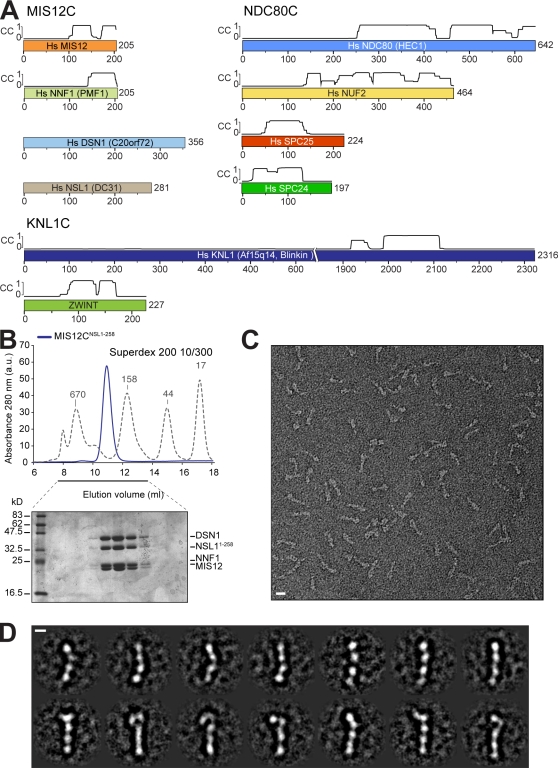
The NSL1 subunit structures interactions between the MIS12, NDC80, and KNL1 kinetochore complexes (see also a related paper by Maskell et al. in this issue).
Abstract
Kinetochores are nucleoprotein assemblies responsible for the attachment of chromosomes to spindle microtubules during mitosis. The KMN network, a crucial constituent of the outer kinetochore, creates an interface that connects microtubules to centromeric chromatin. The NDC80, MIS12, and KNL1 complexes form the core of the KMN network. We recently reported the structural organization of the human NDC80 complex. In this study, we extend our analysis to the human MIS12 complex and show that it has an elongated structure with a long axis of ∼22 nm. Through biochemical analysis, cross-linking–based methods, and negative-stain electron microscopy, we investigated the reciprocal organization of the subunits of the MIS12 complex and their contacts with the rest of the KMN network. A highlight of our findings is the identification of the NSL1 subunit as a scaffold supporting interactions of the MIS12 complex with the NDC80 and KNL1 complexes. Our analysis has important implications for understanding kinetochore organization in different organisms.
Introduction
Equational division of chromosomes to the daughter cells in mitosis is essential for cell viability. Kinetochores, which are protein assemblies built on the centromeric region of chromosomes, are crucial for this process (for reviews see Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). In mammals, ∼100 different proteins populate mitotic kinetochores. A subset of these is required to form end-on, load-bearing attachments between chromosomes and spindle microtubules that are ultimately responsible for sister chromatid separation at anaphase. Kinetochores also regulate a feedback control mechanism, named the spindle assembly checkpoint, which synchronizes cell cycle progression with the progression of chromosome attachment to the spindle. Furthermore, kinetochores are responsible for the correction of improper kinetochore–microtubule attachments. They achieve this by preventing premature stabilization of incorrect attachments, followed by their clearance (for review see Santaguida and Musacchio, 2009).
The core structural components of kinetochores, as well as those responsible for the spindle checkpoint and error correction, are conserved from yeast to humans (for review see Santaguida and Musacchio, 2009), suggesting that the building plan of kinetochores is largely conserved in evolution. However, kinetochores in different organisms display dramatic variations in complexity. The simplest kinetochores are found in Saccharomyces cerevisiae (Westermann et al., 2007). In this organism, centromeres consist of 150 bp of DNA organized in a specialized centromeric nucleosome containing the histone H3 variant CENP-A (Cse4 in S. cerevisiae). These simple centromeres, known as point centromeres, assemble kinetochores that bind a single microtubule. In higher eukaryotes, centromeres usually take the form of regional centromeres: they extend over very large segments of DNA, up to several million base pairs, and display no univocal relationship between DNA sequence and kinetochore assembly. Kinetochores formed on regional centromeres usually bind multiple microtubules, from 3 in Schizosaccharomyces pombe to 15–30 in humans (for review see Santaguida and Musacchio, 2009).
Kinetochores on regional centromeres appear as trilaminar plates, with electron-opaque inner and outer plates and a translucent middle layer. The inner plate rests on compact centromeric heterochromatin containing specialized CENP-A and H3 nucleosomes. Both types of nucleosomes are required for kinetochore assembly, but their reciprocal organization is unclear (Hori et al., 2008). The constitutive centromere-associated network (CCAN; also known as NAC/CAD) is a set of 14–15 proteins originally identified for their physical proximity to CENP-A (Foltz et al., 2006; Okada et al., 2006; Amano et al., 2009; Carroll et al., 2009). However, certain CCAN components, including CENP-T and CENP-W, bind preferentially on H3 nucleosomes (Hori et al., 2008). CENP-C, a conserved kinetochore protein, also associates with H3 and CENP-A nucleosomes, possibly creating a link between them (Ando et al., 2002; Talbert et al., 2004; Cohen et al., 2008; Erhardt et al., 2008; Hori et al., 2008; Milks et al., 2009; Trazzi et al., 2009; Carroll et al., 2010).
The outer kinetochore contains components required for end-on microtubule attachment. The NDC80 complex (abbreviated as NDC80C, where C, for complex, is used to distinguish the complex from one of its subunits, the NDC80 subunit) and the Dam1 complex are crucial for kinetochore–microtubule attachment (for review see Cheeseman and Desai, 2008). The Dam1 complex, which has only been identified in yeasts, may act as a processivity factor at the kinetochore–microtubule interface, possibly but not exclusively through the formation of a ring around microtubules (Miranda et al., 2005; Westermann et al., 2005; Gestaut et al., 2008; Lampert et al., 2010; Tien et al., 2010).
The four-subunit NDC80C is conserved in evolution (Wigge and Kilmartin, 2001; see Fig. 1 A for a schematic representation of its subunits; Table S1 shows the nomenclature of subunits in different species). It has the shape of a 57-nm-long dumbbell, and it connects to the kinetochore through the terminal globular regions of the SPC24 and SPC25 subunits (Ciferri et al., 2005, 2008; Wei et al., 2005, 2006, 2007; Cheeseman et al., 2006; DeLuca et al., 2006; Wang et al., 2008; Wilson-Kubalek et al., 2008). At the opposite end, a tight arrangement of two calponin homology domains in the NDC80 and NUF2 subunits creates an interface for microtubules that is further regulated by a disordered tail in NDC80 (DeLuca et al., 2006; Wei et al., 2007; Ciferri et al., 2008).
Figure 1.
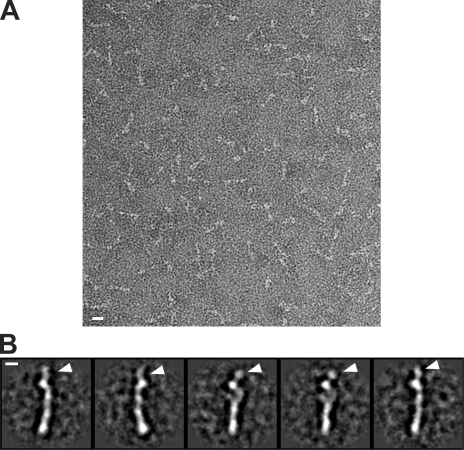
Reconstitution and structural analysis of human MIS12C. (A) Schematic representation of the components of the MIS12C, NDC80C, and KNL1C. Coiled-coil (CC) predictions calculated with program COILS (Lupas et al., 1991) are shown exclusively for subunits with partial or complete coiled-coil content. Alternate names in humans are indicated. Hs, Homo sapiens. (B) Size-exclusion chromatography run of recombinant MIS12CNSL1-258 with corresponding SDS-PAGE separation stained with Coomassie brilliant blue. The molecular mass of the recombinant complex is ∼120 kD, but the protein elutes earlier than expected for a globular protein of equivalent molecular mass, suggesting that it is an oligomer or that it is elongated. The dashed gray line and numbers indicate elution markers in the size-exclusion chromatography experiments and their molecular masses (in kilodaltons), respectively. a.u., arbitrary unit. (C) Negative-stain EM was performed on the recombinant MIS12CNSL1-258. The maximum length of the complex varies between ∼21 and 23 nm depending on the curvature. The maximum thickness of the rodlike structures is ∼3 nm. (D) The class averages represent the characteristic views of the MIS12C and reveal a varying amount of curvature. Bars: (C) 10 nm; (D) 5 nm.
NDC80C is part of a conserved arrangement of interacting subcomplexes, now generally referred to as the KMN network (De Wulf et al., 2003; Desai et al., 2003; Nekrasov et al., 2003; Pinsky et al., 2003; Westermann et al., 2003; Cheeseman et al., 2004, 2006; Obuse et al., 2004; Emanuele et al., 2005; Kops et al., 2005; Liu et al., 2005; Kline et al., 2006; Przewloka et al., 2007; Welburn et al., 2010). Besides NDC80C, the KMN network contains the four-subunit MIS12 complex (MIS12C; also known as the MIND complex or Mtw1 complex; Table S1) and the two-subunit KNL1 complex (KNL1C; Fig. 1 A). KNL1, a subunit of the KNL1C, is required for kinetochore recruitment of checkpoint components such as BUB1 and possibly BUBR1 and interacts stably with ZWINT (Table S1; Desai et al., 2003; Nekrasov et al., 2003; Kiyomitsu et al., 2007; Pagliuca et al., 2009; Schittenhelm et al., 2009).
The KMN network is essential for forming load-bearing, end-on attachments (e.g., Desai et al., 2003; McCleland et al., 2003; DeLuca et al., 2005, 2006; Cheeseman et al., 2006; Vorozhko et al., 2008). Each microtubule-binding site may contain from six to eight KMN complexes (Joglekar et al., 2006, 2008; Johnston et al., 2010). We only have a rudimentary understanding of how these complexes are deposited at kinetochores. Although NDC80C and KNL1 are absent from interphase kinetochores, MIS12C resides there (Obuse et al., 2004; Hemmerich et al., 2008). CENP-C, which partially copurifies with KMN network subunits in proteomic analyses and is required for kinetochore recruitment of MIS12C subunits, may be involved in recruiting MIS12C to interphase kinetochores (e.g., Desai et al., 2003; Westermann et al., 2003; Cheeseman et al., 2004; Liu et al., 2006; Okada et al., 2006; Kwon et al., 2007; Milks et al., 2009). Although MIS12 cycles on interphase kinetochores with a relatively rapid half-time (∼7 s), it becomes stably associated with kinetochores in mitotic prophase (Hemmerich et al., 2008), together with NDC80C and KNL1C (Obuse et al., 2004; Hemmerich et al., 2008). Exactly how these proteins become recruited to kinetochores is currently unclear.
