Folding of gene loci organized in tandem arrays reveals variation of chromatin structure as a function of cell differentiation.
Abstract
Interphase chromatin compaction well above the 30-nm fiber is well documented, but the structural motifs underlying this level of chromatin folding remain unknown. Taking a reductionist approach, we analyzed in mouse embryonic stem (ES) cells and ES-derived fibroblasts and erythroblasts the folding of 10–160-megabase pair engineered chromosome regions consisting of tandem repeats of bacterial artificial chromosomes (BACs) containing ∼200 kilobases of mammalian genomic DNA tagged with lac operator (LacO) arrays. Unexpectedly, linear mitotic and interphase chromatid regions formed from noncontiguously folded DNA topologies. Particularly, in ES cells, these model chromosome regions self-organized with distant sequences segregating into functionally distinct, compact domains. Transcriptionally active and histone H3K27me3-modified regions positioned toward the engineered chromosome subterritory exterior, with LacO repeats and the BAC vector backbone localizing within an H3K9me3, HP1-enriched core. Differential compaction of Dhfr and α- and β-globin transgenes was superimposed on dramatic, lineage-specific reorganization of large-scale chromatin folding, demonstrating a surprising plasticity of large-scale chromatin organization.
Introduction
Work from several laboratories over the past 10–15 yr has revealed several principles for genome organization, which appear to apply in a wide range of species. First is the organization of interphase chromosomes into spatially distinct chromosome territories (Cremer and Cremer, 2001; Cremer et al., 2006). The overall compaction level and intranuclear location varies as a function of gene density for both entire chromosomes (Croft et al., 1999) and specific chromosomal regions (Mahy et al., 2002a; Küpper et al., 2007). Generally decreased compaction and a more interior location within the nucleus is observed with increased gene density, although an interesting inversion of this differential localization has recently been described for retinal cells in the eyes of nocturnal mammals (Solovei et al., 2009).
Second, although transcription occurs throughout the chromosome territory, some genes appear to be preferentially localized toward the chromosome territory periphery (Kurz et al., 1996; Dietzel et al., 1999; Nogami et al., 2000). More generally, transcriptionally active sites within chromosome territories have been reported to localize to the periphery of chromatin subdomains within chromosome territories or to less condensed regions lying between these more condensed subdomains (Visser et al., 1998; Cmarko et al., 1999; Verschure et al., 1999; Mahy et al., 2002b). In mammals, genes that escape X chromosome inactivation are reported to localize at the periphery or loop out from a DAPI-dense Barr body enriched in repetitive sequences and coated with Xist RNA (Chaumeil et al., 2006; Clemson et al., 2006). Third, for highly expressed genes or gene-rich chromosomal regions, extension of these regions within several megabase pair loops well outside the chromosome territory occurs in a significant fraction of cells (Volpi et al., 2000; Mahy et al., 2002a; Chambeyron and Bickmore, 2004). Fourth, this looping outwards from the chromosome territory may be related to the observed association of active genes with “transcription factories,” nuclear regions enriched in RNA polymerase II (Osborne et al., 2004), and/or nuclear speckles (Brown et al., 2008; Hu et al., 2009).
Work aimed at understanding the molecular basis for these phenomena has been difficult because they involve the folding and intranuclear positioning of megabase pairs or larger chromosomal regions. A reductionist approach, although technically challenging, is therefore warranted. Toward this end, we recently demonstrated the use of multicopy insertions of large bacterial artificial chromosome (BAC) transgenes containing 130–220 kbp of mouse or human genomic regions for visualizing the large-scale chromatin folding of inducible genes in CHO cells (Hu et al., 2009). These 1–3 Mbp engineered chromosome regions not only folded into large-scale chromatin structures resembling surrounding euchromatic chromosome regions but also showed a gene-specific localization to nuclear speckles after transcriptional induction.
In this study, we extend this experimental approach to create engineered chromosome regions tens to hundreds of megabase pairs in size formed entirely by tandem arrays of specific, ∼200 kbp gene loci. We apply this approach to compare and contrast the folding of the α-globin, β-globin, and Dhfr gene loci in undifferentiated mouse embryonic stem (ES) cells versus ES cell–derived fibroblasts and erythroblasts. We show that several of the aforementioned properties of genome architecture are recapitulated in the self-assembly of these engineered chromosome regions. We also demonstrate a dramatic plasticity of large-scale chromatin structure, varying as a function of cell differentiation, with linear interphase configurations formed from nonlinearly compacted, topologically complex, looping architectures.
Results
Experimental approach
Previous work in CHO cells analyzed the folding of large transgenes engineered from BACs encompassing three inducible loci, Dhfr, Hsp70, and MT, that were retrofitted by insertion of a 256-mer lac operator (LacO) repeat (Hu et al., 2009). These BAC transgenes assembled into apparently linear large-scale chromatin fibers with mean linear compaction ratios of ∼1,000:1 and ∼330:1 for the transcriptionally uninduced and induced states, respectively, well above the ∼40-fold linear compaction ratio of 30-nm chromatin fibers. The number of GFP foci was two- to fourfold lower than the estimated number of integrated BAC copies, which was interpreted as a result of variable compaction along the large-scale chromatin fiber.
In this study, we followed a similar approach to examine changes in large-scale chromatin structure accompanying cell differentiation. We chose two well-studied developmentally regulated gene loci, the human α- and β-globin gene clusters, and the housekeeping mouse Dhfr locus, which is active in proliferating cells (Fig. 1 A). The β-globin BAC is transcriptionally silent in nonerythroid cells, whereas the α-globin BAC contains several active, housekeeping genes. Differential behavior of α-globin versus β-globin regulatory sequences may be related to their different chromosome environments (Craddock et al., 1995; Brown et al., 2001). The third BAC contains the mouse Dhfr locus and Msh3 genes expressed from a single divergent promoter in proliferating cells.
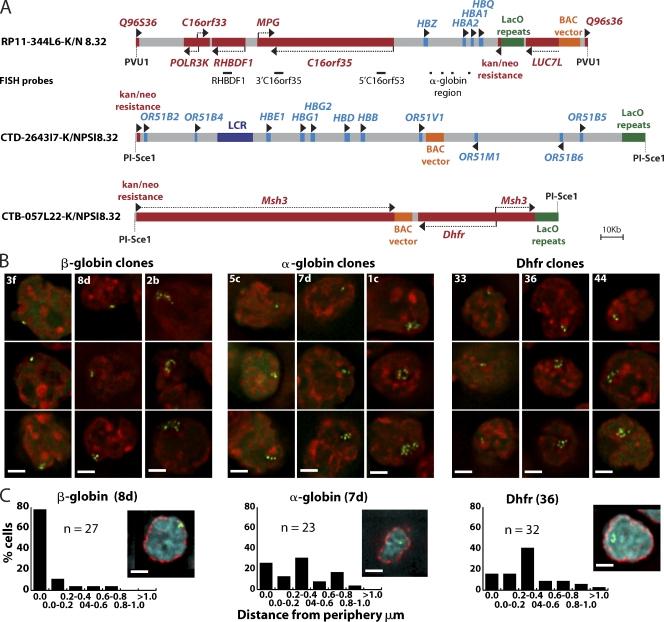
Figure 1.
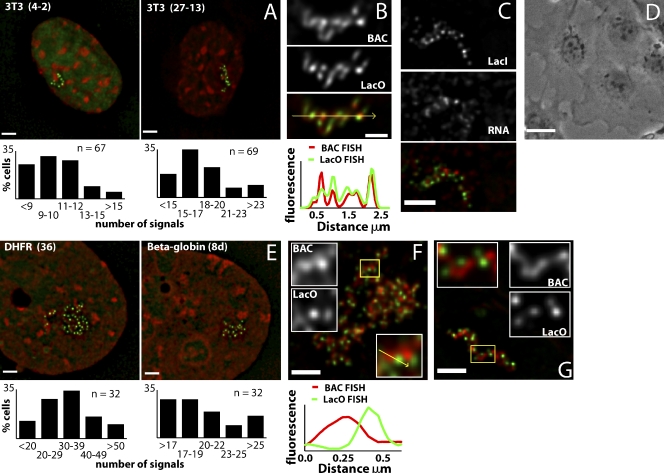
Construction and initial characterization of BAC transgenes. (A) BAC diagrams show positions of genes, regulatory elements, transposons with LacO arrays, vector sequences, and FISH probes (red, active genes; light blue, genes inactive in ES cells). (B) Deconvolved optical sections of EGFP-LacI signals (green) counterstained with DAPI (red) in different transgenic ES cell lines. (C) Deconvolved optical sections of EGFP-LacI (green), anti–nuclear pore immunostaining (red), and DAPI DNA staining (blue). Histograms of the shortest distances between EGFP-LacI spots and the nuclear periphery for one clone of each BAC type. Bars, 2 µm.
BACs with LacO arrays inserted away from the α- and β-globin and Dhfr genes and known regulatory sequences (Fig. 1 A) were linearized before transfection. Stable ES clones were initially screened for the presence of LacO sequences by transient expression of an EGFP-dimeric LacI nuclear localization sequence fusion protein (EGFP-LacI; Robinett et al., 1996). In further experiments, we used the EGFP-LacI lentivirus construct FULGW to transduce ES and differentiated cells, producing lines in which up to 90% of cells expressed EGFP-LacI.
Initial characterization of transgenic cell lines
Signal configurations produced by EGFP-LacI binding to LacO arrays included single spots, clusters of spots, or extended curvilinear strings of spots (Fig. 1 B) in different ES cell clones, similar to those observed for multicopy BAC insertions in CHO cells (Hu et al., 2009).
Interestingly, the intranuclear localization of the β-globin BAC transgene reproduced the extreme peripheral localization of the endogenous β-globin locus in undifferentiated mouse ES cells (Hepperger et al., 2008). In ES clones containing β-globin BACs, the EGFP signal partially overlapped nuclear pore staining in >50% of cells. This peripheral localization was specific to the β-globin BAC (Fig. 1 C, Fig. S1; and Table I).
