 in distilled water in the presence of protease inhibitor cocktail (Roche). To determine GlcNAcT-I activity, ∼50 µg cell extract was incubated at 37°C for 2 h in the presence of 27 µg Man5GlcNAc2Asn (Huang and Montgomery, 1970; Chaney and Stanley, 1986), 62.5 mM 2-(N-morpholino)ethanesulfonate (MES), pH 6.25, 25 mM MnCl2, and 0.75 mM UDP-[3H]-GlcNAc (10,000–20,000 cpm/nmol; PerkinElmer) in a final volume of 50 µl. Reactions were stopped by adding 0.5 ml of Con A buffer (0.1 M sodium acetate, 1.0 M NaCl, 10 mM MgCl2, 10 mM CaCl2, 10 mM MnCl2, and 0.02% sodium azide). After centrifugation in a microfuge for 5 min, the supernatant was added to a 1-ml column of Con A–Sepharose (GE Healthcare). After washing with Con A buffer, product was eluted with 200 mM α-methylmannoside in Con A buffer. Specific activities were determined from 3H-GlcNAc incorporated into Con A–bound product in the presence versus the absence of acceptor Man5GlcNAc2Asn.
GlcNAcT-III activity was assayed in a 40-µl reaction as described previously (Bhaumik et al., 1998) containing 8 μmol GlcNAc, 12.5 mM MnCl2, 6–8 mM UDP-[3H]-GlcNAc (15,000–30,000 cpm/nmol), 1% Triton X-100, and 62.5 mM 1,4-piperazinediethanesulfonic acid sodium salt (Na Pipes; pH 7.0) with or without 40 µg substrate GlcNAc2Man3GlcNAc2 (Seko et al., 1997) and 50–100 µg cell lysate. After 2 h incubation at 37°C, the reaction was stopped by adding 0.5 ml of Con A buffer and 3H-GlcNAc incorporated into Con A–Sepharose-fractionated product in the presence versus the absence of substrate was used to determine specific activity.
β4GalT activity was assayed in 50 µl reaction as described previously (Lee et al., 2001). The reaction contained 5 µmol 2-(N-morpholino) ethanesulfonate buffer, pH 6.5, 3 µmol MnCl2, 1.2% Triton X-100, 35 nmol UDP-[6-3H]-Gal (10,000–30,000 cpm/nmol; PerkinElmer), with or without 10 mM GlcNAc acceptor, and 50–100 µg cell lysate. After 2 h incubation at 37°C, reactions were stopped by adding 1 ml cold water and then passing through a 1-ml column of AG1-X4 (Cl− form; Bio-Rad Laboratories) that was subsequently washed with 2 ml distilled water to obtain 3H-Gal–labeled products. Specific activity was determined from 3H-Gal incorporated in the presence versus the absence of GlcNAc.
Western analysis
Cell lysates were prepared from saline-washed stable cell lines or transient transfectants 1–2 d after transfection in buffer containing 75 µl of 1.5% Triton X-100 (Sigma-Aldrich) and 1× protease inhibitor cocktail in water per 107 cells. Lysate protein concentrations were determined by Dc protein assay (Bio-Rad Laboratories). For electrophoresis, lysates were loaded on 10% Tris-HCl polyacrylamide gels at 10–30 mA for 2 h. Proteins were transferred to polyvinylidene difluoride (PerkinElmer) membrane overnight at 50 mA in a transfer buffer containing 10% methanol. For immunoblotting, primary and secondary antibodies were diluted in Tris-buffered saline (10 mM Tris HCl, pH 7.4, and 150 mM NaCl) containing 0.05% Tween 20 (Sigma-Aldrich) and 3% nonfat dry milk (wt/vol) supplemented with 3% (wt/vol) BSA or 3% nonfat dry milk, respectively. Anti-Myc mAb (9E10) was diluted 1:500, anti-HA mAb (HA.11) 1:1,000, anti-β actin mAb 1:5,000, and HRP-conjugated goat anti–mouse secondary antibody 1:5,000–10,000. The membrane was washed with the same Tris-buffered saline containing 0.05% Tween 20, incubated with Super Signal West Pico chemiluminescence reagent (Thermo Fisher Scientific), and exposed to film (Denville Scientific, Inc.).
