Recruitment of the Crumbs–Stardust polarity complex depends on interactions between Bazooka and the Stardust PDZ domain and is regulated by aPKC-mediated phosphorylation.
Abstract
Apical–basal polarity in Drosophila melanogaster epithelia depends on several evolutionarily conserved proteins that have been assigned to two distinct protein complexes: the Bazooka (Baz)–PAR-6 (partitioning defective 6)–atypical protein kinase C (aPKC) complex and the Crumbs (Crb)–Stardust (Sdt) complex. These proteins operate in a functional hierarchy, in which Baz is required for the proper subcellular localization of all other proteins. We investigated how these proteins interact and how this interaction is regulated. We show that Baz recruits Sdt to the plasma membrane by direct interaction between the Postsynaptic density 95/Discs large/Zonula occludens 1 (PDZ) domain of Sdt and a region of Baz that contains a phosphorylation site for aPKC. Phosphorylation of Baz causes the dissociation of the Baz–Sdt complex. Overexpression of a nonphosphorylatable version of Baz blocks the dissociation of Sdt from Baz, causing phenotypes very similar to those of crb and sdt mutations. Our findings provide a molecular mechanism for the phosphorylation-dependent interaction between the Baz–PAR-3 and Crb complexes during the establishment of epithelial polarity.
Introduction
In Drosophila melanogaster, epithelial differentiation begins with the formation of spot adherens junctions during cellularization (Knust and Bossinger, 2002). By the coalescence of spot adherens junctions, the zonula adherens (ZA) is formed in early gastrulation as a junctional belt in the apical part of the lateral plasma membrane (Tepass and Hartenstein, 1994; McGill et al., 2009). Several proteins essential for the control of apical–basal polarity are localized in the apical plasma membrane domain and are enriched just apical to the ZA (Müller and Bossinger, 2003). The establishment of epithelial polarity and the assembly of the ZA in the ectodermal epithelium are regulated by a complex hierarchy of interacting proteins (Bilder et al., 2003; Johnson and Wodarz, 2003; Müller and Bossinger, 2003; Tanentzapf and Tepass, 2003; Harris and Peifer, 2004, 2005). The first protein complex relevant in this context is the PAR-3 (partitioning defective 3)–PAR-6–atypical PKC (aPKC) complex (Macara, 2004; Suzuki and Ohno, 2006). Its core component PAR-3 (Bazooka [Baz] in Drosophila) serves as a scaffold for aPKC and its regulator PAR-6 (Wodarz et al., 2000; Petronczki and Knoblich, 2001; Macara, 2004; Suzuki and Ohno, 2006). A recent study revealed that Baz localizes to the ZA, whereas PAR-6 and aPKC segregate from Baz and localize slightly more apical (Harris and Peifer, 2005). In the second protein complex required for the establishment of epithelial polarity, the cytoplasmic domain of the transmembrane protein Crumbs (Crb) binds to the Postsynaptic density 95/Discs large (Dlg)/Zonula occludens 1 (PDZ) domain of the membrane-associated guanylate kinase domain protein Stardust (Sdt; Bachmann et al., 2001; Hong et al., 2001). Sdt in turn recruits Pals1-associated tight junction protein (PATJ) and Lin-7 to the complex, which localizes at the apical plasma membrane (Bachmann et al., 2004; Bulgakova and Knust, 2009). Third, a group of proteins consisting of Dlg, Lethal giant larvae (Lgl), and Scribble (Scrib) localizes to the lateral plasma membrane domain and functions as an antagonist to the Crb–Sdt complex, thus restricting the expansion of the apical membrane domain (Bilder et al., 2003; Johnson and Wodarz, 2003; Tanentzapf and Tepass, 2003).
Several molecular interactions between the Crb–Sdt and the PAR-3 (Baz)–PAR-6–aPKC complexes have been uncovered: PAR-6 can bind directly to Crb and to Sdt, although up to now no particular function or mechanism for this binding has been described (Wang et al., 2004; Kempkens et al., 2006). In addition, the cytoplasmic tail of Crb can be phosphorylated by aPKC at two conserved threonine residues, which is required for its proper localization and function (Sotillos et al., 2004).
In this study, we demonstrate a new and functionally important link between both complexes, the transient formation of a complex between Baz and Sdt. The stability of this complex is regulated through phosphorylation of Baz by aPKC, which triggers the dissociation of the Baz–Sdt complex and thus allows the formation of the Crb–Sdt complex. Our results provide mechanistic insight into the molecular interactions between Baz, aPKC, Sdt, and Crb during the establishment of plasma membrane polarity. Because all of the proteins we analyzed in this study are evolutionarily conserved in all higher animals regarding both their structure and function, we expect that this is also true for the mechanisms regulating their interactions that we uncovered in this work.
