Abstract
We report the establishment of a reverse-genetics system for the rescue of recombinant influenza C/JJ/50 virus from seven plasmids. The nucleotide sequence of the whole C/JJ/50 genome was determined and full-length cDNAs were cloned into an RNA pol I/pol II-based bidirectional vector. Transfection of Vero cells and subsequent amplification on MDCK cells yielded viral HA titres of 128. The utility of this bidirectional approach was proved by generating a reassortant virus encoding the NS segment from strain C/JHB/1/66 and a virus with mutations in the noncoding ends of PB1. The latter virus, which has a base-pair mutation within the proposed double-stranded region of the PB1 termini, exhibited impaired replication. In conclusion, our efficient seven-plasmid system for the rescue of recombinant influenza C virus may be used to study the influenza C virus life cycle in more detail and for generation of influenza C virus-based vectors.
Keywords: Influenza C virus, reverse genetics, reassortant virus, noncoding ends, panhandle/corkscrew RNA model
INTRODUCTION
Influenza C virus is a member of the Orthomyxoviridae, which are characterised by a segmented single-stranded RNA genome of negative polarity. Whereas influenza A and B viruses bear eight RNA segments, the genome of influenza C virus comprises seven RNA segments. The two other family members, Thogotovirus and Isavirus, have six and eight RNA segments, respectively.
The first described reverse-genetics system for influenza A viruses was based on in vitro transcribed viral RNA (vRNAs), which were assembled with purified polymerase proteins (PB2, PB1, PA) and nucleoprotein (NP) to form ribonucleoprotein (RNP) complexes. RNPs were transfected into eukaryotic cells, and subsequent infection with a helper virus resulted in generation of recombinant viruses. More recently, systems for the generation of recombinant orthomyxoviruses entirely from cloned cDNAs were established for influenza A virus, influenza B virus, and Thogoto virus. Here, initial studies were based on the transfection of at least 12 plasmids: Eight plasmids (in case of influenza A and B viruses) harbouring the full-length cDNAs cloned between human RNA polymerase I promoter and murine RNA polymerase I terminator for vRNA replication, as well as plasmids for protein expression of at least PB2, PB1, PA, and NP. Subsequently, bidirectional vector systems were established for influenza A and B viruses to restrict the number of plasmids to be transfected to eight: Full-length cDNAs were inserted between RNA polymerase I promoter and terminator as well as RNA polymerase II promoter and termination signal in opposite directions to yield all eight vRNAs and all viral mRNAs simultaneously (reviewed in: Palese and Shaw, 2007). Since then, two groups have developed a system for the generation of recombinant influenza C viruses (Crescenzo-Chaigne and van der Werf, 2007; Muraki et al, 2007), using seven plasmids for production of vRNAs plus protein expression plasmids. Here, we describe the first reverse-genetics system for influenza C virus, which is based on tranfection of only seven bidirectional plasmids.
Having established this efficient and easy to handle system, we generated a reassortant virus harbouring the six large segments from strain C/JJ/50 and the NS segment from C/JHG/66. We also applied our system to study the effect of mutations within the noncoding (nc) ends of one viral segment.
The coding region of each genomic influenza virus RNA is flanked by nc sequences at both ends of the segment. Extensive studies have been carried out to elucidate the nature and function of the nc regions mainly of influenza A viruses. The ultimate nucleotides are highly conserved among the segments and are partially complementary, which allows a base-paired duplex region of four to eight nucleotides between the 3'end (starting from nucleotide 10) and the 5'end (starting from nucleotide 11' – note that nucleotides are counted from both ends, 5' nucleotides are marked with a prime) in case of influenza A virus. This partially double-stranded region was first described as a panhandle/fork structure and was found to be required for vRNA and cRNA promoter activity, endonuclease activity of the viral polymerase complex, and mRNA polyadenylation (reviewed in: Palese and Shaw, 2007). Flick and colleagues (Flick et al, 1996; Flick and Hobom, 1999) extended this model introducing a corkscrew structure, according to which the ultimate nine nucleotides of each end form a local secondary structure. Analysis of the nc ends of influenza C virus revealed that a double-stranded region of at least five base pairs is possible between nucleotides 10 to 14 of the 3'end and nucleotides 11' to 15' of the 5'end, consistent with the panhandle/ corkscrew model for influenza A virus.
In the present study, we introduced a single mutation as well as a base-pair substitution into the nc ends of segment PB1. No recombinant virus could be rescued with the single nucleotide mutation. When the PB1 construct with the double mutation restoring the double-stranded region was employed, we succeeded in rescuing recombinant virus, which exhibited an impaired growth in comparison to wild-type virus.
MATERIALS AND METHODS
Cells and Viruses
Vero and MDCK cells were maintained in Dulbecco's modified Eagle's medium and minimal essential medium, respectively, supplemented with 10% (v/v) fetal calf serum and 2 mM L-glutamine. Cells were grown at 37°C and 5% (v/v) CO2 prior to infection. Wild-type viruses of the strains C/JJ/50 and C/JHB/1/66 were grown in the amniotic cavity of 9-day-old embryonated chicken eggs at 33°C.