Table I.
Size, expression characteristics, and nuclear position of BAC transgenes in ES cells
| BAC clone | Insert copy number/size estimate | Expression relative to the endogenous locusa | Cells with LacI signal touching the nuclear periphery | Mean/median distance of LacI signal from nuclear periphery |
| % | % | µm | ||
| β-Globin 2b | 893 (205 Mb) | NA | 84 (n = 32) | 0.053/0.00 |
| β-Globin 8d | 158 (36.3 Mb) | NA | 78 (n = 27) | 0.075/0.00 |
| β-Globin 3f | 54 (12.4 Mb) | NA | 52 (n = 27) | 0.101/0.00 |
| α-Globin 1c | 609 (115 Mb) | 11.8 | 16 (n = 25) | 0.34/0.35 |
| α-Globin 7d | 105 (20 Mb) | 6.6 | 26 (n = 23) | 0.323/0.312 |
| α-Globin 5c | 56 (12.5 Mb) | 7.3 | 4 (n = 23) | 0.58/0.485 |
| Dhfr 36 | 278 (52.8 Mb) | 52.8 | 16 (n = 32) | 0.359/0.261 |
| Dhfr 44 | 271 (51.5 Mb) | 35.3 | 7 (n = 28) | 0.340/0.377 |
| Dhfr 33 | 111 (21 Mb) | 11.1 | 15 (n = 27) | 0.445/0.384 |
NA, not applicable.
Based on reverse transcription quantitative PCR data for C16orf33 (α-globin clones) and Dhfr (Dhfr clones).
Copy number determination reveals a fundamentally different conformation of BAC transgenes in undifferentiated ES cells
The number, position, and stability of BAC transgene chromosome insertion sites in ES cell clones were determined by hybridization of BAC DNA to metaphase spreads. Copy number estimates for small insertions were made by counting the maximum spot number visualized by FISH on stretched chromatin fibers (fiber-FISH) using LacO DNA as a probe (Fig. 2, A, B, and E; and Table I). Spacing of LacO spots in the fiber-FISH was relatively uniform, which is consistent with tandem BAC arrays. Copy number estimates for larger inserts were derived from ratios of the length of the metaphase chromosome band containing the multicopy BAC to the length of the longest mouse chromosome. Copy number estimates using these ratios were normalized using measurements from β-globin clone 8d, whose copy number was estimated by fiber-FISH (Fig. 2 B and Table I).
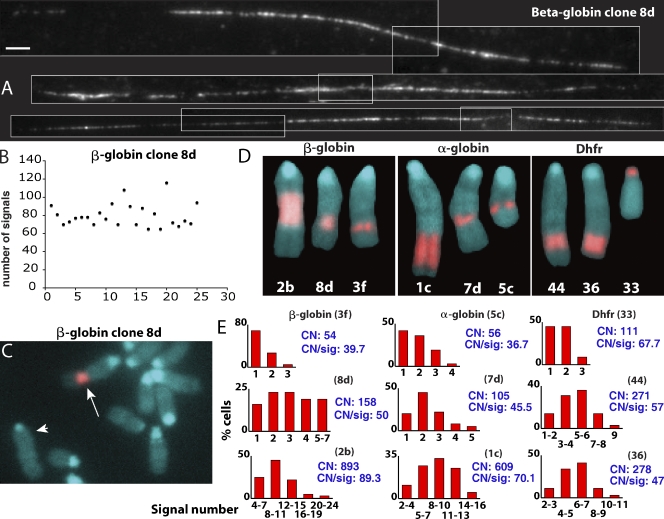
Figure 2.
Transgene copy number estimation. (A) Fiber FISH examples from β-globin clone 8d hybridized with LacO sequences. White lines outline individual images assembled into montages. Bar, 2 µm. (B) Scatter plot showing LacO fiber FISH signal numbers (y axis) in different fiber FISH examples (x axis) for β-globin clone 8d. Variation in signal numbers was related to incomplete stretching, with the maximum signal number used for copy number estimation. (C and D) DAPI (blue) and FISH BAC probe (red) are shown. (C) Partial metaphase spread from β-globin clone 8d, BAC transgene insertion (arrow), longest mouse chromosome used for normalization (arrowhead; see Results). (D) Metaphase chromosomes showing BAC insert size and location for each cell clone selected for further analysis. (E) Histograms showing number of EGFP-LacI signals (x axis) counted in maximum intensity projections of 64 interphase nuclei from each cell clone. Copy number estimates (CN) and mean copy number per GFP signal (CN/sig) are shown in blue.
Three ES clones for each BAC were selected for further analysis (Fig. 1 B, Fig. 2 D, and Table I). All clones were >90% positive for the pluripotency marker Oct-4 and >90% negative for lamin A, a marker for cell differentiation (unpublished data). Copy number per signal was calculated from the BAC copy estimates and the mean number of LacI-EGFP signals counted in maximum intensity projections.
Surprisingly, the BAC copy number per EGFP spot, ranging from 40–89 in these ES cell clones (Fig. 2 E), was >10 fold higher than observed in CHO cells (Hu et al., 2009). With each GFP spot derived from ∼8–16 Mbp of total BAC DNA, the small size of the EGFP-LacI signals in ES cells therefore appeared incompatible with colinear compaction of BAC transgenes along a large-scale chromatin fiber.
Long-distance cis interactions lead to spatial segregation of specific BAC sequences in ES cells within HP1, H3K9me3 enriched regions
To investigate this unexpected coalescence of LacO signals, we used dual-color FISH. In all clones, the LacO FISH signal closely resembled the EGFP-LacI signals, independent of whether the cells expressed EGFP-LacI. In contrast, the BAC FISH signal extended over a much larger area (Fig. 3, A–C; and Fig. S2 A). Control experiments including RNase treatment and hybridization to nondenatured cells excluded significant contribution from residual RNA to the DNA FISH signal (unpublished data).
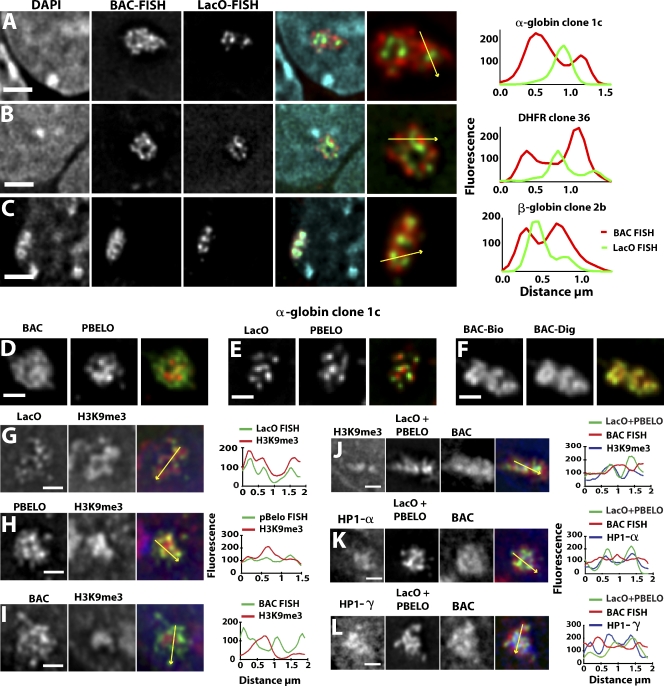
Figure 3.
Positions of LacO and BAC vector sequences relative to other BAC sequences. (A–C) 3D FISH with LacO (green), BAC probes (red), DAPI (blue) for α-globin clone 1c (A), Dhfr clone 36 (B), and β-globin clone 2b (C). (right) Intensity line scans along arrows are plotted. Bars, 2 µm. (D–F) 3D FISH for α-globin clone 1c BAC transgenes hybridized with BAC (green) and PBELO BAC vector backbone (red) probes (D), LacO (green) and PBELO probes (red; E), the same BAC probe labeled with biotin (BAC-Bio; red), or digoxigenin (BAC-Dig; green; F). (G–I) Immuno-3D FISH for α-globin clone 1c BAC transgenes with LacO probe (green) versus H3K9me3 staining (red; G), PBELO vector backbone probe (green) versus H3K9me3 staining (red; H), and BAC probe (green) versus H3K9me3 staining (red; I). (J–L) Three-color immuno-3D FISH with a combined LacO and PBELO vector backbone probe (green), BAC probe (red), and immunostaining (blue) against H3K9me3 (J), HP1-α (K), or HP1-γ (L). (right) Intensity line scans along arrows are plotted. Bars, 1 µm.
In cell clones containing the transcriptionally active α-globin and Dhfr BACs, BAC and LacO hybridization domains were significantly segregated from each other, with BAC staining foci separated by up to 0.5 µm from neighboring LacO signals. Line scans showed that BAC FISH signals peaked in regions with low-intensity LacO FISH signals (Fig. 3, A and B), and red (BAC) and green (LacO) fluorescence correlation coefficients were all <0.6 with means of 0.35 for α-globin and 0.34 for Dhfr clones (Fig. S2 B).
In contrast, although the transcriptionally inactive β-globin BAC sequences still showed separation from LacO foci, there was significantly more overlap of the BAC signal with the LacO foci (Fig. 3 C, right; and Fig. S2 A). Mean BAC-LacO fluorescence correlation coefficient values increased from the 0.34–0.35 values observed for α-globin and Dhfr clones to 0.59 for β-globin clone 2b. Likewise, the distribution of BAC-LacO fluorescence correlation coefficient values for β-globin clone 2b was noticeably skewed to increased values relative to the distributions for the α-globin and Dhfr clones (Fig. S2 B). Moreover, the β-globin BAC transgenes appeared noticeably more condensed than the α-globin and Dhfr BAC transgenes, with the β-globin BAC FISH signal coinciding with a distinctive DAPI-intense region (Fig. 3 C).
