Subsets of 40S ribosomal subunits are required for initiating rRNA processing, rRNA maturation, and nuclear export.
Abstract
Our knowledge of the functions of metazoan ribosomal proteins in ribosome synthesis remains fragmentary. Using siRNAs, we show that knockdown of 31 of the 32 ribosomal proteins of the human 40S subunit (ribosomal protein of the small subunit [RPS]) strongly affects pre–ribosomal RNA (rRNA) processing, which often correlates with nucleolar chromatin disorganization. 16 RPSs are strictly required for initiating processing of the sequences flanking the 18S rRNA in the pre-rRNA except at the metazoan-specific early cleavage site. The remaining 16 proteins are necessary for progression of the nuclear and cytoplasmic maturation steps and for nuclear export. Distribution of these two subsets of RPSs in the 40S subunit structure argues for a tight dependence of pre-rRNA processing initiation on the folding of both the body and the head of the forming subunit. Interestingly, the functional dichotomy of RPS proteins reported in this study is correlated with the mutation frequency of RPS genes in Diamond-Blackfan anemia.
Introduction
The function of ribosomal proteins is a long-standing question that has gained renewed interest with the recent discovery that these proteins play a role in pathologies, especially bone marrow failures (Liu and Ellis, 2006; Ganapathi and Shimamura, 2008). Heterozygous mutations in various genes encoding ribosomal proteins of the small subunit (RPSs) and of the ribosomal protein of the large subunit (RPL) have been linked to Diamond-Blackfan anemia (DBA), a rare congenital disease characterized by deficient production of erythrocyte precursors and a heterogeneous spectrum of congenital abnormalities in different tissues (Draptchinskaia et al., 1999; Gazda et al., 2006, 2008; Cmejla et al., 2007; Farrar et al., 2008; Doherty et al., 2010). Similarly, haploinsufficiency of RPS14 has been identified as a cause of 5q− syndrome, an acquired myelodysplasia, also characterized by defects in erythroid differentiation (Ebert et al., 2008). Beyond DBA and 5q− syndrome, several other rare diseases, such as Treacher-Collins syndrome, Schwachman-Diamond syndrome, and dyskeratosis congenita, have also been linked to mutations in genes encoding proteins involved in ribosome biogenesis (Liu and Ellis, 2006; Ganapathi and Shimamura, 2008), and these pathologies are now designated as ribosomal diseases. The current model, which is supported by experimental data from different animals (Sulic et al., 2005; MacInnes et al., 2008; McGowan et al., 2008; Barkić et al., 2009), postulates that defects in ribosome production may trigger a stress response involving p53, which blocks cell cycle progression and may induce apoptosis. According to this hypothesis, the function of ribosomal proteins in the maturation of the ribosomal subunits, rather than in translation, is central to the pathophysiology of the diseases in which they are involved.
After synthesis, most ribosomal proteins are rapidly transported to the nucleus and integrated into ribosomal particles. It is admitted that a subset of these proteins associate during transcription with the large ribosomal RNA (rRNA) precursor (47S pre-rRNA) along with numerous transacting factors, thus forming a large ribonucleoparticle or the 90S preribosome. Within this particle, the transcribed spacers flanking the 18S, 5.8S, and 28S rRNAs are progressively eliminated through endonucleolytic cleavages and exonucleolytic processing (Hadjiolov, 1985; Eichler and Craig, 1994; Gerbi and Borovjagin, 2004). This leads to the splitting of the 90S particles into pre-40S and pre-60S particles, which are ultimately exported to the cytoplasm where the last maturation steps take place (Fig. 1). Proteomic analyses in Saccharomyces cerevisiae have revealed that ∼150 proteins take part in the maturation process, with putative enzymatic and regulatory functions, such as nucleases, helicases, chaperones, GTPases, and ATPases (Fromont-Racine et al., 2003; Milkereit et al., 2003). In general, the exact role of these proteins in ribosomal production remains unclear. Early in the maturation pathway, pre-rRNAs are also modified by small nucleolar RNAs (sno-RNPs), which catalyze pseudourydilation and 2’-O-methylation of ∼200 nucleotides (Maden, 1990; Kiss, 2002). Additionally, some sno-RNPs, including U3, U8, U14, U17, U22, and E2 in vertebrates, are required for the cleavage of pre-rRNAs. The U3 sno-RNP hybridizes with the 5′- external transcribed spacer (ETS) and with the 18S rRNA and was proposed to chaperone the folding of 18S rRNA (Gerbi and Borovjagin, 2004). snR30, the yeast homologue of U17, hybridizes within the 18S domain in the pre-rRNA and, like U3, is necessary for maturation of this RNA (Fayet-Lebaron et al., 2009).
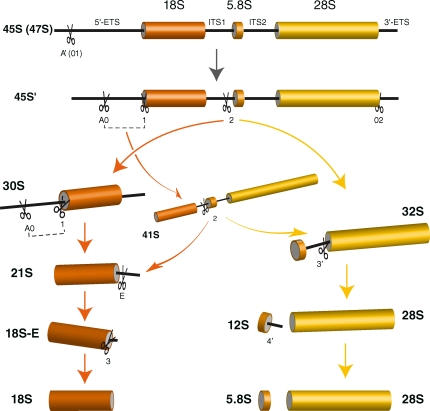
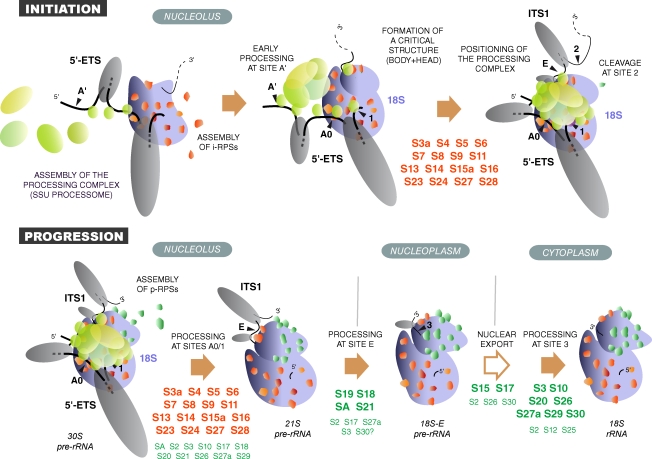
Figure 1.
Pre-rRNA processing pathways in human cells. Two pathways coexist depending on the order of cleavage in the 5′-ETS (sites A0 and 1) and ITS1 (site 2). The major pathway in HeLa cells involves cleavage at site 2 before processing of ITS1, as shown by the higher abundance of 30S pre-rRNA relative to 41S pre-rRNA (see Fig. 3). The 18S-E pre-rRNA is converted to 18S rRNA in the cytoplasm. Nomenclature of the cleavage sites follows Hadjiolova et al. (1993) and Rouquette et al. (2005). The A’ early cleavage site is also designated 01.
Mechanisms of ribosome biogenesis have been most well described in the yeast S. cerevisiae (Warner, 1989; Venema and Tollervey, 1999; Fromont-Racine et al., 2003), including the role of ribosomal proteins in this process (Jakovljevic et al., 2004; Leger-Silvestre et al., 2004, 2005; Ferreira-Cerca et al., 2005, 2007; Zhang et al., 2007; Poll et al., 2009). Although the general scheme of pre-rRNA maturation appears to be conserved in eukaryotes, there are several important differences in vertebrates relative to yeast, including additional cleavage points in the pre-rRNA, supplemental factors involved in ribosome biogenesis, and differential organization of the nucleus and of the rDNA genes, which form clusters on five chromosomes instead of one. Thus, processing of the 5′-ETS in vertebrates includes an early cleavage, which is not found in yeast (Fig. 1; Miller and Sollner-Webb, 1981; Craig et al., 1987). At the other end of the pathway, formation of the 18S RNA 3′ end involves an additional cleavage step in the 5′ part of ITS1 (Rouquette et al., 2005). Additionally, the cleavage order appears to be more flexible in vertebrates (Hadjiolova et al., 1993; Gerbi and Borovjagin, 2004). Functional studies of a limited number of mammalian ribosomal proteins, mostly those linked to pathologies, have revealed that they are involved at various stages in pre-rRNA processing (Rouquette et al., 2005; Choesmel et al., 2007, 2008; Flygare et al., 2007; Idol et al., 2007; Ebert et al., 2008; Farrar et al., 2008; Gazda et al., 2008; Robledo et al., 2008; Doherty et al., 2010).
In this study, we present a global analysis of the function of ribosomal proteins in the formation of the human 40S subunit. Our results show that initiation of the 18S rRNA processing pathway depends strictly on the assembly of a large subset of ribosomal proteins, which probably catalyze the initial steps of the 18S RNA folding, whereas the remaining ribosomal proteins are primarily involved in downstream steps from the nucleolus to the cytoplasm. This functional dichotomy is correlated with distinct timing in the recruitment of these proteins in preribosomes and different positions within the structure of the 40S subunit. Based on these data, we propose that initiation of pre-rRNA processing in mammalian cells requires formation of an assembly intermediate of the entire 40S subunit.
Results
RPS proteins are essential for production of 40S subunits
The vast majority of the 2,000 sequences encoding ribosomal proteins in the human genome are predicted to be pseudogenes, and most ribosomal proteins are presumed to be encoded by a single gene with some exceptions (Kenmochi et al., 1998; Zhang et al., 2002). Among RPS proteins, two are synthesized from several paralogous genes: RPS4, which is encoded by three genes located on the x and y chromosomes (RPS4X, RPS4Y1, and RPSY2; Zhang et al., 2002; Skaletsky et al., 2003) and RPS27, for which there are two paralogous genes, RPS27 on chromosome 1 and RPS27L on chromosome 15 (Zhang et al., 2002; He and Sun, 2007; Li et al., 2007). Additionally, RPS17 is encoded by two genes on chromosome 15, yielding identical mRNAs (Zhang et al., 2002). HeLa cells, which were derived from a female patient, bear only the RPS4X gene.