The mutual arrangement of kinetochore subunits is being actively investigated though super-resolution light microscopy approaches (Schittenhelm et al., 2007; Joglekar et al., 2009; Liu et al., 2009; Maresca and Salmon, 2009; Uchida et al., 2009; Wan et al., 2009; Ribeiro et al., 2010). Supported by this information, and by an increasingly complete census of kinetochore subunits and their interactions, we started an effort of biochemical reconstitution of human kinetochores, aiming to unravel their structure and overall organization. In this study, we report the results of our efforts on the human KMN network.
Results
Reconstitution of MIS12C
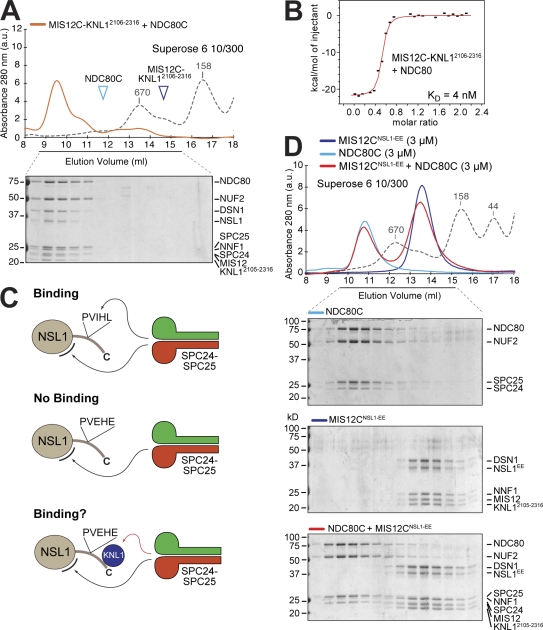
In preliminary experiments, we reconstituted NNF1–MIS12 and NSL1–DSN1 subcomplexes (unpublished data), in agreement with a previous analysis of binary interactions of MIS12C subunits by the yeast two-hybrid method (Kiyomitsu et al., 2007). These initial attempts toward reconstituting MIS12C suggest the existence of tight and discrete binary intersubunit interfaces within the MIS12C. However, the resulting recombinant subcomplexes were only moderately soluble and stable. To reconstitute the entire human MIS12C, we resorted to coexpression in Escherichia coli from the pST39 vector (Tan et al., 2005). Affinity purification from bacterial lysates through a hexahistidine tag on the DSN1 subunit, and additional chromatographic steps, led to the purification of an apparently monodisperse sample (unpublished data). The NSL1 subunit underwent slow spontaneous proteolysis during purification and subsequent storage. By limited proteolysis, we determined that the C-terminal region of NSL1 is unstable (in the absence of binding partners; the tail becomes stabilized in the presence of KNL1, as shown in the next section). Therefore, we created new versions of recombinant MIS12C containing C-terminally deleted NSL1. An MIS12C containing a proteolytically stable segment of NSL1 consisting of residues 1–258 (NSL1258), which we refer to as MIS12CNSL1-258, was stable (Fig. 1 B).
Hydrodynamic analysis by sedimentation velocity analytical ultracentrifugation (AUC) indicated that MIS12CNSL1-258 forms an elongated structure in which each subunit is represented once (Table I). The elongated shape of MIS12C also explains precocious elution of the MIS12CNSL1-258 complex in size-exclusion chromatography, which separates based on the samples’ Stokes radius (Fig. 1 B).
Table I.
Sedimentation velocity AUC
| Construct | Predicted molecular mass | Observed molecular mass | Stoichiometry | Sedimentation coefficient | Frictional coefficient (f/f0) |
| kD | kD | S | |||
| MIS12CNSL1-258 | 116.9 | 115.1 | 1:1:1:1 | 4.08 | 2.079 |
| MIS12CNSL1-258 + SPC2457–197–SPC2570–224 | 151.3 | 160 | 1:1:1:1:1:1 | 4.65 | 2.26 |
| MIS12CNSL1-258 + HP1 | 161.3 | 158 | 1:1:1:1:2 | 4.84 | 2.17 |
| SPC2457–197–SPC2570–224 | 34.4 | 36 | 1:1 | 2.46 | 1.59 |
| HP1 | 44.4 | 45.7 | Homodimer | 2.69 | 1.7 |
Negative-stain EM returned nicely contrasted images of recombinant MIS12CNSL1-258, revealing an elongated, 21–23-nm rod with a diameter of ∼3 nm (Fig. 1 C). Subsequent calculation of class averages by alignment and classification identified four distinct consecutive elements of density (Fig. 1 D), possibly organized in variations of a slightly arched arrangement. In some views, the last element of density, at one end of the rod, displayed two distinct appendices emanating from a central globular structure.
Direct binding of MIS12C to KNL1
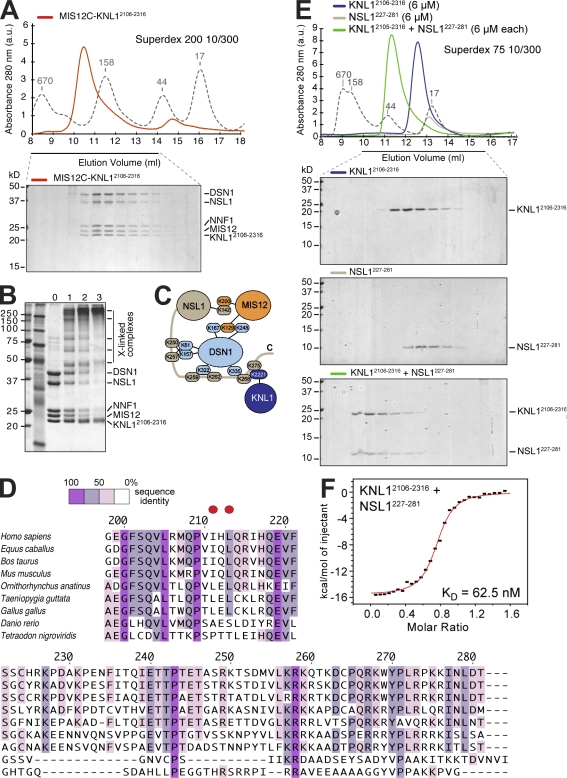
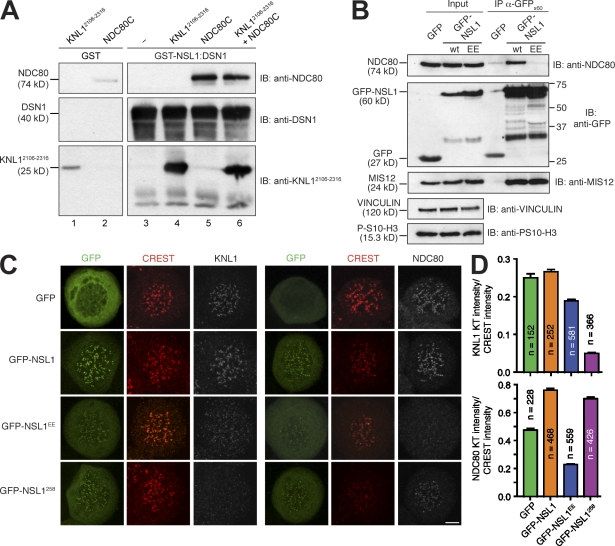
The C-terminal region of KNL1 has been recently implicated in binding to MIS12C and in kinetochore recruitment of KNL1 (Kiyomitsu et al., 2007). To test whether the C-terminal region of KNL1 binds MIS12C directly, we coexpressed full-length MIS12C with residues 2106–2316 of KNL1 (KNL12106–2316). This resulted in the purification of a tight and apparently stoichiometric five-subunit complex (Fig. 2 A). Thus, KNL12106–2316 is sufficient for high-affinity binding to MIS12C.
Figure 2.
The C-terminal domain of KNL1 interacts directly with MIS12C. (A) Size-exclusion chromatography analysis demonstrates that MIS12C and KNL12106–2316 form a stoichiometric complex. (B) Cross-linking analysis. In lanes 1–3, 5 nmol, 11 nmol, or 22 nmol BS2G was added, respectively. (C) Summary of identified intersubunit cross-links. The full list of cross-links is listed in Table S2. (D) Multiple sequence alignment of the C-terminal regions of NSL1 from different species. The two red dots indicate residues that were mutated into glutamic acid (E) in the NSL1EE mutant. (E) Size-exclusion chromatography analysis on the interaction of KNL12106–2316 with a construct encompassing residues 227–281 of the NSL1 subunit of the MIS12C. Elution profile of KNL12106–2316 (top), NSL1227–281 (middle), and of a stoichiometric combination of KNL12106–2316 and NSL1227–281 (bottom). (A and E) Dashed gray lines and numbers indicate elution markers in the size-exclusion chromatography experiments and their molecular masses (in kilodaltons), respectively. (F) ITC on the KNL12106–2316–NSL1227–281 interaction reveals a Kd of ∼130 nM and a stoichiometry of 1:1.
To identify sites of intersubunit contacts, we submitted the five-subunit MIS12C–KNL12106–2316 complex to a previously described cross-linking method (Fig. 2 B; Maiolica et al., 2007). In brief, the method exploits the ability of the isotope-labeled BS2G (bis[sulfosuccinimidyl]glutarate) to cross-link the primary amine of lysines (K) within a distance compatible with the length of the cross-linker (7.7 Å). Besides several intrasubunit cross-links, we also identified several intersubunit cross-links, including contacts between MIS12 and DSN1, MIS12 and NSL1, and between DSN1 or KLN12106–2316 and the C-terminal tail of NSL1 (Fig. 2 C and Table S2). Specifically, cross-links between K268 or K275 in the C-terminal region of NSL1 and K2221 in KNL12106–2316 implicate the proteolytically sensitive C-terminal region of NSL1 in the interaction with KNL1 (Fig. 2 C). This possibility is also consistent with the inability of MIS12CNSL1-258 to bind KNL1 (see Fig. 7 C and not depicted). Moreover, the NSL1 subunit was protected against proteolysis in the complex of MIS12C with KNL12106–2316, further implicating the NSL1 C-terminal tail in this interaction. Collectively, these results suggest that the C-terminal region of NSL1 (whose sequence is displayed in Fig. 2 D) is necessary for high-affinity binding to KNL1.