Endoglycosidase digestions
Endoglycosidase H (Endo H) from Streptomyces plicatus was from Roche and Peptide N-glycosidase F (PNGase F) from Flavobacterium meningosepticum was from New England Biolabs, Inc. PNGase F and Endo H treatments of lysates were performed as suggested by the manufacturers at 37°C for ∼18 h in 20-µl buffers provided by the manufacturers. Lysates of 100–200 µg of protein were treated with 1,000 units of PNGase F or Endo H. Enzyme was replaced by water for control reactions. Reactions were stopped by the addition of gel loading buffer and heating at 95°C for 10 min followed by SDS-PAGE.
Immunofluorescence and confocal microscopy
Cells were plated at 3 × 105 cells/well on poly-l-lysine–coated coverslips on a 6-well plate and incubated for 16 h at 37°C in 5% CO2. After washing with PBS, cells were fixed in 3% (wt/vol) paraformaldehyde, then incubated with blocking buffer supplemented with 0.2% Triton X-100, 1% FBS, and 0.5% (wt/vol) BSA in PBS with Ca2+ and Mg2+ as described previously (Chiu et al., 2002; Mukherjee et al., 2007). After first and secondary antibody incubations, blue DAPI (1 µg/ml; Sigma-Aldrich) was used for nuclear staining. Antibodies and DAPI were diluted in the same blocking buffer containing 0.2% Triton X-100 as described above. Cells were mounted using Fluoromount (SouthernBiotech). For selective permeabilization of plasma membrane, slides were first incubated with 5 µg/ml digitonin (EMD) in 10 mM Hepes buffer containing 0.3 M sucrose, 2.5 mM MgCl2, 0.1 M KCl, and 1 mM EDTA at pH 6.9 for 15 min at 4°C, followed by first and secondary antibody incubations except that no detergent was added to the blocking buffer. Immunofluorescent images were digitally acquired on an inverted microscope (Axiovert 200M; Carl Zeiss, Inc.) coupled to a 12-bit cooled charge-coupled device camera (AxioCam MRm Rev. 3; Carl Zeiss, Inc.) controlled by Axiovision software (AxioVs40, version 4.7.2.0; Carl Zeiss, Inc.), using a 100x/1.3 NA oil immersion objective (EC Plan-NeoFluar; Carl Zeiss, Inc.), and saved as tiff files (1388 × 1040, 8 bit). All pictures were treated identically using Adobe Photoshop to remove background and adjust contrast. Images of confocal microscopy were acquired by capturing Z-series images with a 0.25-µm step size on a confocal microscope (TCS SP2 AOBS; Leica) using a 63x/1.4 NA oil immersion objective (HCX PL APO λBL CS; Leica). Laser lines at 405-, 488-, and 561-nm were provided by 20 mW diode, 100 mW Ar, and 10 mW diode, respectively; sequential excitation by line and detection range settings was used to eliminate cross talk between fluorophores. The images (512 × 512 pixel, 8 bit) were saved as tiff files. The entire Z-series was projected using the sum intensity method provided by ImageJ (NIH).
Brefeldin A treatment
Transfected cells were washed once with PBS containing 1 mM Ca2+ and 1 mM Mg2+ and incubated with DME supplemented with 10% FBS and 100 µg/ml CHX (DMSO stock solution; EMD) for 45 min or 1 h at 37°C in 5% CO2. Cells were then incubated with new medium containing 10 µg/ml BFA (DMSO stock solution; Sigma-Aldrich) with or without 100 µg/ml CHX for another 30 min at 37°C in 5% CO2. Cells were then washed three times with cold PBS containing Ca2+ and Mg2+ and processed for immunofluorescence microscopy.