Coalescence of homologous sequences from different BAC copies was not restricted to the highly repetitive LacO direct repeats. Vector backbone FISH signals were also compact like the LacO signals and showed similar segregation from the BAC signal (Fig. 3 D). Vector backbone and LacO foci did not colocalize but were positioned close to each other (Fig. 3 E). Control experiments with the same BAC probe labeled with two colors produced a significantly higher overlap (Fig. 3 F), which is consistent with the subpixel x–y chromatic shift for these two fluorochromes.
Combined immuno-FISH experiments revealed enrichment of HP1-α and -γ and H3K9me3 immunostaining over the α-globin and Dhfr BAC transgene chromosome subterritories. H3K9me3 and HP1-α immunostaining intensities over the BAC transgene regions were well above nucleoplasmic background levels and of similar intensity to that observed over chromocenters in the same nuclei (Fig. S3, A and B). However, HP1-γ foci, slightly brighter and larger in area than other nuclear HP1-γ foci distributed throughout the nuclei, identified only BAC transgene regions (Fig. S3 C). The HP1, H3K9me3-stained region formed a core within the larger region defined by BAC DNA FISH (Fig. 3, G–L; and Fig. S3), with the combined LacO/BAC vector backbone FISH signal filling most of the H3K9me3/HP1-stained region.
Gene expression of BAC transgenes approaches levels of endogenous genes in ES cells
Despite the extensive, long-distance looping interactions for the LacO direct repeats and vector backbone sequences and their location within a heterochromatin core, we observed relatively normal regulation of the genes contained within these BAC arrays in ES cells. Expression patterns were assessed by RNA FISH using biotin-labeled BAC DNA as probes and by reverse transcription quantitative PCR (Table S1). BAC Dhfr expression per gene copy was 35 and 58% of the expression level of the endogenous Dhfr gene for the two BAC arrays inserted in the middle of chromosome arms versus 11% for the BAC array inserted within the y chromosome centromere (clone 33; Fig. 2 D). Per copy expression of human gene, C16orf33, present on the α-globin BAC, ranged from 11.8 to 7.3% of the endogenous, homologous mouse locus expression level. Expression of human β-globin genes in clone 8d could be detected only after differentiation to erythroblasts.
Peripheral versus interior differential localization of transcriptionally active versus inactive sequences in BAC transgene chromosome subterritory regions
For the transcriptionally active α-globin and Dhfr large transgene arrays, this separation of sequences was frequently characterized by a striking internal localization of LacO and BAC vector signal surrounded by a significant shell of BAC signal (Fig. 3, A and B; and Fig. S2 A). This was particularly pronounced for the large α-globin transgene array containing several housekeeping genes. LacO repeats in a similar size β-globin BAC transgene array did not show such interior partitioning, with similar numbers of LacO foci more uniformly distributed throughout the transgene chromosome subterritory (Fig. 3 C and Fig. S2 A).
In RNA FISH preparations of α-globin and Dhfr BAC arrays, EGFP-LacI foci were observed to colocalize with relatively high-intensity DAPI staining. BAC RNA signals also organized into distinct foci, but these showed significant separation from EGFP-LacI signals and localized predominantly within DAPI-poor channels (Fig. 4 A). In larger BAC arrays (α-globin 1c, Dhfr 36, and Dhfr 44 clones), a distinctive transgene conformation consisting of a central zone of LacO sequences surrounded by peripheral RNA FISH signal was observed in the majority (∼60%) of cells. This peripheral/interior segregation of RNA transcripts versus LacO signals was more pronounced than that observed for BAC versus LacO DNA FISH signals in these clones (Fig. 3, A and B; and Fig. S2 A). Although the DNA BAC FISH signal was typically distributed throughout a disk-shaped area, the RNA FISH signal instead formed more of an external shell. DAPI-depleted channels sited adjacent to LacO foci were also frequently enriched for RNA polymerase II (Fig. 4 B).
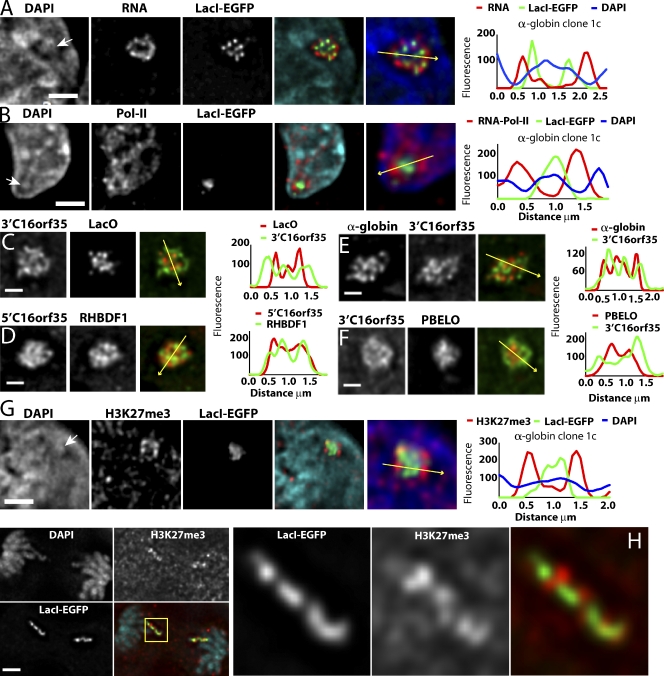
Figure 4.
Peripheral versus interior distribution of transcripts, RNA polymerase II, active versus inactive gene sequences, vector backbone, and LacO repeats relative to BAC transgene territory. (A–H) Undifferentiated, α-globin clone 1c ES cells (DAPI; blue). (right) Intensity line plots along yellow arrows are shown. (A) RNA FISH shows peripheral distribution of BAC transcripts (red) relative to EGFP-LacI spots (green). (B) RNA polymerase II immunostaining (red) relative to EGFP-LacI spots (green). (C–F) 3D DNA FISH are shown. (C) 3′ region of C16orf35 gene (green) and LacO probes are shown (red). (D) 5′ region of C16orf35 (red) and RHBDF1 (green) gene probes are shown. (E) Inactive α-globin region (red) versus active C16orf35 (green) probes are shown. (F) 3′ region of C16orf35 gene (green) versus BAC vector backbone (red) probes are shown. (G and H) Anti-H3K27me3 immunostaining (red) versus EGFP-LacI (green) in interphase (G) versus anaphase nuclei (H). (H) Boxed region indicates region magnified on the right. White arrows mark DAPI-dense regions underlying LacI signals. Bars: (A, B, G, and H) 2 µm; (C–F) 1 µm.
To further examine the spatial segregation of DNA sequences within the BAC chromosome subterritories, we created FISH probes hybridizing to different subregions of the α-globin BAC. DNA FISH signals from 4- to 5-kb probes from active genes Rhbdf1 and C16orf35 (Fig. 1 A) were relatively diffuse and showed minimal overlap with LacO signals (Fig. 4 C). Probes for different active regions clearly showed less spatial segregation from each other than from LacO sequences (Fig. 4 D). This was not simply correlated with the genomic distances separating these probes on the BAC sequence, as the genomic distance between 5′C16orf35 and RHBD1, showing close intermingling of signals (Fig. 4 D), is similar to the distance between 3′C16orf35 and LacO (Fig. 4 C), showing a spatial segregation of signals.
Close localization was observed also between a pool of five 1-kb probes spanning ∼24 kb of the inactive α-globin region with probes from different active gene regions, including 3′C16orf35 (Fig. 4 E) separated by ∼100 kb. However, hybridization of the BAC vector backbone produced signals that were compact and, like the LacO sequences, showed a strong internal localization bias relative to active gene probes (Fig. 4 F).
H3K27me3 domains show a peripheral distribution in the engineered α-globin BAC transgene territories
Although inactive in ES cells, α-globin genes appeared to colocalize with the active genes on the BAC and away from the HP1, H3K9me3-enriched region containing the LacO and vector backbone sequences. In human ES cells, α-globin HBA1 and HBA2 promoter regions are marked by histone H3 trimethyl lysine 27 (H3K27me3) and bind Eed and Suz12, components of polycomb complex PRC2 (Lee et al., 2006). Human α-globin BAC cell clone 1c showed enrichment of H3K27me3 staining peripheral to the DAPI-intense EGFP-LacI signals (Fig. 4 G and Fig. S4 A), which is consistent with FISH data showing intermingling of α-globin genes with active gene sequences (Fig. 4 E). Typically, three to seven H3K27me3 foci were visualized, often adjacent to DAPI-poor channels. Clustered H3K27me3 foci were observed to associate only with engineered chromosome regions containing the α-globin BAC (Fig. S4, B and C), which is consistent with the absence of H3K27me3 marks over the Dhfr promoter or promoters in the β-globin locus (Lee et al., 2006). These H3K27me3 foci were also present adjacent to EGFP-LacI signals within metaphase and anaphase chromosomes, demonstrating that they correspond to histone marks either entirely or largely derived from BAC transgenic sequences (Fig. 4 H). These results suggest that the inactive α-globin genes, marked by the facultative heterochromatin H3K27me3 modification, segregate with active genes and away from the heterochromatic H3K9me3 and HP1-enriched domains.
BAC transgenes assume an entirely different conformation in fibroblasts and fibroblast-like cells
The very different conformation of BAC arrays in mouse ES versus CHO cells (Hu et al., 2009) could be the result of differences in the species origin and/or cell type of these two cell lines or because of differences in the organization of the integrated transgenes. To distinguish between these possibilities, we isolated stable, mouse NIH 3T3 cell clones carrying the Dhfr BAC. Two 3T3 cell clones, with BAC copy number within the range of Dhfr BAC ES clones 33, 36, and 44 were examined in detail.
