The CHD4 helicase is identified as a new component of the genome surveillance machinery in a proteomic screen for factors enriched on chromatin after ionizing radiation (see also related paper by Smeenk et al. in this issue).
Abstract
In response to ionizing radiation (IR), cells delay cell cycle progression and activate DNA repair. Both processes are vital for genome integrity, but the mechanisms involved in their coordination are not fully understood. In a mass spectrometry screen, we identified the adenosine triphosphate–dependent chromatin-remodeling protein CHD4 (chromodomain helicase DNA-binding protein 4) as a factor that becomes transiently immobilized on chromatin after IR. Knockdown of CHD4 triggers enhanced Cdc25A degradation and p21Cip1 accumulation, which lead to more pronounced cyclin-dependent kinase inhibition and extended cell cycle delay. At DNA double-strand breaks, depletion of CHD4 disrupts the chromatin response at the level of the RNF168 ubiquitin ligase, which in turn impairs local ubiquitylation and BRCA1 assembly. These cell cycle and chromatin defects are accompanied by elevated spontaneous and IR-induced DNA breakage, reduced efficiency of DNA repair, and decreased clonogenic survival. Thus, CHD4 emerges as a novel genome caretaker and a factor that facilitates both checkpoint signaling and repair events after DNA damage.
Introduction
DNA double-strand breaks (DSBs) arise as products of stochastic replication failure, reactive oxygen species, or because of environmental clastogens such as ionizing radiation (IR; Löbrich and Jeggo, 2007). DSBs are highly cytotoxic lesions and pose extreme demands on coordinating DNA repair with vital transactions such as transcription, DNA replication, or chromosomal segregation. To safeguard genome integrity challenged by DSBs, cells mobilize repair and signaling pathways, whose activation and coordination involve damaged DNA as well as chromatin composed of histones and histone-binding proteins (Fernandez-Capetillo et al., 2004; Stucki and Jackson, 2006; Jackson and Bartek, 2009; van Attikum and Gasser, 2009).
After DSB generation, the neighboring chromatin undergoes extensive modifications, initiated by the ataxia telangiectasia mutated (ATM)–mediated phosphorylation of the histone H2AX (γ-H2AX) followed by recruitment of the MDC1 adaptor (Stucki et al., 2005) and two ubiquitin ligases, RNF8 and RNF168 (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007; Doil et al., 2009; Stewart et al., 2009). The ensuing chromatin ubiquitylation allows amplification of the ATM signaling and local concentration of repair factors including the BRCA1A complex (van Attikum and Gasser, 2009). In parallel, the DSB sites undergo local histone eviction and enzymatic DNA resection, and the resulting single-stranded DNA generates a structural platform for another signaling module triggered by assembly of the ataxia telangiectasia and Rad3 related (ATR) kinase with its coactivators (Bartek and Lukas, 2007). All of these events are essential for timely initiation and amplification of the DNA damage signaling.
The signal generated at the DSBs must be transmitted to the entire nucleus to delay cell cycle progression (Lukas et al., 2003; Bartek et al., 2004). The key signal transducers are the CHK2 and CHK1 kinases, which propagate and amplify the pathways initiated by ATM and ATR, respectively. Among targets of CHK1/CHK2 is the Cdc25A phosphatase, which, when phosphorylated, undergoes a proteasome-mediated degradation (Mailand et al., 2000). This in turn inhibits Cdk2 and Cdk1, the two major kinases governing cell cycle progression. This checkpoint pathway is rapidly implemented and delays cell cycle for several hours, which in most cases, is sufficient to provide time for repair (Bartek et al., 2004). In parallel, S phase progression can be slowed down also by ATM/ATR-mediated phosphorylation of the cohesin SMC1 (Falck et al., 2002; Kitagawa et al., 2004). Finally, cells possess a mechanism to extend checkpoint activity in cases of complex or extensive DNA damage. This branch depends on p53, which is also targeted by ATM/ATR and CHK2/CHK1 (Bartek and Lukas, 2007). Phosphorylation of p53 leads to its stabilization and transactivation of the p53 targets including the p21Cip1 Cdk inhibitor; p21 then reinforces the cell cycle arrest and can maintain it for an extended period of time (Kastan and Bartek, 2004).
Despite the recent progress in dissecting the pathways involved in DSB repair and signaling, their functional cross talk and coordination are not understood. To elucidate these issues, we performed an unbiased proteomic screen for factors that become specifically enriched on chromatin after IR and report on identification of CHD4 (chromodomain helicase DNA-binding protein 4) as a new component of the genome surveillance machinery.
Results and discussion
Identification of CHD4 as a factor involved in the DNA damage response (DDR)
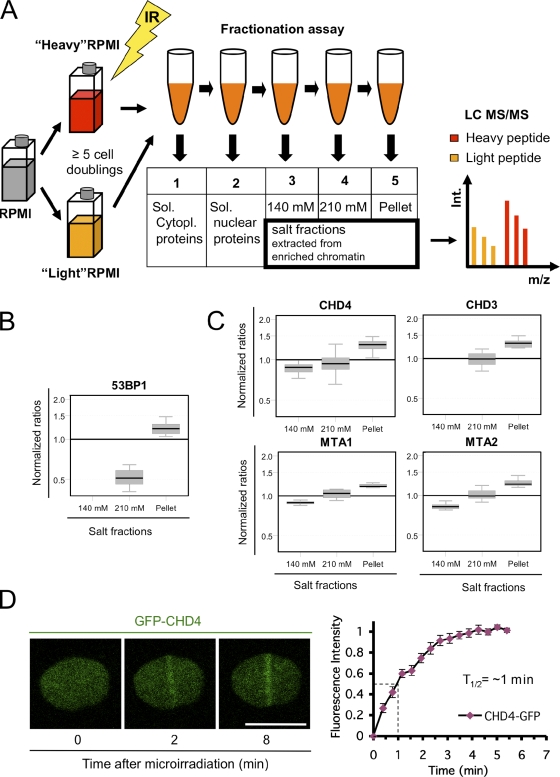
By combining stable isotope labeling with amino acids in cell culture (SILAC) labeling (Ong et al., 2002), cellular fractionation, tandem mass spectrometry (Aebersold and Mann, 2003), and statistical analysis, we screened the nuclear proteome for factors with increased chromatin binding in response to IR. Chromatin-bound proteins were enriched by biochemical fractionation including progressive protein extraction by increasing salt concentrations, and the most tightly bound proteins were solubilized in the pellet fraction (Fig. 1 A). The resulting chromatin fractions were analyzed by tandem mass spectrometry. We used the statistical method analysis of variance (ANOVA) to identify proteins that exhibit DNA damage–induced altered elution from chromatin, resembling the behavior of proteins such as 53BP1 (Fig. 1 B), which is known to bind chromatin in the vicinity of the DSB lesions.