Immunoprecipitation
Anti-Myc mAb (9E10) in 190 mM Hepes (pH 8.9) was incubated with Gammabind G sepharose (GE Healthcare; 5 mg IgG for 1 ml packed beads) for 6 h to overnight at 4°C. Beads were washed with 190 mM Hepes (pH 8.9) and bound antibodies were cross-linked by dimethyl dimelimidate dihydrochloride (11 mg/ml; Sigma-Aldrich) in 190 mM Hepes (pH 9.5) for 1 h at room temperature. The cross-linking reaction was terminated with 0.2 M ethanolamine (pH 8), and beads were washed extensively with PBS and stored in PBS containing 0.01% thimerasol at 4°C. For coimmunoprecipitation, transfected cells were lysed with 800 µl lysis buffer (RIPA buffer [Millipore] containing 150 mM NaCl, 1% NP-40, 0.5% deoxycolate, and 0.1% sodium dodecyl sulfate [SDS] in 50 mM Tris, pH 7.5) containing EDTA-free protease inhibitor cocktail (Roche). Lysates were centrifuged at 10,000 g for 20 min at 4°C. Supernatants were precleaned with 25 µl packed Gammabind G Sepharose for 4–6 h at 4°C, followed by immunoprecipitation with 25 µl anti-Myc beads overnight at 4°C. Anti-Myc beads were washed three times with 1 ml fresh lysis buffer and once with 1 ml PBS, and analyzed by immunoblotting. Two separate aliquots of anti-Myc beads (each 40%) and two aliquots of 1/40th cell lysate were boiled at 95°C for 5 min in gel loading buffer containing 5% 2-mercaptoethanol and 3% SDS. After electrophoresis on two separate 10% Tris-HCl polyacrylamide gels, proteins were transferred to polyvinylpyrolodine (PVDF) membranes that were incubated with the indicated primary antibodies followed by HRP-coupled secondary antibodies. Signals were detected using the Super Signal West Pico chemiluminescence reagent (Thermo Fisher Scientific).
Sertoli cell adhesion assay
TM4 Sertoli cells were plated on a 12-well plate and used at 80–90% confluence. CHO cells were labeled by incubation in warm PBS containing 5 µM carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) reagent (Invitrogen) followed by 15 min incubation at 37°C. Then reagents were removed and cell pellets were resuspended in warm culture medium and incubated at 37°C for 30 min. The culture medium was removed, the cell pellet resuspended with 1x HBSS containing 1 mM Ca2+, Mg2+, and 2% BSA (wt/vol), and 3.5 × 105 cells were added to wells containing fixed TM4 Sertoli cells (3% [wt/vol] paraformaldehyde) and incubated at 37°C for 4 h. After incubation, wells were washed four times with 1.5 ml HBSS containing 1 mM Ca2+, 1 mM Mg2+, and 2% BSA and pictures were taken with a digital camera (10x/0.25 NA; A-Plan Ph1; Carl Zeiss, Inc.) via an inverted microscope (Axiovert 200M; Carl Zeiss, Inc.). Pictures were processed by Adobe Photoshop and quantified by NIH ImageJ.
Online supplemental material
Fig. S1 shows cDNA and deduced protein sequence of GnT1IP-L and -S (GenBank/EMBL/DDBJ accession no. HM067443). Fig. S2 shows that nonmembrane-bound GnT1IP-S was modified with complex N-glycans and secreted, and partially colocalized with GlcNAcT-I. Fig. S3 shows MALDI-TOF mass spectra of N-glycans from CHO cells expressing GnT1Ip-L or Myc-GnT1Ip-S. Fig. S4 shows membrane-bound GnT1IP-S was localized to the ER and early Golgi compartment like GnT1IP-L. Fig. S5 shows that deletion constructs of GnT1IP interacted with medial-Golgi but not trans-Golgi enzymes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004102/DC1.
Supplementary Material
Acknowledgments
The authors thank the late Dennis Shields for help with microscopy, Subha Sundaram for MS experiments, Reto Muller for mouse testes poly(A+) and advice, Paula Cohen for spermatocytes and spermatids, and those mentioned in Materials and methods for reagents.
This work was supported by NIH grant RO1 36434 (to P. Stanley) and partial support was provided by the Albert Einstein Cancer Center grant NCI PO1 13330.