The six to seven mean number of BAC copies per EGFP-LacI spot in these two 3T3 clones was about two- to threefold higher than in CHO cells but ∼10-fold lower than in mouse ES cells. Moreover, GFP spots in the 3T3 clones were frequently arranged in extended, fiber-like conformations (Fig. 5 A), as observed previously in CHO cells (Hu et al., 2009), and BAC and LacO DNA FISH signals showed a much higher colocalization in 3T3 (Fig. 5 B) than in ES cells. RNA FISH signals partially overlapped or were closely adjacent to EGFP-LacI signals (Fig. 5 C), and both signals were associated with DAPI-poor regions of the nucleus.
Figure 5.
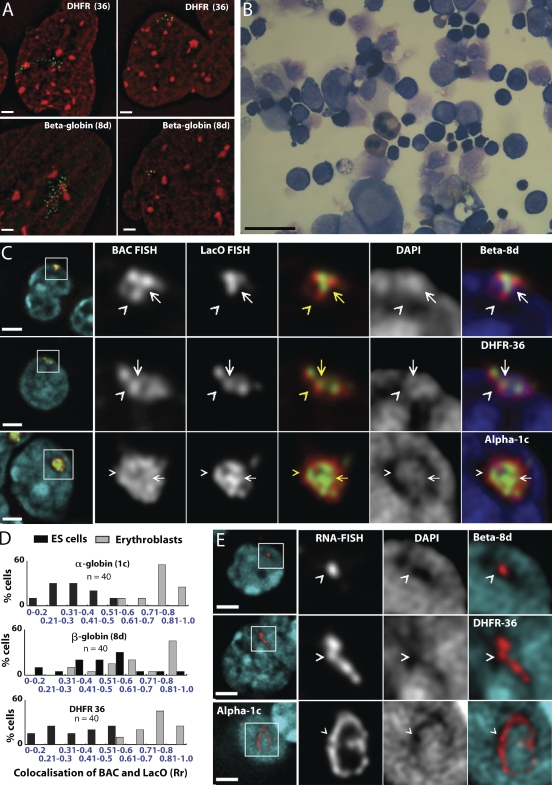
Different organization of BAC transgenes in fibroblasts. (A) 3T3 Dhfr BAC clones 4–2 (left) or 27–13 (right) are shown for EGFP-LacI (green) and DAPI (red). (bottom) Histograms show numbers of EGFP-LacI signals in nuclear maximum intensity projections (n = 67–69). (B) 3D DNA FISH using whole BAC (red) or LacO (green) probes in 3T3 clone 27–13. (bottom) Signal intensities along the arrow are shown. (C) 3T3 clone 27–13 for BAC RNA FISH (red) and EGFP-LacI (green). (D) ES cells differentiated to fibroblast-like cells (phase contrast). (E) ES clones Dhfr 36 (left) or β-globin 8d (right) differentiated to fibroblast-like cells for EGFP-LacI (green) and DAPI (red). Histograms show numbers of EGFP-LacI signals in maximum intensity projections. (F) 3D DNA FISH using BAC (red) or LacO (green) probes on fibroblast-like cells differentiated from ES Dhfr clone 36. Clusters of similar-size units show segregation of BAC and LacO sequences. Enlargement of individual units (of yellow boxed area) and intensity line drawing (bottom) shows the spatial distribution of BAC and LacO signals with relative fluorescence intensity along arrow. (G) 3D DNA FISH on fibroblast-like cells differentiated from ES β-globin clone 8d using BAC (red) and LacO (green) probes. LacO foci are offset from the central axis of a continuous fiber formed by BAC sequence. Enlargements of the yellow boxed area show a segment of fiber-like structure. DAPI counterstaining in E has been γ processed (factor of 0.7) to enhance chromatin substructure. Bars: (B) 1 µm; (D) 25 µm; (A, C, and E–G) 2 µm.
We next directly examined changes in chromatin conformation of specific ES cell BAC transgene clones accompanying cell differentiation to fibroblast-like cells (Fig. 5 D). These cells stained negatively for Oct-4, a marker for pluripotent ES cells, and positively for lamin A, a marker for differentiated cells, and mitotic cells were present in the population, indicating continued cell proliferation (unpublished data).
Mean BAC copies per GFP spot values in these cells were very similar to those observed in 3T3 cells (Fig. 5 E), with similar numbers for both Dhfr and β-globin BACs (Dhfr clone 36, 7.3; β-globin clone 8d, 7.9). In many cells, distinct linear arrangements of GFP spots or BAC and LacO FISH signals could be traced for up to 5 µm, which was quite similar to the predominant pattern in 3T3 cells.
However, a much wider continuum of morphologies was observed in the fibroblast-like cells differentiated from ES cells. Although in some cells, LacO and BAC FISH signals showed the linear colocalizing patterns characteristic of both proliferating and quiescent 3T3 cells, other cells showed a more open network or chain-like pattern, sometimes with no visible connection between subunits (Fig. 5 F). Distinct subunits were observed consisting of a single LacO FISH spot associated with one end of a short fiber-like BAC signal. Subunit lengths averaged 0.7 µm and contained ∼1–2 Mb DNA or 5–10 copies of engineered BAC. Between these two extremes, in about one third of cells, BAC sequences merged to form continuous, fiber-like structures, but the LacO repeats segregated into distinct foci localizing to the edges of these apparent fibers. The number of BAC copies per LacO foci was similar to that in fibroblasts and other fibroblast-like cells.
Large-scale chromatin conformation of BAC transgene arrays in retinoic acid (RA)–differentiated and erythroblast cells
To address whether these differences in large-scale chromatin conformation between ES and fibroblast cells represented the difference between an undifferentiated pluripotent versus differentiated state versus a lineage-specific conformation, we examined the BAC transgene arrays in other differentiated cell types.
RA-induced differentiation of ES cells leads to a mixture of differentiated cell types, particularly neuroprogenitors (Guan et al., 2001). After 10–14 d in RA without leukemia inhibitory factor (LIF), ∼100% of cells lost the Oct4 pluripotent marker (unpublished data). In these cells, we saw a range of EGFP-LacI staining patterns, including flattened, fibroblast-like cells that showed either the aforementioned linear string of spots or network patterns (Fig. 6 A, left). Other differentiated cell types showed GFP spot patterns similar to undifferentiated ES cells (Fig. 6 A, right).
Figure 6.
Transgene conformation in RA-differentiated and erythroid cells. (A) High GFP spot number, linear fibroblast-like (left) versus low GFP spot number, compact ES-like (right) BAC conformations in RA-differentiated Dhfr 36 or β-globin 8d cells (red, DAPI; green, GFP-LacI) (B) Erythroblasts derived from ES cells after 13 d of differentiation (May-Giemsa staining). (C) 3D DNA FISH of transgene arrays in ES clones β-globin 8d, Dhfr 36, and α-globin1c cells after differentiation to erythroblasts: DAPI (blue), LacO (green), and BAC probes (red). Whole cells are shown in first left panels with enlargements (marked by white boxes) in right panels. Fringes of BAC signals (carets) overlap the edges of DAPI-poor regions but not LacO signal. Overlapping BAC and LacO signals coincide with high-intensity DAPI staining (arrows). (D) Histograms showing degree of colocalization of LacO and BAC FISH signals in individual cells (as measured by Pearson’s correlation coefficients [Rr]) before (black) or after (gray) differentiation to erythroblasts. Dhfr and α-globin BAC transgenes show clear transition from low to high overlap during differentiation. (E) RNA FISH BAC (red) signal and DAPI (blue) in clones β-globin 8d, Dhfr 36, and α-globin 1c after erythroid differentiation. (right) Enlargements of the boxed areas (left) show that RNA signals (carets) coincide with DAPI-poor regions. DAPI signal in A has been γ processed (factor of 0.7). Bars: (A, C, and E) 2 µm; (B) 20 µm.
We next turned to a differentiation protocol producing nearly pure populations of erythroid cells (Nakano et al., 1996). For the three clones showing successful differentiation at days 12–14 of this protocol, 94% of harvested cells were mixed early- and late-stage definitive erythroblasts, as determined by May-Giemsa staining (Fig. 6 B), and were diamino fluorine positive, indicating that hemoglobin was expressed.
The large-scale chromatin structure after differentiation to erythroid cells was similar for all BAC transgene arrays yet quite different from undifferentiated ES cells, fibroblasts, or RA-differentiated ES cells. Analysis of BAC transgene morphology was restricted to DNA and RNA FISH because EGFP-LacI expression could be achieved in very few erythrocytes. Compact chromatin domains showed a significantly higher colocalization of LacO and BAC sequences than observed in other cell types (Fig. 6, C and D). Histograms of intensity correlation coefficients showed near-complete separation between values for undifferentiated ES versus erythroid cells (Fig. 6 D). The β-globin BAC array reached similar intensity colocalization correlation levels in erythroid cells, but the net increase from levels in undifferentiated ES cells was smaller because of its more compact structure in these cells.
BAC transgene arrays in erythroid cells showed an outer fringe of BAC FISH signal depleted of LacO hybridization (Fig. 6 C, carets). Erythroblast nuclei were dominated by large clumps of DAPI-intense staining, interspersed with low-intensity staining, a pattern that survived both DNA and RNA FISH procedures. The outer fringe of BAC FISH signal bordered or marginally overlapped DAPI-poor nuclear regions, whereas overlapping BAC and LacO signals localized to regions of high DAPI intensity (Fig. 6 C, arrows). Segregation of BAC from LacO signals rarely exceeded 0.2 µm.
RNA FISH signals, seen in a varying proportion of cells in erythroblasts derived from all three transgenes, predominantly colocalized with DAPI-poor channels (Fig. 6 E). In the largest clone, α-globin 1c, and to a lesser degree in Dhfr 36, RNA signals often appeared as circular or curved linear configurations (Fig. 6 E, bottom). In contrast with BAC DNA FISH signals, RNA signals in all clones tended to be located in the center rather than at the edges of DAPI-poor regions, suggesting that most of the BAC array chromosome subterritory consists of condensed chromatin regions, with transcribed RNA localizing to the surrounding peripheral DAPI-poor regions. These RNA FISH signals may represent sites of transcription of a subset of expressed BAC transgenes that are highly decompacted and not detected by DNA FISH under the experimental conditions used. Alternatively, these RNA FISH signals may represent accumulations of transcript spatially offset from the actual sites of transcription.