Results and discussion
Phosphorylation of S980 is required for proper subcellular localization of Baz
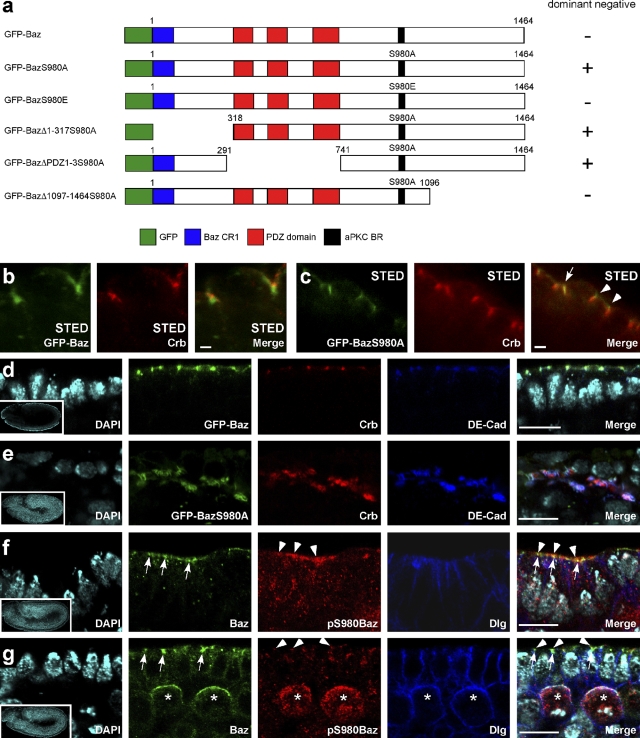
In mammalian epithelial cells, the overexpression of a version of PAR-3 that cannot be phosphorylated by aPKC-λ/ζ (PAR-3S827A) causes defects in the formation of tight junctions and in the establishment of apical–basal cell polarity after calcium switch (Nagai-Tamai et al., 2002). S980 of Drosophila Baz, which corresponds to S827 of PAR-3, is also phosphorylated by aPKC (Kim et al., 2009), but no particular function has been described for this phosphorylation event so far. Therefore, we investigated whether phosphorylation of Baz by aPKC at S980 might be required for the proper subcellular localization and function of Baz. Using stimulated emission depletion (STED) microscopy, we have been able to determine the exact subcellular localization of Baz, Crb, and Sdt relative to each other with a resolution <50 nm, in contrast to the resolution limit of ∼200 nm set by conventional confocal microscopy (Hell, 2009). Consistent with published data (Harris and Peifer, 2005), endogenous Baz as well as GFP-Baz (Fig. 1 a) always localized slightly basal to Crb (Fig. 1 b) and Sdt (not depicted), with a mean distance between the peaks of GFP-Baz and Crb of 268 ± 69 nm (n = 17). GFP-BazS980E (Fig. 1 a), which mimics constitutive phosphorylation of S980 of Baz, showed the same localization basal to Crb as wild-type Baz and GFP-Baz (Fig. S1 g). Staining with a phospho-specific antibody raised against a Baz peptide phosphorylated at S980 (Kim et al., 2009; Krahn et al., 2009) showed that this phosphorylated form of Baz only partially colocalized with the bulk of Baz and was concentrated in the most apical part of the region where Baz is localized (Fig. 1, f and g). In contrast, GFP-BazS980A (Fig. 1 a) did not have a defined localization with respect to Crb and Sdt and could frequently be found colocalized with or even apical of Crb and Sdt (Fig. 1 c). Collectively, these data indicate that phosphorylation of Baz at S980 is essential for the segregation of Baz at the ZA from the Crb–Sdt complex in the apical plasma membrane. This is consistent with our observation that GFP-Baz and GFP-BazS980E but not GFP-BazS980A rescued the lethality of embryos lacking maternal and zygotic baz expression.
Figure 1.
GFP-BazS980A does not localize properly and causes the formation of protein aggregates when overexpressed. (a) GFP-tagged versions of Baz used in this study. + or − indicate whether overexpression of these variants of Baz causes the dominant-negative phenotype described in Overexpression of nonphosphorylatable Baz phenocopies mutations in crb and sdt. (b) STED imaging reveals localization of GFP-Baz basal to Crb in the embryonic ectoderm. (c) STED imaging reveals colocalization of GFP-BazS980A with Crb (arrow) and localization of GFP-BazS980A apical to Crb (arrowheads) in the embryonic ectoderm. (d) GFP-Baz localizes in dots at the apex of the lateral plasma membrane. (e) Overexpressed GFP-BazS980A localizes to aggregates containing additional proteins, including Crb and DE-cadherin, that are mislocalized to the cytosol. (f and g) Subcellular localization of Baz phosphorylated at S980. Embryos at stage 9 were stained with an antibody that recognizes Baz irrespective of its phosphorylation status (Baz) and with an antibody that recognizes Baz only when it is phosphorylated at S980 (pS980Baz). Note that in the epithelium, both signals overlap only partially, with pS980Baz staining (arrowheads) being enriched at the apical border of larger spots stained with the conventional Baz antibody (arrows). (g) pS980Baz staining is also detectable in the apical cortex of mitotic neuroblasts (asterisks). (d–g) Insets show overviews of the embryos from which the high magnification images were taken. Baz transgenes were overexpressed with da::GAL4. See also Fig. S1. Bars: (b and c) 1 µm; (d–g) 10 µm.
Overexpression of nonphosphorylatable Baz phenocopies mutations in crb and sdt
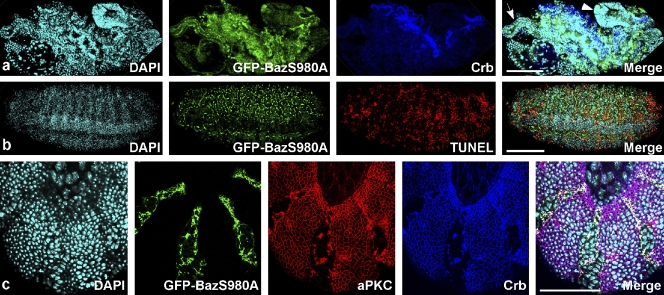
To determine whether the failure of Baz to segregate from the Crb–Sdt complex affects embryonic development, we overexpressed GFP-BazS980A with the UAS-GAL4 system (Brand and Perrimon, 1993). Strong overexpression of GFP-BazS980A triggered the formation of mislocalized aggregates that contained all proteins of the apical junctional complexes that we investigated (Drosophila epithelial cadherin [DE-cadherin], Armadillo, α-catenin, PAR-6, aPKC, Crb, Sdt, PATJ, and Lin-7; Fig. 1 e, Fig. S1, and not depicted). In contrast, Dlg as a marker for the lateral plasma membrane domain was excluded from these aggregates and localized normally at the cortex (unpublished data). We do not think that the formation of aggregates upon GFP-BazS980A overexpression is caused by nonspecific segregation of apical components because we observed these aggregates only in epithelia that express Sdt and Crb and not in neuroblasts and oocytes, although in these cell types some apical components are present, including aPKC and PAR-6. Moreover, we did not observe the formation of aggregates in embryos overexpressing GFP-Baz or GFP-BazS980E, which are both fully functional and rescue baz loss of function mutations. Upon GFP-BazS980A overexpression, the morphology of the epithelial monolayer was disrupted (Fig. 2 a), the cells rounded up, and most of the cells died by apoptosis in late embryogenesis (Fig. 2 b and compare Video 1 with Video 2). These dominant-negative effects of GFP-BazS980A overexpression were cell autonomous because upon overexpression in stripes using the en::GAL4 driver, only cells within the stripes showed mislocalization of aPKC and Crb (Fig. 2 c). Deletion of the N-terminal CR1 domain or the three PDZ domains did not affect the dominant-negative phenotype of GFP-BazS980A overexpression (Fig. 1 a and not depicted). In contrast, overexpression of a GFP-BazS980A version lacking the region from aa 1097–1464, which is required for membrane targeting of Baz (Krahn et al., 2010), did not cause dominant-negative effects (Fig. 1 a and not depicted). Thus, we conclude that GFP-BazS980A has to be localized to the plasma membrane to induce a dominant-negative phenotype.