KNL12106–2316 and a construct encompassing the C-terminal tail of NSL1 (NSL1227–281) coeluted in a complex from a size-exclusion chromatography column (Fig. 2 E). By isothermal titration calorimetry (ITC), we determined that these species bind each other with a Kd of ∼63 nM. Nevertheless, the KNL12106–2316–NSL1227–281 complex does not fully recapitulate the interaction of KNL12106–2316 with MIS12C, as we have evidence that the overall binding affinity of KNL12106–2316 for MIS12C is significantly higher (although the instability of the C-terminal tail of NSL1 in the absence of binding partners prevented us from measuring it directly). For instance, the KNL12106–2316–NSL1227–281 complex dissociates when loaded onto a strong anion-exchange column, probably because of the strongly electrostatic character of the interaction (NSL1227–281 has an isoelectric point [pI] of ∼10.5, whereas KNL12106–2316 has a pI of ∼4.5). Conversely, the KNL12106–2316–MIS12C complex resists this treatment (unpublished data). As our cross-linking analysis suggests that the C-terminal region of NSL1 makes several contacts with the DSN1 subunit (Fig. 2 C), additional binding contacts from DSL1 to KNL1 may increase the overall binding affinity.
Direct binding of MIS12C to NDC80C
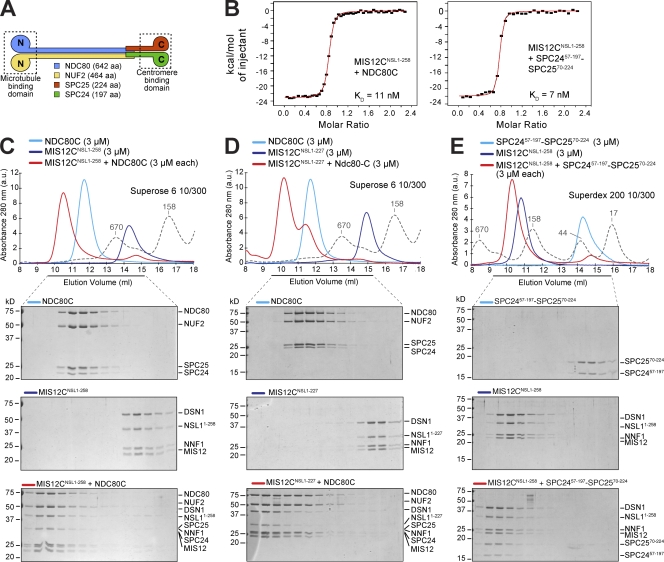
Next, we asked whether MIS12C interacts directly with NDC80C or whether this interaction requires KNL1. We purified a homogeneous form of the four-subunit NDC80C (whose organization is schematically presented in Fig. 3 A) after coexpression of its subunits in insect cells (Fig. 3 C). We then mixed MIS12CNSL1-258 with NDC80C in approximately stoichiometric amounts and separated the resulting species by size-exclusion chromatography (Fig. 3 C). We observed the formation of a high–molecular weight complex containing all eight subunits, indicating that MIS12CNSL1-258 and NDC80C enter a tight binary complex in the absence of KNL1. A more extensive C-terminal deletion of NSL1 (MIS12CNSL1-227) was also compatible with the interaction with NDC80C (Fig. 3 D). Thus, the KNL1-binding region of NSL1 is not required for high-affinity binding of MIS12C to NDC80C.
Figure 3.
Interaction of MIS12C with NDC80C. (A) Schematic representation of the NDC80C. (B) Binding curves were measured by ITC by titrating NDC80C (left) or SPC2457–197–SPC2570–224 (right) in a cell containing MIS12CNSL1-258. (C) Size-exclusion chromatography elution profiles and SDS-PAGE analysis of recombinant NDC80C expressed in, and purified from, insect cells (top), MIS12CNSL1-258 (middle), and their stoichiometric combination (bottom). (D) Elution profiles and SDS-PAGE analysis of NDC80C (top), MIS12CNSL1-227 (middle), and their stoichiometric combination (bottom). The MIS12CNSL1-227 construct binds NDC80C. (E) As in C, but with the kinetochore-binding portion of NDC80C, the SPC2457–197–SPC2570–224 construct. (C–E) Dashed gray lines and numbers indicate elution markers in the size-exclusion chromatography experiments and their molecular masses (in kilodaltons), respectively.
In agreement with previous analyses showing that the kinetochore-binding domain of NDC80C resides in the SPC24–SPC25 subcomplex (see Introduction), the dimeric constructs SPC2457–197–SPC2570–224 and SPC24104–197–SPC2599–224 also bound MIS12C tightly (Fig. 3 E and Fig. S1 A). By ITC, we found similar binding constants for the interaction of MIS12C with NDC80C or the SPC2457–197–SPC2570–224 subcomplex, both ranging near 10 nM (Fig. 3 B). As determined by AUC, the MIS12C–SPC2457–197–SPC2570–224 complex contains one copy of each subunit (Table I). Recombinant MIS12C did not cosediment with taxol-stabilized microtubules, nor did it increase the affinity of NDC80C for microtubules (Fig. S1 B).
HP1-α binds NSL1
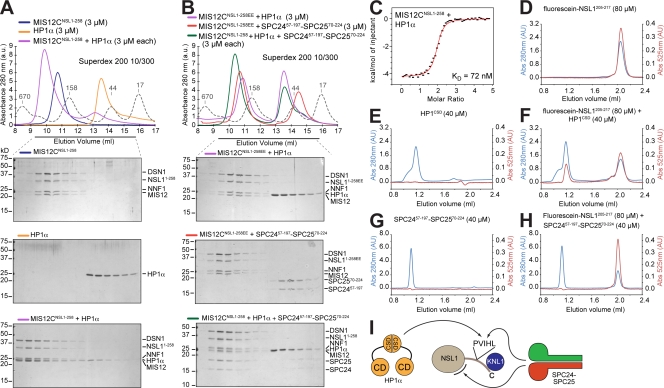
HP1 (heterochromatin protein 1), a crucial component of centromeric heterochromatin (for reviews see Pidoux and Allshire, 2005; Lomberk et al., 2006), has been identified as a binding partner of the KMN network in humans and flies and may participate in directing MIS12C to the centromere in interphase (Obuse et al., 2004; Przewloka et al., 2007; Kiyomitsu et al., 2010). HP1 is recruited to centromeric heterochromatin in an Suv39h-dependent manner (Peters et al., 2001) and may have a role in the recruitment of MIS12 to the centromere during interphase (Obuse et al., 2004). To test whether HP1 binds MIS12C directly, we performed a size-exclusion chromatography analysis on stoichiometric combinations of these proteins and found that HP1-α coeluted with MIS12C (Fig. 4 A).
Figure 4.
Interaction of MIS12C with HP1. (A) Size-exclusion chromatography elution profiles and SDS-PAGE analysis of MIS12CNSL1-258 (top), HP1-α (middle), and their stoichiometric combination (bottom). (B) The MIS12CNSL1-258EE mutant does not bind HP1-α (top). SPC2457–197–SPC2570–224 is also unable to bind MIS12CNSL1-258EE, indicating that the binding sites for HP1-α and SPC2457–197–SPC2570–224 overlap, at least in part (middle). When HP1-α and SPC2457–197–SPC2570–224 were combined in stoichiometric amounts with MIS12CNSL1-258, HP1-α did not coelute with MIS12CNSL1-258, whereas SPC2457–197–SPC2570–224 was incorporated in a complex with MIS12CNSL1-258, suggesting that SPC2457–197–SPC2570–224 binds MIS12CNSL1-258 with higher affinity (bottom). (A and B) Dashed gray lines and numbers indicate elution markers in the size-exclusion chromatography experiments and their molecular masses (in kilodaltons), respectively. (C) ITC binding curve for the interaction of MIS12CNSL1-258 with HP1-α. (D) Elution profile from a size-exclusion chromatography Superdex 75 PC 3.2/30 column of a fluorescein-labeled synthetic peptide encompassing residues 205–217 of NSL1 (fluorescein-NSL1205–217). The red trace reports absorbance (Abs) at 525 nm. Panels D through H were run under the same experimental conditions. (E) Elution profile of the chromoshadow domain of HP1-α. (F) Elution profile of a mixture of the chromoshadow domain of HP1-α and fluorescein-NSL1205–217 demonstrating a shift in the elution profile of the fluorescein-NSL1205–217 peptide. (G) Elution profile of SPC2457–197–SPC2570–224. (H) The SPC2457–197–SPC2570–224 construct does not coelute with the fluorescein-NSL1205–217 peptide, suggesting that this region of NSL1 is insufficient for high-affinity binding to NDC80C. (I) Summary of interactions presented in the figure. The position of the second binding site for SPC24–SPC24, indicated by a black curved segment, is actually on MIS12C, but not necessarily on the NSL1 subunit as shown. CD, chromodomain; CSD, chromoshadow domain.
HP1-α forms homodimers. Each HP1 subunit consists of a chromodomain, which binds to methylated lysine 9 of histone H3, and of a chromoshadow domain, which is responsible for dimerization and for the interactions with binding partners containing motifs conforming to the PXVXL consensus, where X is any amino acid (for review see Lomberk et al., 2006). In their dimeric configuration, the chromoshadow domain forms a shared groove that hosts a single PXVXL motif in either of two equivalent orientations (Brasher et al., 2000). It has been noted that residues 209–213 of NSL1 contain a PVIHL motif that largely conforms to the PXVXL consensus (residues 209–213, Fig. 2 D; Kiyomitsu et al., 2010). To confirm the role of this motif in HP1-α binding, we mutated it into PVEHE within the framework of the MIS12CNSL1-258 complex (MIS12CNSL1-258EE). In agreement with the hypothesis that the PVIHL motif of NSL1 is important for HP1-α binding, MIS12CNSL1-258EE was unable to bind HP1-α (Fig. 4 B, top gel). This result provides a direct biochemical confirmation of the recently described interaction of HP1 with NSL1 (Kiyomitsu et al., 2010). By AUC, we found that HP1 binds to the MIS12CNSL1-258 complex as a dimer, which is as expected for an interaction mediated by the chromoshadow domain of HP1 (Table I).