Despite the more compact BAC transgene conformation present in erythroid cells, Dhfr transgene expression per copy number in clone 36 cells differentiated into erythroid cells remained at ∼40% the expression level of the endogenous gene. α- and β-globin transgenes showed considerably decreased expression levels relative to the endogenous genes, and the endogenous gene expression was decreased relative to control cells without transgenes (Table II), suggesting that this decrease was because of titration of specific transcription factors.
Table II.
Expression characteristics of transgenic clones after erythroid differentiation
| BAC clone | Gene analyzed | Expression/copy from transgenea | Total expression from transgenea | Expression from endogenous locusb |
| % | % | % | ||
| β-Globin 8d | HBB/Hbb-b1 | 0.0089 | 1.4 | 10.3 |
| α-Globin 1c | HBA/Hba-a1 | 0.26 | 160 | 4.3 |
| Dhfr 36 | Dhfr | 40 | 5,560 | ND |
Based on reverse transcription quantitative PCR data.
Relative to the endogenous locus in cells without the transgene.
Relative to expression in cells without the transgene.
Discussion
In this study, we demonstrate the ability to create engineered chromosome regions tens to hundreds of megabase pairs in size, consisting of high copy number repeats of individual cloned, ∼200-kb genomic fragments. These chromosome regions range from the size of chromosome bands to longer than the arms of some mouse chromosomes yet remain stable through multiple cell divisions. By several criteria, these engineered chromosome regions recapitulate at least partially several functional features of endogenous wild-type chromosomes: expression of housekeeping genes on the BAC transgenes was maintained per copy within severalfold of the endogenous genes in ES and fibroblast-like cells, positioning of the mouse β-globin locus to the nuclear periphery in ES cells (Hepperger et al., 2008) was mimicked by multicopy human β-globin BAC transgenes independent of chromosome insertion site, and appearance of H3K27me3-modified foci only within the α-globin BAC transgene array suggests that this epigenetic marking of inactive α-globin genes is correctly reproduced by the transgenes.
Using these engineered chromosome regions, we demonstrate a surprising plasticity of large-scale chromatin organization, both in terms of the chromatin folding within a single chromosome territory and changes in folding patterns after differentiation. In the following paragraphs, we summarize our major results and describe their implications for understanding higher-level chromatin folding within native chromosomes.
Chromosome territory formation
Conceptually, formation of compact chromosome territories could require complex, long-range interactions between specific sequences on native chromosomes. Alternatively, chromosome territory formation might emerge from the intrinsic folding properties of local chromatin regions just hundreds of kilobases in size. Our results support the second possibility, as all BAC transgene arrays in all cell types formed compact spatial domains, which was similar to endogenous chromosome territories.
Similarly, the characteristic variability in compaction for different megabase pair subterritory regions correlating with difference in gene density (Goetze et al., 2007) could be influenced mainly by the type and number of regulatory elements and activity of genes distributed throughout these regions. Alternatively, this variability might be because of the presence of a small number of specific, sparsely distributed cis sequences, which modulate gene activity only indirectly by controlling subterritory compaction. The more condensed subterritory domains formed in ES cells by the transcriptionally inactive versus active arrays support the former explanation. Thus, our results extend these statistical correlations between gene density and chromosome subterritory compaction to direct experimental manipulation and measurement.
Sequence segregation and functional polarization within ES cell chromosome territories
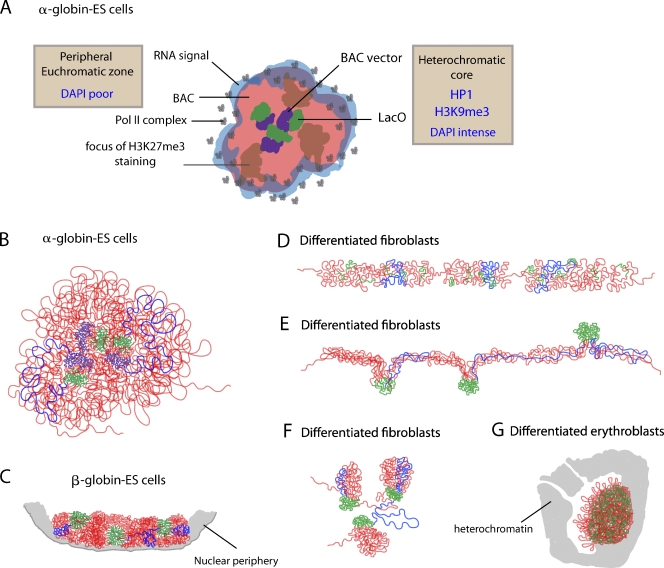
In ES cells, there was a spatial separation between LacO and vector backbone foreign DNA sequences embedded within a heterochromatic domain and the remainder of the BAC sequences corresponding to mammalian DNA (Fig. 7, A and B). This segregation was accompanied by a polarized distribution of the LacO and vector backbone sequences to the interior of the engineered chromosome subterritories with other BAC sequences preferentially localized to the subterritory exterior (Fig. 7 A). This polarized sequence segregation appeared to be functionally driven because BAC DNA of the inactive β-globin region in ES cells was apparently as condensed as the LacO foci, showed no preferential localization to the exterior of chromosome subterritories, and had substantially more spatial overlap with the LacO sequences (Fig. 7 C). Furthermore, for both the Dhfr and α-globin BAC transgenes, RNA transcripts and RNA polymerase II signals showed an even more extreme peripheral distribution, not overlapping with LacI-stained regions but coinciding with DAPI-poor channels surrounding the subterritories (Fig. 7 A).
Figure 7.
Summary of BAC transgene conformations in ES and differentiated cells. (A) Cartoon showing spatial segregation of DNA sequences and nascent transcripts relative to distributions of HP1, H3K9me3, H3K27me3, and RNA polymerase II within α-globin BAC transgene arrays in ES cells. (B–G) Models for changes in DNA topology within BAC transgene arrays as a function of cell differentiation: LacO repeats (green), BAC vector sequence (purple), and remainder of the BAC (red) are shown, with blue lines tracing the path of a single BAC copy.
This pattern of segregation strikingly parallels the proposed localization of repetitive DNA to the DAPI-condensed Barr body of the mammalian inactive X chromosome, external distribution of genes that escape x inactivation, and external bias relative to the Barr body of inactive X chromosome–linked genes (Chaumeil et al., 2006; Clemson et al., 2006).
Global changes in chromosome territories and large-scale chromatin plasticity during development
Nuclear size, shape, and DAPI staining texture showed pronounced differences between ES, fibroblast-like, and erythroid-differentiated cells paralleled by changes in the overall compaction and organization of the engineered chromosome regions, independent of the transcription status of the BAC transgenes. Our ability to control and manipulate the sequence content of these 10–180-Mbp chromosome regions highlights the origin of this variability as intrinsic, tissue-specific differences in chromatin compaction as opposed to differentiation-induced changes in gene expression.
Plasticity in overall chromosome subterritory geometry of the engineered chromosome regions was paralleled by significant differences in the local folding and topology of each BAC repeat (Fig. 7, B–G). Despite quite different folding trajectories and overall compaction in different cell types, expression levels of the Dhfr BAC transgenes were remarkably constant relative to the endogenous Dhfr gene, even in erythroid cells. Normal transcriptional activity is therefore compatible with high compaction. Moreover, these results suggest that regulation of gene expression may not depend on the exact patterns of large-scale chromatin folding but, rather, on segregation of particular sequences within chromosome territories and chromatin fiber–fiber aggregation properties.
A common theme emerging in both ES and erythroid cells was the striking localization of nascent RNA transcripts to the engineered chromosome subterritory periphery surrounding a more condensed DNA core (Fig. 7 A). These RNA FISH signals may represent sites of transcription of a subset of expressed BAC transgenes at the edge of the BAC transgene territory. Alternatively, these RNA FISH signals may represent accumulations of transcript spatially offset from the actual sites of transcription.
Linear large-scale chromatin structures from noncontiguous DNA topologies
Several classes of models for large-scale chromatin folding above the 10- and 30-nm chromatin fibers have been proposed, including hierarchical, looping, and network models (Belmont, 2002; Swedlow and Hirano, 2003; Kireeva et al., 2004; Branco and Pombo, 2007). However, what has generally not been questioned is the implicit assumption that there is a single folding motif for chromatin organization.
Our analysis of the large-scale chromatin-folding patterns within these engineered chromosome regions instead suggests that different folding motifs dominate for the same chromosome region depending on the cell type. Furthermore, the combination of different folding motifs observed within the same cell type reveal surprising aspects of chromatin organization.
In ES cells, extensive looping mediated by LacO and vector backbone interactions was observed (Fig. 7 B). Additional looping was implied by the foci of H3K27me3 immunostaining in α-globin BAC transgene arrays. A relatively low level of chromatin compaction was inferred by a noticeable separation of up to ∼0.5 µm between BAC and LacO FISH signals, implying linear chromatin compaction ratios in maximally extended loops of ∼70:1, just about twofold more condensed than extended 30-nm chromatin fibers. The persistence within mitotic chromosomes of these GFP-LacI– and H3K27me3-modified foci indicates that folding motifs underlying mitotic chromosome compaction are superimposed on top of these long-range looping interactions.
In contrast, in fibroblasts (Fig. 7 D), large-scale chromatin fibers were observed. These were similar to previous observations in CHO cells of large-scale chromatin fibers with estimated linear compaction ratios of 200–1,000:1 (Tumbar et al., 1999; Hu et al., 2009), which were explained by hierarchical folding models (Belmont and Bruce, 1994; Kireeva et al., 2004). Remarkably, an alternative pattern in about one third of ES cells differentiated into fibroblast-like cells showed continuous, large-scale chromatin fibers but with LacO repeats segregating into distinct foci localizing at the fiber periphery (Fig. 7 E). An intermediate pattern was observed in ES cell–differentiated fibroblast-like cells in which BAC hybridization showed short, apparent large-scale chromatin fiber segments connected in a network pattern. However, localization of LacO foci to the tips of these apparent fiber segments again indicated the presence of extensive looping of BAC DNA within these fiber-like segments (Fig. 7 F). In all cases, a similar number (about seven) of estimated BAC copies per LacO foci was observed.