We designed two siRNAs to inhibit expression of each of the 33 genes encoding 40S subunit proteins in HeLa cells, except for RPS28 and RPS29, in which case a single siRNA was used as a result of their very small ORFs. Regarding RPS15, RPS19, and RPS24, the most efficient siRNA was selected based on our previous studies (Rouquette et al., 2005; Choesmel et al., 2007, 2008). Using quantitative RT-PCR, we checked the efficiency of a large number of these siRNAs and found a >90% drop in the amount of the corresponding mRNA 48 h after transfection with little or no influence on the levels of other RPS mRNAs (Fig. S1), which is consistent with previous studies (Rouquette et al., 2005; Choesmel et al., 2007, 2008; Robledo et al., 2008). All pairs of siRNAs were efficient at destabilizing the targeted mRNA, except for RPS15a, for which only one of three siRNAs tested proved to be functional.
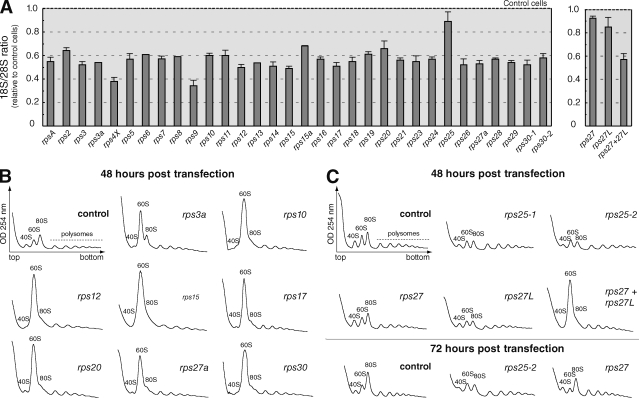
Knockdown of all the RPS proteins presumed to be encoded by a single gene (including RPS4X) resulted in a significant drop in the amount of 18S rRNA relative to 28S rRNA (Fig. 2 A), indicating that RPS proteins are essential for production of the 40S ribosomal subunit. Analysis of cytoplasmic ribosomes on a sucrose gradient for a large subset of these siRNAs confirmed the inhibition of 40S ribosomal subunit production (Fig. 2 B). The only exception was RPS25, the depletion of which had only a mild effect on 18S rRNA levels, even though the amount of RPS25 mRNA was reduced by 95% (Fig. S1). However, the polysome profile of RPS25-depleted cells revealed a decrease in the amount of free 40S subunits (Fig. 2 C), suggesting that this ribosomal protein may be required for efficient production or stability of the 40S subunit. Knockdown of either RPS27 or RPS27L expression had a limited effect on the levels of 18S rRNA. However, codepletion of RPS27 and RPS27L resulted in a dramatic decrease in both the amount of 18S rRNA (Fig. 2 A, right) and on the ribosome profile (Fig. 2 C), suggesting redundant functions for the two isoforms in ribosome biogenesis.
Figure 2.
Knockdown of each RPS protein affects 40S particle production. (A) Total RNAs were extracted from HeLa cells 48 h after transfection with siRNAs targeting RPS mRNAs. The 18S/28S ratio was calculated from Northern blot analysis to estimate the impact of each siRNA on 40S subunit production compared with control cells. Means of three to eight independent experiments with two different siRNAs ± SD are shown. (B) Cytoplasmic fractions were prepared in the presence of cycloheximide from cells transfected for 48 h with rps siRNAs or with a scrambled siRNA. Depletion of RPS proteins leads to the loss of free 40S particles and accumulation of free 60S particles, indicating a strong alteration in 40S subunit production. For each target protein, similar results were obtained with both siRNAs. (C) Knockdown of RPS25, RPS27, or RPS27L for 72 h, instead of 48 h, had no additional effect.
Two functional groups of ribosomal proteins in pre-rRNA maturation
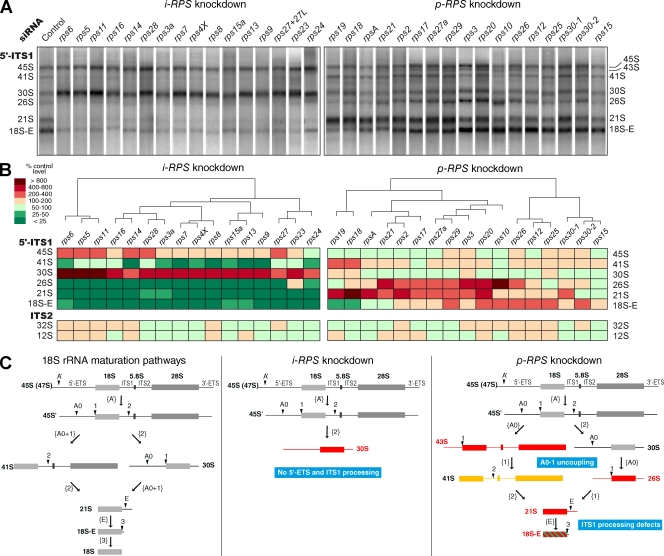
To study the role of the RPS proteins in pre-rRNA maturation, total RNAs extracted from siRNA-transfected cells were analyzed by Northern blotting with probes hybridizing to ITS1 or ITS2. Variations in pre-rRNA levels relative to control cells in multiple independent experiments were quantified by phosphoimaging and analyzed by principal component analysis and hierarchical clustering (Fig. 3, A and B). Strong variations were detected in the levels of the 18S rRNA precursors, whereas the precursors of the 60S subunit rRNAs (12S and 32S pre-rRNAs) were rather stable (Fig. 3 B), which is consistent with a specific defect in 40S subunit production.
Figure 3.
Analysis of pre-rRNA profiles upon individual knockdown of RPS proteins reveals their sequential role during 40S particle formation. Total RNAs from HeLa cells were extracted 48 h after transfection, resolved on a 1% agarose gel, and transferred to a nylon membrane, which was hybridized with various 32P-labeled probes (5′-ITS1, ITS2, 18S, or 28S). (A) A representative result with the 5′-ITS1 probe is shown for each RPS protein. The proteins fall into two categories: i-RPSs act in the early steps of processing (left), whereas p-RPSs impact a broad range of 18S rRNA maturation steps (right). (B) Hierarchical clustering of the pre-rRNA processing phenotypes after principal component analysis. Pre-rRNAs were quantified on Northern blots by phosphoimaging and normalized to the amount of 28S. The relative abundance of the precursors detected either with the 5′-ITS1 probe (top) or the ITS2 probe (bottom) relative to control cells is indicated by the color code (percentage of control level). Each column in the table corresponds to the mean of two to eight experiments and mixes the data obtained with two different siRNAs for most genes. siRNAs rps30-1 and rps30-2 yielded slightly different results but were very close in the clustering. (C) Alteration of the 18S rRNA maturation pathways in mammalian cells on knockdown of i-RPSs or p-RPSs. (left) Processing of the large 47S primary transcript can follow two alternative paths. (middle) Upon depletion of i-RPSs, all cleavages beyond that at site 2 were no longer taking place, leading to major accumulation of 30S pre-rRNAs. (right) When p-RPSs were knocked down, the 30S pre-rRNAs were at least partly processed, leading to 18S-E pre-rRNA production, and a wider range of phenotypes was observed: blocked or delayed processing at sites A0, 1, E, and 2 (accumulation of 30S, 21S, and 18S-E) and uncoupling of cleavage at sites A0 and 1 (accumulation of 26S and 45S). Red, strong accumulation; yellow, moderate accumulation; green, under accumulation.
This analysis led us to classify RPS proteins into two functional groups relative to pre-rRNA processing. The first class, designated initiation RPS (i-RPS), consisted of 16 of the 32 proteins. Knockdown of these proteins resulted in the accumulation of the 45S and 30S pre-rRNAs, whereas the other precursors almost disappeared. This phenotype indicated the failure of all the processing steps in the 5′-ETS and in the ITS1 necessary to generate 18S rRNA beyond cleavage of ITS1 at site 2 (Fig. 3 C). This group of proteins comprises RPS3a, RPS4, RPS5, RPS6, RPS7, RPS8, RPS9, RPS11, RPS13, RPS14, RPS16, RPS15a, RPS23, RPS24, and RPS28. The two RPS27 isoforms can also be classified into this group: single depletion of either resulted in mild accumulation of the 45S and 30S pre-RNAs, whereas depletion of both led to very strong accumulation of these intermediates (Fig. S2).
Depletion of the 16 remaining RPS proteins was characterized by partial or complete 5′-ETS maturation and various defects in ITS1 processing after cleavage at site 2. Thus, these were designated as progression RPS (p-RPS). The phenotypes in this group were heterogeneous and could be distinguished according to the extent of ITS1 processing. In the case of RPS19, RPS18, RPSA, and RPS21, low levels of 18S-E were detected, indicating that maturation of the 21S pre-rRNA was very strongly affected. This was most marked with RPS18 and RPS19, two proteins that play a crucial role in ITS1 processing in yeast (Leger-Silvestre et al., 2004, 2005). Knockdown of RPS2, RPS3, RPS17, RPS20, RPS27a, and RPS29 also yielded higher amounts of 21S pre-rRNA, but production of 18S-E pre-rRNA indicated efficient processing at site E. Depletion of RPS10, RPS12, RPS15, RPS26, and RPS25 was associated with higher levels of 18S-E pre-rRNA but did not alter conversion of the 21S species to 18S-E RNA. Although the two rps30 siRNAs induced accumulation of 18S-E pre-rRNA, one resulted in accumulation of 21S pre-rRNA, whereas the other did not. Given the short size of the RPS30 coding sequence, we were not able to design a third siRNA to refine this result.