HP1 and NDC80 bind MIS12C competitively
Binding of HP1-α to MIS12C was compatible with the presence of the KNL12106–2316 segment (unpublished data). However, when HP1-α and SPC2457–197–SPC2570–224 were combined, most if not all HP1-α was displaced from MIS12CNSL1-258, and SPC2457–197–SPC2570–224 concomitantly shifted into the fast-eluting MIS12CNSL1-258-containing peak (Fig. 4 B, middle gel). Thus, HP1-α and NDC80C are competitive binders of MIS12C, suggesting that their binding sites on MIS12C partly coincide. To corroborate this idea, we combined SPC2457–197–SPC2570–224 with MIS12CNSL1-258EE. No complex formed, which is in agreement with the idea that HP1-α and NDC80 share their binding site on MIS12 and that the NSL1 subunit is decisively implicated in this interaction (Fig. 4 B, bottom gel).
By ITC, we determined that MIS12CNSL1-258 binds HP1-α with a Kd of 72 nM (Fig. 4 C), a sevenfold lower binding affinity relative to that measured for the interaction with NDC80C or SPC2457–197–SPC2570–224 (Fig. 3 B). This may explain why SPC2457–197–SPC2570–224 effectively competes the binding of HP1-α to MIS12C. We next asked whether the NSL1 segment containing the PVIHL motif was sufficient for HP1-α or NDC80C binding. To this end, we generated fluorescein-labeled synthetic peptides encompassing residues 205–217 or 202–219 of NSL1 (fluorescein-NSL1205–217 or fluorescein-NSL1202–219). Using fluorescence anisotropy–based assays, we determined that these peptides bound the chromoshadow domain of HP1-α with an affinity around 400–500 nM (Fig. S2 A), six- to sevenfold lower than that measured by ITC for the MIS12CNSL1-258–HP1-α interaction. Overall, these observations indicate that the MIS12C–HP1-α interaction is largely, although not exclusively, determined by the chromoshadow domain of HP1-α and the NSL1205–217 segment.
In contrast, there was no binding of fluorescein-NSL1205–217 or fluorescein-NSL1202–219 to SPC24104–197–SPC2599–224 (Fig. S2 B), suggesting that the binding site for NDC80C on MIS12C cannot be recapitulated by the isolated fluorescein-labeled NSL1205–217 peptide. This conclusion was corroborated by size-exclusion chromatography coelution experiments. The fluorescein-NSL1205–217 peptide, whose absorbance could be monitored at 525 nm, eluted as expected for its size from a size-exclusion chromatography column (Fig. 4 D). When combined with the HP1-α chromoshadow domain (whose profile is shown in Fig. 4 E), part of fluorescein-NSL1205–217 eluted with it, which is indicative of complex formation (Fig. 4 F). When the same experiment was performed with SPC2457–197–SPC2570–224 or SPC24104–197–SPC2599–224, no shift in the elution profile of fluorescein-NSL1205–217 was observed, suggesting that the interaction is not strong enough for coelution from a size-exclusion chromatography column (Fig. 4, G and H; and Fig. S2, C and D). Because the SPC2457–197–SPC2570–224 or SPC24104–197–SPC2599–224 constructs bind tightly to MIS12C, we conclude that NSL1205–217 is necessary but not sufficient to bind SPC24–SPC25 and that other residues in MIS12C are required. Unfortunately, cross-linking analysis has been so far unable to identify such additional residues.
The C-terminal tail of NSL1 is required for kinetochore binding of NDC80 and KNL1
Collectively, our results point to the DSN1–NSL1 dimer as a crucial binding site for HP1, NDC80C, and KNL1C. Indeed, a recombinant DSN1–NSL1 dimer was sufficient to bind NDC80C or the C-terminal tail of KNL1 in vitro (Fig. 5 A). The NSL1 C-terminal tail is crucially involved in the binding of NDC80C and KNL1, and NDC80C and HP1-α are competitive binding partners of NSL1. Recently, it has been proposed that the MIS12C–HP1 interaction may be important for centromeric cohesion and for centromere recruitment of the chromosome–passenger complex (Kiyomitsu et al., 2010). Our observation that HP1 and NDC80 bind to MIS12 competitively predicts that mutations in the HP1-binding site of NSL1 that prevent HP1 binding also prevent NDC80 binding. To test this prediction in cells, we expressed the NSL1EE mutant in HeLa cells as a fusion to GFP. In agreement with our in vitro observations, the GFP-NSL1EE mutant was unable to interact with endogenous NDC80, contrarily to wild-type GFP-NSL1 (Fig. 5 B). However, the mutant localized normally to kinetochores (Fig. 5 C). As expected, its expression there largely reduced the amount of NDC80 at kinetochores (Fig. 5, C and D). Therefore, we suspect that the recently described mitotic phenotype caused by the expression of the NSL1EE mutant in HeLa cells (Kiyomitsu et al., 2010) may also reflect a defect in the localization of NDC80, rather than being exclusively a consequence of the mislocalization of HP1.
Figure 5.
Role of the NSL1 C-terminal tail. (A) The DSN1–NSL1 subcomplex is sufficient to bind KNL1 and NDC80C. GST-NSL1–DSN1 complex was purified by glutathione Sepharose affinity purification and used as an affinity bait for pull-down assays. Copurification of DSN1 with GST-tagged NSL1 was assessed by immunoblotting. GST-NSL1–DSN1 specifically binds KNL12106–2316 (lane 4) and NDC80C (as judged by immunoblotting on the NDC80 subunit; lane 5), and the binding is not mutually exclusive (lane 6). The levels of unspecific binding to GST are shown in lanes 1 and 2. (B) HeLa cells were transiently transfected with plasmids containing GFP alone, GFP-tagged NSL1, and GFP-tagged NSL1EE. After 48 h, cells were washed once and treated with 5 µM S-trityl-l-cysteine for 16 h. Mitotic cells were harvested by vigorous shake-off. Immunoprecipitates (IPs) with an anti-GFP antibody were examined by immunoblotting (IB) with the indicated antibodies. Only GFP-NSL1 was able to bind endogenous NDC80. The band indicated by the asterisk may represent a degradation product of the GFP fusion proteins. wt, wild type. (C) HeLa cells were transiently transfected with plasmids containing GFP alone, GFP-tagged NSL1, GFP-tagged NSL1EE, and GFP-tagged NSL1258. After 48 h, cells were washed once, treated with 3.3 µM nocodazole for 14 h, and analyzed by immunofluorescence. Bar, 5 µm. (D) The means of KNL1 or NDC80 kinetochore (KT) intensities normalized to CREST signal in cells transiently expressing GFP alone, GFP-tagged NSL1, GFP-tagged NSL1EE, and GFP-tagged NSL1258 from the experiment in C were calculated and plotted. Error bars represent the SEM. For each condition, at least four different cells were used in the quantification.
We also expressed a truncated form of NSL1 (NSL1258). Like NSL1EE, this mutant also localized normally to kinetochores. Upon expression of NSL1258, the kinetochore levels of KNL1 were substantially reduced (Fig. 5, C and D), as expected based on our in vitro analysis. Overall, this analysis confirms that the C-terminal tail of NSL1 is a crucial determinant of kinetochore localization of KNL1 and NDC80.
Testing additional interactions within the KMN network
As a further step in the analysis of the KMN assembly, we asked whether the MIS12C–KNL12106–2316 complex interacted directly with NDC80C. Indeed, when mixed at equimolar concentrations, MIS12C–KNL12106–2316 and NDC80C became stoichiometrically engaged in a high–molecular weight complex (Fig. 6 A). By ITC, we measured a Kd of 4 nM for this interaction (Fig. 6 B), a value in range with those measured for the NDC80C–MIS12C interaction in the absence of KNL1 (Fig. 3 B). Thus, KNL1, when bound to MIS12C, may not contribute significantly to the interaction with NDC80C.
Figure 6.
Interaction of MIS12C with HP1. (A) Elution profile and SDS-PAGE analysis of a stoichiometric mixture of MIS12C–KNL12106–2316 and NDC80C. The elution position of individual complexes in indicated. (B) ITC analysis of the interaction of MIS12C–KNL12106–2316 with NDC80C. (C) SPC24–SPC25 binds the C-terminal tail of NSL1 and an additional site on MIS12 (see legend to Fig. 4 I). If the PVIHL motif is mutated, SPC24–SPC25 does not bind. If SPC24–SPC24 bound KNL1 (the red arrow indicates this interaction is hypothetical), additional binding energy may be recovered, and SPC24–SPC25 would be expected to bind even with a mutated PVIHL motif. (D) Addition of KNL12106–2316 to MIS12C does not rescue the lack of interaction of NDC80C to the PVIHL mutant, suggesting that SPC24–SPC25 and KNL12106–2316 do not interact. (A and D) Dashed gray lines and numbers indicate elution markers in the size-exclusion chromatography experiments and their molecular masses (in kilodaltons), respectively.
A possible problem arising from using ITC to measure the very tight interactions discussed in the previous paragraph is that relatively high concentrations of binding species are necessary, possibly leading to an underestimation of the binding affinity. Thus, although our ITC experiments clearly demonstrate that the interaction of MIS12C and NDC80C is very high affinity, they may be unsuitable to distinguish an additional positive contribution from KNL1. Such a contribution may be predicted on the basis of a previous study of the Caenorhabditis elegans KMN network showing that KNL1 is required for the establishment of a tight interaction of NDC80C with MIS12C (Cheeseman et al., 2006).
To test a possible contribution of KNL1, we resorted to two additional experiments. First, we asked whether KNL12106–2316 and NDC80C were able to coelute from a size-exclusion chromatography column. No shift was observed, suggesting that if an interaction between NDC80C and KNL12106–2316 exists, it is low affinity (Fig. S1 C). Nevertheless, these species may be able to form a low-affinity interaction, insufficient to allow the detection in a binary binding assay but sufficient to cause a dramatic increase of the overall binding affinity within the context of a positively cooperative binding mechanism in which multiple surfaces interact (Whitty, 2008). (The observation that the PVIHL motif of NSL1 is necessary but not sufficient for high-affinity binding of MIS12C and NDC80C is an excellent example for how a relatively small contribution to the binding energy can dramatically increase the overall binding affinity.) To expose such a contribution from KNL1, we asked whether the presence of KNL1 rescued lack of NDC80C binding by the MIS12CNSL1-258EE mutant. The rationale for this, illustrated in Fig. 6 C, is that if KNL1 contacts NDC80C directly, the binding energy provided by this additional interaction, even if relatively small, may compensate for the decrease in binding affinity caused by mutations in the PVIHL motif of NSL1. Contradicting the hypothesis, the presence of KNL12106–2316 did not rescue the inability of NDC80C to bind MIS12CNSL1-258EE (Fig. 6 D). We conclude that if a connection between KNL1 and NDC80 exists in the human KMN, it must be of very modest strength, at least under the in vitro conditions of our experiments.