Footnotes
Abbreviations used in this paper:
- β4GalT
- β-1,4-galactosyltransferase
- BFA
- brefeldin A
- CHX
- cycloheximide
- Endo H
- endoglycosidase H
- GlcNAcT
- N-acetylglucosaminyltransferase
- GNA
- G. nivalus agglutinin
- GnT1IP
- GlcNAcT-I inhibitory protein
- Hygro
- hygromycin
- L-PHA
- P. vulgaris leukoagglutinin
- ManII/Iix
- α-mannosidase II/IIx
- PNGase F
- peptide-N-glycosidase F
References
- Akama T.O., Nakagawa H., Sugihara K., Narisawa S., Ohyama C., Nishimura S., O’Brien D.A., Moremen K.W., Millan J.L., Fukuda M.N.. 2002. Germ cell survival through carbohydrate-mediated interaction with Sertoli cells. Science. 295:124–127. 10.1126/science.1065570 [DOI] [PubMed] [Google Scholar]
- Akama T.O., Nakagawa H., Wong N.K., Sutton-Smith M., Dell A., Morris H.R., Nakayama J., Nishimura S., Pai A., Moremen K.W., et al. 2006. Essential and mutually compensatory roles of alpha-mannosidase II and alpha-mannosidase IIx in N-glycan processing in vivo in mice. Proc. Natl. Acad. Sci. USA. 103:8983–8988. 10.1073/pnas.0603248103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka-Manya K., Manya H., Nakajima A., Kawakita M., Endo T.. 2006. Physical and functional association of human protein O-mannosyltransferases 1 and 2. J. Biol. Chem. 281:19339–19345. 10.1074/jbc.M601091200 [DOI] [PubMed] [Google Scholar]
- Aryal R.P., Ju T., Cummings R.D.. 2010. The endoplasmic reticulum chaperone Cosmc directly promotes in vitro folding of T-synthase. J. Biol. Chem. 285:2456–2462. 10.1074/jbc.M109.065169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R., Bhaumik M., Raju T.S., Stanley P.. 2002. Truncated, inactive N-acetylglucosaminyltransferase III (GlcNAc-TIII) induces neurological and other traits absent in mice that lack GlcNAc-TIII. J. Biol. Chem. 277:26300–26309. 10.1074/jbc.M202276200 [DOI] [PubMed] [Google Scholar]
- Bhaumik M., Harris T., Sundaram S., Johnson L., Guttenplan J., Rogler C., Stanley P.. 1998. Progression of hepatic neoplasms is severely retarded in mice lacking the bisecting N-acetylglucosamine on N-glycans: evidence for a glycoprotein factor that facilitates hepatic tumor progression. Cancer Res. 58:2881–2887. [PubMed] [Google Scholar]
- Breton C., Imberty A.. 1999. Structure/function studies of glycosyltransferases. Curr. Opin. Struct. Biol. 9:563–571. 10.1016/S0959-440X(99)00006-8 [DOI] [PubMed] [Google Scholar]
- Campbell C., Stanley P.. 1984. A dominant mutation to ricin resistance in Chinese hamster ovary cells induces UDP-GlcNAc:glycopeptide beta-4-N-acetylglucosaminyltransferase III activity. J. Biol. Chem. 259:13370–13378. [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B.. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37(Database issue):D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P. 2001. Improving the efficacy of antibody-based cancer therapies. Nat. Rev. Cancer. 1:118–129. 10.1038/35101072 [DOI] [PubMed] [Google Scholar]
- Chalmel F., Rolland A.D., Niederhauser-Wiederkehr C., Chung S.S., Demougin P., Gattiker A., Moore J., Patard J.J., Wolgemuth D.J., Jégou B., Primig M.. 2007. The conserved transcriptome in human and rodent male gametogenesis. Proc. Natl. Acad. Sci. USA. 104:8346–8351. 10.1073/pnas.0701883104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney W., Stanley P.. 1986. Lec1A Chinese hamster ovary cell mutants appear to arise from a structural alteration in N-acetylglucosaminyltransferase I. J. Biol. Chem. 261:10551–10557. [PubMed] [Google Scholar]
- Chen W., Stanley P.. 2003. Five Lec1 CHO cell mutants have distinct Mgat1 gene mutations that encode truncated N-acetylglucosaminyltransferase I. Glycobiology. 13:43–50. 10.1093/glycob/cwg003 [DOI] [PubMed] [Google Scholar]
- Chiu R., Novikov L., Mukherjee S., Shields D.. 2002. A caspase cleavage fragment of p115 induces fragmentation of the Golgi apparatus and apoptosis. J. Cell Biol. 159:637–648. 10.1083/jcb.200208013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R.D., Esko J.D.. 2009. Principles of glycan recognition. In Essentials of Glycobiology. Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etlzler M.E., editors. Cold Spring Harbor Laboratory Press, New York. 387–402. [PubMed] [Google Scholar]