Interestingly, with the exception of RPS12, RPS15, RPS19, and RPS25, alteration of ITS1 processing upon depletion of p-RPSs was always paralleled by accumulation of 26S and 43S pre-rRNAs (Fig. 3, A and B), two intermediate species that start at site A0 in the 5′-ETS (Rouquette et al., 2005). The presence of the 26S and 43S pre-rRNAs, which were barely detectable in control cells, indicated that the 5′-ETS was cleaved at site A0, independent of processing at site 1 (5′ extremity of the 18S rRNA; Fig. 3 C). Analysis of the 5′-ETS cleavage products in mouse cells has shown that these two processing steps are normally strongly coupled (Kent et al., 2008). Thus, some p-RPSs could be required early in maturation for coordinated processing at sites A0 and 1 in the 5′-ETS.
In summary, these data lead us to distinguish two functional categories of RPSs. In the first (i-RPSs), protein depletion blocked initiation of all the processing steps specific to the 18S pre-rRNA maturation pathway. Proteins of the second group (p-RPSs) were required for progression of the processing pathway, but in most cases, did not prevent production of the 18S-E pre-rRNA.
Early cleavage of the 5′-ETS is insensitive to depletion of RPS proteins
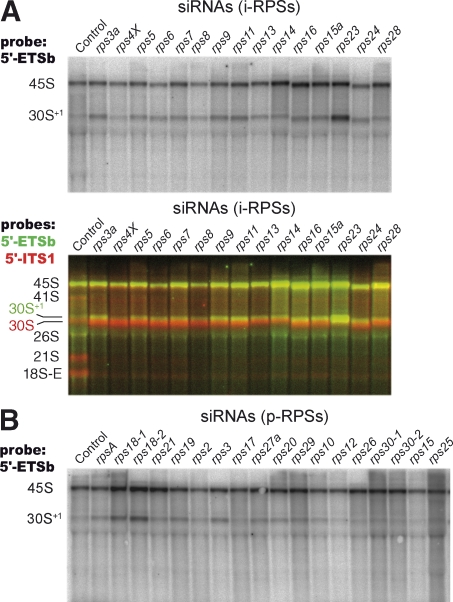
The primary event of mammalian pre-rRNA processing is the cleavage of the 5′-ETS at the so-called early cleavage site (site A’; also known as 01) positioned around nucleotide 414 in human pre-rRNA (Kass et al., 1987). This step, which does not occur in S. cerevisiae, requires nucleolin (Ginisty et al., 1998) and U3 sno-RNA (Kass and Sollner-Webb, 1990). To evaluate the requirement of ribosomal proteins in this particular step, we probed Northern blots of RNAs extracted from RPS-depleted cells with an oligonucleotide hybridizing upstream of site A’. When compared with controls, we generally noticed a slight increase in the amount of 45S/47S pre-rRNA upon knockdown of i-RPSs as well as an extended form of the 30S RNA unprocessed at site A’ that we called 30S+1 (Fig. 4, top). However, the majority of the 30S pre-rRNA accumulating on treatment with these siRNAs was processed at site A’, as indicated by a probe hybridizing downstream of site A’ (Fig. 4, middle). With the rps23-1 siRNA, we observed a stronger accumulation of unprocessed 30S+1 pre-rRNA, but this result was not reproduced with two other siRNAs against the RPS23 mRNA, which otherwise efficiently affected maturation at sites A0 and 1 (unpublished data). Regarding p-RPSs, we found that siRNAs against RPS18 partially inhibited processing at site A’ (Fig. 4, bottom).
Figure 4.
Processing of the early cleavage site in the 5′-ETS of the primary transcript is still taking place upon depletion of RPS proteins. Pre-rRNAs uncleaved at site A’ were detected with probe 5′-ETSb, spanning nucleotides 301–324. This probe predominantly revealed the 45S pre-rRNA together with an extended form of the 30S pre-rRNA that we called 30S+1. (A) The 30S+1 species accumulated slightly on depletion of i-RPS proteins. However, the majority of the 30S RNA detected with probe 5′-ITS1 corresponds to a faster migrating band (overlay), indicating that it is processed at site A’. (B) Mild accumulation of 30S+1 RNA was observed on depletion of some p-RPSs only, especially RPS18.
These data indicate that processing at the early cleavage site, which is not universal in eukaryotes, is much less sensitive to ribosomal protein depletion than the other processing steps in the pre-40S particle maturation pathway. We conclude that this early cleavage step is independent of the initial steps of the pre-40S particle formation and is uncoupled from the other cleavages of the 5′-ETS.
Depletion of RPS proteins affects organization of nucleolar chromatin
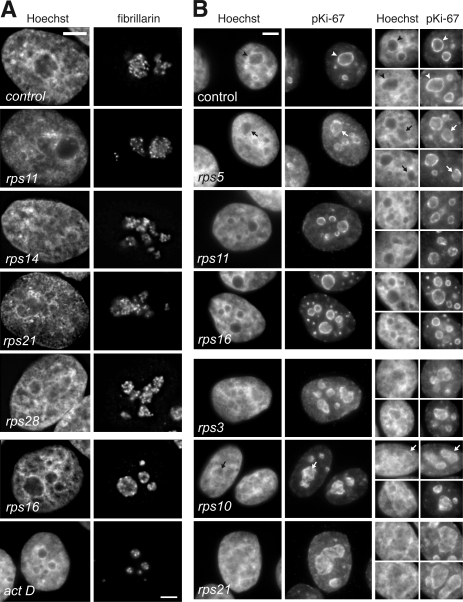
Knockdown of a large subset of RPS proteins induced strong changes in nuclear morphology, as revealed by localization of fibrillarin, a core component of C/D box sno-RNPs and a marker of the dense fibrillar component, a subdomain of the nucleolus. In most cases, the overall nucleolar morphology was irregular, although fibrillarin was still distributed in bead-like structures corresponding to fibrillar centers surrounded by the dense fibrillar component, as in the controls (Fig. 5 A, top). Depletion of RPS16 resulted in a distinct phenotype with smaller, rounded nucleoli (Fig. 5 A, bottom); fibrillarin appeared as sparser dots, clustered at the periphery of the nucleoli, a pattern strongly reminiscent of the stepwise nucleolar segregation observed upon inhibition of rDNA transcription by actinomycin D (Dousset et al., 2000). This suggests that knockdown of RPS16 severely interfered with rDNA transcription.
Figure 5.
Nucleolar morphology and chromatin condensation are modified by the depletion of specific RPSs. (A) Nucleolar morphology visualized with mAb 72B9. Fibrillarin is distributed in the dense fibrillar component, which outlines the fibrillar centers (small dots). Depletion of RPSs leads to changes in the number and shape of the nucleoli. Upon depletion of RPS16, nucleoli show partial segregation of the fibrillar components, similar to what is observed on RNA polymerase I inhibition for 4 h with 5 nM actinomycin D. (B) Knockdown of RPSs affected chromatin condensation and localization. Perinucleolar chromatin, visualized with antibodies to pKi-67, appeared as a rim around nucleoli in control HeLa cells (arrowheads). Upon knockdown of some proteins, like RPS5, RPS11, or RPS16, this rim was interrupted and disorganized (arrows show some examples). Depletion of other RPSs, like RPS3, RPS10, or RPS21, resulted in the redistribution of chromatin within nucleoli, as also seen by Hoechst staining of nucleoli, which was as intense as in the nucleoplasm. Bars, 5 µm.
To further understand the changes in nucleolar morphology, we looked at the distribution of chromatin around and within nucleoli. After depletion of i-RPSs RPS5, RPS11, RPS16 (Fig. 5 B, top), RPS23, or RPS28 (not depicted), nucleoli were clearly distinguishable as dark regions in nuclei counterstained with Hoechst, but perinucleolar heterochromatin tended to be poorly detected. Accordingly, antibodies directed against pKi-67, a proliferation marker that binds to heterochromatic regions (Scholzen et al., 2002), showed disruptions in the heterochromatin surrounding nucleoli in i-RPS–treated cells, whereas it appeared as a continuous cord around control nucleoli. On knockdown of RPS16 and RPS23, pKi-67–positive clusters of highly condensed chromatin were also found in the nucleoplasm. Disorganization of perinucleolar chromatin was even more striking in cells depleted of p-RPSs RPS3, RPS10, RPS21 (Fig. 5 B, bottom), RPS12, RPS20, RPS29, or RPS30 (not depicted); anti–pKi-67 labeling showed invasion of the nucleoli by chromatin, whereas perinucleolar heterochromatin was reduced to several sparse patches, as if perinucleolar heterochromatin unwound into the nucleolus. These rearrangements of nucleolar chromatin are likely to strongly contribute to the structural changes of nucleoli in RPS-depleted cells. These results argue for a tight relationship between pre-rRNA maturation and nucleolar chromatin organization.
p-RPS protein assembly controls nucleolar release, nuclear export, and cytoplasmic maturation of pre-40S particles
We next examined the impact of RPS protein knockdown on preribosome transport from the nucleolus to the cytoplasm by fluorescence in situ hybridization (Fig. 6) and cell fractionation experiments (Fig. 7). In control cells, FISH experiments performed using a probe spanning site 1 (junction of the 5′-ETS and 18S RNA) detected early preribosomes in the nucleoli, and the 5′-ITS1 probe also revealed pre-40S particles in the cytoplasm (Fig. 6 A). Depletion of any i-RPS produced bright FISH signals strictly restricted to the nucleolus, which is consistent with interruption of pre-rRNA maturation at an early stage (Fig. 6 B). Early 5′-ETS–containing precursors, detected with the-ETS1-18S probe, filled the nucleolus, as expected from the accumulation of 45S- and 30S-containing preribosomes. In contrast, labeling with this probe was weaker than in the control upon depletion of most p-RPSs. This low abundance of early precursors, also observed on Northern blots (Fig. 3 A), could indicate a decrease in RNA polymerase I activity in response to pre-rRNA–processing defects.