Figure 2.
Overexpression of GFP-BazS980A disrupts epithelial morphogenesis and induces apoptosis. (a) A late stage embryo overexpressing GFP-BazS980A with da::GAL4 shows a severely disrupted morphology caused by the degeneration of the epidermis. The embryo is dorsally open, and internal organs including the foregut (arrow) and midgut (arrowhead) protrude to the outside. (b) Upon overexpression of GFP-BazS980A with da::GAL4, many epidermal cells die by apoptosis, marked by TUNEL labeling. GFP-BazS980A puncta correspond to aggregates such as those shown at higher magnification in Fig. 1 e. (c) The dominant-negative effect of GFP-BazS980A overexpression is cell autonomous. GFP-BazS980A was overexpressed under control of en::GAL4 in segmentally repeated stripes. Note that aPKC and Crb are lost from the plasma membrane in cells that overexpress GFP-BazS980A but localize to the cell outlines in the region between stripes. See also Fig. S2 and Videos 1 and 2. Bars: (a and b) 100 µm; (c) 50 µm.
In embryonic neuroblasts, GFP-BazS980A localized to the apical cortex like wild-type Baz (Kuchinke et al., 1998), without affecting the localization of cell fate determinants, spindle orientation, asymmetric cell division, or viability of the flies (Fig. S2 a and not depicted). Oocyte polarity was not affected upon GFP-BazS980A overexpression, and the mutant protein localized correctly to the anterior cortex of the oocyte (Fig. S2 b).
Genetic interactions of nonphosphorylatable Baz with cell polarity regulators
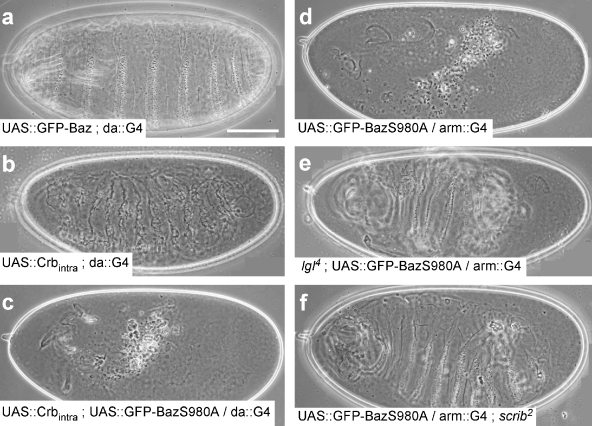
Proper cell polarity is the prerequisite for the secretion of a contiguous cuticle by the epidermis at the end of embryogenesis. Embryos homozygous for loss of function mutations in the cell polarity regulators crb, sdt, baz, aPKC, and PAR-6 secrete only little scraps of cuticle due to the degeneration of the epidermis (Wieschaus et al., 1984; Tepass et al., 1990; Bachmann et al., 2001; Hong et al., 2001; Kim et al., 2009). Whereas overexpression of full-length wild-type GFP-Baz did not affect viability of embryos and allowed secretion of a normal cuticle (Fig. 3 a), overexpression of GFP-BazS980A caused a cuticle phenotype very similar to crb or sdt mutant embryos (Fig. 3 d). In contrast, overexpression of Crbintra, consisting of the signal peptide, the transmembrane domain, and the cytoplasmic tail of Crb, resulted in ectopic secretion of cuticle (Fig. 3 b; Wodarz et al., 1995). Overexpression of GFP-BazS980A together with Crbintra resulted in the same phenotype as GFP-BazS980A overexpression alone, demonstrating that GFP-BazS980A overexpression is epistatic over Crbintra overexpression (Fig. 3 c). The cuticle phenotype of crb and sdt mutations can be strongly suppressed by simultaneous reduction in the levels of lgl, dlg, and scrib gene function (Bilder et al., 2003; Tanentzapf and Tepass, 2003), which has led to the model that the functions of Crb and Sdt are mainly required to suppress the activities of Lgl, Dlg, and Scrib. We observed a strong suppression of the cuticle phenotype caused by overexpression of GFP-BazS980A when these embryos lacked one or both wild-type copies of lgl (Fig. 3 e) or scrib (Fig. 3, compare d with f). Together, we conclude from our data that overexpression of GFP-BazS980A inhibits the activity of the Crb–Sdt complex and mimics crb and sdt loss of function phenotypes. This hypothesis also explains why the overexpression of GFP-BazS980A does not affect the polarity of neuroblasts and oocytes because Crb and Sdt are not expressed in these two cell types.
Figure 3.
Overexpression of GFP-BazS980A causes cuticle phenotypes similar to crb and sdt loss of function mutations. (a) Overexpression of GFP-Baz does not affect embryonic development and allows the formation of a normal cuticle. (b) Overexpression of the membrane-bound intracellular domain of Crb (Crbintra) causes the formation of an ectopic cuticle. (c and d) Co-overexpression of Crbintra and GFP-BazS980A (c) causes the same phenotype as overexpression of GFP-BazS980A alone (d). (e and f) Cuticle defects upon GFP-BazS980A overexpression are strongly suppressed when the embryo is at the same time mutant for lgl (e) or scrib (f). Genotypes are given in each panel. Bar, 100 µm.