- Cummings R.D., Liu F.T.. 2009. Galectins. In Essentials of Glycobiology. Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etlzler M.E., editors. Cold Spring Harbor Laboratory Press, New York. 475–487. [PubMed] [Google Scholar]
- Dennis J.W., Nabi I.R., Demetriou M.. 2009. Metabolism, cell surface organization, and disease. Cell. 139:1229–1241. 10.1016/j.cell.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H.. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2:953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Fink J.L., Aturaliya R.N., Davis M.J., Zhang F., Hanson K., Teasdale M.S., Kai C., Kawai J., Carninci P., Hayashizaki Y., Teasdale R.D.. 2006. LOCATE: a mouse protein subcellular localization database. Nucleic Acids Res. 34(Database issue):D213–D217. 10.1093/nar/gkj069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo C.G., Daniotti J.L., Maccioni H.J.. 2001. Physical and functional association of glycolipid N-acetyl-galactosaminyl and galactosyl transferases in the Golgi apparatus. Proc. Natl. Acad. Sci. USA. 98:1625–1630. 10.1073/pnas.031458398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassinen A., Rivinoja A., Kauppila A., Kellokumpu S.. 2010. Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. J. Biol. Chem. 285:17771–17777. 10.1074/jbc.M110.103184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson U., Pulkkinen V., Dixon M., Peyrard-Janvid M., Rehn M., Lahermo P., Ollikainen V., Salmenkivi K., Kinnula V., Kere J., et al. 2006. ELMOD2 is a candidate gene for familial idiopathic pulmonary fibrosis. Am. J. Hum. Genet. 79:149–154. 10.1086/504639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe M.H., Slusarewicz P., Misteli T., Watson R., Warren G.. 1995. Evidence for recycling of the resident medial/trans Golgi enzyme, N-acetylglucosaminyltransferase I, in ldlD cells. J. Biol. Chem. 270:25057–25063. 10.1074/jbc.270.42.25057 [DOI] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W.. 1993. TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler. 374:166. [Google Scholar]
- Huang C.C., Montgomery R.. 1970. Microheterogeneity and paucidispersity of glycoproteins. I. The carbohydrate of chicken ovalbumin. Carbohydr. Res. 13:127–137. 10.1016/S0008-6215(00)84902-2 [DOI] [PubMed] [Google Scholar]
- Iguchi N., Tobias J.W., Hecht N.B.. 2006. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc. Natl. Acad. Sci. USA. 103:7712–7717. 10.1073/pnas.0510999103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioffe E., Stanley P.. 1994. Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc. Natl. Acad. Sci. USA. 91:728–732. 10.1073/pnas.91.2.728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.J., Morrison C.A., Stoddart R.W.. 1992. Histochemical analysis of rat testicular glycoconjugates. 1. Subsets of N-linked saccharides in seminiferous tubules. Histochem. J. 24:319–326. 10.1007/BF01046163 [DOI] [PubMed] [Google Scholar]
- Ju T., Cummings R.D.. 2002. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc. Natl. Acad. Sci. USA. 99:16613–16618. 10.1073/pnas.262438199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju T., Aryal R.P., Stowell C.J., Cummings R.D.. 2008. Regulation of protein O-glycosylation by the endoplasmic reticulum-localized molecular chaperone Cosmc. J. Cell Biol. 182:531–542. 10.1083/jcb.200711151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius K., Mølgaard A., Gupta R., Brunak S.. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 15:153–164. 10.1093/glycob/cwh151 [DOI] [PubMed] [Google Scholar]
- Kanda Y., Yamada T., Mori K., Okazaki A., Inoue M., Kitajima-Miyama K., Kuni-Kamochi R., Nakano R., Yano K., Kakita S., et al. 2007. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology. 17:104–118. 10.1093/glycob/cwl057 [DOI] [PubMed] [Google Scholar]
- Kudo T., Iwai T., Kubota T., Iwasaki H., Takayma Y., Hiruma T., Inaba N., Zhang Y., Gotoh M., Togayachi A., Narimatsu H.. 2002. Molecular cloning and characterization of a novel UDP-Gal:GalNAc(alpha) peptide beta 1,3-galactosyltransferase (C1Gal-T2), an enzyme synthesizing a core 1 structure of O-glycan. J. Biol. Chem. 277:47724–47731. 10.1074/jbc.M205839200 [DOI] [PubMed] [Google Scholar]