This segregation of LacO repeats in the latter two patterns described in fibroblast-like cells (Fig. 7, E and F) implies that extensive topological looping of distant DNA fragments exist within fiber-like, large-scale chromatin structures (Fig. 7 D), challenging predictions of hierarchical folding models for large-scale chromatin structure, which assume linear folding of adjacent sequences. Presumably, to form mitotic chromosomes, these fiber-like structures themselves undergo a higher level of folding as a unit, thereby combining both looping and hierarchical folding motifs.
The extent to which linear chromosome structures form through aggregation of nonlinear sequences in native chromosomes remains unknown. Our results, together with the demonstration by RNA-TRAP and 3C-derived methods of long-range cis interactions between distant sequences (Carter et al., 2002; Tolhuis et al., 2002; Simonis et al., 2006), suggest that this may be a more fundamental feature of higher-order chromatin structure than previously imagined. Our findings also resolve the apparent contradiction between hierarchical models of chromatin folding and frequent long-range DNA interactions demonstrated by 3C-like molecular methods. Specifically, our results show a plasticity of large-scale chromatin folding, which allows the formation of large-scale chromatin fibers that can still incorporate complex DNA-looping topologies.
Increased association of LacO repeats and BAC vector sequences may reflect extreme plasticity of large-scale chromatin structure in ES cells, facilitating long-range interactions
In mouse ES cells, we observed a high degree of association of LacO arrays from neighboring BAC copies. LacO repeats separated by several megabase pairs have shown pairing in Arabidopsis thaliana, which is dependent on DNA methylation (Watanabe et al., 2005) and association with chromocenters; however, such pairing has not been previously appreciated in Drosophila melanogaster or mammalian cells, and we did not observe LacO association with chromocenters.
This phenomenon was not restricted to high copy number direct repeats, as the BAC vector backbone sequences in ES cells present at only one copy per BAC coalesced into foci that were similar and adjacent to LacO foci. We also observed clustering of H3K27me3-modified regions of the α-globin BAC, and similar-sized H3K27me3 foci were seen throughout the nucleus.
Previous experiments have demonstrated association of transgenic repeats in other systems (Assaad et al., 1993; Dorer and Henikoff, 1994). Whether the association of LacO and BAC vector sequences represents similar, possibly RNAi-mediated association of foreign DNA, transgenic repeats, true homologous pairing, or coalescence of sequences with common epigenetic marks is unclear. Despite common epigenetic marks, LacO and the single-copy vector backbone sequences still segregated separately. In contrast, single-copy housekeeping genes on the α-globin BAC showed little evidence of self-association.
Whatever their origin, these cis sequence long-range associations in the engineered BAC transgene arrays were most pronounced in undifferentiated ES cells. This may be related either to the high frequency of homologous recombination (Sedivy and Dutriaux, 1999), and therefore possible increased pairing of homologous sequences in ES cells, or to a more plastic, flexible, and/or dynamic large-scale chromatin structure in these cells. Structural characteristics attributed to mouse ES but not differentiated cell chromatin include pools of highly mobile chromosomal proteins (Meshorer and Misteli, 2006) and global, low-level transcription (Efroni et al., 2008). Similar association of distantly separated, endogenous chromatin may occur in mouse ES cells facilitated through the formation of comparable decondensed, looped configurations, possibly mediated through repeat sequences.
Future directions
In this study, we have constructed chromosome regions up to hundreds of megabase pairs in size. Several fundamental features of natural chromosomes are recapitulated by the self-assembly of these engineered chromosome regions. We also demonstrate a surprising plasticity of large-scale chromatin folding and chromosome territory architecture as a function of cell differentiation. The ability to generate model chromosome territories from multicopy BAC sequences and to manipulate these BAC sequences using BAC recombineering should allow future dissection of cis elements controlling large-scale chromatin folding and chromosome territory organization.
Materials and methods
BAC modification
BAC modifications were performed as previously described (Hu et al., 2009). p[Kan/Neo-PSI-8.32] was created from p[Kan/Neo-8.32] by addition of a PI–SceI homing endonuclease site between the kanomycin/neomycin resistance genes and LacO array. Tn5 transposition was used to insert the transposon from p[Kan/Neo-8.32] into BAC RP11-344L6 containing the human α-globin–like genes and the transposon from p[K/N-PSI-8.32] into BACs CTD-2643I7 and CTB-057L22 containing the human β-globin–like genes and the mouse Dhfr locus, respectively. CTB-057L22 modified to contain LacO repeats and a zeocin resistance gene was used to create 3T3 clones. Restriction endonuclease digest of resistant colonies was used to screen for presence of the full-length LacO array, and the position of inserts was determined by DNA sequence analysis. BACs were obtained from Invitrogen.
Creation of ES and 3T3 cell lines carrying BAC insertions
Mouse ES HM1 cells (provided by A. Smith, University of Cambridge, Cambridge, England, UK) were maintained on a feeder layer of mitomycin C–treated mouse embryonic fibroblasts or in flasks coated with 0.1% gelatin in DME (Invitrogen) supplemented with 15% ES cell–screened FCS (Invitrogen), 1% MEM nonessential amino acids, 0.001% 2-mercaptoethanol, and 1,000 U/ml LIF (Millipore). HM1 cells were used between passages 27 and 33. Mouse 3T3 cells and embryonic fibroblasts were obtained from the American Type Culture Collection and maintained in DME with 10% FCS (Hyclone).
DNA was prepared from BAC clones using a large construct kit (QIAGEN). 10 µg purified BAC DNA was linearized by PI–SceI (CTD-2643I7 K/N PSI 8.32 and CTB-057L22 K/N PSI 8.32) or PvuI digestion (RP11-344L6 K/N 8.32), extracted with phenol/chloroform, checked by pulse field gel electrophoresis to confirm linearization, and used immediately to transfect ES or 3T3 cells. ES cells were transfected at 70–90% confluence in T25 tissue culture flasks using 30 µl Lipofectamine 2000 (Invitrogen), incubated overnight, plated onto four 94-mm hybridoma dishes (Greiner Bio-One) coated with 0.1% gelatin, and incubated for 24 h before selection with 320 µg/ml G418. Transfected 3T3 cells were selected with 75 µg/ml zeocin. 10 d after plating, colonies were washed twice with PBS, immersed in 0.05% trypsin and 0.02% EDTA for 5 min, and replated into 96-well plates. Individual 3T3 subclones were selected by serial dilution of a mixed clonal population obtained after 3 wk selection. Expanded ES clones retaining an undifferentiated ES cell colony morphology and 3T3 cell clones were screened for BAC inserts by transient expression of GFP-LacI.
Expression of EGFP-LacI
1.6 µg plasmid p3′SS-dimer-EGFP (Robinett et al., 1996) and 4 µl Lipofectamine 2000 were used to transiently transfect 50–70% confluent ES cells in 12-well plates. 24 h after transfection, cells were washed in PBS, fixed for 10 min in 1.6% formaldehyde in PBS, permeabilized with 0.1% Triton X-100 (Thermo Fisher Scientific) in PBS, stained in 0.2 µg/ml DAPI in PBS, and mounted in ProLong antifade (Invitrogen).
Lentivirus FUPW, obtained from T. Brennan (University of California, San Francisco, San Francisco, CA), was constructed from FUGW (Lois et al., 2002) by excision of EGFP at XbaI and EcoRI sites. FULGW was created by ligating the 9,200-bp fragment generated by XbaI and HpaI digestion of FUPW with the EGFP-LacI fragment created by digesting p3′SS-dimer-EGFP-LacI with MunI, filling in the ends with Klenow, and digesting with XbaI.
Infectious lentivirus particles were produced by cotransfection of 293T cells with FULGW, CMVΔR8.2 HIV-1 packaging, and vesicular stomatitis virus G envelope glycoprotein plasmids. Lentivirus particles were harvested after 48 h and concentrated in 1 ml PBS after ultracentrifugation. 50 µl lentivirus was added to 104 − 2 × 105 ES cells or ES cells differentiated to fibroblasts. 24 h after transduction, cells were washed twice in PBS, resuspended in medium, and either fixed or passaged.
RNA and DNA FISH
ES and 3T3 cells were grown overnight on coverslips coated for 4 h with 0.1% fibronectin (Sigma-Aldrich). Erythroblasts differentiated from ES cells were plated for 1 h on coverslips coated for 30 min with 1 mg/ml poly-l-lysine. Methanol/acetic acid–fixed metaphase spreads were prepared according to standard cytogenetic procedures (Beatty and Scherer, 2002). Stretched chromatin fibers were prepared by cytospin as described using 100 µl cells per slide at 3 × 106 cells/ml (Heng, 2002). Cells were prepared for 3D DNA and RNA FISH by fixing in freshly prepared 4% calcium magnesium-free (CMF) PBS buffer (CMF-PBS) for 10 min at room temperature. Cells were permeabilized with 0.5% Triton X-100 in CMF-PBS for 5 min. Coverslips were equilibrated in 20% glycerol in CMF-PBS for 30 min before being immersed in liquid nitrogen for 2–3 s, then thawed at room temperature and replaced briefly in 20% glycerol. After an additional three freeze thaw cycles, cells were immersed in 0.1 M HCl for 7.5 min, washed in CMF-PBS, then stored at 4°C in 50% deionized formamide/2× SSC (Solevei et al., 2002).
FISH probes included the three BACs used to create transgenic cell lines without LacO inserts. In addition, small probes were amplified by PCR from subregions of RP11-344L6 and subcloned into pUC18; XbaI or EcoRI sequences were included at the 5′ ends of primers to facilitate cloning (Table S1). Plasmids p[Kan/Neo- 8.32] and pBeloBAC11 were used to detect LacO arrays and the BAC vector backbone sequences (Shockett et al., 1995).