Baz is required for recruitment of Sdt to the plasma membrane
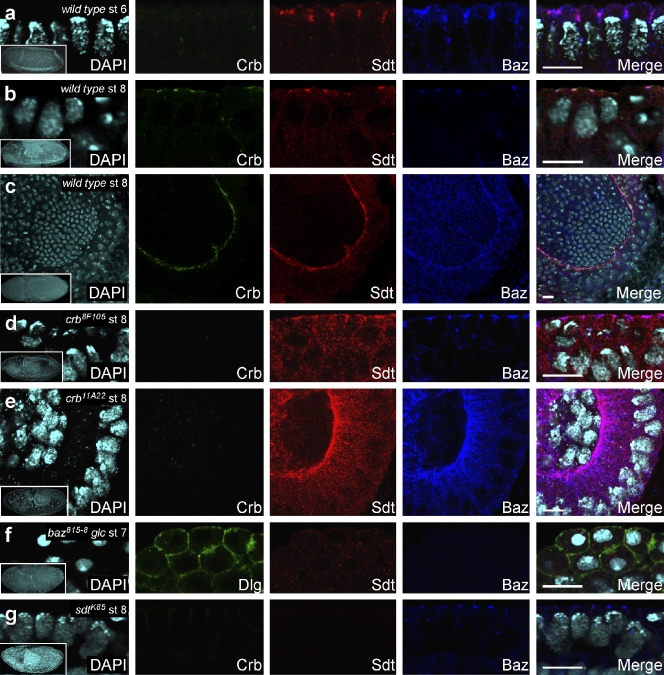
To investigate the functional interactions between Baz and the Crb–Sdt complex, we analyzed the subcellular localization of Baz, Crb, and Sdt in wild-type embryos and in embryos mutant for crb, sdt, and baz. In wild-type embryos at stage 6, Crb staining just started to become detectable, whereas Sdt was already robustly expressed and colocalized with Baz in the apical region of the lateral plasma membrane of the blastoderm epithelium (Fig. 4 a). From stage 7 onward, Sdt colocalized with Crb and partially also with Baz in all ectodermal epithelia (Fig. 4, b and c; Bachmann et al., 2001; Hong et al., 2001). In crb8F105 and crb11A22 mutant embryos at stage 8, a significant amount of Sdt remained colocalized with Baz in the apical region of the lateral plasma membrane (Fig. 4, d and e). In baz815-8 mutant embryos lacking both maternal and zygotic Baz at stage 7, neither Crb nor Sdt were detectable at the plasma membrane (Fig. 4 f and Fig. S3). In sdtK85 mutant embryos at stage 8, Crb was completely mislocalized, whereas Baz was still detectable in apicolateral spots at the membrane (Fig. 4 g). Together, our data show that Baz is necessary for membrane localization of Sdt in the complete absence of Crb, whereas both Crb and Sdt are dispensable for the apical membrane localization of Baz at early stages of embryonic development.
Figure 4.
Sdt colocalizes with Baz in the absence of Crb. (a) In a wild-type embryo at stage (st) 6, Crb is very weakly expressed, but Sdt and Baz colocalize in the apical portion of the lateral plasma membrane. (b) In the epidermis of a wild-type embryo at stage 8, Crb and Sdt colocalize slightly apical to Baz. (c) In the proctodeal invagination of a wild-type embryo at stage 8, Crb and Sdt colocalize slightly apical to Baz. (d and e) In embryos mutant for crb8F105 (d) and crb11A22 (e), Sdt and Baz colocalize in the absence of Crb. Note that the C-terminally truncated Crb8F105 protein is diffusely localized in the epidermis, and thus, the fluorescence signal is below the detection level. (f) In a baz815-8 mutant embryo derived from a germline clone (glc), Sdt is completely delocalized, whereas Dlg still localizes to the cortex. (g) In a sdtK85 mutant embryo, Crb is completely delocalized, but Baz still localizes to apical spots at the membrane. See also Fig. S3. (a–g) Insets show an overview of the embryo from which the respective high magnification images were taken. Bars, 10 µm.
Baz binds directly to the PDZ domain of Sdt
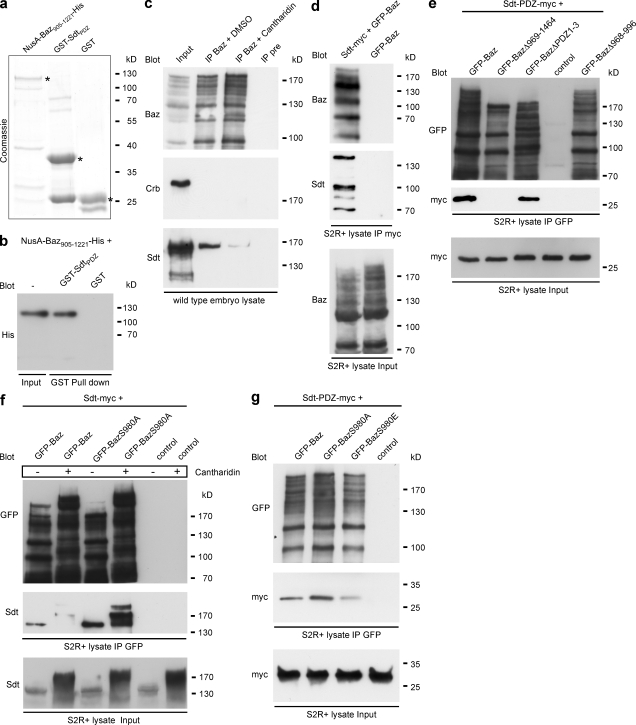
These data pointed to a function of Baz in the recruitment of Sdt to the membrane, independent of Crb. To test whether Baz directly binds to Sdt, we performed pull-down experiments with recombinant proteins expressed in Escherichia coli (Fig. 5 a). A His-tagged fragment of Baz comprising aa 905–1221 (NusA-Baz905–1221-His) was efficiently pulled down by a fusion protein of the PDZ domain of Sdt with GST (GST-SdtPDZ) but not by GST alone (Fig. 5 b). To check whether this interaction occurs also in vivo in wild-type embryos, we immunoprecipitated Baz and probed Western blots for the presence of Sdt and Crb in the Baz immunocomplex. We did not detect Crb in a complex with Baz, but Sdt immunoprecipitated together with Baz (Fig. 5 c). Myc-tagged full-length Sdt and a fragment of Sdt consisting only of the myc-tagged PDZ domain also coimmunoprecipitated with GFP-Baz in S2R+ cells (Fig. 5, d and e). These findings exclude the possibility that in vivo Baz and Sdt interact only indirectly via PAR-6, which can bind to both Baz (Petronczki and Knoblich, 2001) and to a region at the N terminus of Sdt (Wang et al., 2004). We used the same assay to narrow down the region of Baz that is required for binding to Sdt in vivo. Deletion of the three PDZ domains of Baz did not affect binding of Baz to Sdt-PDZ-myc, whereas deletion of the C terminus of Baz (aa 969–1464) or internal deletion of the so-called aPKC-binding region (aa 968–996; Nagai-Tamai et al., 2002) completely abolished the binding of Sdt-PDZ-myc to Baz (Fig. 5 e). Together, these data show that Baz and Sdt bind directly to each other, both in vitro and in vivo. The interaction is mediated by the PDZ domain of Sdt and depends on the presence of the region between aa 968 and 996 of Baz, which contains the aPKC phosphorylation site S980.
Figure 5.