- Lee J., Sundaram S., Shaper N.L., Raju T.S., Stanley P.. 2001. Chinese hamster ovary (CHO) cells may express six beta 4-galactosyltransferases (beta 4GalTs). Consequences of the loss of functional beta 4GalT-1, beta 4GalT-6, or both in CHO glycosylation mutants. J. Biol. Chem. 276:13924–13934. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Yuan L.C., Bonifacino J.S., Klausner R.D.. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 56:801–813. 10.1016/0092-8674(89)90685-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather J.P. 1980. Establishment and characterization of two distinct mouse testicular epithelial cell lines. Biol. Reprod. 23:243–252. 10.1095/biolreprod23.1.243 [DOI] [PubMed] [Google Scholar]
- Metzler M., Gertz A., Sarkar M., Schachter H., Schrader J.W., Marth J.D.. 1994. Complex asparagine-linked oligosaccharides are required for morphogenic events during post-implantation development. EMBO J. 13:2056–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward T.A., Heitzmann M., Bill K., Längle U., Schumacher P., Forrer K.. 2008. Effect of constant and variable domain glycosylation on pharmacokinetics of therapeutic antibodies in mice. Biologicals. 36:41–47. 10.1016/j.biologicals.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Mori K., Iida S., Yamane-Ohnuki N., Kanda Y., Kuni-Kamochi R., Nakano R., Imai-Nishiya H., Okazaki A., Shinkawa T., Natsume A., et al. 2007. Non-fucosylated therapeutic antibodies: the next generation of therapeutic antibodies. Cytotechnology. 55:109–114. 10.1007/s10616-007-9103-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Chiu R., Leung S.M., Shields D.. 2007. Fragmentation of the Golgi apparatus: an early apoptotic event independent of the cytoskeleton. Traffic. 8:369–378. 10.1111/j.1600-0854.2007.00542.x [DOI] [PubMed] [Google Scholar]
- Nilsson T., Hoe M.H., Slusarewicz P., Rabouille C., Watson R., Hunte F., Watzele G., Berger E.G., Warren G.. 1994. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 13:562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T., Rabouille C., Hui N., Watson R., Warren G.. 1996. The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase I in retention, kin recognition and structural maintenance of the Golgi apparatus in HeLa cells. J. Cell Sci. 109:1975–1989. [DOI] [PubMed] [Google Scholar]
- Niu T.K., Pfeifer A.C., Lippincott-Schwartz J., Jackson C.L.. 2005. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell. 16:1213–1222. 10.1091/mbc.E04-07-0599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North S.J., Huang H.H., Sundaram S., Jang-Lee J., Etienne A.T., Trollope A., Chalabi S., Dell A., Stanley P., Haslam S.M.. 2010. Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem. 285:5759–5775. 10.1074/jbc.M109.068353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opat A.S., Houghton F., Gleeson P.A.. 2000. Medial Golgi but not late Golgi glycosyltransferases exist as high molecular weight complexes. Role of luminal domain in complex formation and localization. J. Biol. Chem. 275:11836–11845. 10.1074/jbc.275.16.11836 [DOI] [PubMed] [Google Scholar]
- Opat A.S., Houghton F., Gleeson P.A.. 2001. Steady-state localization of a medial-Golgi glycosyltransferase involves transit through the trans-Golgi network. Biochem. J. 358:33–40. 10.1042/0264-6021:3580033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik S.K., Stanley P.. 2006. Lectin-resistant CHO glycosylation mutants. Methods Enzymol. 416:159–182. 10.1016/S0076-6879(06)16011-5 [DOI] [PubMed] [Google Scholar]
- Patnaik S.K., Potvin B., Carlsson S., Sturm D., Leffler H., Stanley P.. 2006. Complex N-glycans are the major ligands for galectin-1, -3, and -8 on Chinese hamster ovary cells. Glycobiology. 16:305–317. 10.1093/glycob/cwj063 [DOI] [PubMed] [Google Scholar]
- Rodeheffer C., Shur B.D.. 2002. Targeted mutations in beta1,4-galactosyltransferase I reveal its multiple cellular functions. Biochim. Biophys. Acta. 1573:258–270. [DOI] [PubMed] [Google Scholar]
- Russo R.N., Shaper N.L., Shaper J.H.. 1990. Bovine beta 1----4-galactosyltransferase: two sets of mRNA transcripts encode two forms of the protein with different amino-terminal domains. In vitro translation experiments demonstrate that both the short and the long forms of the enzyme are type II membrane-bound glycoproteins. J. Biol. Chem. 265:3324–3331. [PubMed] [Google Scholar]