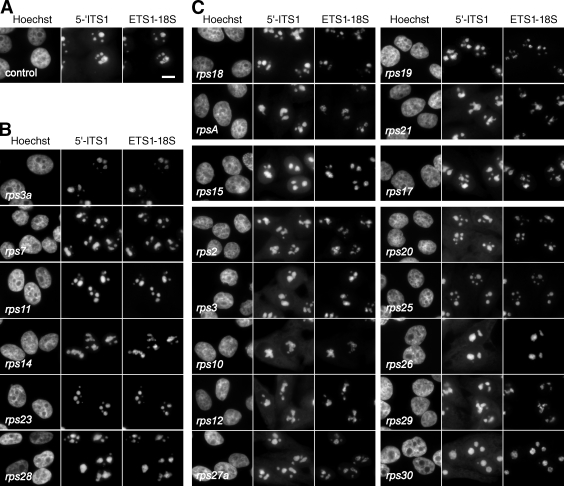
Figure 6.
Intracellular localization of the precursors to the 40S subunit in RPS-depleted cells. Pre-rRNAs of the 18S rRNA maturation pathway were detected by FISH with probes 5′-ITS1 (Cy3) and ETS1-18S (Cy5). (A–C) Control cells (A), representative examples of cells in which i-RPSs have been depleted (B), and cells in which p-RPSs have been knocked down (C). These FISH experiments were systematically performed in parallel with Northern blot analyses. Images were captured in identical conditions, and gray levels were scaled within the same lower and upper limits, except for RPS23-depleted cells for which the upper limit was twice that of the other images because of a much lower signal. The 5′-ITS1 labeling is displayed with a γ value of 1.5 to enhance the lowest gray levels. Bar, 10 µm.
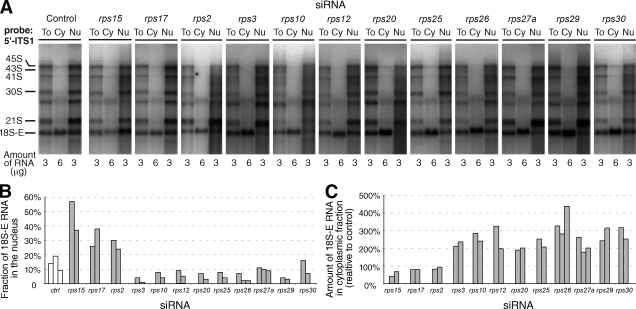
Figure 7.
Nucleocytoplasmic distribution of the 18S rRNA precursors in p-RPS–depleted cells. (A) At 48 h after transfection with siRNAs, cytoplasmic and nuclear RNAs were isolated and analyzed by Northern blotting. A representative experiment shows the patterns obtained after hybridization with the 5′-ITS1 probe for total extracts (To), the cytoplasmic fractions (Cy), and nuclear fractions (Nu). The amount of RNA loaded on the gel was 3 µg/well for total and nuclear extracts and 6 µg/well for cytoplasmic extracts. (B) Percentage of 18S-E pre–rRNA found in the nuclear fraction, as calculated from the balance of the whole fractionation procedure. The levels of 18S-E were measured with a phosphoimager. (C) Amount of 18S-E rRNA in the cytoplasmic fraction relative to control. The amount of 18S-E rRNA in the cytoplasmic fractions was normalized according to the amount of 28S rRNA (measured after hybridization with a specific probe on the phosphoimager), and siRNA-treated cells were compared with control cells. (B and C) The results of two or three independent experiments are shown for each RPS.
In cells deficient in 18S-E RNA production after treatment with rps18, rps19, rpsa, or rps21 siRNAs (Fig. 6 C, top), pre-40S particles, detected with the 5′-ITS1 probe, were also retained in the nucleolus, indicating that preribosomes containing the 21S pre-rRNA or earlier precursors were not allowed to leave the nucleolus or were very rapidly degraded in the nucleoplasm.
Further along the pathway leading to the cytoplasm, depletion of RPS15 and RPS17 resulted in accumulation of pre-40S particles in the nucleoplasm and lower levels in the cytoplasm, indicating a defect in nuclear export (Fig. 6 C, middle). A high proportion of 18S-E pre-rRNA was found consistently in nuclear fractions, especially in the case of RPS15 (Fig. 7, A and B).
Finally, depletion of the other p-RPSs resulted in high levels of cytoplasmic signals by FISH with the 5′-ITS1 probe (Fig. 6 C, bottom). Accumulation of 18S-E RNA in the cytoplasm was confirmed by semiquantitative measurements of 18S-E RNA in the cytoplasmic fraction, except in the case of RPS2 (Fig. 7, A and C). We noticed that on knockdown of RPS2, RPS26, and RPS30, FISH labeling also increased in the nucleus, suggesting delayed nuclear export in addition to processing defects. For RPS2, this observation was supported by a higher proportion of 18S-E RNA in the nuclear fraction (Fig. 7 B). Depletion of RPS25 yielded contradictory results because the fluorescence levels in the cytoplasm with the 5′-ITS1 probe were comparable with control cells (Fig. 6 C), whereas higher levels of 18S-E RNA in the cytoplasmic fraction were observed on fractionation (Fig. 7 C). However, for most p-RPSs, FISH and cell fractionation data consistently indicated that accumulation of the 18S-E pre-rRNA was primarily caused by a defect in cytoplasmic processing at site 3, except in the case of RPS15 and RPS17, the depletion of which hampers nuclear export of the pre-40S subunits.
Discussion
Mammalian RPS proteins fall into two functional classes that match with their timing of assembly
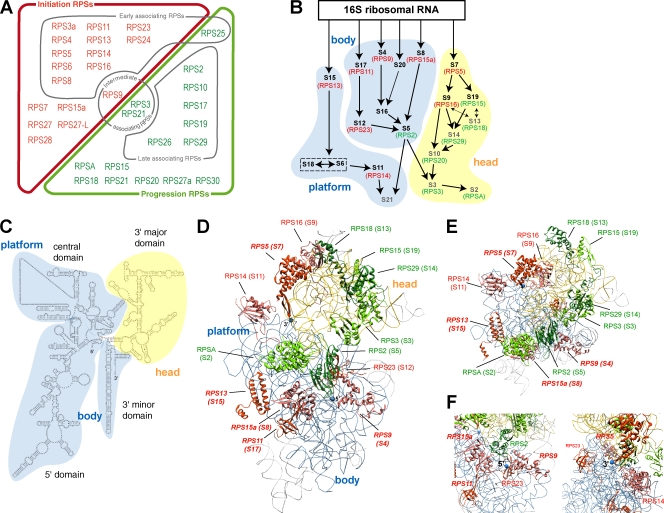
Our results show that human RPS proteins fall into two main functional categories according to their action in pre-rRNA processing (Fig. 8 A). Pioneering studies in the 1970s–1980s classified a large fraction of the mammalian ribosomal proteins according to their order of incorporation into preribosomes (Prestayko et al., 1974; Auger-Buendia et al., 1979; Lastick, 1980; Todorov et al., 1983; Hadjiolov, 1985). Comparison of these two complementary datasets (Fig. 8 A) shows that RPS proteins found to assemble early into 90S preribosomes are all included within the i-RPS set, which is consistent with a role at the very beginning of the maturation process. In contrast, proteins incorporated into preribosomes at later time points correspond to p-RPSs. The only discrepancy concerns RPS25, found to assemble early, but the function of which in pre-rRNA processing remains unclear. Thus, the timing of RPS incorporation into preribosomes is in very good agreement with our functional data. Along the same lines, i-RPSs include the eukaryotic homologues of the bacterial ribosomal proteins identified as primary binders in in vitro reconstitution of the 30S subunits, as well as homologues of some secondary binders (Mizushima and Nomura, 1970; Held et al., 1973; Fig. 8 B and Table S2). In contrast, the homologues of the other bacterial secondary binders and of the tertiary binders are found among p-RPSs. This correlation strongly suggests that 40S subunit formation follows common rules in mammalian cells and in bacteria. It is interesting that the 17 eukaryotic RPS proteins that lack bacterial homologues are equally distributed among i-RPS and p-RPSs. Thus, the emergence of additional ribosomal proteins in archaea and eukaryotes during evolution has contributed to an increase in complexity along the entire maturation pathway.
Figure 8.