Direct binding of Sdt to Baz is regulated by phosphorylation of BazS980. (a) Purified fusion proteins used for in vitro binding assays stained with Coomassie. (b) Western blot of GST pull-down experiment. (c) Coimmunoprecipitation of endogenous embryonic proteins in the absence or presence of the phosphatase inhibitor cantharidin. IP, immunoprecipitation. (d) Coimmunoprecipitation after transfection of GFP-Baz and Sdt-myc into S2R+ cells. (e) Mapping of the binding site of Baz for Sdt. GFP-Baz and deletion versions of Baz were cotransfected into S2R+ cells with Sdt-PDZ-myc. (f) Coimmunoprecipitation after cotransfection of GFP-Baz or GFP-BazS980A with Sdt-myc into S2R+ cells in the absence (−) or presence (+) of cantharidin. (g) Coimmunoprecipitation after cotransfection into S2R+ cells of Sdt-PDZ-myc with GFP-tagged versions of Baz.
Phosphorylation of S980 of Baz weakens the binding between Baz and Sdt
To test whether the phosphorylation status of Baz might affect the interaction between Baz and Sdt, we treated embryo lysates with the phosphatase inhibitor cantharidin, which prevents dephosphorylation of several sites in the Baz protein, including S980 (Krahn et al., 2009). Compared with the DMSO-treated control, the coimmunoprecipitation of Sdt with Baz was strongly reduced upon cantharidin treatment (Fig. 5 c), which suggested that phosphorylation of Baz diminishes the binding affinity between Baz and Sdt.
To investigate whether phosphorylation of S980 of Baz specifically affects the binding between Baz and Sdt, we cotransfected GFP-Baz and GFP-BazS980A together with myc-tagged full-length Sdt into S2R+ cells. GFP-Baz coimmunoprecipitated with Sdt, and the amount of coprecipitated Sdt was strongly reduced upon treatment of the cell lysates with cantharidin (Fig. 5 f). Compared with GFP-Baz, a significantly higher amount of Sdt coimmunoprecipitated with GFP-BazS980A, which did not decrease upon cantharidin treatment (Fig. 5 f), confirming that phosphorylation of S980 of Baz reduces the binding affinity between Baz and Sdt. To investigate the effect of S980 phosphorylation on the binding between Baz and Sdt by an additional approach, we compared the amounts of Sdt-PDZ-myc that coimmunoprecipitated with GFP-Baz, GFP-BazS980A, and the phosphomimetic version GFP-BazS980E. Consistent with our results obtained by phosphatase treatment, GFP-BazS980E bound much less Sdt-PDZ-myc than GFP-BazS980A and slightly less than GFP-Baz (Fig. 5 g).
Conclusions
Based on our results, we propose the following model for the interaction between the PAR-3 (Baz)–PAR-6–aPKC complex and the Crb–Sdt complex during the establishment of apical–basal cell polarity in early embryogenesis. During cellularization, shortly before Crb expression starts, Baz and Sdt form a complex that localizes to the apical region of the lateral plasma membrane. In this complex, the PDZ domain of Sdt binds to the region surrounding S980 of Baz, which is a phosphorylation target of aPKC. As long as S980 is not phosphorylated by aPKC, this complex is stable, and the PDZ domain of Sdt is not available for binding to the C terminus of Crb. Upon phosphorylation of S980 of Baz by aPKC, the binding between Baz and Sdt becomes weaker, causing the dissociation of the Baz–Sdt complex at the ZA and releasing Sdt for binding to Crb. This mechanism provides an explanation for the enrichment of the Crb–Sdt complex in the immediate vicinity of the ZA because Baz initially recruits Sdt to the ZA and then releases it locally for binding to Crb, which localizes to the apical plasma membrane domain. Whether there is direct competition between Baz and Crb for binding to Sdt remains to be further investigated. The separation of the ZA from the adjacent apical membrane domain may then be achieved by the recruitment of aPKC and PAR-6 to the Crb–Sdt complex via binding of PAR-6 to Sdt or directly to Crb (Wang et al., 2004; Kempkens et al., 2006). When the dissociation of the Baz–Sdt complex is blocked, for instance by overexpression of GFP-BazS980A, the Crb–Sdt complex cannot form, which results in phenotypes very similar to those of crb or sdt loss of function mutations (Tepass et al., 1990; Bachmann et al., 2001; Hong et al., 2001; Kim et al., 2009). This hypothesis is supported by the observation that the cuticle phenotype of GFP-BazS980A overexpression was strongly suppressed by concomitant reduction of Lgl or Scrib activity, as was reported for crb and sdt mutant phenotypes (Bilder et al., 2003; Tanentzapf and Tepass, 2003). Also consistent with this model is our observation that aPKC mutant embryos derived from germline clones, in which phosphorylation of S980 of Baz cannot occur, exhibit a very similar epithelial phenotype as embryos mutant for crb or sdt (Kim et al., 2009). We do not think that the dominant-negative phenotype of GFP-BazS980A overexpression is primarily caused by altered binding of aPKC to Baz or misregulation of aPKC kinase activity because, in this case, we would also expect dominant-negative effects upon overexpression of GFP-BazS980A in neuroblasts, in which the regulation of aPKC kinase activity is crucial for asymmetric cell division (Betschinger et al., 2003; Wirtz-Peitz et al., 2008).
Two recently published manuscripts also reported dominant-negative phenotypes in epithelial development similar to the ones we describe in this study upon overexpression of GFP-BazS980A (Morais-de-Sá et al., 2010; Walther and Pichaud, 2010). However, these phenotypes were attributed predominantly to the effect of phosphorylation of BazS980 by aPKC on the binding affinity between aPKC and Baz. According to their model, phosphorylation of Baz by aPKC leads to the dissociation of the Baz–aPKC complex, which triggers the segregation of Baz to the ZA and of aPKC and PAR-6 to the apical membrane domain. Although our data also underline the importance of the phosphorylation of Baz at S980 by aPKC, our model goes beyond the one proposed by Morais-de-Sá et al. (2010) and Walther and Pichaud (2010) by showing that all of the phenotypes observed upon overexpression of GFP-BazS980A can be explained by the phosphorylation-dependent binding of Sdt to Baz.
It has recently been proposed that phosphorylation of mammalian PAR-3 by aPKC-λ/ζ is required for separation of PAR-3 from aPKC and PAR-6, which is the prerequisite for apical domain formation in mammalian epithelia (Horikoshi et al., 2009; McCaffrey and Macara, 2009). One of these studies furthermore proposed that binding of aPKC and PAR-6 to PAR-3 may be an important intermediate step to recruit aPKC and PAR-6 to the membrane before they dissociate from PAR-3 and bind to other apical membrane–anchoring factors such as Cdc42 or the Crb–PALS-1 complex (Horikoshi et al., 2009). Our results are conceptually similar but further extend this model by demonstrating for the first time the direct interaction between Baz and Sdt. These findings represent an important advancement in our understanding of the molecular mechanisms that control the establishment of apical–basal cell polarity in the Drosophila ectoderm. To fully understand this process, it will be important to know how aPKC is activated in early embryogenesis and how the phosphorylation of Baz changes the binding interface between Baz, Sdt, and aPKC. Because all of the components of the molecular mechanism that we describe in this study are conserved in evolution, we are eager to see whether their interactions are regulated in a similar way during the polarization of mammalian epithelia.