Figure 1.
Identification of CHD4 as a factor involved in the DDR. (A) Proteomic screening procedure. GM00130 lymphocytes were grown in heavy or light SILAC media, exposed to 10 Gy of IR, fractionated, and analyzed by tandem mass spectrometry (MS/MS). LC, liquid chromatography. (B) Box plot showing quantitative tandem mass spectrometry data for 53BP1 (positive control). Y axis, normalized ratios (IR peptide/control peptide) showing protein elution by progressive salt fractionation of irradiated lymphocytes relative to control lymphocytes. The box represents the central 50% of the distributions, and the whiskers approximate the 95% interval. (C) Tandem mass spectrometry data for NuRD subunits are shown. Box plots are as in B. (D) Accumulation of GFP-CHD4 at laser-generated DSBs (left) and real-time recruitment of GFP-CHD4 derived from 10 independent cells (right). Error bars indicate SEM. Bar, 10 µm.
Among the statistically highest scoring proteins (99.9% confidence level) were several subunits of the nucleosome-remodeling and histone deacetylase (NuRD) complex. NuRD uniquely couples two chromatin-directed enzymatic functions: ATP-dependent nucleosome remodeling and histone deacetylation. The NuRD complex is composed of the chromatin-remodeling subunit CHD4/3, HDAC1/2 (histone deacetylase 1 and 2), RbAp (retinoblastoma-associated protein 46 and 48), MBD (methyl-CpG–binding domain–containing protein 2 and 3), and MTA1–3 (metastasis-associated proteins 1, 2, and 3; Tong et al., 1998; Wade et al., 1998; Xue et al., 1998; Zhang et al., 1998; Bowen et al., 2004). The NuRD subunits CHD4, CHD3, MTA1, and MTA2 all showed a statistically significant increase in chromatin binding in response to IR (Fig. 1 C).
CHD4 physically interacts with ATR (Schmidt and Schreiber, 1999), and it was recently identified as an ATM/ATR phosphotarget on S1349Q in a proteomic screen (Matsuoka et al., 2007). Consistent with this previously described involvement of CHD4 in the main DDR pathways and the mass spectrometry data reported herein, we obtained evidence that the IR-induced chromatin enrichment of CHD4 can be explained at least in part by its retention directly at the sites of DNA damage. Thus, after microirradiating cells expressing GFP-CHD4 by a laser under conditions that induce DNA strand breaks (Lukas et al., 2003; Bekker-Jensen et al., 2006), we observed rapid accumulation of GFP-CHD4 at the sites of DNA damage (Fig. 1 D, left). Of note, the accumulation of CHD4 at DSBs represented only a fraction of the nuclear pool of the protein, the bulk of which remained dispersed in the nucleus, indicating a dynamic exchange between DSBs and the undamaged parts of the nucleus. Time-lapse analysis revealed that the accumulation of GFP-CHD4 at the DSB sites reached half-maximum within 1 min after microirradiation (Fig. 1 D, right), a value reminiscent of the earliest DSB interactors (Lukas et al., 2004; Mailand et al., 2007; Doil et al., 2009). Collectively, these data are consistent with a role of CHD4 in the initial stages of the DDR, and we set out to investigate the physiological relevance of these findings.
Knockdown of CHD4 sensitizes cells to IR and slows down cell cycle progression
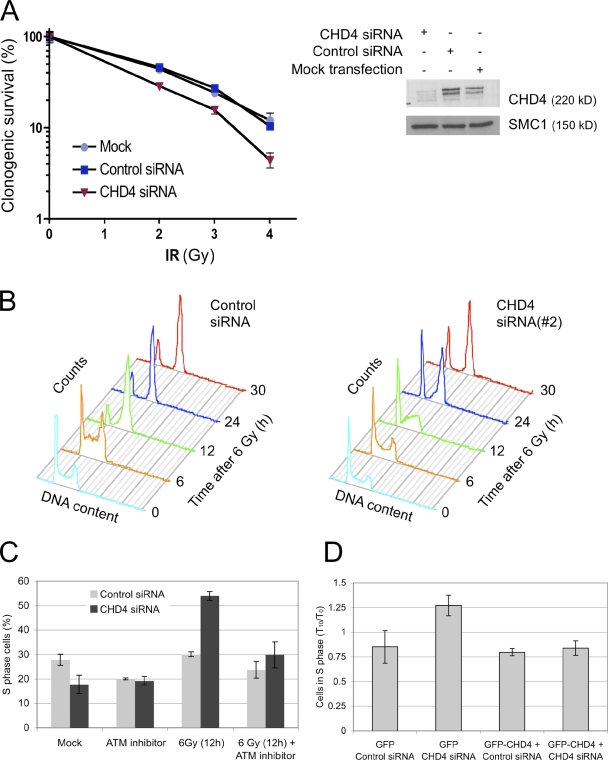
We started by examining the impact of CHD4 on cellular fitness and observed that knockdown of CHD4 reduced colony formation of cells exposed to IR (Fig. 2 A). To gain insight into the impaired survival, we first followed cell cycle progression. Although treating cells with CHD4 siRNAs moderately reduced S and G2 compartments in otherwise unstressed cells, combined irradiation and CHD4 depletion had a much more pronounced impact on cell cycle progression. This was manifested by a marked S phase delay (Fig. 2 B, 12 h) followed by accumulation of cells in G2 (Fig. 2 B, 24 and 30 h). Furthermore, a fraction of CHD4-deficient cells was also clearly arrested in G1, an effect that was virtually absent in a control cell population treated with an identical dose of IR (Fig. 2 B, compare the matching 12- and 24-h time points). Similar consequences of CHD4 knockdown were observed after treating irradiated cells with an independent siRNA (Fig. S1 A). The extended cell cycle checkpoints were further substantiated by a more pronounced inhibition of the cyclin A–associated kinase activity (Fig. S1 B), decreased DNA replication measured by BrdU incorporation (Fig. S1 C), and delayed mitotic entry (Fig. S1 D). Importantly, the impact of CHD4 knockdown on cell cycle progression involved bona fide DNA damage signaling, demonstrated by the reversal of the S phase accumulation by treating the cells with a specific inhibitor of ATM (Fig. 2 C). Finally, reintroducing siRNA-resistant GFP-CHD4 into cells depleted of endogenous CHD4 reversed the S phase accumulation, arguing against off-target effects of the siRNA treatment (Fig. 2 D and Fig. S1 E).