Probes were labeled with biotin or digoxigenin and used in single- or dual-color FISH experiments as described previously (Hu et al., 2009). For analysis of BAC and LacO sequences, 75 ng digoxigenin-labeled BAC and 50 ng biotin-labeled K/N 8.32 DNA in a total volume of 5 µl of hybridization buffer were used. For analyzing RP11-344L6 subregions, 62.5 ng each of pairs of differentially labeled PCR-derived probes in a total volume of 5 µl were used. For RNA FISH, 60 ng probe was hybridized in 6 µl of hybridization buffer.
For 3D DNA FISH, hybridization mix (50% deionized formamide, 10% dextran sulfate, and 2× SSC) was applied before mounting coverslips to glass slides using rubber cement. Denaturation at 75°C for 2 min on a heat block was followed by hybridization overnight at 37°C in a humidified chamber followed by washes in 0.4× SSC at 70°C for 2 min and SSCT (2× SSC/0.2% Tween 20) at room temperature. RNA FISH was performed as previously described (Chaumeil et al., 2004). DNA probes were resuspended in formamide, denatured at 75°C, and immediately brought to a final concentration of 50% formamide, 1% BSA, 2× SSC, and 2 mM vanadyl ribonucleoside complex (New England Biolabs, Inc.). Hybridization was performed at 37°C overnight in a wet chamber followed by three washes in 50% formamide, 2× SSC, pH 7.2, and three washes in 2× SSC at 42°C. Detection used streptavidin Alexa Fluor 594 (1:200; Invitrogen) or antidigoxigenin fluorescein antibodies (1:200; Roche) in 4% BSA/SSCT (3D FISH) or 1% BSA/4× SSC/2 mM vanadyl ribonucleoside complex (RNA FISH).
Immunofluorescence
Cells for immunostaining were permeabilized in 0.1% Triton X-100 in PBS* (5 mM MgCl2 and 0.1 mM EDTA in CMF-PBS) for 5 min before fixation with 1.6% formaldehyde in PBS* for 10 min and washing three times with CMF-PBS. Before application of primary antibodies, a 1-h block with 5% normal goat or donkey serum diluted in CMF-PBS was used, with the exception of monoclonal RL1, which was blocked with 5% BSA in CMF-PBS. Primary antibodies were applied for 24 h at 4°C at the following dilutions in PBS*/0.1% Triton X-100: mouse monoclonal antibody RL1 against nuclear pore O-linked glycoprotein, 1:750 (ABR-Affinity BioReagents); rabbit anti-H3-K27me3, 1:1,000 (Millipore); rabbit anti-RNA polymerase II (N-20), 1:500; rabbit anti–Oct-4 monoclonal antibody, 1:200 (Santa Cruz Biotechnology, Inc.); and anti–lamin A antibody, 1:1,000 (obtained from B. Goldman, Northwestern University, Evanston, IL). Secondary donkey or goat anti–rabbit and anti–mouse antibodies conjugated to FITC or Texas red (Jackson ImmunoResearch Laboratories) were used at a 1:500 dilution and applied for 2 h at room temperature after three 5-min washes in CMF-PBS.
Immunostaining followed by 3D DNA FISH
Immunostaining on mouse ES cells was performed as described in the previous section. Rabbit anti–histone H3K9me3 (Millipore) was diluted at 1:500, mouse anti–HP1-α (Millipore) was diluted at 1:500, and mouse anti–HP1-γ (Millipore) was diluted at 1:1,000. Secondary goat anti–rabbit or anti–mouse antibodies conjugated with Texas red or Alexa Fluor 350 (Jackson ImmunoResearch Laboratories) were used at 1:500 dilutions. After immunostaining, cells were postfixed in 3% paraformaldehyde in PBS for 10 min at room temperature and permeabilized in 0.1 M HCl/0.7% Triton X-100 in 2× SSC for 10 min on ice. DNA FISH was performed as described previously. Probes prepared with plasmids p[K/N-PSI-8.32], pBeloBAC11, and BAC RP11-344L6 were used to detect LacO arrays, BAC vector backbone, and BAC DNA, respectively.
Quantitative RT-PCR
RNA was prepared using an RNAeasy mini kit with on-column DNase digestion (QIAGEN) according to the manufacturer’s protocol. cDNA was prepared from 0.2–1.0 µg RNA using avian myeloblastosis virus reverse transcription (Invitrogen) and oligo (dT) primers (Sigma-Aldrich). PCR reactions used SYBR green PCR master mix (Applied Biosystems), 0.1 µm of each primer (Table S2), and 1 µl cDNA in 25 µl total vol in sealed 96-well PCR plates (ABgene) on a thermocycler (iCycler; Bio-Rad Laboratories). Denaturation was performed at 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 30 s. Log dilutions (1–10−4) were prepared for each cDNA, and reactions were performed in triplicate. Normalization to the β-actin housekeeping gene was used for comparisons of gene expression between different cell lines. Comparisons of expression of transgenes versus their endogenous counterparts assumed that both endogenous alleles were transcribed. To compare expression from human transgenes versus mouse endogenous loci, we used the comparative Ct (2-Δ Δ Ct) method (Wong and Medrano, 2005) with primer pairs with similar PCR efficiencies (<0.1 differences in slope).
ES differentiation
ES cells were differentiated to fibroblast-like cells by growth in DME with 10% FCS, 1% MEM nonessential amino acids, and 0.001% 2-mercaptoethanol without LIF for four passages in gelatin-coated flasks followed by four to seven passages in uncoated flasks. After about six passages in uncoated flasks, a monolayer of fibroblast-like cells supporting ES-like colonies formed. Washing twice with PBS 1 h after passage before addition of new medium increased the proportion of fibroblast-like cells. Cells were plated onto fibronectin-coated coverslips for 24 h before FULGW transduction or FISH.
ES cells were differentiated with RA as previously described (Chaumeil et al., 2004). Briefly, ES cells were plated at 10,000 cells/cm2 and, after attachment, washed in CMF-PBS, and incubated in DME with 10% FCS, 1% MEM nonessential amino acids, 100 nM all trans-RA (Sigma-Aldrich), and 0.1 µM 2-mercaptoethanol for 7–14 d. Media were changed daily.
Erythroblasts were derived from ES cells essentially as previously described (Nakano et al., 1996). OP9 cells (American Type Culture Collection) were maintained, and ES cells differentiated in Alpha minimum essential medium without ribonucleosides and deoxyribonucleosides (Invitrogen) supplemented with 2 mM l-glutamine (Hyclone), 1 mM sodium pyruvate (Sigma-Aldrich), and 20% fetal bovine serum (Hyclone). Dispersed ES cells were plated at 104 cells/well into 6-well plates of ∼90% confluent OP9 cells. 3 d after plating, half of the medium was replaced, and 4 IU recombinant human erythropoietin-α (EPO; ProSpec) was added to each well. After 5 d, cells were dispersed with 0.5 ml 0.25% trypsin and 0.53 mM EDTA (Sigma-Aldrich) at 37°C for 5 min and transferred to fresh plates of ∼90% confluent OP9 cells at 1–2 × 105 cells/well in 2 ml medium with 4 IU EPO. 5 d later, cells were removed by vigorous pipetting, centrifuged, resuspended in medium with EPO, and transferred to fresh OP9 cells. 3–4 d after replating, detached erythroblasts were harvested. For RNA or DNA FISH, cells were resuspended in 200 µl medium and plated onto coverslips coated with 0.2 mg/ml poly-l-lysine for 1 h at 37°C.
DAPI (Sigma-Aldrich) staining was performed as previously described (Kaiho and Mizuno, 1985). Stained cells were plated onto poly-l-lysine–coated coverslips for 10 min before washing in PBS and mounting in glycerol.
Microscopy and image analysis
3D optical section datasets of fixed cells were collected using a 60× NA 1.4 objective on an inverted light microscope (IMT-2; Olympus) equipped with a slow-scan cooled charge-coupled device camera unit (CE200A; Photometrics), motorized filter wheels, and a microstepping motor for z focus, all driven by the Resolve3D software data collection program (Applied Precision) running on an SGI 4D/35TG. This system duplicates that described previously by Hiraoka et al. (1991). Alternatively, for data presented in Fig. 3 (G–L) and Fig. S3, a personal deconvolution microscope system (DeltaVision; Applied Precision) was used with a 60× NA 1.4 lens. Deconvolution used an enhanced ratio iterative-constrained algorithm (Agard et al., 1989). XY and Z optical displacement between different filter sets was determined experimentally using fluorescent microspheres (Tetraspeck; Invitrogen). Distances between EGFP-LacI signals and the nuclear periphery, defined by nuclear pore staining, were made from signal edges, and measurements were taken from the EGFP-LacI signal closest to the periphery. Corrections for optical displacement, maximum intensity projections of EGFP-LacI signals, γ adjustments, distance and fluorescence intensity measurements, and colocalization analysis of BAC and LacO FISH signals were performed with ImageJ (National Institutes of Health) open-source software. ImageJ software was also used to measure colocalization between BAC and LacO FISH signals through calculation of the Pearson’s correlation coefficient (Rr) on single optical sections after subtracting background and outlining the signal area using the masking tool. Figures were prepared using Photoshop and Illustrator software (Adobe).