- Seko A., Yamashita K.. 2008. Activation of beta1,3-N-acetylglucosaminyltransferase-2 (beta3Gn-T2) by beta3Gn-T8. Possible involvement of beta3Gn-T8 in increasing poly-N-acetyllactosamine chains in differentiated HL-60 cells. J. Biol. Chem. 283:33094–33100. 10.1074/jbc.M806933200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seko A., Koketsu M., Nishizono M., Enoki Y., Ibrahim H.R., Juneja L.R., Kim M., Yamamoto T.. 1997. Occurence of a sialylglycopeptide and free sialylglycans in hen’s egg yolk. Biochim. Biophys. Acta. 1335:23–32. [DOI] [PubMed] [Google Scholar]
- Shur B.D., Evans S., Lu Q.. 1998. Cell surface galactosyltransferase: current issues. Glycoconj. J. 15:537–548. 10.1023/A:1006951407168 [DOI] [PubMed] [Google Scholar]
- Song Y., Aglipay J.A., Bernstein J.D., Goswami S., Stanley P.. 2010. The bisecting GlcNAc on N-glycans inhibits growth factor signaling and retards mammary tumor progression. Cancer Res. 70:3361–3371. 10.1158/0008-5472.CAN-09-2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess A.N., Feig C., Schulze W., Chalmel F., Cappallo-Obermann H., Primig M., Kirchhoff C.. 2007. Cross-platform gene expression signature of human spermatogenic failure reveals inflammatory-like response. Hum. Reprod. 22:2936–2946. 10.1093/humrep/dem292 [DOI] [PubMed] [Google Scholar]
- Sprenger J., Lynn Fink J., Karunaratne S., Hanson K., Hamilton N.A., Teasdale R.D.. 2008. LOCATE: a mammalian protein subcellular localization database. Nucleic Acids Res. 36(Database issue):D230–D233. 10.1093/nar/gkm950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P. 1983. Selection of lectin-resistant mutants of animal cells. Methods Enzymol. 96:157–184. 10.1016/S0076-6879(83)96015-9 [DOI] [PubMed] [Google Scholar]
- Stanley P., Sundaram S., Tang J., Shi S.. 2005. Molecular analysis of three gain-of-function CHO mutants that add the bisecting GlcNAc to N-glycans. Glycobiology. 15:43–53. 10.1093/glycob/cwh136 [DOI] [PubMed] [Google Scholar]
- Stanley P., Schachter H., Taniguchi N.. 2009. N-glycans. In Essentials of Glycobiology. Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etlzler M.E., editors. Cold Spring Harbor Laboratory Press, New York. 101–114. [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 101:6062–6067. 10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale R.D., Jackson M.R.. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu. Rev. Cell Dev. Biol. 12:27–54. 10.1146/annurev.cellbio.12.1.27 [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg D., Thierry-Mieg J.. 2006. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 7:S12-1–14. 10.1186/gb-2006-7-s1-s12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Lowe J.. 2009. Biological roles of glycans. In Essentials of Glycobiology. Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etlzler M.E., editors. Cold Spring Harbor Laboratory Press, New York. 75–88 [PubMed] [Google Scholar]
- Ward T.H., Polishchuk R.S., Caplan S., Hirschberg K., Lippincott-Schwartz J.. 2001. Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155:557–570. 10.1083/jcb.200107045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Shekar S.C., Flinn R.J., El-Sibai M., Jaiswal B.S., Sen K.I., Janakiraman V., Seshagiri S., Gerfen G.J., Girvin M.E., Backer J.M.. 2009. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110alpha and are disrupted in oncogenic p85 mutants. Proc. Natl. Acad. Sci. USA. 106:20258–20263. 10.1073/pnas.0902369106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Sato Y., Isaji T., Fukuda T., Matsumoto A., Miyoshi E., Gu J., Taniguchi N.. 2008. Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J. 275:1939–1948. 10.1111/j.1742-4658.2008.06346.x [DOI] [PubMed] [Google Scholar]
- Zhou Q., Shankara S., Roy A., Qiu H., Estes S., McVie-Wylie A., Culm-Merdek K., Park A., Pan C., Edmunds T.. 2008. Development of a simple and rapid method for producing non-fucosylated oligomannose containing antibodies with increased effector function. Biotechnol. Bioeng. 99:652–665. 10.1002/bit.21598 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.