Figure 2.
Knockdown of CHD4 sensitizes cells to IR and deregulates cell cycle progression. (A) Clonogenic survival assay. U2OS cells were treated with control or CHD4 siRNAs (SMARTpool) for 72 h as indicated, irradiated, and colonies with >50 cells were counted. CHD4 down-regulation was monitored by immunoblotting. SMC1, loading control. (B) U2OS cells were treated with control or CHD4 siRNA (#2) for 72 h, irradiated (6 Gy), and analyzed at the indicated time points by flow cytometry. (C) U2OS cells were treated with control or CHD4 siRNAs (SMARTpool), treated with ATM inhibitor for 1.5 h, irradiated, and analyzed by flow cytometry. The efficiency of CHD4 siRNAs in B and C is shown in Fig. S3 B. (D) U2OS cell lines conditionally expressing GFP or GFP-CHD4 resistant to siRNA (#3) were treated with control or CHD4 siRNA (#3) as indicated. After 48 h, the transgenes were induced by addition of doxycycline after an additional 24 h, irradiated, and analyzed by flow cytometry. To compensate for minor differences in the starting S phase content in the two cell lines, the data were normalized and are presented as the ratios between the S phase content measured 10 h after IR (T10) and that in unirradiated cells (T0). The GFP-CHD4 cell line and the efficiency of siRNA (#3) are characterized in Fig. S1 E. Error bars indicate SEM.
Elevated checkpoint signaling in the absence of CHD4
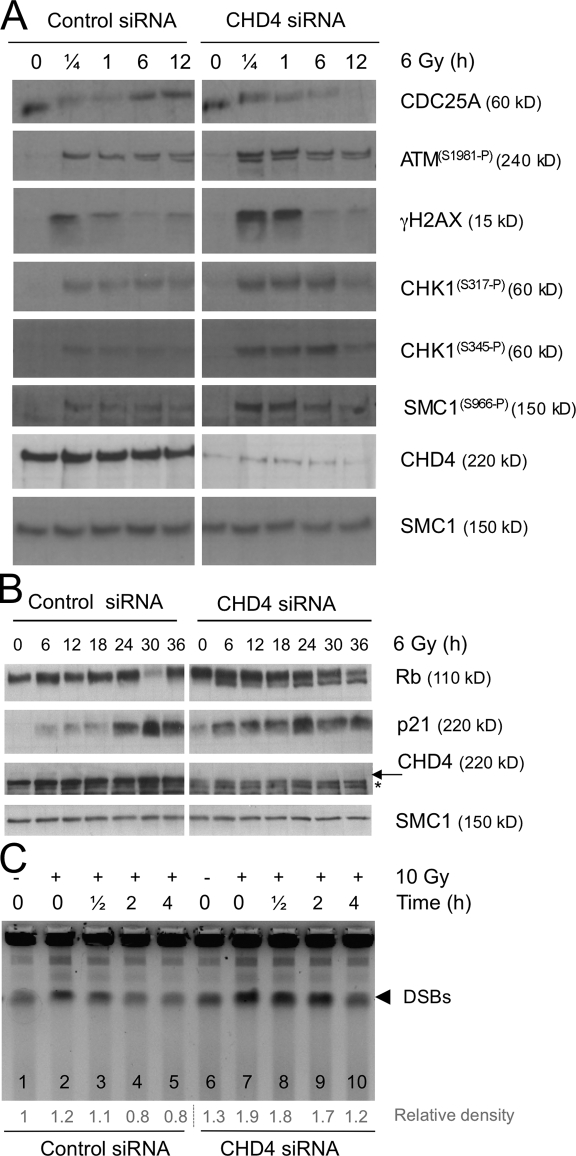
To understand the reasons for these cell cycle aberrations, we examined the critical components of the genome surveillance pathways. Consistent with the aforementioned cell cycle delay, we observed that the Cdc25A phosphatase, the key effector of rapidly deployed cell cycle checkpoints, became degraded and remained low in the absence of CHD4, even at stages when Cdc25A in control cells recovered to predamage levels (Fig. 3 A). We could confirm that the absence of Cdc25A recovery in CHD4-deficient cells was caused by ongoing protein degradation and not by reduced mRNA expression (Fig. S2, A and B). Together, these data indicate that in the absence of CHD4, the rapid checkpoint response lasts longer than in CHD4-proficient cells.
Figure 3.
Extended checkpoint signaling in the absence of CHD4. (A) U2OS cells were treated with control or CHD4 siRNAs (SMARTpool) for 72 h, irradiated, and analyzed by immunoblotting with antibodies and at the specified time points. (B) U2OS cells were treated with siRNAs as in A, irradiated, and analyzed by immunoblotting. Asterisk, nonspecific band; arrow, CHD4. (A and B) Total SMC1, loading control. (C) U2OS were treated with control or CHD4 siRNAs (SMARTpool) for 72 h and irradiated. The DNA breakage was analyzed by PFGE at the indicated time points (top). The relative densities of the DSB bands (bottom) were normalized to the value measured in nonirradiated cells treated with control siRNA (lane 1).
Because CHD4-depleted cells responded to IR also by a G1 and G2 accumulation (Fig. 2 B), we examined the p53–p21 axis that plays an important role in sustaining the cell cycle arrest at these crucial transitions. Interestingly, knocking down CHD4 triggered p21 accumulation already in unstressed cells, and this further increased after exposing the cells to IR (Fig. 3 B and Fig. S2 C). Such p21 elevation was associated with a progressive dephosphorylation of pRb, an established surrogate of Cdk inhibition (Fig. 3 B). In addition, the IR-induced induction of p21 was p53 dependent (Fig. S2 D), and it was consistently more robust and occurred earlier in CHD4-depleted cells (Fig. 3 B and Fig. S2 C). We conclude that the sustained branch of the DNA damage–induced checkpoint signaling is more robust in CHD4-depleted cells and that it is partially activated already before exposing cells to external genotoxic stress.