The binding site for ZWINT
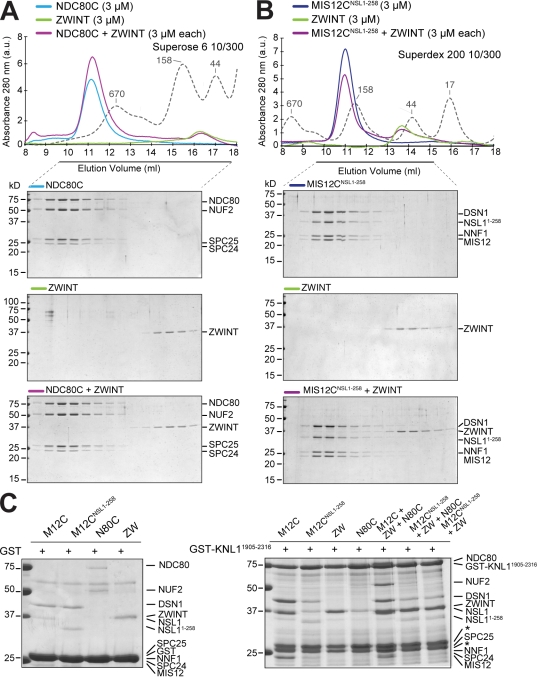
It was found by the yeast two-hybrid method that ZWINT interacts with the C-terminal region of KNL1 (Kiyomitsu et al., 2007). In this position, ZWINT is believed to act as a receptor for the ROD–ZW10–ZWILCH (RZZ) complex, a kinetochore complex required for kinetochore recruitment of dynein and of the spindle checkpoint proteins MAD1 and MAD2 (for review see Musacchio and Salmon, 2007). We expressed and characterized a recombinant version of ZWINT and tested its ability to bind MIS12C and NDC80C in size-exclusion chromatography coelution experiments. No binding of ZWINT to any of these constructs was observed, suggesting that ZWINT does not bind directly to MIS12C or NDC80C (Fig. 7, A and B). We then tested whether ZWINT bound KNL12106–2316 in a size-exclusion chromatography coelution experiment. Once again, we did not observe binding between these species (Fig. S3 A). Therefore, we tested binding of ZWINT to a GST fusion of a larger KNL1 segment (GST-KNL11904–2316), which also contains the predicted coiled-coil of KNL1 (Fig. 1 A and Fig. S4). MIS12C readily bound to this construct, whereas MIS12CNSL1-258 did not (Fig. 7 C, right, first and second lanes). Thus, the presence of the coiled-coil of KNL1 does not provide strong additional binding contacts to MIS12C, suggesting that the interaction of the KNL1 C-terminal domain with MIS12C is crucial for the KNL1–MIS12 interaction even in the context of a larger KNL1 segment. In agreement with our findings, only the C-terminal region of KNL1 was found to interact with the MIS12C in a yeast three-hybrid experiment (Kiyomitsu et al., 2007). ZWINT also readily bound to GST-KNL11904–2316, forming an apparently stoichiometric complex in the absence of other proteins (Fig. 7 C, right, third lane). In agreement with our observation that ZWINT and MIS12CNSL1-258 do not bind tightly, ZWINT was unable to rescue the inability of MIS12CNSL1-258 to bind GST-KNL11904–2316 (Fig. 7 C, right, seventh lane). No direct binding of NDC80 to GST-KNL11904–2316 was observed (Fig. 7 C, right, fourth lane). However, the addition of MIS12C and ZWINT led to NDC80C binding, presumably through the mediation of MIS12C (Fig. 7 C, right, fifth lane). In agreement with this hypothesis, MIS12CNSL1-258 was unable to promote the retention of NDC80C even in the presence of ZWINT (Fig. 7 C, right, sixth lane). Additional binding assays, supporting the same conclusions, are shown in Fig. S3 (B–D). Collectively, these results suggest that ZWINT does not establish tight contacts with NDC80C or MIS12C and that its interaction with the KMN network complex is primarily mediated by the interaction with the C-terminal domain of KNL1.
Figure 7.
Interactions of ZWINT. (A) Elution profile and SDS-PAGE analysis of NDC80 and recombinant ZWINT. No complex with NDC80C was observed. (B) As in A, but with MIS12C. (A and B) Dashed gray lines and numbers indicate elution markers in the size-exclusion chromatography experiments and their molecular masses (in kilodaltons), respectively. (C, left) Only background binding to GST bound to glutathione Sepharose beads was observed. (right) A GST-KNL11904–2316 fusion protein was used as an affinity bait on glutathione Sepharose beads. This construct was then incubated with the indicated proteins. Asterisks mark two bands that copurify with GST-KNL11904–2316 on the glutathione Sepharose beads. Additional controls and input proteins are shown in Fig. S3.
Overall organization of the KMN network
EM analysis in Fig. 1 suggests the possibility that the four subunits of MIS12C are arranged sequentially along the MIS12C rod. We sought to extend our understanding of the subunit organization within the MIS12C by decorating it with additional KMN subunits. Specifically, we attempted to visualize a complex of MIS12C with SPC2457–197–SPC2570–224, as the latter forms an extended structure expected to significantly alter the appearance of MIS12C in EM images (Wei et al., 2006; Ciferri et al., 2008). We used negative-stain EM, under conditions similar to those already used for the isolated MIS12C, to visualize the complex of MIS12C with SPC24–SPC25. As shown previously for MIS12C (Fig. 1), we observed good contrast and were able to calculate class averages from the raw images. After image alignment (Sander et al., 2003) and multivariate statistical analysis (van Heel and Frank, 1981), additional density corresponding to the SPC24–SPC25 component was visible as an extension to the arched arrangement of density corresponding to the MIS12C rod (Fig. 8, indicated by arrowheads). These results strongly suggest that the NDC80C emanates from one end of the MIS12C and are consistent with the increase in the hydrodynamic radius of the MIS12C–SPC24–SPC25 complex relative to both individual complexes (Table I).
Figure 8.
EM analysis of MIS12C–SPC24–SPC25. (A) Negative-stain EM was performed on the recombinant MIS12CNSL1-258–SPC2457–197–SPC2570–224 complex. (B) Class averages from negative-stain EM images shown in A. The arrowheads point to an element of density that is not present in the class averages of the MIS12CNSL1-258 complex shown in Fig. 1 D and that likely represents the SPC24–SPC25 complex. Bars: (A) 10 nm; (B) 5 nm.
Discussion
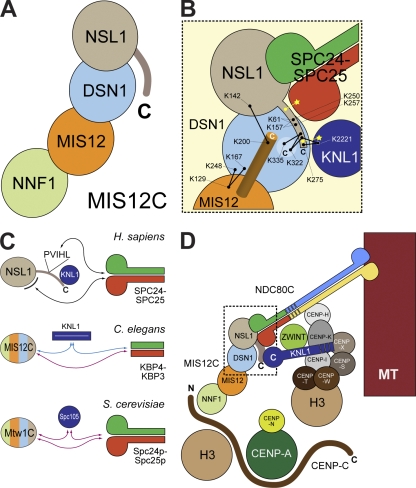
We propose that the MIS12C rod is a concatenation of the NNF1, MIS12, DSN1, and NSL1 subunits, with NNF1 and NSL1 at the two opposite ends of the ∼22-nm rod (Fig. 9 A). MIS12 forms cross-links with NSL1 and DSN1, suggesting that it neighbors these proteins. Because MIS12 forms at least one cross-link with residues within the predicted globular region of DSN1 (K129MIS12 with K167DSN1 or K248DSN1; Fig. 1 C), we suspect that it directly contacts DSN1. The NSL1 C-terminal tail probably meanders on a surface contributed by DSN1 and possibly by its neighboring MIS12 and NSL1 subunits (Fig. 9 B). Both SPC24–SPC25 and KNL1 are likely to form additional contacts with this surface: the C-terminal tail of NSL1 is necessary for both interactions, but we observed an increase in binding energy when the interactions were measured with the entire MIS12C rather than the isolated C-terminal tail of NSL1.
Figure 9.
Organization of the KMN network. (A) Schematic illustration of the organization of the human MIS12C complex. (B) Summary of interactions identified in this study. The black dots connected by a line represent cross-links. Yellow dots represent defined binding sites. Yellow stars represent undefined binding sites. The orange cylinder represents a predicted C-terminal helix in the MIS12 subunit. (C) Summary of interactions in KMN complexes in different species. Black arrows represent established interactions. Blue arrows represent established binding requirements that have not been mapped at the molecular level. Purple arrows represent putative interactions. In C. elegans, KBP-4 and KBP-3 (SPC24 and SPC25 homologues, respectively) are 97- and 134-residue proteins lacking the globular domains of SPC24 and SPC25 in other species. A globular domain at the C-terminal end of KNL1 is also missing in this organism (see alignment in Fig. S4). In S. cerevisiae, Mtw1 (MIS12 homologue) does not have a PVIHL motif or a positively charged C-terminal domain, and the binding site for Spc24p–Spc24p and Spc105p (KNL1 homologue) is therefore unknown. The four-color scheme for MIS12C in C. elegans and S. cerevisiae conveys that the binding sites have not been mapped and could therefore be anywhere on these structures. (D) An extension of the two-hand model (Cheeseman et al., 2008) for kinetochore recruitment of the KMN network based on our experiments. MT, microtubule.
The model in Fig. 9 is consistent with previous yeast two- and three-hybrid analyses (Kiyomitsu et al., 2007, 2010). It is further supported by our observation that the SPC24–SPC25 dimer appears to emerge from one end of the MIS12C rod (Fig. 8). For technical reasons, we were unable to achieve EM images of the MIS12C–KNL1C of sufficient quality. MIS12C and NDC80C may add their lengths in series, at least partially, thus potentially creating a structure up to 80–90 nm in length. Future studies will have to address whether the structures created by the MIS12C, NDC80C, and KNL1C can be reconciled with the fibrous material revealed by recent electron tomography studies of the kinetochore (Dong et al., 2007; McIntosh et al., 2008). In an accompanying paper in this issue, Maskell et al. describe an analysis of the organization and interactions of the yeast Mtw1 complex, homologous to the MIS12C, that is largely consistent with ours.
The globular C-terminal region of KNL1, which mediates the interaction with MIS12C, is conserved in KNL1 sequences from vertebrates and invertebrates (Fig. S4). Notable exceptions are C. elegans and Drosophila melanogaster, whose sequences in this region are not recognizably related to those of other species (and are therefore not present in the alignment in Fig. S4). In contrast, the preceding coiled-coil is essentially ubiquitous in KNL1 (Fig. S4). In humans, this region of KNL1 binds to ZWINT (Fig. 7). In C. elegans, where the C-terminal globular region is missing, it may also bind NDC80C and MIS12C.