Materials and methods
Fly stocks and genetics
The following mutant alleles were used in this study: crb11A22 (Jürgens et al., 1984), crb8F105 (Wodarz et al., 1993), sdtK85 (provided by E. Knust, Max Planck Institute of Cell Biology and Genetics, Dresden, Germany; Berger et al., 2007), lgl4 (provided by D. Bilder, University of California, Berkeley, Berkeley, CA; Mechler et al., 1985; Bilder et al., 2003; Tanentzapf and Tepass, 2003), and scrib2 (Bilder et al., 2003; Tanentzapf and Tepass, 2003). baz815-8 (McKim et al., 1996; Krahn et al., 2010) germline clone embryos were obtained using the Flippase recombination target–dominant female sterile method (Chou and Perrimon, 1992). UAS::GFP-Baz transgenes were generated using standard germline transformation. da::GAL4, en::GAL4, arm::GAL4, nos::GAL4, and wor::GAL4 driver lines were obtained from the Bloomington Drosophila Stock Center.
Immunohistochemistry
Embryos were fixed in a 1:1 mixture of 4% formaldehyde, phosphate buffer, pH 7.4, and heptane for 20 min. After removal of the vitelline envelope by vigorous shaking in a 1:1 mixture of methanol and heptane, embryos were rehydrated in PBS and 0.1% Tween 20 (PBT) for 20 min and then incubated with primary antibodies in PBT and 5% normal horse serum. The primary antibodies used were rabbit anti–PKC-ζ C20 (1:1,000; Santa Cruz Biotechnology, Inc.), rabbit anti-Baz (1:1,000; Wodarz et al., 1999), rabbit anti–Baz phospho-S980 (1:100; Krahn et al., 2009), mouse anti-Crb Cq4 (1:50; Developmental Studies Hybridoma Bank; Tepass and Knust, 1993), mouse anti-Sdt (1:20; provided by E. Knust; Berger et al., 2007), rabbit anti–Lin-7 (1:500; provided by E. Knust; Bachmann et al., 2004), rabbit anti-PATJ (1:1,000; provided by E. Knust; Richard et al., 2006), guinea pig anti-Mira (1:1,000; Kim et al., 2009), rat anti–DE-cadherin DCAD2 (1:20; Developmental Studies Hybridoma Bank; Oda et al., 1994), mouse anti-Dlg 4F3 (1:50; Developmental Studies Hybridoma Bank), rabbit anti-Staufen (1:1,000; provided by D. St Johnston, Gurdon Institute, Cambridge, England, UK; St Johnston et al., 1991), mouse anti-Gurken 1D12 (1:10; Developmental Studies Hybridoma Bank), and mouse anti-GFP 3E6 (1:1,000; Invitrogen). DNA was stained with DAPI (Invitrogen). Secondary antibodies conjugated to Cy2 and Cy3 were obtained from Jackson ImmunoResearch Laboratories, Inc. Secondary antibodies conjugated to Alexa Fluor 647 were obtained from Invitrogen. After repeated washing in PBT, embryos were mounted in Mowiol 4-88 (Polysciences Europe) supplemented with 1,4-diazabicyclo (2.2.2) octane (DABCO). TUNEL assays, for detection of cell death in situ, were performed with an in situ cell death detection kit (Roche) according to the manufacturer’s instructions (Wang et al., 1999). Images were taken on a confocal microscope (LSM 510 Meta; Carl Zeiss, Inc.) using 25× NA 0.8 Plan-Neofluar and 63× NA 1.40 Plan-Apochromat objectives and processed using Photoshop (Adobe).
STED microscopy
Embryos were fixed and incubated with primary antibodies as described in the previous section before being incubated with the following secondary antibodies: ATTO 594 (ATTO-TEC GmbH) goat anti–rabbit IgG (dianova GmbH) and KK 114 (provided by K. Kolmakov and V. Belov, Max-Planck-Institut für Biophysikalische Chemie, Göttingen, Germany) sheep anti–mouse IgG (dianova GmbH). Two-color STED images were recorded with a custom-built STED microscope that combined two pairs of excitation and STED laser beams all derived from a single supercontinuum fiber laser source similar to the one described previously (Wildanger et al., 2008). Excitation wavelengths were 570 ± 5 nm (ATTO 594) and 650 ± 5 nm (KK 114), and STED wavelengths were 720 ± 20 nm (ATTO 594) and 755 ± 20 nm (KK 114). The fluorescence was detected in the spectral ranges of 600–640 nm for ATTO 594 and 660–690 nm for KK 114. Apart from the elimination of the cross talk between the two detection channels by means of linear unmixing, all image data shown in this study and used for subsequent analysis are raw data; in particular, no deconvolution algorithms were used.
Antibodies and Western blotting
Primary antibodies were used for Western blotting according to standard procedures as follows: mouse anti-Sdt (1:100; provided by E. Knust; Berger et al., 2007), mouse anti-Crb Cq4 (1:20; Developmental Studies Hybridoma Bank; Tepass and Knust, 1993), rabbit anti-Baz (1:2,000; Wodarz et al., 1999), rabbit anti–PKC-ζ C20 (1:2,000; Santa Cruz Biotechnology, Inc.), rabbit anti-phospho–PKC-ζ T410 (1:1,000; Santa Cruz Biotechnology, Inc.), rabbit anti-phospho–PKC-ι T560 (1:500; Abcam), rabbit antiactin (1:1,000; Sigma-Aldrich), mouse anti-GFP (1:1,000; Roche), mouse anti-His 4A12E4 (1:1,000; Invitrogen), and mouse anti-myc 9E10 (1:100; Developmental Studies Hybridoma Bank).