Distribution of i-RPSs and p-RPSs in the 40S ribosomal subunit. (A) Functional classification of RPSs according to their role in pre-rRNA processing. This classification is compared with the association order established in previous experiments for a large number of RPSs (see Discussion; Hadjiolov, 1985). RPSs shown to associate early belong to the i-RPS set (red), except for RPS25, whereas late associating proteins correspond to p-RPSs (green). (B) Position of i-RPSs and p-RPSs bacterial homologues in the assembly map of the 30S subunit (adapted from Holmes and Culver, 2004). Arrows between ribosomal proteins indicate their mutual dependency for association. Primary and secondary binding ribosomal proteins are shown in black and tertiary ribosomal proteins in gray. Human homologues are indicated by brackets (red, i-RPSs; green, p-RPSs). Correspondence of the ribosomal protein nomenclatures in Homo sapiens, S. cerevisiae, and Escherichia coli is provided in Table S2. (C) Secondary structure of the murine 18S rRNA. (D) Position of the i-RPS (in red or salmon red) and p-RPS (in dark and light green) proteins in the model of the canine mammalian ribosome (Protein Data Bank accession no. 2ZKQ) based on cryoelectron microscopy (Chandramouli et al., 2008). The 18S rRNA head domain is colored in yellow, whereas the body is in blue. Because localization of ribosomal proteins in eukaryotic models relies on the crystal structure of bacterial ribosomal subunits, only the 15 RPS proteins having a homologue in bacteria are positioned. The names of the corresponding bacterial proteins are indicated in parentheses. Proteins homologous to bacterial primary binders are in bold, italic characters. Pictures were produced using Chimera (Pettersen et al., 2004). (E) Top view of the head showing arrangement of p-RPSs. (F) Closer view of the 18S rRNA 5′ and 3′ ends (blue dots) shows that they are surrounded by i-RPSs.
Initiation of pre-rRNA processing depends on assembly of a small subunit intermediate
Ribosomal proteins are thought to stabilize secondary structures in the rRNA, promote formation of tertiary structures, and prevent misfolding (Adilakshmi et al., 2008). In particular, the bacterial counterparts of i-RPS proteins, which include all primary binding proteins, are known to participate in the initial folding steps of the 16S rRNA subdomains. The 30S pre-rRNA accumulating on i-RPS knockdown is not converted to unstable 21S pre-rRNA, as previously shown by pulse-chase experiments in RPS24-depleted cells (Choesmel et al., 2008). Thus, our data suggest that assembly of i-RPSs is necessary to reach a critical conformational state in the pre-rRNA before the initiation of processing. Distribution of the bacterial homologues of i-RPSs in the small ribosomal subunit (Fig. 8 C) indicates that this assembly intermediate primarily concerns the body of the subunit, which is mostly formed of the first 1,200 nucleotides of the 18S rRNA. The i-RPSs RPS24, RPS7, and RPS3a were also localized on the body of the rat 40S subunits by immunoelectron microscopy (Lutsch et al., 1990). However, initiation of pre-rRNA processing also strongly requires RPS5 or RPS16, which are located on the head (Fig. 8 C). Considering the role of their bacterial counterparts S7 and S9, in 30S subunit formation (Fig. 8 B), RPS5 and RPS16 are expected to initiate folding of the head domain. i-RPSs RPS5, RPS14, and RPS16 surround the 3′ end of the 18S rRNA, whereas the 5′ end is framed by i-RPSs RPS9, RPS11, and RPS23 (Fig. 8 F), which may be important for correct positioning of the 18S rRNA extremities during the initial folding steps.
Thus, minimal folding of both the body and the head of the small subunit appears to be a prerequisite for initiating the cleavage of the transcribed spacers in the human 40S subunit biogenesis pathway (Fig. 9). Depletion of i-RPSs phenocopies depletion of the U3 and snR30/U17 sno-RNAs in yeast or vertebrates (Beltrame and Tollervey, 1995; Borovjagin and Gerbi, 2001; Fayet-Lebaron et al., 2009) and early assembly of i-RPSs should be envisioned as part of the stepwise assembly of the small subunit processome components (Fig. 9; Gallagher et al., 2004; Pérez-Fernández et al., 2007). The function provided by i-RPSs may be necessary for hybridization to the 18S rRNA of sno-RNAs involved in pre-rRNA cleavage. Along this line, uncoupling of cleavages at sites A0 and 1, observed upon depletion of most p-RPSs, could also indicate improper hybridization of U3 in the 5′-ETS and the 18S rRNA because this anomaly was observed upon mutation of U3 in Xenopus laevis oocytes (Borovjagin and Gerbi, 2001). Processing at the early cleavage site A’ in the 5′-ETS, which depends on binding of U3 to the 5′-ETS in mouse somatic cells (Kass et al., 1987), still takes place upon depletion of i-RPSs. This suggests that in mammalian cells, sno-RNP U3, and by extension the small subunit processome, can act in two distinct RPS-independent (site A’) and RPS-dependent (sites A0, 1, and perhaps E) processing steps (Fig. 9).
Figure 9.
Model of RPS protein activity in ribosome biogenesis. i-RPS proteins (red) assemble with the nascent pre-rRNA, most likely cotranscriptionally (Chooi and Leiby, 1981), together with early preribosomal factors (blue), in particular UTP proteins and sno-RNPs, like U3 and U17. i-RPS proteins participate in the folding of the 18S rRNA domain, whereas large secondary structures form in the transcribed spacers (Renalier et al., 1989; Michot and Bachellerie, 1991), potentially involving the binding of preribosomal factors. This allows formation of an assembly intermediate in which the processing machinery is correctly positioned, thus initiating cleavage of the 5′-ETS. p-RPSs (green) are mostly involved in downstream steps, although they may be necessary for coordination of processing at sites A0 and 1. The pre-40S particles are released from the nucleolus after cleavage at site E and are exported to the cytoplasm where the final processing step occurs. The RPS proteins required at each processing step are indicated.
p-RPSs are involved in nuclear export and cytoplasmic processing of the pre-40S particles
After initiation of pre-rRNA processing, p-RPSs are required in the maturation pathway in both nuclear and cytoplasmic steps (Fig. 9). In contrast to i-RPSs, p-RPSs are located on the head of the subunit, which formed from a large domain in the 3′ half of the 18S rRNA (Fig. 8, D and E). This observation is consistent with results in yeast showing that head proteins are linked to nuclear export and cytoplasmic processing (Ferreira-Cerca et al., 2005, 2007).
Our data show that release of the pre-40S particles from the nucleolus required processing of the 21S pre-rRNA into 18S-E rRNA, as observed upon depletion of RPS18, RPS19, RPS21, and RPSA. Recent positioning, by template matching, of RPS19 in close proximity to RPS18 on the head of the 40S subunit (Taylor et al., 2009) correlates with the very similar effects of the knockdown of these proteins in ITS1 processing, in both humans (Choesmel et al., 2008; this study) and in yeast (Leger-Silvestre et al., 2004, 2005). Remarkably, Rps0p and Rps21p, the yeast orthologues of RPSA and RPS21, were also found to be required for ITS1 cleavage at site A2 in yeast (Tabb-Massey et al., 2003). These converging results in yeast and mammalian cells argue for a conserved role of these proteins in the 90S to pre-40S particle transition, potentially through recruitment of pre-40S–specific preribosomal factors (Leger-Silvestre et al., 2005). Our data do not support the direct involvement of RPS18 and RPS19 in nuclear export of the pre-40S particles.
In contrast, the particular role of RPS15 in nuclear export of the pre-40S particles downstream of nucleolar release is confirmed in this study (Leger-Silvestre et al., 2004; Rouquette et al., 2005). Additionally, RPS17 has a function similar to that of RPS15, although accumulation of 26S and 21S pre-rRNAs on RPS17 knockdown indicates that this protein is also required upstream of nuclear export. Although not precisely positioned on the 40S subunit, RPS17 was localized on the head by immunoelectron microscopy (Lutsch et al., 1990). Other p-RPSs, especially RPS2 (Perreault et al., 2008), may contribute to nuclear export, albeit less critically. As previously proposed for yeast pre-40S particles (Oeffinger et al., 2004; Ferreira-Cerca et al., 2005), several ribosomal and nonribosomal proteins may be required to shield the pre-rRNA for passage through the hydrophobic environment of the nuclear pore complex. RPS15 and RPS17 may be more directly involved in the recruitment of the yet to be described export factors.
Most other p-RPSs are required for processing of the 18S-E pre-rRNA in the cytoplasm. These proteins may be required for ensuring and regulating the association of nonribosomal factors with pre-40S particles in the cytoplasm, such as the RIO2 kinase or the Nob1 endonuclease (Rouquette et al., 2005; Zemp et al., 2009). In this respect, we cannot exclude that the accumulation of 26S pre-rRNA observed on depletion of p-RPSs may be secondary to the trapping in unprocessed pre-40S particles of nonribosomal factors necessary for cleavage of the 5′-ETS. Conversely, it seems likely that some i-RPSs also participate in late maturation steps in addition to their early role in pre-rRNA processing, as already shown for yeast RPS14 (Jakovljevic et al., 2004) and RPS5 (Neueder et al., 2010), which are located in the neighborhood of the 18S rRNA 3′ end.
Comparison with formation of the yeast 40S subunit
Our results show a substantial conservation of the function of the ribosomal proteins between yeast and mammals, but they also highlight several differences. In S. cerevisiae, Rps12p and Rps25p were described as nonessential proteins involved in translation (Ferreira-Cerca et al., 2005). Although the mild phenotype observed on knockdown of RPS25 does not exclude a similar role in mammals, we found that RPS12 was essential for production or stability of the 40S subunits in human cells. Also, knockdown of proteins RPS7 and RPS28 in HeLa cells unambiguously blocks 5′-ETS and ITS1 processing, confirming previous results (Gazda et al., 2008; Robledo et al., 2008). In contrast, it has been reported that depletion of yeast Rps28p and Rps7p only delayed 5′-ETS and ITS1 processing (Ferreira-Cerca et al., 2005, 2007). Determining how much of the differences stressed in this study between yeast and human ribosomal proteins may be ascribed to evolution versus technical considerations requires further work. Finally, this study provides, to our knowledge, the first description of the function of eukaryotic RPS4, RPS15a (RPS22 in yeast), and RPS29 in pre-rRNA maturation.