CHD4-deficient cells transiently hyperactivate ATM and ATR
Consistent with the extended Cdc25A degradation, both ATM- (ATMS1981-P, γ-H2AX, and SMC1S966-P) and ATR-mediated (CHK1S317-P and CHK1S345-P) signaling were elevated to supraphysiological levels in the absence of CHD4, an effect that was most evident on increased γ-H2AX (Fig. 3 A and Fig. S2 C). This spike of ATM/ATR phosphorylations was restricted to the early stages of DDR (≤1 h after IR) and was consistently observed after knocking down CHD4 with independent siRNAs targeting distinct regions of the CHD4 transcript (Fig. 3 A and Fig. S2 C). Two scenarios could explain the enhanced ATM/ATR activities. First, depletion of CHD4 may cause local chromatin changes that increase the signaling from a similar amount of DNA breaks as generated in control cells. Second, CHD4 depletion may result in more widespread chromatin modifications that poise it to accumulate more breaks when exposed to IR. Although these scenarios are not mutually exclusive, we obtained evidence that the latter might represent the source of the transient spike of the ATM/ATR-mediated phosphorylations in CHD4-deficient conditions. Thus, using pulsed field gel electrophoresis (PFGE; Hanada et al., 2007), we detected an increased amount of DSBs in CHD4-deficient cells immediately after IR exposure (Fig. 3 C, compare lane 2 with lane 7; and Fig. S2 E). Of note, the elevated amount of DSBs in CHD4-depleted cells remained apparent during the first 2 h after IR (Fig. 3 C, compare lanes 3 and 4 with lanes 8 and 9, respectively), indicating that CHD4-deficient cells have reduced ability to repair DSBs. Finally, the PFGE assay revealed reproducible DSB generation even in nonirradiated CHD4-depleted cells, an effect evident especially after quantitatively knocking down CHD4 by the most efficient siRNA (#2; Fig. S2 E). This is consistent with a recent study reporting that the physiological loss of the NuRD complex in senescent cells is accompanied by a progressive increase of spontaneous DNA damage (Pegoraro et al., 2009).
Knockdown of CHD4 impairs retention of repair proteins at the sites of DNA damage
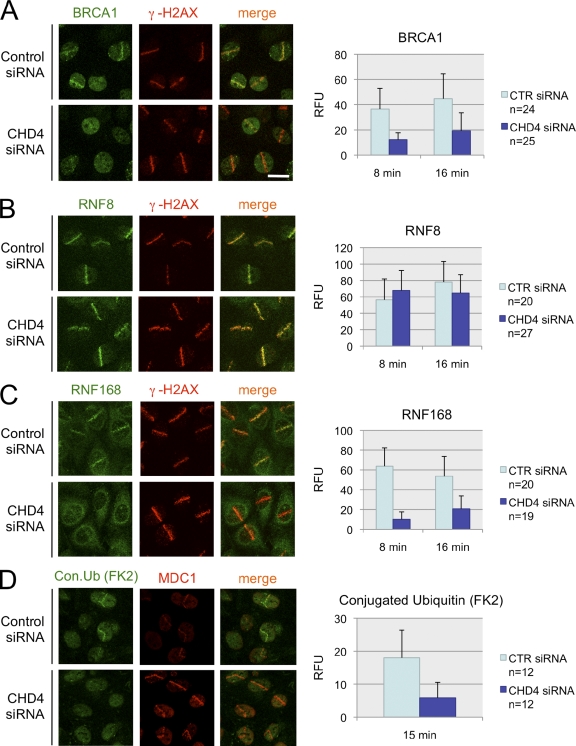
To gain more insight into mechanisms that might be subverted by NuRD disruption, we turned to the earlier observation that a fraction of CHD4 accumulates directly at the DSB sites (Fig. 1 D) and asked whether this influences accumulation of proteins in this compartment. Interestingly, accumulation of BRCA1, a key genome caretaker involved in DSB signaling and repair, was consistently impaired in early phases of the DSB response in CHD4-depleted cells (Fig. 4 A). This was unexpected because the extent of H2AX phosphorylation (an upstream prerequisite for BRCA1 retention on damaged chromatin) was more pronounced in CHD4-deficient cells (Fig. 3 A and Fig. S2 C). Importantly, these observations were not restricted to the laser-induced DNA lesions; the transient impairment of BRCA1 focus formation was also observed in irradiated cells treated with two independent siRNAs against CHD4 (Fig. S3 A).
Figure 4.
Impaired ubiquitylation and delayed accumulation of BRCA1 at the site of DSBs in the absence of CHD4. (A–D) U2OS cells were treated with control (CTR) or CHD4 siRNAs (SMARTpool) for 72 h, microirradiated by the laser, and immunostained with antibodies to BRCA1 (A), RNF8 (B), RNF168 (C), and conjugated ubiquitin (FK2 antibody; D). Cells were coimmunostained with antibodies to γ-H2AX (A–C) or MDC1 (D) to mark the DSB-containing tracks. (left) Representative fields for each DSB regulator (A–C, acquired 8 min after microirradiation; D, acquired 15 min after microirradiation) are shown. (right) Graphs show quantification of relative fluorescence intensities in the microirradiated areas subtracted by the background fluorescence in the undamaged parts of the nucleus. The efficiency of CHD4 siRNAs in A–D is shown in Fig. S3 B. RFU, relative fluorescence units. Error bars indicate SD. Bar, 10 µm.
Because the BRCA1 requires binding to conjugated ubiquitin for its accumulation at DSBs, we tested the impact of CHD4 knockdown on RNF8 and RNF168, the two key ubiquitin ligases involved in this process. Strikingly, although RNF8 remained stable and robustly accumulated at DSBs regardless of the CHD4 status (Fig. 4 B and Fig. S3 B), RNF168 was partially destabilized, and its retention of RNF168 was attenuated in CHD4-depleted cells (Fig. 4 C and Fig. S3 B). Correspondingly, generation of ubiquitin conjugates at the microlaser-generated DSBs was reduced (Fig. 4 D). Together, these data suggest that the transition from the RNF8- to RNF168-controlled step in the DSB-induced chromatin response might be more complex than originally thought (Doil et al., 2009; Stewart et al., 2009). For instance, we can envisage that the transient recruitment of CHD4 to the DSB sites (and the ensuing chromatin remodeling) allows more efficient recognition of the nascent ubiquitin chains (generated by RNF8) by the ubiquitin-binding domains of RNF168, which would in turn facilitate RNF168 recruitment and amplification of the local ubiquitin reaction.