However, the sequence of NSL1 has considerably diverged in evolution (Meraldi et al., 2006). This protein has a conserved structural core, roughly corresponding to residues 1–205 of the human sequence (Fig. S5). The NSL1 C-terminal tail, which is required for the interaction of MIS12C with NDC80C and KNL1C, extends this conserved core and may not be present outside vertebrates. The interaction of a basic NSL1 stretch (residues 257–281) in this tail with the C-terminal region of KNL1 may therefore be limited to vertebrates. Also, the PVIHL motif mediating the interaction of NSL1 with NDC80C does not appear to be conserved outside vertebrates. However, in C. elegans, KNL1 and MIS12C bind in vitro in the absence of NDC80C (Cheeseman et al., 2006), like their human counterparts. As neither segment mediating the interaction of the human proteins is recognizably homologous to sequences of C. elegans, distinct binding determinants must be present in worms. In contrast, although human NDC80C and MIS12C interact with very high affinity in the absence of KNL1 (this study), the C. elegans NDC80C and MIS12C do not bind tightly without KNL1 (Cheeseman et al., 2006). Thus, it appears that KNL1 contributes to creating a high-affinity binding site for NDC80C on the C. elegans KMN network, although this may not be the case within the human KMN.
In summary, some aspects of KMN network assembly may differ in evolution. A speculative comparative analysis of KMN assembly in three species is discussed in Fig. 9 C. If confirmed, this flexibility in the assembly plan of the KMN may indicate that MIS12C acts as a localization scaffold for the assembly of the KMN network but that the exact geometry of the arrangement of intersubunit contacts may be less crucial for successful chromosome segregation.
Our data are consistent with super-resolution microscopy analyses placing the SPC24–SPC25 dimer near MIS12C and the C-terminal tail of KNL1/Spc105 (Schittenhelm et al., 2007, 2009; Joglekar et al., 2009; Wan et al., 2009). In agreement with a direct interaction between MIS12C and KNL1, MIS12C is required for kinetochore recruitment of KNL1 in different organisms (Cheeseman et al., 2004, 2008). Conversely, mitotic kinetochore localization of MIS12 subunits is strongly affected upon depletion of KNL1 by RNAi (Cheeseman et al., 2004, 2008). Similarly, recruitment of the NDC80C is also severely affected by reductions in the levels of MIS12C subunits (Cheeseman et al., 2004; Kline et al., 2006; Liu et al., 2006).
Proteins within the CCAN subcomplex, including CENP-C, CENP-H, CENP-K, CENP-I, CENP-T/W, and CENP-X, have been implicated in kinetochore recruitment of KNL1 (Hori et al., 2003; Mikami et al., 2005; Liu et al., 2006; Cheeseman et al., 2008; Amano et al., 2009). Super-resolution light microscopy analyses suggest that CENP-C, CENP-I, and CENP-T/W are positioned near MIS12C, SPC24–SPC25, and the C-terminal region of KNL1 (Schittenhelm et al., 2007; Joglekar et al., 2009; Wan et al., 2009). The recently discussed two-hand model of outer kinetochore assembly (Fig. 9 D; Cheeseman et al., 2008) suggests that in vertebrates, CENP-C and CENP-H/I/K may be scaffolds that recruit different outer kinetochore proteins to centromeric chromatin. Specifically, CENP-C may be important for kinetochore recruitment of MIS12C (Cheeseman et al., 2004; Liu et al., 2006; Kwon et al., 2007; Milks et al., 2009), and the CENP-H/I/K complex may contribute to the recruitment of KNL1 but not MIS12C (Liu et al., 2006; Okada et al., 2006; Kwon et al., 2007; Cheeseman et al., 2008). Although KNL1 may not bind NDC80C in humans (this study), the CENP-H/I/K complex may also directly contribute to NDC80C recruitment, which is in agreement with the reported interaction of NUF2 with CENP-H (Mikami et al., 2005). In C. elegans, where the CCAN, with the exclusion of CENP-C, is probably missing, KNL1 may be recruited exclusively through the CENP-C–MIS12 pathway (Cheeseman et al., 2004). Because KNL1 is required to stabilize the interaction of NDC80C with MIS12C in C. elegans, both MIS12C and KNL1 are absolutely required for kinetochore localization of NDC80C (Cheeseman et al., 2006).
How CENP-C and the CENP-H/K/I/L complex interact with centromeric chromatin is less clear. CENP-C may bridge H3- and CENP-A–containing nucleosomes in the centromere with its N- and C-terminal regions, respectively (Ando et al., 2002; Talbert et al., 2004; Cohen et al., 2008; Hori et al., 2008; Milks et al., 2009; Trazzi et al., 2009; Carroll et al., 2010). In contrast, CENP-H/K/I may be recruited through the CENP-T/W complex and probably reside on a distinct H3-containing nucleosome relative to that hosting CENP-C (Fig. 9 D; Hori et al., 2008). However, at least in Xenopus laevis extracts, CENP-C has been recently implicated in kinetochore recruitment of CENP-K, possibly suggesting that the two hands are not completely independent (Milks et al., 2009). In summary, the network of interactions in the kinetochore is unlikely to be linear, and species-specific variation may accompany an otherwise conserved building plan.
Materials and methods
Plasmids
Genes encoding MIS12C subunits were PCR amplified from a human cDNA library. Silent mutations were introduced to remove internal restriction sites. The genes were then subcloned into the pST39 polycistronic expression vector, according to the cloning strategy described previously (Tan et al., 2005). Site-directed mutagenesis, performed with the QuikChange Mutagenesis kit (Agilent Technologies), was used to generate premature stop codons in the NSL1 gene, thus generating shorter MIS12C complex constructs (MIS12CNSL1-227 and MIS12CNSL1-258). Sequences encoding C-terminal KNL1 fragments (1904–2316 and 2106–2316) were PCR amplified from a human cDNA library and cloned in the first cassette of pGEX-6P-2rbs, a dicistronic derivative of pGEX-6P vector generated in-house. For coexpression experiments with NSL1 fragments, sequences encoding KNL12106–2316 and NSL1227–281 were PCR amplified and subcloned in the first and second cassette of pGEX-6P-2rbs, respectively. The sequence encoding KNL12106–2316 was PCR amplified from the pGEX-6P-2rbs vector and subcloned in the pET28a vector and used subsequently for coexpression with MIS12C using a two-plasmid strategy (see next section). SPC2457–197–SPC2570–224 and SPC24104–197–SPC2599–224 constructs were created by subcloning in dicistronic vector pGEX-6P-2rbs as described previously (Ciferri et al., 2005). HP1-α and HP1110–191 (chromoshadow domain) were PCR amplified from the full-length HP1-α gene (gift from B. Amati, European Institute of Oncology, Milan, Italy) and subcloned into pGEX-6P-1 (GE Healthcare). cDNAs encoding full-length human NDC80, NUF2, SPC24, and SPC25 were PCR amplified and subcloned in the pFL (NDC80 and NUF2) and pUCDM (SPC24 and SPC25, the latter in frame with a C-terminal hexahistidine tag) vectors to support insect cell expression (Bieniossek et al., 2008). The vectors were fused and processed as described previously (Bieniossek et al., 2008). The resulting baculovirus was used for expression of NDC80C in Hi5 insect cells (Invitrogen). The sequence encoding human ZWINT was subcloned in pGEX-6P-1 for bacterial expression. All constructs were verified by sequencing.
Protein expression and purification
For expression of MIS12C (and its deletion mutants) BL21(DE3)pLysS cells containing the pST39 plasmid were grown in Terrific broth at 37°C to an OD600 of ∼0.8. Protein expression was induced by the addition of 0.1 mM ITPG at 18°C, and cells were incubated overnight. Cell pellets were resuspended in buffer A (20 mM Tris/HCl, pH 8.0, 300 mM NaCl, 10% [vol/vol] glycerol, and 2 mM 2-mercaptoethanol) supplemented with protease inhibitor cocktail (Protease Inhibitor Set III; EMD), lysed by sonication, and cleared by centrifugation at 45,000 g for 60 min. The cleared lysate was applied to Ni-NTA agarose beads (QIAGEN) preequilibrated in buffer A and incubated for 2 h. Beads were washed with 30 vol of buffer A containing 10 mM imidazole, and the bound protein was eluted with buffer A supplemented with 250 mM imidazole. The eluate was dialyzed against ion-exchange buffer A (20 mM Tris/HCl, pH 8.0, 30 mM NaCl, 1 mM EDTA, and 1 mM DTT) and applied to a Resource Q (ResQ; GE Healthcare) column preequilibrated in the same buffer. Elution of bound protein was achieved by a linear gradient from 30 to 300 mM NaCl in 20 bed column volumes. Relevant fractions were concentrated in 10-kD molecular mass cut-off Vivaspin concentrators (Sartorius) and applied to a Superdex 200 10/300 column (GE Healthcare) equilibrated in size-exclusion chromatography buffer (20 mM Tris/HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT). Size-exclusion chromatography was performed under isocratic conditions at a flow rate of 0.4 ml/min, and the relevant fractions containing MIS12C complex were pooled, concentrated, flash-frozen in liquid N2, and stored at −80°C.
MIS12C–KNL12106–2316 was expressed in BL21(DE3)Rosetta cells containing the pST39-MIS12C full-length and pET28a-GST-KNL12106–2316 fragment plasmids. Cell growth and protein expression and the initial steps of purification were performed as for MIS12C. After elution from Ni-NTA agarose beads with 250 mM imidazole, the eluate was dialyzed against GST-binding buffer (20 mM Tris/HCl, pH 8, 300 mM NaCl, 10% [vol/vol] glycerol, 1 mM EDTA, and 1 mM DTT) and applied to glutathione Sepharose 4 Fast Flow beads (GE Healthcare) preequilibrated in the GST-binding buffer for a further 3 h. Beads were washed in 30 vol of GST-binding buffer, and GST was cleaved by overnight incubation with PreScission protease (GE Healthcare) at 4°C. The eluate containing the MIS12C–KNL12106–2316 complex was dialyzed against ion-exchange buffer A and applied to a ResQ column preequilibrated in the same buffer. After elution from the ResQ column, further purification by size-exclusion chromatography and subsequent storage were performed as described above for the isolated MIS12C.