It was proposed that early pre-rRNA processing in yeast involves a stable assembly intermediate of the body of the small subunit, whereas assembly of the head would be required for nuclear export and cytoplasmic processing (Ferreira-Cerca et al., 2007). In contrast, we find that RPS5 and RPS16, two proteins whose bacterial homologues initiate folding of the head of the small subunit, are necessary for cleavage of the 5′-ETS, which strongly argues for a folding intermediate of the entire subunit being a prerequisite for initiation of pre-rRNA processing in mammalian cells. This difference between yeast and mammalian cells may correspond to changes in the coordination between pre-rRNA synthesis, folding, processing, and protein assembly. In this respect, it is interesting that 18S rRNA production, as observed in pulse-chase analyses, is slower in HeLa cells (30–60 min) than in yeast (2–5 min). In yeast, pre-rRNA processing was recently shown to be cotranscriptional (Osheim et al., 2004; Kos and Tollervey, 2010), which remains to be demonstrated in mammalian cells. Furthermore, ribosomal protein assembly in yeast and mammalian cells may have coevolved with several variations in pre-rRNA processing, such as the conversion of 21S into 18S-E pre-rRNA, which represents an additional cleavage step in mammalian nuclei.
p-RPSs are more frequently mutated in DBA
Systematic sequencing of ribosomal protein genes has revealed that ∼50% of DBA patients bear hemizygous mutations in ribosomal protein genes, especially RPS genes: RPS19, 25–30% of DBA patients; RPS26, 6%; RPS10, 3%; RPS17, 1–2%; RPS24, 1–2%; and RPS7, <1%; Gazda et al., 2006, 2008; Doherty et al., 2010). The functions of these genes do not appear to particularly overlap during pre-rRNA maturation. Additionally, RPL genes (RPL5, RPL11, and RPL35) are also mutated in DBA. This makes it difficult to pinpoint a particular maturation step in relation to the disease and, rather, suggests that the common denominator is primarily ribosomal stress, resulting from altered pre-rRNA maturation and nucleolar disorganization. Nevertheless, it is remarkable that DBA patients affected in p-RPS genes (RPS19, RPS10, RPS26, and RPS17) far outnumber those with mutations in i-RPS genes (RPS7 and RPS24). Among several hypotheses, we favor the idea that because of their role at the onset of pre-rRNA processing, haploinsufficiency of iRPSs would have a stronger impact on ribosome biogenesis and would lead to embryonic lethality. In contrast, ribosomal stress levels induced by p-RPS mutations would remain tolerable in most physiological processes, with the exception of erythropoiesis and some developmental mechanisms. Similar to p-RPSs, the two most frequently mutated RPLs in DBA, RPL5 (7–10%) and RPL11 (5%), are not essential for the early pre-rRNA processing steps (Zhang et al., 2007; Gazda et al., 2008). However, this hypothesis does not explain the very limited number of ribosomal protein genes linked to the disease (including among p-RPSs) or the strong prevalence of RPS19 mutations. The possibility remains that ribosomal proteins involved in DBA do indeed share specific functions still to be uncovered.
Materials and methods
Cell culture and siRNAs
Human cervical carcinoma HeLa cells were grown in DME supplemented with 10% FCS, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen). Specific inhibition of RNA polymerase I transcription was achieved in the presence of either 5 or 10 ng/ml actinomycin D (Sigma-Aldrich) in the cell culture medium. Sequences of siRNAs used to knockdown RPS proteins are listed in Table S1. All siRNAs were purchased from Eurogentec and diluted to 100 µM. After washing the cells once in DME (without FCS and antibiotics), 10 µl of the siRNA solution was added on ice to 107 HeLa cells in suspension in 200 µl of the same medium. Electrotransformation was performed at 250 V and at 975 µF with a gene pulser (Bio-Rad Laboratories) in a cuvette with a 4-mm interelectrode distance. The cells were diluted in 20 ml DME, supplemented with FCS and antibiotics, and plated in a 140-cm2 Petri dish. Control samples were electrotransformed with a scrambled siRNA or without siRNA, which lead to similar results.
Quantitative RT-PCR
Knockdown efficiency of siRNAs was assessed by quantitative PCR, using GAPDH as an internal control to normalize expression of the target genes. Total RNAs were isolated with Trizol reagent (Invitrogen). First stand synthesis of cDNAs was performed with 2 µg DNA and avian myeloblastosis virus reverse transcriptase (Promega). Real-time PCR was performed with Platinum SYBR green quantitative PCR SuperMix-UDG (Invitrogen) using an ICyclerQIM (Bio-Rad Laboratories) or a Mastercycler ep Realplex (Eppendorf). After denaturation at 95°C for 3 min, 40 cycles were performed as follows: 95°C for 30 s, 56°C for 20 s, and 72°C for 30 s. The fluorescent signals were measured at 78°C after each extension step. Triplicate reactions were prepared for each sample, and two independent experiments were run. Data analysis was performed using the IQCycler or the Realplex software. The concentration of each gene-specific mRNA in treated cells relative to untreated cells was obtained by subtracting the normalized Ct values obtained for untreated cells from those obtained from treated samples (ΔΔCt = ΔCttreated − ΔCtuntreated), and the relative concentration was determined (2−ΔΔCt).
Cell fractionation and RNA extractions
After treatment with siRNAs for 48 h, the cells were successively washed at 4°C with DME, PBS, and buffer A (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, and 10 mM KCl). 10% of the suspension was kept for total RNA extraction. The cells were mechanically disrupted with a dounce homogenizer in buffer A containing 0.5 mM DTT. After centrifugation (1,000 g at 4°C for 10 min), the supernatant (i.e., the cytoplasmic fraction) was frozen. The pellet containing the nuclei was washed with 10 mM Tris-HCl, pH 7.5, 3.3 mM MgCl2, and 250 mM sucrose. After centrifugation, nuclei were put in suspension in a solution containing 10 mM MgCl2 and 250 mM sucrose and further purified by centrifugation (500 g for 10 min) on a sucrose cushion (0.5 mM MgCl2 and 350 mM sucrose). The pellet was lysed in 10 vol 50 mM Na acetate, pH 5.1, 140 mM NaCl, and 0.3% SDS, and the nuclear RNAs were extracted twice with phenol. Total and cytoplasmic RNAs were purified with Trizol reagent. After alcohol precipitation, RNA pellets were dissolved in formamide, quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific), and diluted to 1 mg/ml.
Analysis of ribosomes by sucrose density gradient centrifugation
At 48 or 72 h after transfection, HeLa cells were treated with 100 µg/ml cycloheximide (Sigma-Aldrich) for 10 min. The cytoplasmic fractions were prepared on ice as described above, except that cycloheximide was added to all buffers. 1 mg of proteins was loaded on a 10–50% (wt/wt) sucrose gradient prepared with a Gradient Master former (BioComp Instruments). The tubes were centrifuged (36,000 rpm at 4°C for 105 min) with an SW41 rotor (Optima L100XP ultracentrifuge; Beckman Coulter). The gradient fractions were collected at OD254 nm with a Foxy Jr. gradient collector (Teledyne Isco).
Northern blot analysis of pre-rRNA species
RNAs (3 µg/well for total and nuclear extracts and 6 µg/well for cytoplasmic extracts) were separated on a 1% agarose gel prepared with Tri/Tri buffer (30 mM triethanolamine and 30 mM tricine, pH 7.9) containing 1.2% formaldehyde and run in Tri/Tri buffer at 140 V. RNAs were transferred to a Hybond N+ nylon membrane (GE Healthcare) and fixed by UV cross-linking. Membranes were prehybridized for 1 h at 45°C in 6× SSC, 5× Denhardt’s solution, 0.5% SDS, and 0.9 µg/ml tRNA. The 32P-labeled oligodeoxynucleotide probe was added and incubated overnight at 45°C. The probes used in this study were 5′-ETSb (5′-AGACGAGAACGCCTGACACGCACGGCAC-3′), 18S (5′-TTTACTTCCTCTAGATAGTCAAGTTCGACC-3′), 5′-ITS1 (5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′), ITS2b (5′-CTGCGAGGGAACCCCCAGCCGCGCA-3′), ITS2d/e (5′-GCGCGACGGCGGACGACACCGCGGCGTC-3′), and 28S (5′-CCCGTTCCCTTGGCTGTGGTTTCGCTAGATA-3′). For detection of ITS2, probes ITS2b and ITS2d/e were mixed in equal amounts. After hybridization, the membranes were washed twice for 10 min at room temperature in 2× SSC with 0.1% SDS and once in 1× SSC with 0.1% SDS. Labeled RNA signals were acquired with a phosphorimager (FLA2000; Fujifilm) and quantified with the Image Gauge software. The data were analyzed by principal component analysis and hierarchical clustering as described below.
Fluorescence in situ hybridization
Precursors to the 18S rRNA were localized with the 5′-ITS1 probe conjugated to Cy3 (GE Healthcare) on amino-modified deoxythymidine or with a probe spanning the 5′-ETS–18S junction (ETS1–18S probe: 5′-GATCAACCAGGTAGGTAAGGTAGAATC-3′) conjugated to Cy5 (Sigma-Aldrich). Cells grown on glass coverslips were washed twice in PBS and fixed for 30 min with 4% paraformaldehyde in PBS (EMS). The cells were washed twice in PBS and permeabilized for 18 h in 70% ethanol at 4°C. After two washes in 2× SSC containing 10% formamide, the following steps were performed in the dark: hybridization at 37°C for ≥5 h in a buffer containing 10% formamide, 2.1× SSC, 0.5 µg/ml tRNA, 10% dextran sulfate, 250 µg/ml BSA, 10 mM ribonucleoside vanadyl complexes, and 0.5 ng/µL of each probe. After two washes at 37°C with 2× SSC containing 10% formamide, the cells were rinsed in PBS, counter stained with Hoechst 33342 (Invitrogen) to visualize DNA, and mounted in Mowiol (version 4.88; Polyscience, Inc.). Observations were made with an inverted microscope (IX-81; Olympus) equipped with a camera (CoolSNAP HQ; Photometrics). Image analysis was performed with MetaMorph software (MDS Analytical Technologies).