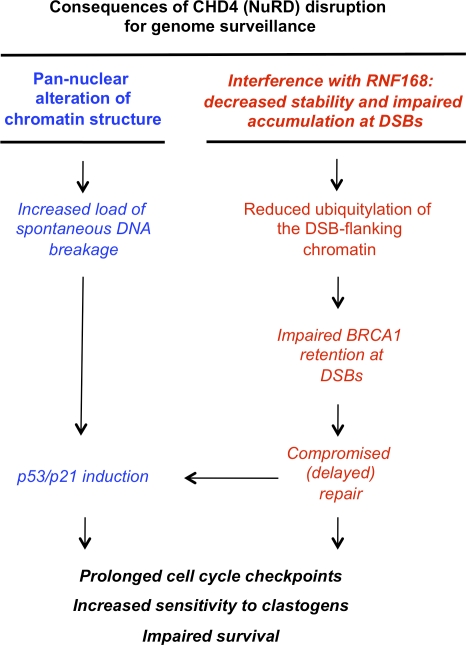
Model of the CHD4 involvement in protecting the genome against chromosomal breakage
Large-scale genetic and biochemical surveys suggested a role of the NuRD chromatin-remodeling complex in the DDR (van Haaften et al., 2006; Matsuoka et al., 2007). In this study, we provide additional and complementary evidence from an unbiased proteomic screen in which we identified four distinct subunits of the NuRD complex among proteins that gain affinity to chromatin damaged by IR. In addition, our study and an accompanying paper in this issue by Smeenk et al. report the first set of functional analyses that delineate the consequences of CHD4 disruption for genome surveillance. Among the salient alterations in CHD4-deficient cells are decreased clonogenic survival, supraphysiological increase of ATM/ATR signaling, protracted cell cycle checkpoints after IR, and delayed assembly of a subset of repair factors at the sites of DNA damage. In addition, we consistently observed that reduction of cellular levels of CHD4 is accompanied by spontaneous DNA damage. Molecular explanations of these diverse defects are likely complex, and our data indicate that disruption of CHD4 may deregulate DDR at multiple levels (Fig. 5).
Figure 5.
A proposed model of CHD4 involvement in genome maintenance. See Results and discussion for details.
First, the decreased stability and impaired accumulation of RNF168 at DSBs, the reduced local chromatin ubiquitylation, and the impaired accumulation of BRCA1 on damaged chromosomes likely attenuate the DNA repair efficiency and thereby contribute to the increased IR sensitivity in CHD4-deficient cells. The compromised DNA repair under such conditions might also contribute to the extended cell cycle checkpoints caused by continuous presence of unrepaired DNA and chromatin intermediates that amplify ATR and ATM signaling. Such a protracted cell cycle arrest, when extended over certain threshold, may undermine viability for instance by inducing cell death or allowing checkpoint adaptation followed by mitotic entry with unrepaired DSBs (Syljuåsen et al., 2006).
However, recent development in the field and results in this study indicate that the local events at the DSB sites cannot explain the entire complexity of phenotypes observed in CHD4-deficient cells. Most notably in this regard, the prolonged S phase, transient spike of ATM/ATR signaling after IR, and spontaneous DNA damage were not readily observed in experiments analyzing the currently known factors associated with the DSB-flanking chromatin. Based on our PFGE results, we propose that the latter consequences of CHD4 depletion reflect at least in part more global alterations of higher-order chromatin structure. This in turn can render chromatin vulnerable and prone to accumulate more breaks. In support of such a scenario, a recent study (Pegoraro et al., 2009) showed that loss of NuRD components during premature and physiological ageing induced alterations of the higher order chromatin structure accompanied by increased spontaneous DNA damage. Moreover, the genome-protective role of heterochromatin seems to be conserved throughout evolution, indicated by another recent study showing that loss of heterochromatin-associated histone methylations in Drosophila melanogaster also leads to spontaneous DNA damage, checkpoint activation, and chromosomal instability (Peng and Karpen, 2009). Interestingly, the NuRD complex (specifically its MBD3 subunit) was shown to facilitate deposition and stability of epigenetic marks including the heterochromatin-associated histone methylations (Morey et al., 2008). Thus, in addition to facilitating local assembly of repair factors directly at DSBs (and thereby directly contributing to repair efficiency), the NuRD complex may contribute to genome maintenance by organizing potentially vulnerable segments of eukaryotic genomes (such as the heterochromatin-associated repetitive sequences; Peng and Karpen, 2009) to a state that makes them more resilient to adverse effects of stochastic or clastogen-induced DNA breakage.
Materials and methods
Peptide preparation and tandem mass spectrometry
Proteins extracted from chromatin preparations were dissolved in LDS sample buffer, heated for 10 min at 70°C, reduced with DTT, and alkylated with iodoacetamide. Proteins were separated by electrophoresis on NuPAGE Bis-Tris 4–12% gradient gels (Invitrogen) and stained with Coomassie blue. Gel slices (10 slices) were cut into small pieces, washed several times with 20 mM ammonium bicarbonate and 50% acetonitrile, and incubated with 12.5 ng/µl endoprotease trypsin in 20 mM ammonium bicarbonate at 37°C overnight. The resulting peptides were extracted with 1% trifluoroacetic acid desalted on STAGE tips, and eluted into 96-well plates for mass spectrometric analysis.
Mass spectrometry and peptide identification and quantitation
Mass spectrometry was performed by liquid chromatography mass spectrometry using an HP1100 system (Agilent Technologies) and a linear ion-trap Fourier-transform ion cyclotron resonance mass spectrometer (Thermo Fisher Scientific). Peptides were separated by a 120-min linear gradient of 95% buffer A (0.5% acetic acid in water) to 50% buffer B (80% acetonitrile and 0.5% acetic acid in water). The linear ion-trap Fourier-transform ion cyclotron resonance instrument was operated in the data-dependent mode to acquire high-resolution precursor ion spectra (m/z 300–1,500; resolution 50,000; and ion accumulation to a target value of 3 × 106 ions) in the ion cyclotron resonance cell. The three most intense ions were sequentially isolated for accurate mass measurements by selected ion monitoring (SIM) scans (10-D mass window; resolution 50,000; and a target accumulation value of 50,000). The ions were simultaneously fragmented in the linear ion trap with a normalized collision energy setting of 27% and a target value of 10,000. Peak lists were extracted using MSQuant, an in-house developed open source application (http://msquant.sourceforge.net), and used for searches in the International Protein Index sequence database using Mascot (Matrix Science). MSQuant was also used to calculate peptide isotope ratios and to evaluate the certainty in peptide identification and quantitation based on Mascot score and MS3 scoring or by manual inspection. Initially, all peptides with a Mascot score of ≥20 were quantified automatically.
Data analysis
The quantified peptide ratios were analyzed through the use of a statistical analysis in which each individual peptide ratio was considered to be an independent estimate of the relative abundance of the corresponding protein. Raw individual peptide ratios were normalized by dividing by the median value, calculated across all three chromatin fractions; the final value is therefore the abundance of a peptide in the treated samples, relative to the untreated, in which a value of 1 corresponds to equivalent amounts. For each individual protein, ANOVA was used as a large-scale screening technique to identify proteins that showed a statistically significant variation in the elution profile, i.e., under the null hypothesis of each chromatin fraction in the elution profile having the same mean. Normalized peptide ratios were assumed to be log-normally distributed about the mean, and were therefore log (ln)-transformed before analysis: applying the Shapiro-Wilks test for normality on the log-transformed ratios showed that this assumption could not be rejected at the 95% level for the majority (92%) of proteins. The ANOVA was then performed using the R statistical package (http://www.r-project.org). Proteins in which the null hypothesis could be rejected at the 99% level were identified as candidates.