Immunoprecipitation
For immunoprecipitations, wild-type embryos from an overnight collection were dechorionated and lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, and 50 mM Tris-HCl, pH 7.5) supplemented with protease inhibitors. After centrifugation, 2 µl of rabbit anti-Baz (Wodarz et al., 1999) or 2 µl of the corresponding preimmune serum was added to cell lysate. Immunocomplexes were harvested using protein A/G–conjugated agarose (Roche), washed five times in lysis buffer, and boiled in 2× SDS sample buffer before SDS-PAGE and Western blotting. Lysates from S2R+ cells were processed accordingly, and Sdt-myc was immunoprecipitated with anti-myc antibody 9E10 (Developmental Studies Hybridoma Bank). GFP-tagged versions of Baz were immunoprecipitated with GFP-Binder (ChromoTek).
GST pull-down
The PDZ domain of Sdt fused to GST (provided by E. Knust) was expressed in BL-21–competent bacterial cells and purified using glutathione beads (GE Healthcare). A fragment of Baz encompassing the aPKC phosphorylation site S980 (Baz905–1221) was cloned into the NGWA vector (provided by D. Busso, Université Louis Pasteur, Illkirch, France; Busso et al., 2005). After expression in BL-21–competent bacterial cells, NusA-Baz905–1221-His was purified using Protino-Ni-TED columns (MACHEREY-NAGEL). For pull-down experiments, ∼10 µg NusA-Baz905–1221-His was incubated with equal amounts of either GST-SdtPDZ or GST bound to glutathione beads in lysis buffer for 2 h at 4°C.
Online supplemental material
Fig. S1 is related to Fig. 1 and shows the mislocalization of PATJ, Sdt, and Lin-7 upon GFP-BazS980A overexpression and also the subcellular localization of GFP-BazS980E in the embryonic epidermis. Fig. S2 is related to Fig. 2 and shows that GFP-BazS980A localizes like wild-type Baz in embryonic neuroblasts and in oocytes. Fig. S3 is related to Fig. 4 and shows that Crb and Sdt are not localized to the plasma membrane in embryos derived from baz germline clones. Video 1 is related to Fig. 2 and shows the normal development of an embryo overexpressing GFP-Baz. Video 2 is related to Fig. 2 and shows the dramatically abnormal development of an embryo overexpressing GFP-BazS980A. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006029/DC1.
Acknowledgments
We thank E. Knust for providing numerous antibodies, fly stocks, and DNAs. We also thank D. Bilder, D. Busso, V. Belov, K. Kolmakov, D. St Johnston, U. Tepass, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank for sending reagents. We thank D. St Johnston, F. Pichaud, S. Hell, and members of the Wodarz laboratory for discussion. M. Honemann-Capito and K. Curth provided technical assistance.
This work was supported by grants of the Deutsche Forschungsgemeinschaft to A. Wodarz (priority program SPP 1111 “Cell Polarity”; Forschungszentrum der Deutschen Forschungsgemeinschaft für Molekularphysiologie des Gehirns [CMPB]).
Footnotes
Abbreviations used in this paper:
- aPKC
- atypical PKC
- Baz
- Bazooka
- Crb
- Crumbs
- DE-cadherin
- Drosophila epithelial cadherin
- Dlg
- Discs large
- Lgl
- Lethal giant larvae
- PATJ
- Pals1-associated tight junction protein
- PDZ
- Postsynaptic density 95/Dlg/Zonula occludens 1
- Scrib
- Scribble
- Sdt
- Stardust
- STED
- stimulated emission depletion
- ZA
- zonula adherens
References
- Bachmann A., Schneider M., Theilenberg E., Grawe F., Knust E. 2001. Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature. 414:638–643 10.1038/414638a [DOI] [PubMed] [Google Scholar]
- Bachmann A., Timmer M., Sierralta J., Pietrini G., Gundelfinger E.D., Knust E., Thomas U. 2004. Cell type-specific recruitment of Drosophila Lin-7 to distinct MAGUK-based protein complexes defines novel roles for Sdt and Dlg-S97. J. Cell Sci. 117:1899–1909 10.1242/jcs.01029 [DOI] [PubMed] [Google Scholar]
- Berger S., Bulgakova N.A., Grawe F., Johnson K., Knust E. 2007. Unraveling the genetic complexity of Drosophila stardust during photoreceptor morphogenesis and prevention of light-induced degeneration. Genetics. 176:2189–2200 10.1534/genetics.107.071449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 422:326–330 10.1038/nature01486 [DOI] [PubMed] [Google Scholar]
- Bilder D., Schober M., Perrimon N. 2003. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 5:53–58 10.1038/ncb897 [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415 [DOI] [PubMed] [Google Scholar]
- Bulgakova N.A., Knust E. 2009. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J. Cell Sci. 122:2587–2596 10.1242/jcs.023648 [DOI] [PubMed] [Google Scholar]
- Busso D., Delagoutte-Busso B., Moras D. 2005. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 343:313–321 10.1016/j.ab.2005.05.015 [DOI] [PubMed] [Google Scholar]
- Chou T.B., Perrimon N. 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 131:643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J., Peifer M. 2004. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 167:135–147 10.1083/jcb.200406024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J., Peifer M. 2005. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J. Cell Biol. 170:813–823 10.1083/jcb.200505127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell S.W. 2009. Microscopy and its focal switch. Nat. Methods. 6:24–32 10.1038/nmeth.1291 [DOI] [PubMed] [Google Scholar]
- Hong Y., Stronach B., Perrimon N., Jan L.Y., Jan Y.N. 2001. Drosophila Stardust interacts with Crumbs to control polarity of epithelia but not neuroblasts. Nature. 414:634–638 10.1038/414634a [DOI] [PubMed] [Google Scholar]
- Horikoshi Y., Suzuki A., Yamanaka T., Sasaki K., Mizuno K., Sawada H., Yonemura S., Ohno S. 2009. Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J. Cell Sci. 122:1595–1606 10.1242/jcs.043174 [DOI] [PubMed] [Google Scholar]
- Johnson K., Wodarz A. 2003. A genetic hierarchy controlling cell polarity. Nat. Cell Biol. 5:12–14 10.1038/ncb0103-12 [DOI] [PubMed] [Google Scholar]
- Jürgens G., Wieschaus E., Nüsslein-Volhard C., Kluding H. 1984. Mutations affecting the pattern of the larval cuticle of Drosophila melanogaster. II. Zygotic loci on the third chromosome. Rouxs Arch. Dev. Biol. 193:283–295 10.1007/BF00848157 [DOI] [PubMed] [Google Scholar]