Immunofluorescence
mAb 72B9 (antifibrillarin) was provided by J. Cavaillé (Université de Toulouse, Toulouse, France), and the anti–pKi-67 (MIB-1) antibody was purchased from Dako. Cells grown on coverslips were fixed for 10 min with 4% paraformaldehyde in PBS, rinsed, and permeabilized with 0.4% Triton X-100 in PBS for 5 min. After rinsing, cells were incubated first with 1% BSA in PBS for 1 h and with the primary antibody in the same buffer for 3 h. Dilution of the antibody was 1:75 for mAb MIB-1 and 1:200 for mAb 72B9. Cells were washed three times in PBS, and nonspecific sites were further saturated with 1% BSA in PBS for 30 min. Cells were incubated for 1 h with a goat anti–mouse secondary antibody conjugated to Alexa Fluor 555 (Invitrogen) at a 1:1,000 dilution. After rinsing twice in PBS and counter staining with Hoechst 33342, coverslips were mounted in Mowiol.
Principal component analysis and hierarchical clustering
Principal component analysis (Pearson, 1901; Gordon, 1981) was used to classify RPS proteins according to pre-rRNA maturation defects induced by siRNAs. It was applied independently on both sets of RPSs, i-RPSs and p-RPSs. The two datasets were organized in tables, the rows of which correspond to RPS proteins, and the columns to pre-rRNAs detected by Northern blotting with the 5′-ITS1 or the ITS2 probes. The tables each contain the logarithm of the ratio of the pre-rRNA level in the knockdown context to its level in the control. Each column corresponds to the mean of two to eight experiments, depending on the RPS. The data were further centered and divided by the SD. Only the four first principal components were considered because they explained 86% and 87% of the total data variability for the i-RPS and p-RPS datasets, respectively. Hierarchical classification was next applied on the coordinates of the RPS proteins corresponding to these first four principal components. Classification on this subvector space, instead of the total vector space, is expected to eliminate most of the background noise caused by random fluctuations, which often correspond to the variability explained by the last principal components. An agglomerative approach, which begins with each element as a separate cluster and merges them into successively larger clusters, was performed using Euclidean distances. At each step, Ward’s criterion was used to merge two clusters to minimize the loss of inertia between classes. Principal component analysis and hierarchical clustering were performed with the R freeware, using the ade4 package (http://www.r-project.org/).
Online supplemental material
Fig. S1 shows the quantitative RT-PCR analysis of RPS mRNA knockdown with siRNAs. Fig. S2 shows the effect of combining rps27 and rps27A siRNAs on pre-rRNA processing, as visualized by Northern blotting with the 5′-ITS1 probe. Table S1 displays the RPS gene accession numbers and the corresponding siRNA sequences. Table S2 shows the correspondence between bacterial, yeast, and human RPSs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005117/DC1.
Acknowledgments
We thank our colleagues in the Gleizes group and the LBME members for their scientific input through meetings and discussions. We are indebted to Yves Henry and Michael O’Donohue for critical reading of the manuscript.
P.E. Gleizes is a fellow of the Institut Universitaire de France. This work was supported by the Agence Nationale de la Recherche (RIBODBA and RIBEUC projects) and by the Association pour la Recherche sur le Cancer (grant 1058).
Footnotes
Abbreviations used in this paper:
- DBA
- Diamond-Blackfan anemia
- ETS
- external transcribed spacer
- i-RPS
- initiation RPS
- p-RPS
- progression RPS
- RPL
- ribosomal protein of the large subunit
- RPS
- ribosomal protein of the small subunit
- rRNA
- ribosomal RNA
- sno-RNA
- small nucleolar RNA
References
- Adilakshmi T., Bellur D.L., Woodson S.A. 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 455:1268–1272 10.1038/nature07298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger-Buendia M.A., Longuet M., Tavitian A. 1979. Kinetic studies on ribosomal proteins assembly in preribosomal particles and ribosomal subunits of mammalian cells. Biochim. Biophys. Acta. 563:113–128 [DOI] [PubMed] [Google Scholar]
- Barkić M., Crnomarkovic S., Grabusić K., Bogetić I., Panić L., Tamarut S., Cokarić M., Jerić I., Vidak S., Volarević S. 2009. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol. Cell. Biol. 29:2489–2504 10.1128/MCB.01588-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 14:4350–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovjagin A.V., Gerbi S.A. 2001. Xenopus U3 snoRNA GAC-box A’ and box A sequences play distinct functional roles in rRNA processing. Mol. Cell. Biol. 21:6210–6221 10.1128/MCB.21.18.6210-6221.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli P., Topf M., Ménétret J.F., Eswar N., Cannone J.J., Gutell R.R., Sali A., Akey C.W. 2008. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 16:535–548 10.1016/j.str.2008.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choesmel V., Bacqueville D., Rouquette J., Noaillac-Depeyre J., Fribourg S., Crétien A., Leblanc T., Tchernia G., Da Costa L., Gleizes P.E. 2007. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 109:1275–1283 10.1182/blood-2006-07-038372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choesmel V., Fribourg S., Aguissa-Touré A.H., Pinaud N., Legrand P., Gazda H.T., Gleizes P.E. 2008. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum. Mol. Genet. 17:1253–1263 10.1093/hmg/ddn015 [DOI] [PubMed] [Google Scholar]
- Chooi W.Y., Leiby K.R. 1981. An electron microscopic method for localization of ribosomal proteins during transcription of ribosomal DNA: a method for studying protein assembly. Proc. Natl. Acad. Sci. USA. 78:4823–4827 10.1073/pnas.78.8.4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmejla R., Cmejlova J., Handrkova H., Petrak J., Pospisilova D. 2007. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum. Mutat. 28:1178–1182 10.1002/humu.20608 [DOI] [PubMed] [Google Scholar]
- Craig N., Kass S., Sollner-Webb B. 1987. Nucleotide sequence determining the first cleavage site in the processing of mouse precursor rRNA. Proc. Natl. Acad. Sci. USA. 84:629–633 10.1073/pnas.84.3.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty L., Sheen M.R., Vlachos A., Choesmel V., O’Donohue M.F., Clinton C., Schneider H.E., Sieff C.A., Newburger P.E., Ball S.E., et al. 2010. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 86:222–228 10.1016/j.ajhg.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset T., Wang C., Verheggen C., Chen D., Hernandez-Verdun D., Huang S. 2000. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell. 11:2705–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N., Gustavsson P., Andersson B., Pettersson M., Willig T.N., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H., et al. 1999. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21:169–175 10.1038/5951 [DOI] [PubMed] [Google Scholar]
- Ebert B.L., Pretz J., Bosco J., Chang C.Y., Tamayo P., Galili N., Raza A., Root D.E., Attar E., Ellis S.R., Golub T.R. 2008. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 451:335–339 10.1038/nature06494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler D.C., Craig N. 1994. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49:197–239 10.1016/S0079-6603(08)60051-3 [DOI] [PubMed] [Google Scholar]
- Farrar J.E., Nater M., Caywood E., McDevitt M.A., Kowalski J., Takemoto C.M., Talbot C.C.J., Jr., Meltzer P., Esposito D., Beggs A.H., et al. 2008. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 112:1582–1592 10.1182/blood-2008-02-140012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet-Lebaron E., Atzorn V., Henry Y., Kiss T. 2009. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J. 28:1260–1270 10.1038/emboj.2009.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S., Pöll G., Gleizes P.E., Tschochner H., Milkereit P. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 20:263–275 10.1016/j.molcel.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S., Pöll G., Kühn H., Neueder A., Jakob S., Tschochner H., Milkereit P. 2007. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 28:446–457 10.1016/j.molcel.2007.09.029 [DOI] [PubMed] [Google Scholar]
- Flygare J., Aspesi A., Bailey J.C., Miyake K., Caffrey J.M., Karlsson S., Ellis S.R. 2007. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 109:980–986 10.1182/blood-2006-07-038232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. 2003. Ribosome assembly in eukaryotes. Gene. 313:17–42 10.1016/S0378-1119(03)00629-2 [DOI] [PubMed] [Google Scholar]
- Gallagher J.E., Dunbar D.A., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506–2517 10.1101/gad.1226604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi K.A., Shimamura A. 2008. Ribosomal dysfunction and inherited marrow failure. Br. J. Haematol. 141:376–387 10.1111/j.1365-2141.2008.07095.x [DOI] [PubMed] [Google Scholar]
- Gazda H.T., Grabowska A., Merida-Long L.B., Latawiec E., Schneider H.E., Lipton J.M., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A., et al. 2006. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 79:1110–1118 10.1086/510020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda H.T., Sheen M.R., Vlachos A., Choesmel V., O’Donohue M.F., Schneider H., Darras N., Hasman C., Sieff C.A., Newburger P.E., et al. 2008. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 83:769–780 10.1016/j.ajhg.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi S.A., Borovjagin A.V. 2004. Pre-ribosomal RNA processing in multicellular organisms. The Nucleolus. Olson M.O.J, editor Kluwer Academic/Plenum Publishers, New York: 170–198 [Google Scholar]