Plasmids and transfections
The expression plasmids for CHD4 were generated by inserting PCR-amplified CHD4 cDNA in frame into either pcDNA3.1-HA or pcDNA4TO-GFP. siRNA-resistant form of CHD4 was generated using the QuickChange site-directed mutagenesis kit (Agilent Technologies) with the oligonucleotides (forward) 5′-CGGCCAGAGCGGCAATTTTTCGTGAAATGGCAAGGC-3′ and (reverse) 5′-GCCTTGCCATTTCACGAAAAATTGCCGCTCTGGCCG-3′. Plasmid transfections were performed using FuGENE 6 according to the manufacturer’s recommendations (Roche).
RNA interference
The siRNA oligonucleotides were obtained from Thermo Fisher Scientific (SMARTpool of four oligonucleotides) and MWG Biotech (individual siRNAs). The annotations and sequences of the siRNA oligonucleotides were as follows (sense strands): (#1) 5′-GAAUAAAUUUCUAGCUCGAUU-3′, (#2) 5′-GGUGUUAUGUCUUUGAUUCUU-3′, (#3) 5′-GAGCGGCAGUUCUUUGUGAUU-3′, and (#4) 5′-AAGAAGAUCUAGCCCGAAAUU-3′.
All siRNA transfections were performed with 100 nM siRNA duplexes using Lipofectamine RNAiMAX (Invitrogen). Cells were transfected by siRNAs 24 and 48 h after plating. Transfection reagents and remaining oligonucleotides were washed off 6 h after treatment. Samples were harvested 72 h after initiation of transfection unless stated otherwise. Control siRNA (5′-GGGAGGACAAGACGUUCUA-3′) was against HSP70B (Leung et al., 1990), a variant of the human heat shock protein that is not expressed in U2OS cells. For complementation assays, oligonucleotide duplex #3 was used by transient transfection.
Cell culture
Human U2OS osteosarcoma cells and 293T human embryonic kidney cells were grown in DME containing 10% fetal bovine serum (Invitrogen), 100 U penicillin, and 100 µg/ml streptomycin. GM00130 (Coriell Cell Repositories) Epstein-Barr virus–transformed B-lymphocytes were cultured in RPMI1640 and GlutaMAX (Invitrogen) containing 15% serum, 100 U penicillin, and 100 µg/ml streptomycin. Where indicated, the culture medium was supplied with 5 mM of the proteasome inhibitor MG132 (EMD) or 10 µM ATM inhibitor (KU55933; Kudos Pharmaceuticals). For SILAC experiments, cells were grown for at least five cell divisions in l-lysine– and l-arginine–deficient RPMI1640 and GlutaMAX containing 15% dialyzed serum and either normal isotopes or 2H4-lysine/13C6-arginine, also referred to as light and heavy, respectively. U2OS derivative cell lines expressing GFP-CHD4 protein in a doxycycline-responsive manner were isolated by cotransfecting U2OS cells with pcDNA6/TR (Invitrogen) and pcDNA4/TO-GFP-CHD4 constructs and selecting productively transfected cells with 400 µg/ml zeocin and 5 µg/ml blasticidin S (Invitrogen).
Generation of DNA damage
IR was delivered by an x-ray generator (150 kV; 15 mA; 2.18 Gy/min dose rate; HF160; Pantak). Laser microirradiation and real-time recording was performed as described previously (Lukas et al., 2003, 2004). Typically, a mean of 150 cells were microirradiated for each experiment.
BrDU incorporation
Cells were labeled for 10 min with 25 µM BrdU (Sigma-Alrich), fixed in 4% formaldehyde solution (VWR) and permeabilized with 0.2% Triton X-100. Samples were treated with DNase (Roche) for 30 min at room temperature and immunostained according to the manufacturer’s instruction.
Antibodies
Rabbit polyclonal antibodies used in this study included CHK1 phospho-S317 (2344; Cell Signaling Technology), CHK1 phospho-S345 (2341; Cell Signaling Technology), SMC1 (ab9262; Abcam), phospho–SMC1-S966 (ab1276; Abcam), phospho-ATM S1981 (200-301-400; Rockland), γ-H2AX (2577; Cell Signaling Technology), cyclin A (sc-751; Santa Cruz Biotechnology, Inc.), HA (sc-7392; Santa Cruz Biotechnology, Inc.), histone 3 phospho-Ser10 (06-570; Millipore), p53 (sc-6243; Santa Cruz Biotechnology, Inc.), and RNF8 (Mailand et al., 2007). Mouse monoclonal antibodies included CHD4 (H00001108-MO1; Abnova), Rb (#554136; BD), Cdc25A (sc-7389; Santa Cruz Biotechnology, Inc.), MDC1 (DCS-380), γ-H2AX (05-636; Millipore), BrdU (RPN20AB; APBiotech), BRCA1 (sc-6954; Santa Cruz Biotechnology, Inc.), RNF168 (Doil et al., 2009), p21 (DCS-60, -61, -62 in combination), and conjugated ubiquitin, FK2 (PW 8810; Enzo Life Sciences, Inc.). Goat antibody to tubulin was obtained from Santa Cruz Biotechnology, Inc. (sc-7396). Additional antibodies to MDC1, RNF8, and RNF168 were provided by S. Jackson (The Gurdon Institute, University of Cambridge, Cambridge, England, UK), J. Chen (MD Anderson Cancer Center, University of Texas, Houston, TX), and D. Durocher (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada), respectively. Secondary antibodies included peroxidase-linked secondary antibodies (GE Healthcare) and donkey anti–goat antibody (Santa Cruz Biotechnology, Inc.) for immunoblotting and Alexa Fluor for immunofluorescence.
Immunochemical techniques
Total cell lysates were prepared in Laemmli sample buffer (50 mM Tris, pH 6.8, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol). Alternatively, cell extracts were prepared by the following buffer (50 mM Tris, pH 7.5, 120 mM NaCl, 0.5% NP-40 [IGEPAL CA-630; Sigma-Aldrich], and 1 mM EDTA). Protease inhibitors 5 µg/ml leupeptin, 2 µg/ml aprotinin, and 0.1 mM PMSF, phosphatase inhibitors 10 mM β-glycerophosphate, 0.1 mM Na3VO4, and 1 mM NaF, and 1 mM DTT (to reduce disulfide bonds) were added before use. Immunoprecipitation, immunoblotting, and immunofluorescence procedures were described previously (Lukas et al., 2003, 2004; Mailand et al., 2006).