KNL12106–2316–NSL1227–281 expression was performed in BL21(DE3)pLysS cells containing pGEX-6P-2rbs-KNL12106–2316–NSL1227–281 vector. Cell pellets (obtained as described above for other constructs) were resuspended in GST-binding buffer supplemented with protease inhibitor cocktail, lysed by sonication, and cleared by centrifugation at 45,000 g for 60 min. The cleared lysate was applied to glutathione Sepharose 4 Fast Flow beads preequilibrated in the GST-binding buffer and treated as described above for equivalent GST fusion construct. The eluate was dialyzed against ion-exchange buffer A and applied to a ResQ column preequilibrated in the same buffer. The NSL1227–281 peptide was collected in flow-through fractions, and the bound KNL12106–2316 protein was eluted by a linear gradient from 30 to 350 mM NaCl in 20 bed column volumes. The fractions containing the target protein were concentrated in 3-kD (NSL1227–281) or 10-kD (KNL12106–2316) molecular mass cut-off Vivaspin concentrators and applied to a Superdex 75 10/300 column equilibrated in size-exclusion chromatography buffer as described above.
For SPC2457–197–SPC2570–224, SPC24104–197–SPC2599–224, HP1-α, and HP1110–191 (chromoshadow domain) expression, BL21(DE3)pLysS cells containing the relevant plasmids were grown in Luria-Bertani or Terrific broth at 37°C to an OD600 of ∼0.6–0.8. Protein expression was induced with 0.3 mM ITPG at 20°C, and cells were incubated overnight. Cell pellets were resuspended in GST-binding buffer supplemented with protease inhibitor cocktail, lysed by sonication, and cleared by centrifugation. The cleared lysate was applied to, and eluted from, glutathione Sepharose 4 Fast Flow beads as described above for equivalent GST constructs. The supernatants containing the proteins of interest were concentrated in Vivaspin concentrators and applied to a Superdex 75 10/300 column equilibrated in size-exclusion chromatography buffer. Size-exclusion chromatography and subsequent storage were performed as described above for other constructs.
Hi5 insect cells infected with the pFL derivative driving NDC80C expression were harvested by centrifugation and lysed by sonication in buffer A supplemented with protease inhibitors. The lysate was cleared by centrifugation at 45,000 g for 60 min. The cleared lysate was applied to Ni-NTA agarose beads preequilibrated in buffer A and incubated for 2 h. Beads were washed three times in buffer B (20 mM Tris/HCl, pH 8.0, 1 M NaCl, 5% [vol/vol] glycerol, 20 mM imidazole, and 2 mM 2-mercaptoethanol), and the bound protein was eluted in buffer C (20 mM Tris/HCl, pH 8.0, 300 mM NaCl, 5% [vol/vol] glycerol, 250 mM imidazole, and 2 mM 2-mercaptoethanol). The eluate was supplemented with 1 mM EDTA, concentrated in 10-kD molecular mass cut-off Vivaspin concentrators, and applied to a Superose 6 10/300 column equilibrated in size-exclusion chromatography buffer. Size-exclusion chromatography and subsequent storage were performed as described above.
Analytical size-exclusion chromatography
Analytical size-exclusion chromatography experiments were performed on calibrated Superdex 75 or Superdex 200 10/300 columns or on Superdex 75 PC 3.2/30 columns. All samples were eluted under isocratic conditions at 4°C in size-exclusion chromatography buffer at a flow rate of 0.4 ml/min. Elution of proteins was monitored at 280 nm. To detect complex formation, proteins were mixed at the indicated concentrations (in 200 µl for runs in 10/300 columns or 50 µl for runs in 75 PC 3.2/30 column), incubated for 1–3 h on ice, and then subjected to chromatography. Fractions were collected and analyzed by SDS-PAGE and Coomassie staining.
GST pull-down experiments
GST fusion proteins were expressed and purified as described in Protein expression and purification. Purified GST-KNL1 C-terminal fragments were incubated with their interacting partners at 4°C for 1 h in a total volume of 50 µl. After incubation and extensive washing, the products remaining associated with the beads were separated by SDS-PAGE and detected by Coomassie staining or immunoblotting.
Fluorescence anisotropy
Fluorescence anisotropy measurements were performed with an Infinite F200 instrument (Tecan) at 20°C. Fixed concentrations (10 nM) of fluorescein-labeled NSL1 peptides (synthesized by Mimotopes) were mixed with increasing concentrations of the indicated SPC24–SPC25 or HP1 constructs in PBS buffer, and reaction mixtures were allowed to equilibrate overnight at 4°C. Fluorescein was excited with polarized light at 485 nm, and the emitted light was detected at 535 nm through both horizontal and vertical polarizers. The Kd was determined by fitting the fluorescence polarization data to the equation Pobs = Pmax[C]/([C] + Kd), where Pobs is the observed fluorescence polarization signal, Pmax is the saturation value of polarization (with all the peptide in complex with the protein), and [C] is the protein concentration.
Microtubule cosedimentation assay
Tubulin was purchased from Cytoskeleton, Inc., and microtubules were polymerized according to the producer’s instructions. For microtubule-binding reactions, 2 µM of microtubules was diluted in general tubulin buffer (80 mM Pipes, pH 6.8, 1 mM MgCl2, 1 mM EGTA, and 50 µM paclitaxel) supplemented with 50 mM NaCl. 1 µM NDC80C, 1 µM BSA, and the indicated concentrations of MIS12CNSL1-258 were added to a final reaction volume of 50 µl. Reactions were incubated at room temperature for 10 min, transferred onto 100 µl of cushion buffer (80 mM Pipes, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 50 µM paclitaxel, and 50% glycerol), and ultracentrifuged for 10 min at 90,000 rpm in a TLA100 rotor (Beckman Coulter) at 25°C. Pellets and supernatants were analyzed by SDS-PAGE.
Negative-stain EM
MIS12 complexes were prepared for EM by the GraFix protocol (Kastner et al., 2008), which combines mild chemical stabilization and glycerol gradient centrifugation. The fractionated complexes were adsorbed to a thin film of carbon and then transferred to an electron microscopic grid covered with a perforated carbon film. The bound MIS12C or MIS12C–SPC2457–197–SPC2570–224 and particles were stained with 2% uranyl formate, blotted, and air dried. Images were recorded at a magnification of 155,000 on a 4k × 4k charge-coupled device camera (TVIPS GmbH) using twofold pixel binning (1.8 Å/pixel) in an electron microscope (CM200 FEG; Philips/FEI) operated at 160 kV acceleration voltage. 5,389 particle images (4,969 particles for the complex of MIS12C with SPC24–SPC25) were interactively selected and subjected to several rounds of alignment (Sander et al., 2003) and classification (van Heel and Frank, 1981) until stable class averages were obtained.
Sedimentation velocity AUC
Sedimentation velocity experiments were performed on an analytical ultracentrifuge (Optima XL-A; Beckman Coulter) using conventional charcoal-filled epon double-sector quartz cells in an An-60 Ti rotor (Beckman Coulter). The rotor speed was 40,000 rpm, and the temperature was maintained at 20°C. Before centrifugation, samples were prepared by dialysis against the buffer blank solution (20 mM Tris/HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT). During experiments, radial absorbance scans (λ = 280 nm) were collected at time intervals of 5 min, and 150 scans were used in the analysis. The data recorded from moving boundaries were analyzed using the software SEDFIT (Schuck, 2000) in terms of continuous distribution function of sedimentation coefficients (c(S)).
ITC
All samples were extensively dialyzed into fresh buffer (20 mM Tris/HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT). ITC measurements were performed at 25°C on a VP-ITC MicroCalorimeter (MicroCal). In each titration, the protein in the cell (at a 4–5-µM concentration) was titrated with 35 10-µl injections (at 240-s intervals) of protein ligand (at 10-fold higher molar concentration). The injections were continued beyond saturation levels to allow for determination of heats of ligand dilution. Data were fitted by least-square procedures to a single-site binding model using ORIGIN 5.0 software package (MicroCal).
Cell culture and plasmids
HeLa cells were grown in DME (EuroClone) supplemented with 10% fetal bovine serum (HyClone) and 2 mM l-glutamine. Nocodazole and S-trityl-l-cysteine (Sigma-Aldrich) were used at 3.3 µM and 5 µM, respectively. HeLa cells were transfected with FuGENE 6 Transfection Agent (Roche). For GFP-NSL1 cloning, GFP cDNA was cloned into pCDNA5-FRT/TO vector previously modified to carry an internal ribosomal entry site (IRES) sequence to have pCDNA5FRT/TOGFP-IRES vector. Human NSL1 cDNA was cloned into pCDNA5FRT/TOGFP-IRES vector to express all GFP-NSL1 constructs. Empty pCDNA5FRT/TOGFP-IRES was used for GFP expression as a control. All constructs were checked by DNA sequencing.
Immunoprecipitation and immunoblotting
For immunoprecipitation, mitotic cells were harvested by shake-off and lysed in lysis buffer (150 mM KCl, 75 mM Hepes, pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 15% glycerol, 0.1% NP-40, and protease inhibitors [EMD]). Protein extracts were incubated with rabbit anti-GFP polyclonal antibody (produced at the Italian Foundation for Cancer Research Institute of Molecular Oncology–European Institute of Oncology [IFOM–IEO] campus antibody facility) cross-linked to protein–A beads (GE Healthcare) for 2 h at 4°C. The following antibodies were used for immunoblotting: mouse antivinculin (working dilution 1:100,000; Sigma-Aldrich), rabbit anti–P-S10-H3 (working dilution 1:1,000; Abcam), rabbit anti-GFP (working dilution 1:5,000; produced at the IFOM–IEO campus antibody facility), mouse anti-HEC1 (human NDC80; working dilution 1:1,000; clone 9G3.23; GeneTex, Inc.), and rabbit anti-hMIS12 (working dilution 1:1,500; Abcam).
Immunofluorescence and quantification
Immunofluorescence microscopy was performed according to Santaguida et al. (2010). The following antibodies were used: anticentromeric antibody (working dilution 1:100; Antibodies Inc.) and mouse anti-HEC1 (human NDC80; working dilution 1:1,000; clone 9G3.23; GeneTex, Inc.). Cy3- and Cy5-labeled secondary antibodies for immunofluorescence were purchased from Jackson ImmunoResearch Laboratories, Inc. and used at 1:100 dilution. Cells were imaged using a confocal microscope (TCS SP2; Leica) equipped with a 63× NA 1.4 objective lens using the LCS 3D software (Leica). Images shown in Fig. 5 C were acquired as z sections at 0.25 µm and converted into maximal intensity projections using ImageJ (National Institutes of Health). Measurements of kinetochore intensities were performed using intensity projections of images with Imaris software (Bitplane AG). Kinetochore intensity was calculated by subtracting the mean background signal of the chromosome area of a cell and normalized for the CREST signal of the same kinetochore.