- Kempkens O., Médina E., Fernandez-Ballester G., Ozüyaman S., Le Bivic A., Serrano L., Knust E. 2006. Computer modelling in combination with in vitro studies reveals similar binding affinities of Drosophila Crumbs for the PDZ domains of Stardust and DmPar-6. Eur. J. Cell Biol. 85:753–767 10.1016/j.ejcb.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Kim S., Gailite I., Moussian B., Luschnig S., Goette M., Fricke K., Honemann-Capito M., Grubmüller H., Wodarz A. 2009. Kinase-activity-independent functions of atypical protein kinase C in Drosophila. J. Cell Sci. 122:3759–3771 10.1242/jcs.052514 [DOI] [PubMed] [Google Scholar]
- Knust E., Bossinger O. 2002. Composition and formation of intercellular junctions in epithelial cells. Science. 298:1955–1959 10.1126/science.1072161 [DOI] [PubMed] [Google Scholar]
- Krahn M.P., Egger-Adam D., Wodarz A. 2009. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev. Cell. 16:901–908 10.1016/j.devcel.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Krahn M.P., Klopfenstein D.R., Fischer N., Wodarz A. 2010. Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 20:636–642 10.1016/j.cub.2010.01.065 [DOI] [PubMed] [Google Scholar]
- Kuchinke U., Grawe F., Knust E. 1998. Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr. Biol. 8:1357–1365 10.1016/S0960-9822(98)00016-5 [DOI] [PubMed] [Google Scholar]
- Macara I.G. 2004. Parsing the polarity code. Nat. Rev. Mol. Cell Biol. 5:220–231 10.1038/nrm1332 [DOI] [PubMed] [Google Scholar]
- McCaffrey L.M., Macara I.G. 2009. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 23:1450–1460 10.1101/gad.1795909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M.A., McKinley R.F., Harris T.J. 2009. Independent cadherin–catenin and Bazooka clusters interact to assemble adherens junctions. J. Cell Biol. 185:787–796 10.1083/jcb.200812146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S., Dahmus J.B., Hawley R.S. 1996. Cloning of the Drosophila melanogaster meiotic recombination gene mei-218: a genetic and molecular analysis of interval 15E. Genetics. 144:215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechler B.M., McGinnis W., Gehring W.J. 1985. Molecular cloning of lethal(2)giant larvae, a recessive oncogene of Drosophila melanogaster. EMBO J. 4:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais-de-Sá E., Mirouse V., St Johnston D. 2010. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 141:509–523 10.1016/j.cell.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.A., Bossinger O. 2003. Molecular networks controlling epithelial cell polarity in development. Mech. Dev. 120:1231–1256 10.1016/j.mod.2003.06.001 [DOI] [PubMed] [Google Scholar]
- Nagai-Tamai Y., Mizuno K., Hirose T., Suzuki A., Ohno S. 2002. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 7:1161–1171 10.1046/j.1365-2443.2002.00590.x [DOI] [PubMed] [Google Scholar]
- Oda H., Uemura T., Harada Y., Iwai Y., Takeichi M. 1994. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev. Biol. 165:716–726 10.1006/dbio.1994.1287 [DOI] [PubMed] [Google Scholar]
- Petronczki M., Knoblich J.A. 2001. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3:43–49 10.1038/35050550 [DOI] [PubMed] [Google Scholar]
- Richard M., Grawe F., Knust E. 2006. DPATJ plays a role in retinal morphogenesis and protects against light-dependent degeneration of photoreceptor cells in the Drosophila eye. Dev. Dyn. 235:895–907 10.1002/dvdy.20595 [DOI] [PubMed] [Google Scholar]
- Sotillos S., Díaz-Meco M.T., Caminero E., Moscat J., Campuzano S. 2004. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J. Cell Biol. 166:549–557 10.1083/jcb.200311031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Beuchle D., Nüsslein-Volhard C. 1991. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 66:51–63 10.1016/0092-8674(91)90138-O [DOI] [PubMed] [Google Scholar]
- Suzuki A., Ohno S. 2006. The PAR-aPKC system: lessons in polarity. J. Cell Sci. 119:979–987 10.1242/jcs.02898 [DOI] [PubMed] [Google Scholar]
- Tanentzapf G., Tepass U. 2003. Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5:46–52 10.1038/ncb896 [DOI] [PubMed] [Google Scholar]
- Tepass U., Hartenstein V. 1994. The development of cellular junctions in the Drosophila embryo. Dev. Biol. 161:563–596 10.1006/dbio.1994.1054 [DOI] [PubMed] [Google Scholar]
- Tepass U., Knust E. 1993. Crumbs and stardust act in a genetic pathway that controls the organization of epithelia in Drosophila melanogaster. Dev. Biol. 159:311–326 10.1006/dbio.1993.1243 [DOI] [PubMed] [Google Scholar]
- Tepass U., Theres C., Knust E. 1990. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 61:787–799 10.1016/0092-8674(90)90189-L [DOI] [PubMed] [Google Scholar]
- Walther R.F., Pichaud F. 2010. Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr. Biol. 20:1065–1074 10.1016/j.cub.2010.04.049 [DOI] [PubMed] [Google Scholar]
- Wang S.L., Hawkins C.J., Yoo S.J., Müller H.A., Hay B.A. 1999. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 98:453–463 10.1016/S0092-8674(00)81974-1 [DOI] [PubMed] [Google Scholar]
- Wang Q., Hurd T.W., Margolis B. 2004. Tight junction protein Par6 interacts with an evolutionarily conserved region in the amino terminus of PALS1/stardust. J. Biol. Chem. 279:30715–30721 10.1074/jbc.M401930200 [DOI] [PubMed] [Google Scholar]
- Wieschaus E., Nüsslein-Volhard C., Jürgens G. 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. III. Zygotic loci on the X chromosome and fourth chromosome. Rouxs Arch. Dev. Biol. 193:296–307 10.1007/BF00848158 [DOI] [PubMed] [Google Scholar]
- Wildanger D., Rittweger E., Kastrup L., Hell S.W. 2008. STED microscopy with a supercontinuum laser source. Opt. Express. 16:9614–9621 10.1364/OE.16.009614 [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., Knoblich J.A. 2008. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 135:161–173 10.1016/j.cell.2008.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Grawe F., Knust E. 1993. CRUMBS is involved in the control of apical protein targeting during Drosophila epithelial development. Mech. Dev. 44:175–187 10.1016/0925-4773(93)90066-7 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Hinz U., Engelbert M., Knust E. 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 82:67–76 10.1016/0092-8674(95)90053-5 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., Knust E. 1999. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 402:544–547 10.1038/990128 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Grimm A., Knust E. 2000. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150:1361–1374 10.1083/jcb.150.6.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]