Microscopy
Confocal microscopy was performed essentially as described previously (Bekker-Jensen et al., 2006; Doil et al., 2009) using a confocal microscope (LSM 510; Carl Zeiss, Inc.) mounted on an inverted microscope (Axiovert 100M; Carl Zeiss, Inc.) equipped with a Plan Neofluar 40× 1.3 NA oil immersion objective. Dual-color images were acquired using laser lines 488-nm and 543-nm for excitation of Alexa Fluor 488 and Alexa Fluor 568 dyes, respectively. Band-pass filters 505–530 nm and 560–615 nm were used to collect the emitted fluorescence signals. For quantification of protein accumulations at laser-generated DSBs, mean nondamaged nuclear fluorescence intensity was subtracted from mean fluorescence intensity of damaged regions in each cell. All quantified images of the same antibody staining were captured with the same microscope settings (laser intensity, detector gain, and detector offset). Quantification of IR-induced foci per nucleus was preformed as described previously (Doil et al., 2009).
Flow cytometry
Cells were harvested by trypsinization, fixed in 70% ethanol, and resuspended in 200 µl propidium iodide (PI) buffer (Facsflow [BD], 0.1 mg/ml PI, and 0.02% NaN3). Samples were incubated for 30 min at 37°C before analysis. Cell cycle analysis was performed by flow cytometry on a flow cytometer (FACSCalibur; BD). Mitotic entry was examined by staining with primary antibody for histone 3 phospho-S10 followed by FITC-conjugated secondary antibody and PI. The method was previously described (Syljuåsen et al., 2004). Data were quantified by Modfit LT (version 3.1; BD).
Chromatin fractionation
GM00130 suspension cells (3 × 107 cells) were harvested by centrifugation and resuspended in 1 ml of buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, and 10% glycerol + full inhibitor range [1 mM DTT, 5 µg/ml aprotinin, 5 µg/ml leupeptin, 0.1 mM PMSF, 0.1 mM vanadate, 10 mM β-glycerolphosphate, okadeic acid, and 1 mM NAF]). Cells were lysed in 0.02% Triton X-100 for 1 min, and soluble cytoplasmic proteins were collected. The pellet was washed twice in buffer A and incubated for 30 min at 4°C in no-salt buffer (3 mM EDTA, pH 7.9, and 0.2 mM EGTA + full inhibitor range). Cells were spun for 5 min at 1,500 g, and soluble nuclear proteins were collected. The pellet was washed twice in no-salt buffer. The chromatin-enriched pellet was resuspended in buffer S (20 mM Hepes, pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol + 0.5 mM DTT, vanadate, okadeic acid, and NAF) with 120 mM NaCl. Samples were incubated for 15 min at 4°C and spun at 2,000 g for 5 min. Extracted proteins were collected in the supernatant. This was repeated with an NaCl concentration of 210 mM, and finally, samples were resuspended in loading sample buffer (50 mM Tris, pH 6.8, 100 mM DTT, 2% SDS, 0.1% Bromophenol blue, and 10% glycerol), boiled for 5 min, and sonicated.
Kinase assay
After standard immunoprecipitation, beads were washed two times in kinase buffer (50 mM Hepes, pH 7.5, 10 mM MgCl2, 2.5 mM EGTA, 10 mM β-glycerolphosphate, 1 mM NAF, 1 mM DTT, and 0.1 mM Na3VO4). 30 µl kinase reaction mix (18 µl kinase buffer, 75 µM cold ATP, 2 µg protein H1, and 10 µCi γ-[32P] ATP) was added and incubated for 30 min at 30°C with interval shaking. Samples were analyzed using 12% SDS-PAGE followed by Coomassie staining and drying, and the signal was quantified using a phosphoimager.
Clonogenic survival assay
U2OS cells were untreated or transfected with control or CHD4 siRNA. 2 d after transfection, between 100 and 2,000 cells (depending on radiation dose to yield 30–200 colonies per dish) were seeded to 6-cm-diameter dishes, incubated for 20–24 h, and treated with IR (0, 2, 3, or 4 Gy). Subsequently, cells were incubated for an additional 10 d and stained with crystal violet. Colonies containing >50 cells were scored as survivors. Survival fractions were calculated in each experiment as the mean cloning efficiency (from at least two parallel dishes) after treatment corrected for plating efficiency.
PFGE
Optimized PFGE protocol allowing quantitative detection of DSBs already in a range of 10 Gy of IR was performed as described previously (Hanada et al., 2007). In brief, U2OS cells were treated with indicated siRNAs on two sequential days. After 72 h, cells were either irradiated or left untreated, trypsinized, and agarose plugs 5 × 105 cells were prepared with a CHEF disposable plug mold (Bio-Rad Laboratories). Plugs were incubated in lysis buffer (100 mM EDTA, 0.2% sodium deoxycholate, 1% sodium laurylsarcosine, and 1 mg/ml proteinase K) for ∼40 h at 37°C. After washing with TE buffer (10 mM Tris, pH 8.0, and 100 mM EDTA), plugs were embedded into a 0.9% agarose gel. Electrophoresis was performed in TBE buffer with the following parameters: (block I) 9 h, 120° angle, 5.5 V/cm voltage, 30–18-s interval; (block II) 6 h, 117° angle, 4.5 V/cm voltage, 18–9-s interval; (block III) 6 h, 112° angle, 4 V/cm voltage, 9–5-s interval. The gel was stained with ethidium bromide.
Real-time PCR
Real-time PCR was performed as described previously (Löffler et al., 2003) with the following primer sequences: Cdc25A, (sense) 5′-ACCGTCACTATGGACCAGC-3′ and (antisense) 5′-TTCAGAGCTGGACTACATCC-3′; and porphobilinogen deaminase, (sense) 5′-TCCAAGCGGAGC-CATGTCTG-3′ and (antisense) 5′-AGAATCTTGTCCCCTGTGGTGGA-3′.
Online supplemental material
Fig. S1 provides additional evidence for extended cell cycle delay in CHD4-depleted cells exposed to IR. Fig. S2 shows impact of CHD4 depletion on checkpoint signaling and DNA repair. Fig. S3 shows impaired BRCA1 retention at IR-induced nuclear foci and partial destabilization of RNF168 in CHD4-deficient cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912135/DC1.