- Ginisty H., Amalric F., Bouvet P. 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17:1476–1486 10.1093/emboj/17.5.1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A.D. 1981. Classification: Methods for the Exploratory Analysis of Multivariate Data. CRC Monographs on Statistics & Applied Probability; Chapman and Hall, London/New York: 193 [Google Scholar]
- Hadjiolov A.A. 1985. The Nucleolus and Ribosome Biogenesis. Cell Biology Monographs. Springer-Verlag, New York: 268 [Google Scholar]
- Hadjiolova K.V., Nicoloso M., Mazan S., Hadjiolov A.A., Bachellerie J.P. 1993. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 212:211–215 10.1111/j.1432-1033.1993.tb17652.x [DOI] [PubMed] [Google Scholar]
- He H., Sun Y. 2007. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 26:2707–2716 10.1038/sj.onc.1210073 [DOI] [PubMed] [Google Scholar]
- Held W.A., Mizushima S., Nomura M. 1973. Reconstitution of Escherichia coli 30 S ribosomal subunits from purified molecular components. J. Biol. Chem. 248:5720–5730 [PubMed] [Google Scholar]
- Holmes K.L., Culver G.M. 2004. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat. Struct. Mol. Biol. 11:179–186 10.1038/nsmb719 [DOI] [PubMed] [Google Scholar]
- Idol R.A., Robledo S., Du H.Y., Crimmins D.L., Wilson D.B., Ladenson J.H., Bessler M., Mason P.J. 2007. Cells depleted for RPS19, a protein associated with Diamond Blackfan anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol. Dis. 39:35–43 10.1016/j.bcmd.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Jakovljevic J., de Mayolo P.A., Miles T.D., Nguyen T.M., Léger-Silvestre I., Gas N., Woolford J.L., Jr 2004. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell. 14:331–342 10.1016/S1097-2765(04)00215-1 [DOI] [PubMed] [Google Scholar]
- Kass S., Sollner-Webb B. 1990. The first pre-rRNA-processing event occurs in a large complex: analysis by gel retardation, sedimentation, and UV cross-linking. Mol. Cell. Biol. 10:4920–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. 1987. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol. Cell. Biol. 7:2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenmochi N., Kawaguchi T., Rozen S., Davis E., Goodman N., Hudson T.J., Tanaka T., Page D.C. 1998. A map of 75 human ribosomal protein genes. Genome Res. 8:509–523 [DOI] [PubMed] [Google Scholar]
- Kent T., Lapik Y.R., Pestov D.G. 2009. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA. 15:14–20 10.1261/rna.1384709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 109:145–148 10.1016/S0092-8674(02)00718-3 [DOI] [PubMed] [Google Scholar]
- Kos M., Tollervey D. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 37:809–820 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastick S.M. 1980. The assembly of ribosomes in HeLa cell nucleoli. Eur. J. Biochem. 113:175–182 10.1111/j.1432-1033.1980.tb06152.x [DOI] [PubMed] [Google Scholar]
- Léger-Silvestre I., Milkereit P., Ferreira-Cerca S., Saveanu C., Rousselle J.C., Choesmel V., Guinefoleau C., Gas N., Gleizes P.E. 2004. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 23:2336–2347 10.1038/sj.emboj.7600252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léger-Silvestre I., Caffrey J.M., Dawaliby R., Alvarez-Arias D.A., Gas N., Bertolone S.J., Gleizes P.E., Ellis S.R. 2005. Specific role for yeast homologs of the Diamond Blackfan anemia-associated Rps19 protein in ribosome synthesis. J. Biol. Chem. 280:38177–38185 10.1074/jbc.M506916200 [DOI] [PubMed] [Google Scholar]
- Li J., Tan J., Zhuang L., Banerjee B., Yang X., Chau J.F., Lee P.L., Hande M.P., Li B., Yu Q. 2007. Ribosomal protein S27-like, a p53-inducible modulator of cell fate in response to genotoxic stress. Cancer Res. 67:11317–11326 10.1158/0008-5472.CAN-07-1088 [DOI] [PubMed] [Google Scholar]
- Liu J.M., Ellis S.R. 2006. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 107:4583–4588 10.1182/blood-2005-12-4831 [DOI] [PubMed] [Google Scholar]
- Lutsch G., Stahl J., Kärgel H.J., Noll F., Bielka H. 1990. Immunoelectron microscopic studies on the location of ribosomal proteins on the surface of the 40S ribosomal subunit from rat liver. Eur. J. Cell Biol. 51:140–150 [PubMed] [Google Scholar]
- MacInnes A.W., Amsterdam A., Whittaker C.A., Hopkins N., Lees J.A. 2008. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc. Natl. Acad. Sci. USA. 105:10408–10413 10.1073/pnas.0805036105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B.E. 1990. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 39:241–303 10.1016/S0079-6603(08)60629-7 [DOI] [PubMed] [Google Scholar]
- McGowan K.A., Li J.Z., Park C.Y., Beaudry V., Tabor H.K., Sabnis A.J., Zhang W., Fuchs H., de Angelis M.H., Myers R.M., et al. 2008. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 40:963–970 10.1038/ng.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J.P. 1991. Secondary structure of the 5′ external transcribed spacer of vertebrate pre-rRNA. Presence of phylogenetically conserved features. Eur. J. Biochem. 195:601–609 10.1111/j.1432-1033.1991.tb15743.x [DOI] [PubMed] [Google Scholar]
- Milkereit P., Kühn H., Gas N., Tschochner H. 2003. The pre-ribosomal network. Nucleic Acids Res. 31:799–804 10.1093/nar/gkg165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.G., Sollner-Webb B. 1981. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 27:165–174 10.1016/0092-8674(81)90370-6 [DOI] [PubMed] [Google Scholar]
- Mizushima S., Nomura M. 1970. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 226:1214 10.1038/2261214a0 [DOI] [PubMed] [Google Scholar]
- Neueder A., Jakob S., Pöll G., Linnemann J., Deutzmann R., Tschochner H., Milkereit P. 2010. A local role for the small ribosomal subunit primary binder rpS5 in final 18S rRNA processing in yeast. PLoS One. 5:e10194 10.1371/journal.pone.0010194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Dlakic M., Tollervey D. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 18:196–209 10.1101/gad.285604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y.N., French S.L., Keck K.M., Champion E.A., Spasov K., Dragon F., Baserga S.J., Beyer A.L. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 16:943–954 10.1016/j.molcel.2004.11.031 [DOI] [PubMed] [Google Scholar]
- Pearson K. 1901. On lines and planes of closest fit to systems of points in space. Philos. Mag. 2:559–572 [Google Scholar]
- Pérez-Fernández J., Román A., De Las Rivas J., Bustelo X.R., Dosil M. 2007. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 27:5414–5429 10.1128/MCB.00380-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault A., Bellemer C., Bachand F. 2008. Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2. Nucleic Acids Res. 36:6132–6142 10.1093/nar/gkn625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pöll G., Braun T., Jakovljevic J., Neueder A., Jakob S., Woolford J.L.J., Jr., Tschochner H., Milkereit P. 2009. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One. 4:e8249 10.1371/journal.pone.0008249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestayko A.W., Klomp G.R., Schmoll D.J., Busch H. 1974. Comparison of proteins of ribosomal subunits and nucleolar preribosomal particles from Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Biochemistry. 13:1945–1951 10.1021/bi00706a026 [DOI] [PubMed] [Google Scholar]
- Renalier M.H., Mazan S., Joseph N., Michot B., Bachellerie J.P. 1989. Structure of the 5′-external transcribed spacer of the human ribosomal RNA gene. FEBS Lett. 249:279–284 10.1016/0014-5793(89)80641-6 [DOI] [PubMed] [Google Scholar]
- Robledo S., Idol R.A., Crimmins D.L., Ladenson J.H., Mason P.J., Bessler M. 2008. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 14:1918–1929 10.1261/rna.1132008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J., Choesmel V., Gleizes P.E. 2005. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 24:2862–2872 10.1038/sj.emboj.7600752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen T., Endl E., Wohlenberg C., van der Sar S., Cowell I.G., Gerdes J., Singh P.B. 2002. The Ki-67 protein interacts with members of the heterochromatin protein 1 (HP1) family: a potential role in the regulation of higher-order chromatin structure. J. Pathol. 196:135–144 10.1002/path.1016 [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 423:825–837 10.1038/nature01722 [DOI] [PubMed] [Google Scholar]
- Sulic S., Panic L., Barkic M., Mercep M., Uzelac M., Volarevic S. 2005. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 19:3070–3082 10.1101/gad.359305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb-Massey A., Caffrey J.M., Logsden P., Taylor S., Trent J.O., Ellis S.R. 2003. Ribosomal proteins Rps0 and Rps21 of Saccharomyces cerevisiae have overlapping functions in the maturation of the 3′ end of 18S rRNA. Nucleic Acids Res. 31:6798–6805 10.1093/nar/gkg899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.J., Devkota B., Huang A.D., Topf M., Narayanan E., Sali A., Harvey S.C., Frank J. 2009. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 17:1591–1604 10.1016/j.str.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov I.T., Noll F., Hadjiolov A.A. 1983. The sequential addition of ribosomal proteins during the formation of the small ribosomal subunit in Friend erythroleukemia cells. Eur. J. Biochem. 131:271–275 10.1111/j.1432-1033.1983.tb07259.x [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261–311 10.1146/annurev.genet.33.1.261 [DOI] [PubMed] [Google Scholar]
- Warner J.R. 1989. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev. 53:256–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I., Wild T., O’Donohue M.F., Wandrey F., Widmann B., Gleizes P.E., Kutay U. 2009. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185:1167–1180 10.1083/jcb.200904048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Harrison P., Gerstein M. 2002. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 12:1466–1482 10.1101/gr.331902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M.P., Hiley S.L., Hughes T., Woolford J.L., Jr 2007. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21:2580–2592 10.1101/gad.1569307 [DOI] [PMC free article] [PubMed] [Google Scholar]