Cross-linking
The protein cross-linking method has been described in detail previously (Maiolica et al., 2007).
Online supplemental material
Fig. S1 shows additional properties of MIS12C–NDC80C–KNL1. Fig. S2 shows that NDC80C does not bind tightly to the PVIHL motif of NSL1. Fig. S3 shows additional characterization of the role of ZWINT. Fig. S4 shows multiple sequence alignment of the C-terminal region of KNL1. Fig. S5 shows multiple sequence alignment of NSL1. Table S1 lists the nomenclature of KMN subunits in different species. Table S2 shows a summary of cross-linking data analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002070/DC1.
Acknowledgments
The AUC data were collected by the late J. Eccleston (National Institute for Medical Research, London, England, UK), and A. Petrovic would like to dedicate this work to his memory.
We thank T. Kiyomitsu, M. Yanagida, and members of the Musacchio laboratory for helpful discussions and S. Ramachandra and F. Çivril for generating initial expression constructs for ZWINT.
Work in the Musacchio laboratory is funded by the Association for International Cancer Research, the Telethon Foundation, the European Union’s Sixth Framework Program 3D-Repertoire, the European Union’s Seventh Framework Program European Research Council grant KINCON, the Italian Association for Cancer Research, the Fondo di Investimento per la Ricerca di Base, the Cariplo Foundations, the Human Frontier Science Program, and Programmi Integrati di Oncologia Strategici 7/07. Work in the Stark laboratory is supported by the Bundesministerium für Bildung und Forschung and the European Union’s Sixth Framework Program 3D-Repertoire. S. Pasqualato and S. Santaguida are supported by fellowships from the Italian Foundation for Cancer Research.
Footnotes
Abbreviations used in this paper:
- AUC
- analytical ultracentrifugation
- CCAN
- constitutive centromere-associated network
- IRES
- internal ribosomal entry site
- ITC
- isothermal titration calorimetry
References
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., Fukagawa T. 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186:173–182 10.1083/jcb.200903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S., Yang H., Nozaki N., Okazaki T., Yoda K. 2002. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22:2229–2241 10.1128/MCB.22.7.2229-2241.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C., Richmond T.J., Berger I. 2008. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr. Protoc. Protein Sci. Chapter 5:Unit 5.20. [DOI] [PubMed] [Google Scholar]
- Brasher S.V., Smith B.O., Fogh R.H., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19:1587–1597 10.1093/emboj/19.7.1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., Straight A.F. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., Straight A.F. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Niessen S., Anderson S., Hyndman F., Yates J.R., III, Oegema K., Desai A. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268 10.1101/gad.1234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C., De Luca J., Monzani S., Ferrari K.J., Ristic D., Wyman C., Stark H., Kilmartin J., Salmon E.D., Musacchio A. 2005. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 280:29088–29095 10.1074/jbc.M504070200 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., Simons K.T. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491 10.1091/mbc.E08-03-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A.D., Sorger P.K. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17:2902–2921 10.1101/gad.1144403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., McEwen B.F. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531 10.1091/mbc.E04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K.J., Meng X., Khodjakov A., McEwen B.F. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9:516–522 10.1038/ncb1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., McCleland M.L., Satinover D.L., Stukenberg P.T. 2005. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol. Biol. Cell. 16:4882–4892 10.1091/mbc.E05-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., Straight A.F. 2008. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 183:805–818 10.1083/jcb.200806038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., III, Cleveland D.W. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Gestaut D.R., Graczyk B., Cooper J., Widlund P.O., Zelter A., Wordeman L., Asbury C.L., Davis T.N. 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10:407–414 10.1038/ncb1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180:1101–1114 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347–3362 10.1242/jcs.00645 [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D.C., Molk J.N., Bloom K.S., Salmon E.D. 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8:581–585 10.1038/ncb1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E.D., Bloom K.S. 2008. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 181:587–594 10.1083/jcb.200803027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., Salmon E.D. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston K., Joglekar A., Hori T., Suzuki A., Fukagawa T., Salmon E.D. 2010. Vertebrate kinetochore protein architecture: protein copy number. J. Cell Biol. 189:937–943 10.1083/jcb.200912022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B., Fischer N., Golas M.M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., et al. 2008. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 5:53–55 10.1038/nmeth1139 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Iwasaki O., Obuse C., Yanagida M. 2010. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J. Cell Biol. 188:791–807 10.1083/jcb.200908096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline S.L., Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173:9–17 10.1083/jcb.200509158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R., III, Tagaya M., Cleveland D.W. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60 10.1083/jcb.200411118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., Fukagawa T. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168 10.1091/mbc.E07-01-0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Hornung P., Westermann S. 2010. The Dam1 complex confers microtubule plus end–tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189:641–649 10.1083/jcb.200912021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Rattner J.B., Jablonski S.A., Yen T.J. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J.R., III, He X. 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24:2919–2930 10.1038/sj.emboj.7600762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G., Wallrath L., Urrutia R. 2006. The Heterochromatin Protein 1 family. Genome Biol. 7:228 10.1186/gb-2006-7-7-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., Rappsilber J. 2007. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics. 6:2200–2211 10.1074/mcp.M700274-MCP200 [DOI] [PubMed] [Google Scholar]
- Maresca T.J., Salmon E.D. 2009. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 184:373–381 10.1083/jcb.200808130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell D.P., Hu X.-W., Singleton M.R. 2010. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 190:823–834 10.1083/jcb.201002059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., Gardner R.D., Kallio M.J., Daum J.R., Gorbsky G.J., Burke D.J., Stukenberg P.T. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114 10.1101/gad.1040903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., Grishchuk E.L., Morphew M.K., Efremov A.K., Zhudenkov K., Volkov V.A., Cheeseman I.M., Desai A., Mastronarde D.N., Ataullakhanov F.I. 2008. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 135:322–333 10.1016/j.cell.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., McAinsh A.D., Rheinbay E., Sorger P.K. 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7:R23 10.1186/gb-2006-7-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami Y., Hori T., Kimura H., Fukagawa T. 2005. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol. Cell. Biol. 25:1958–1970 10.1128/MCB.25.5.1958-1970.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks K.J., Moree B., Straight A.F. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 20:4246–4255 10.1091/mbc.E09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.J., De Wulf P., Sorger P.K., Harrison S.C. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nekrasov V.S., Smith M.A., Peak-Chew S., Kilmartin J.V. 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:4931–4946 10.1091/mbc.E03-06-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135–1141 10.1038/ncb1187 [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., III, Desai A., Fukagawa T. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Pagliuca C., Draviam V.M., Marco E., Sorger P.K., De Wulf P. 2009. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS One. 4:e7640 10.1371/journal.pone.0007640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 107:323–337 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- Pidoux A.L., Allshire R.C. 2005. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:569–579 10.1098/rstb.2004.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B.A., Tatsutani S.Y., Collins K.A., Biggins S. 2003. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell. 5:735–745 10.1016/S1534-5807(03)00322-8 [DOI] [PubMed] [Google Scholar]
- Przewloka M.R., Zhang W., Costa P., Archambault V., D’Avino P.P., Lilley K.S., Laue E.D., McAinsh A.D., Glover D.M. 2007. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One. 2:e478 10.1371/journal.pone.0000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S.A., Vagnarelli P., Dong Y., Hori T., McEwen B.F., Fukagawa T., Flors C., Earnshaw W.C. 2010. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA. 107:10484–10489 10.1073/pnas.1002325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander B., Golas M.M., Stark H. 2003. Corrim-based alignment for improved speed in single-particle image processing. J. Struct. Biol. 143:219–228 10.1016/j.jsb.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Heeger S., Althoff F., Walter A., Heidmann S., Mechtler K., Lehner C.F. 2007. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 116:385–402 10.1007/s00412-007-0103-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Chaleckis R., Lehner C.F. 2009. Intrakinetochore localization and essential functional domains of Drosophila Spc105. EMBO J. 28:2374–2386 10.1038/emboj.2009.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Bryson T.D., Henikoff S. 2004. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3:18 10.1186/jbiol11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Kern R.C., Selleck W. 2005. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40:385–395 10.1016/j.pep.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Tien J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., Davis T.N. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S., Perini G., Bernardoni R., Zoli M., Reese J.C., Musacchio A., Della Valle G. 2009. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS One. 4:e5832 10.1371/journal.pone.0005832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K.S., Takagaki K., Kumada K., Hirayama Y., Noda T., Hirota T. 2009. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 184:383–390 10.1083/jcb.200811028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M., Frank J. 1981. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 6:187–194 [DOI] [PubMed] [Google Scholar]
- Vorozhko V.V., Emanuele M.J., Kallio M.J., Stukenberg P.T., Gorbsky G.J. 2008. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma. 117:169–179 10.1007/s00412-007-0135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.W., Long S., Ciferri C., Westermann S., Drubin D., Barnes G., Nogales E. 2008. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J. Mol. Biol. 383:894–903 10.1016/j.jmb.2008.08.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Sorger P.K., Harrison S.C. 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 102:5363–5367 10.1073/pnas.0501168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Schnell J.R., Larsen N.A., Sorger P.K., Chou J.J., Harrison S.C. 2006. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 14:1003–1009 10.1016/j.str.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Wei R.R., Al-Bassam J., Harrison S.C. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14:54–59 10.1038/nsmb1186 [DOI] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I.M., Anderson S., Yates J.R., III, Drubin D.G., Barnes G. 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163:215–222 10.1083/jcb.200305100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H.W., Niederstrasser H., Wong J., Drubin D.G., Nogales E., Barnes G. 2005. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290 10.1016/j.molcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Westermann S., Drubin D.G., Barnes G. 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76:563–591 10.1146/annurev.biochem.76.052705.160607 [DOI] [PubMed] [Google Scholar]
- Whitty A. 2008. Cooperativity and biological complexity. Nat. Chem. Biol. 4:435–439 10.1038/nchembio0808-435 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kilmartin J.V. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360 10.1083/jcb.152.2.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Kubalek E.M., Cheeseman I.M., Yoshioka C., Desai A., Milligan R.A. 2008. Orientation and structure of the Ndc80 complex on the microtubule lattice. J. Cell Biol. 182:1055–1061 10.1083/jcb.200804170 [DOI] [PMC free article] [PubMed] [Google Scholar]