Online supplemental material
Fig. S1 shows transgene position relative to nuclear periphery. Fig. S2 shows LacO and BAC FISH in ES cells. Fig. S3 shows localization of LacO, vector backbone, and BAC sequences relative to H3K9me3, HP1-α, and HP1-γ marks. Fig. S4 shows H3K27me3 versus LacO distribution in ES cells. Table S1 shows primers for BAC RP11-344L6 FISH probes, and Table S2 shows primers for reverse transcription quantitative PCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912167/DC1.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (grant GM58460 to A.S. Belmont) and the Human Frontier Science Program (grant HFSP RGP0019/200 to A.S. Belmont and E. Heard). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Abbreviations used in this paper:
- BAC
- bacterial artificial chromosome
- CMF
- calcium magnesium free
- EPO
- erythropoietin-α
- ES
- embryonic stem
- LacO
- lac operator
- LIF
- leukemia inhibitory factor
- RA
- retinoic acid
References
- Agard D.A., Hiraoka Y., Shaw P., Sedat J.W. 1989. Fluorescence microscopy in three dimensions. Methods Cell Biol. 30:353–377 10.1016/S0091-679X(08)60986-3 [DOI] [PubMed] [Google Scholar]
- Assaad F.F., Tucker K.L., Signer E.R. 1993. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol. Biol. 22:1067–1085 10.1007/BF00028978 [DOI] [PubMed] [Google Scholar]
- Beatty B.G., Scherer S.W. 2002. Human chromosome mapping of single copy genes. FISH: A Practical Approach. Beatty B., Mai S., Squire J., Oxford University Press, Oxford: 29–53 [Google Scholar]
- Belmont A.S. 2002. Mitotic chromosome scaffold structure: new approaches to an old controversy. Proc. Natl. Acad. Sci. USA. 99:15855–15857 10.1073/pnas.262672799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A.S., Bruce K. 1994. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 127:287–302 10.1083/jcb.127.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco M.R., Pombo A. 2007. Chromosome organization: new facts, new models. Trends Cell Biol. 17:127–134 10.1016/j.tcb.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Brown K.E., Amoils S., Horn J.M., Buckle V.J., Higgs D.R., Merkenschlager M., Fisher A.G. 2001. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3:602–606 10.1038/35078577 [DOI] [PubMed] [Google Scholar]
- Brown J.M., Green J., das Neves R.P., Wallace H.A.C., Smith A.J.H., Hughes J., Gray N., Taylor S., Wood W.G., Higgs D.R., et al. 2008. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 182:1083–1097 10.1083/jcb.200803174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D., Chakalova L., Osborne C.S., Dai Y.F., Fraser P. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623–626 10.1038/ng1051 [DOI] [PubMed] [Google Scholar]
- Chambeyron S., Bickmore W.A. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119–1130 10.1101/gad.292104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J., Okamoto I., Heard E. 2004. X-chromosome inactivation in mouse embryonic stem cells: analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol. 376:405–419 10.1016/S0076-6879(03)76027-3 [DOI] [PubMed] [Google Scholar]
- Chaumeil J., Le Baccon P., Wutz A., Heard E. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20:2223–2237 10.1101/gad.380906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., Hall L.L., Byron M., McNeil J., Lawrence J.B. 2006. The x chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl. Acad. Sci. USA. 103:7688–7693 10.1073/pnas.0601069103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D., Verschure P.J., Martin T.E., Dahmus M.E., Krause S., Fu X.D., van Driel R., Fakan S. 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell. 10:211–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock C.F., Vyas P., Sharpe J.A., Ayyub H., Wood W.G., Higgs D.R. 1995. Contrasting effects of alpha and beta globin regulatory elements on chromatin structure may be related to their different chromosomal environments. EMBO J. 14:1718–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301 10.1038/35066075 [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer M., Dietzel S., Müller S., Solovei I., Fakan S. 2006. Chromosome territories—a functional nuclear landscape. Curr. Opin. Cell Biol. 18:307–316 10.1016/j.ceb.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131 10.1083/jcb.145.6.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel S., Schiebel K., Little G., Edelmann P., Rappold G.A., Eils R., Cremer C., Cremer T. 1999. The 3D positioning of ANT2 and ANT3 genes within female x chromosome territories correlates with gene activity. Exp. Cell Res. 252:363–375 10.1006/excr.1999.4635 [DOI] [PubMed] [Google Scholar]
- Dorer D.R., Henikoff S. 1994. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 77:993–1002 10.1016/0092-8674(94)90439-1 [DOI] [PubMed] [Google Scholar]
- Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D., Buetow K.H., et al. 2008. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2:437–447 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze S., Mateos-Langerak J., Gierman H.J., de Leeuw W., Giromus O., Indemans M.H., Koster J., Ondrej V., Versteeg R., van Driel R. 2007. The three-dimensional structure of human interphase chromosomes is related to the transcriptome map. Mol. Cell. Biol. 27:4475–4487 10.1128/MCB.00208-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K., Chang H., Rolletschek A., Wobus A.M. 2001. Embryonic stem cell-derived neurogenesis. Retinoic acid induction and lineage selection of neuronal cells. Cell Tissue Res. 305:171–176 10.1007/s004410100416 [DOI] [PubMed] [Google Scholar]
- Heng H.H.Q. 2002. High resolution FISH mapping using chromatin and DNA fibre. FISH: A Practical Approach. Beatty B., Mai S., Squire J., Oxford University Press, Oxford: 77–92 [Google Scholar]
- Hepperger C., Mannes A., Merz J., Peters J., Dietzel S. 2008. Three-dimensional positioning of genes in mouse cell nuclei. Chromosoma. 117:535–551 10.1007/s00412-008-0168-2 [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Swedlow J.R., Paddy M.R., Agard D.A., Sedat J.W. 1991. Three-dimensional multiple-wavelength fluorescence microscopy for the structural analysis of biological phenomena. Semin. Cell Biol. 2:153–165 [PubMed] [Google Scholar]
- Hu Y., Kireev I., Plutz M.J., Ashourian N., Belmont A.S. 2009. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J. Cell Biol. 185:87–100 10.1083/jcb.200809196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiho S., Mizuno K. 1985. Sensitive assay systems for detection of hemoglobin with 2,7-diaminofluorene: histochemistry and colorimetry for erythrodifferentiation. Anal. Biochem. 149:117–120 10.1016/0003-2697(85)90483-X [DOI] [PubMed] [Google Scholar]
- Kireeva N., Lakonishok M., Kireev I., Hirano T., Belmont A.S. 2004. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 166:775–785 10.1083/jcb.200406049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper K., Kölbl A., Biener D., Dittrich S., von Hase J., Thormeyer T., Fiegler H., Carter N.P., Speicher M.R., Cremer T., Cremer M. 2007. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 116:285–306 10.1007/s00412-007-0098-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz A., Lampel S., Nickolenko J.E., Bradl J., Benner A., Zirbel R.M., Cremer T., Lichter P. 1996. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 135:1195–1205 10.1083/jcb.135.5.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. 2006. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 125:301–313 10.1016/j.cell.2006.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C., Hong E.J., Pease S., Brown E.J., Baltimore D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 295:868–872 10.1126/science.1067081 [DOI] [PubMed] [Google Scholar]
- Mahy N.L., Perry P.E., Bickmore W.A. 2002a. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753–763 10.1083/jcb.200207115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy N.L., Perry P.E., Gilchrist S., Baldock R.A., Bickmore W.A. 2002b. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 157:579–589 10.1083/jcb.200111071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Misteli T. 2006. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 7:540–546 10.1038/nrm1938 [DOI] [PubMed] [Google Scholar]
- Nakano T., Kodama H., Honjo T. 1996. In vitro development of primitive and definitive erythrocytes from different precursors. Science. 272:722–724 10.1126/science.272.5262.722 [DOI] [PubMed] [Google Scholar]
- Nogami M., Kohda A., Taguchi H., Nakao M., Ikemura T., Okumura K. 2000. Relative locations of the centromere and imprinted SNRPN gene within chromosome 15 territories during the cell cycle in HL60 cells. J. Cell Sci. 113:2157–2165 [DOI] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Fraser P. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065–1071 10.1038/ng1423 [DOI] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy J.M., Dutriaux A. 1999. Gene targeting and somatic cell genetics—a rebirth or a coming of age? Trends Genet. 15:88–90 10.1016/S0168-9525(98)01689-8 [DOI] [PubMed] [Google Scholar]
- Shockett P., Difilippantonio M., Hellman N., Schatz D.G. 1995. A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc. Natl. Acad. Sci. USA. 92:6522–6526 10.1073/pnas.92.14.6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38:1348–1354 10.1038/ng1896 [DOI] [PubMed] [Google Scholar]
- Solevei I., Walter M., Cremer M., Habermann F., Schermelleh L., Cremer T. 2002. FISH on three-dimensionally preserved nuclei. FISH: A Practical Approach. Beatty B., Mai S., Squire J., Oxford University Press, Oxford: 119–154 [Google Scholar]
- Solovei I., Kreysing M., Lanctôt C., Kösem S., Peichl L., Cremer T., Guck J., Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 137:356–368 10.1016/j.cell.2009.01.052 [DOI] [PubMed] [Google Scholar]
- Swedlow J.R., Hirano T. 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell. 11:557–569 10.1016/S1097-2765(03)00103-5 [DOI] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 10:1453–1465 10.1016/S1097-2765(02)00781-5 [DOI] [PubMed] [Google Scholar]
- Tumbar T., Sudlow G., Belmont A.S. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354 10.1083/jcb.145.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschure P.J., van Der Kraan I., Manders E.M., van Driel R. 1999. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 147:13–24 10.1083/jcb.147.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser A.E., Eils R., Jauch A., Little G., Bakker P.J.M., Cremer T., Aten J.A. 1998. Spatial distributions of early and late replicating chromatin in interphase chromosome territories. Exp. Cell Res. 243:398–407 10.1006/excr.1998.4144 [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Chevret E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Pecinka A., Meister A., Schubert I., Lam E. 2005. DNA hypomethylation reduces homologous pairing of inserted tandem repeat arrays in somatic nuclei of Arabidopsis thaliana. Plant J. 44:531–540 10.1111/j.1365-313X.2005.02546.x [DOI] [PubMed] [Google Scholar]
- Wong M.L., Medrano J.F. 2005. Real-time PCR for mRNA quantitation. Biotechniques. 39:75–85 10.2144/05391RV01 [DOI] [PubMed] [Google Scholar]