Acknowledgments
We thank Dr. Per Guldberg for reagents and assistance with real-time PCR experiments and Haico van Attikum for sharing unpublished results.
This work was supported by grants from the Danish Cancer Society, the Danish National Research Foundation, the Lundbeck Foundation (projects R-44-A4400 and R-13-A1287), the European Commission (projects GENICA, DNA Repair, and CZ.1.05/2.1.00/01.0030), the Danish Research Council, and the John and Birthe Meyer Foundation.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3 related
- DDR
- DNA damage response
- DSB
- double-strand break
- IR
- ionizing radiation
- NuRD
- nucleosome remodeling and deacetylase
- PFGE
- pulsed field gel electrophoresis
- PI
- propidium iodide
- SILAC
- stable isotope labeling with amino acids in cell culture
References
- Aebersold R., Mann M. 2003. Mass spectrometry-based proteomics. Nature. 422:198–207 10.1038/nature01511 [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas J. 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19:238–245 10.1016/j.ceb.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas C., Lukas J. 2004. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5:792–804 10.1038/nrm1493 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J. 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173:195–206 10.1083/jcb.200510130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen N.J., Fujita N., Kajita M., Wade P.A. 2004. Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta. 1677:52–57 [DOI] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Falck J., Petrini J.H.J., Williams B.R., Lukas J., Bartek J. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30:290–294 10.1038/ng845 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. 2004. H2AX: the histone guardian of the genome. DNA Repair (Amst.). 3:959–967 10.1016/j.dnarep.2004.03.024 [DOI] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., Kanaar R. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14:1096–1104 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- Huen M.S.Y., Grant R., Manke I., Minn K., Yu X.C., Yaffe M.B., Chen J.J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B., Bartek J. 2004. Cell-cycle checkpoints and cancer. Nature. 432:316–323 10.1038/nature03097 [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Bakkenist C.J., McKinnon P.J., Kastan M.B. 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 18:1423–1438 10.1101/gad.1200304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T.K., Rajendran M.Y., Monfries C., Hall C., Lim L. 1990. The human heat-shock protein family. Expression of a novel heat-inducible HSP70 (HSP70B’) and isolation of its cDNA and genomic DNA. Biochem. J. 267:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löbrich M., Jeggo P.A. 2007. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 7:861–869 10.1038/nrc2248 [DOI] [PubMed] [Google Scholar]
- Löffler H., Syljuåsen R.G., Bartkova J., Worm J., Lukas J., Bartek J. 2003. Distinct modes of deregulation of the proto-oncogenic Cdc25A phosphatase in human breast cancer cell lines. Oncogene. 22:8063–8071 10.1038/sj.onc.1206976 [DOI] [PubMed] [Google Scholar]
- Lukas C., Falck J., Bartkova J., Bartek J., Lukas J. 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5:255–260 10.1038/ncb945 [DOI] [PubMed] [Google Scholar]
- Lukas C., Melander F., Stucki M., Falck J., Bekker-Jensen S., Goldberg M., Lerenthal Y., Jackson S.P., Bartek J., Lukas J. 2004. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 23:2674–2683 10.1038/sj.emboj.7600269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N., Falck J., Lukas C., Syljuâsen R.G., Welcker M., Bartek J., Lukas J. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science. 288:1425–1429 10.1126/science.288.5470.1425 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Bartek J., Lukas J. 2006. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 23:307–318 10.1016/j.molcel.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z.M., Solimini N., Lerenthal Y., et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316:1160–1166 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- Morey L., Brenner C., Fazi F., Villa R., Gutierrez A., Buschbeck M., Nervi C., Minucci S., Fuks F., Di Croce L. 2008. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol. Cell. Biol. 28:5912–5923 10.1128/MCB.00467-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 1:376–386 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- Pegoraro G., Kubben N., Wickert U., Göhler H., Hoffmann K., Misteli T. 2009. Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol. 11:1261–1267 10.1038/ncb1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.C., Karpen G.H. 2009. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 5:e1000435 10.1371/journal.pgen.1000435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D.R., Schreiber S.L. 1999. Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry. 38:14711–14717 10.1021/bi991614n [DOI] [PubMed] [Google Scholar]
- Smeenk G., Wiegant W.W., Vrolijk H., Solari A.P., Pastink A., van Attikum H. 2010. The NuRD chromatin–remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 190:741–749 10.1083/jcb.201001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136:420–434 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Stucki M., Jackson S.P. 2006. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst.). 5:534–543 10.1016/j.dnarep.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 123:1213–1226 10.1016/j.cell.2005.09.038 [DOI] [PubMed] [Google Scholar]
- Syljuåsen R.G., Sørensen C.S., Nylandsted J., Lukas C., Lukas J., Bartek J. 2004. Inhibition of Chk1 by CEP-3891 accelerates mitotic nuclear fragmentation in response to ionizing radiation. Cancer Res. 64:9035–9040 10.1158/0008-5472.CAN-04-2434 [DOI] [PubMed] [Google Scholar]
- Syljuåsen R.G., Jensen S., Bartek J., Lukas J. 2006. Adaptation to the ionizing radiation-induced G2 checkpoint occurs in human cells and depends on checkpoint kinase 1 and Polo-like kinase 1 kinases. Cancer Res. 66:10253–10257 10.1158/0008-5472.CAN-06-2144 [DOI] [PubMed] [Google Scholar]
- Tong J.K., Hassig C.A., Schnitzler G.R., Kingston R.E., Schreiber S.L. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 395:917–921 10.1038/27699 [DOI] [PubMed] [Google Scholar]
- van Attikum H., Gasser S.M. 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19:207–217 10.1016/j.tcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- van Haaften G., Romeijn R., Pothof J., Koole W., Mullenders L.H., Pastink A., Plasterk R.H., Tijsterman M. 2006. Identification of conserved pathways of DNA-damage response and radiation protection by genome-wide RNAi. Curr. Biol. 16:1344–1350 10.1016/j.cub.2006.05.047 [DOI] [PubMed] [Google Scholar]
- Wade P.A., Jones P.L., Vermaak D., Wolffe A.P. 1998. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8:843–846 10.1016/S0960-9822(98)70328-8 [DOI] [PubMed] [Google Scholar]
- Wang B., Elledge S.J. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA. 104:20759–20763 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong J., Moreno G.T., Young M.K., Côté J., Wang W. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 2:851–861 10.1016/S1097-2765(00)80299-3 [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy G., Seelig H.P., Lane W.S., Reinberg D. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 95:279–289 10.1016/S0092-8674(00)81758-4 [DOI] [PubMed] [Google Scholar]