Sequencing of 3'- and 5'-noncoding ends of viral RNA segments
In order to determine the 3'- and 5'-nc regions of the influenza C/JJ/50 virus, first strand cDNA synthesis was performed using the SMART technology (Clontech) with modifications according to Matz (Matz, 2003), and nc ends were amplified by 3' and 5' RACE-PCR. First, virus was purified by ultracentrifugation through a 20% (w/v) sucrose cushion for 90min at 116,000xg and 4°C, and then treated with benzonase (Sigma) at room temperature for 30min to eliminate host cell-derived nucleic acids. Then, viral RNA was isolated with the QiaAmp viral RNA mini kit (Qiagen) largely according to the manufacturer's instructions, except that no carrier-RNA was added, and vRNA was added to a polyadenylation reaction using the Poly(A) Tailing Kit from Ambion. After polyadenylation, vRNA was again purified with the QiaAmp viral RNA mini Kit (with the addition of carrier-RNA). First strand cDNA was prepared using RevertAidTM H Minus M-MuLV Reverse Transcriptase (Fermentas), using the primer TRsa, 5'CGCAGTCGG TACTTTTTTTTTTTTTTTTTTCA, which base-pairs with the oligo(A) tail of the vRNA and adds a known sequence to the 5'end of the cDNA, and the primer TS, 5'AAGCAGT GGTATCAACGCAGAGTACGCrGrGrG. The oligo(rG) of the latter primer can base-pair with CCC added to the 3'end of the cDNA by M-MuLV Reverse Transcriptase, and the enzyme switches the templates and continues replication using the TS-oligo as template. The viral 5'-nc ends were then amplified with internal forward primers and primer TS-PCR, 5'AAGCAGTGGTATCA ACGCAGAGT, while the viral 3'ends were manifolded with primer TRsa and internal reverse primers. Finally, PCR products were purified with the Wizard SV Gel and PCR Clean-Up System from Promega and were sequenced by Eurofins MWG Operon (Germany). Oligonucleotide primers were obtained from VBC-Biotech (Austria). Sequences of internal primers and PCR programmes can be provided on request.
Construction of a bidirectional plasmid and insertion of viral segments
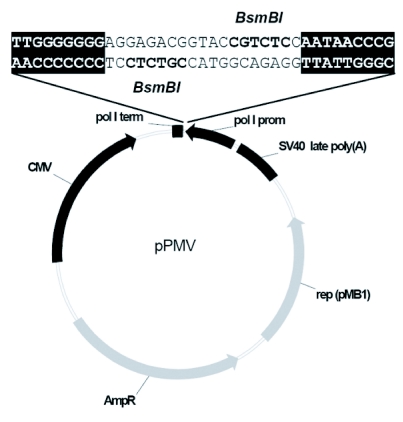
In the first step to construct the bidirectional expression plasmid pPMV, the CMV promoter, the multiple cloning site and the SV40 polyadenylation signal from the pCI vector (Promega) were inserted into the EcoO109I and SapI restriction sites of pUC18. Then, 236 nucleotides of the human RNA polymerase I promoter, two BsmBI restriction sites and 33 nucleotides of the murine RNA polymerase I terminator sequence were inserted into the XbaI/XhoI window between the SV40 poly(A) signal and the CMV promoter (Figure 1). The sequence of vector pPMV is available in the GenBank Database. Viral RNA from egg-grown influenza C/JJ/50 virus was prepared with the QiaAmp viral RNA mini kit (Qiagen) according to the manufacturer's instructions, vRNA was reverse-transcribed with RevertAidTM H Minus M-MuLV Reverse Transcriptase (Fermentas), and the seven full-length viral segments were amplified with primers complementary to the last twelve nucleotides using Herculase II Fusion DNA Polymerase (Stratagene). Four silent mutations were cloned into segment P3 by site-directed mutagenesis. The seven segments were inserted into pPMV via BsmBI restriction sites yielding the seven bidirectional plasmids pPMV-PB2, -PB1, -P3m, -HEF, -NP, -M, and -NS. To generate a bidirectional vector encoding the NS segment from strain C/JHB/1/66, vRNA from this strain was extracted, and the segment was amplified as described above. For cloning of pPMV-PB1-5'm, where the wild-type A at position 13' of the viral 5'-nc sequence is substituted by a G, cDNA was amplified with the 12-nucleotide primer complementary to the 3'end as for wild-type PB1 together with a 13-nucleotides long primer harbouring the nucleotide substitution for the 5'end. Plasmid pPMV-PB1-5'-3'm has the nucleotide substitution at position 13' of the 5'end as described above and an additional substitution at position 12 of the 3'end: The wild-type U was replaced by a C, again by site-directed mutagenesis. At least three clones of every construct were sequenced. Primer sequences and PCR programmes are available upon request.
Figure 1.
The cloning vector pPMV allows transcription of inserted cDNA in both orientations by opposed promoters and termination signals. The seven influenza C virus cDNA segments were ligated into the two BsmBI restriction sites leading to nucleotide-exact insertion between polymerase I terminator and promoter for vRNA replication. mRNA is synthesised by CMV promoter and SV40 poly(A) signal. (In bold: BsmBI restriction sites. Shaded: End sequences of murine polymerase I terminator and human polymerase I promoter.)
Transfection and rescue of recombinant viruses
Ninty percent confluent Vero cells, seeded on 35mm plates, were transfected with 0.5μg per bidirectional plasmid and 10μl Lipofectamine 2000 (Invitrogen). Transfection was performed in Opti-MEM with Glutamax (Invitrogen) according to the manufacturer's instructions. Six hours post transfection, cells were washed twice with Opti-MEM and incubated with 3ml Opti-MEM supplemented with Glutamax and 1μg/ml TPCK-trypsin (Sigma) at 33°C. TPCK-trypsin was added to the medium every second day. Seven days after transfection, supernatants were collected, and MDCK cells or fresh Vero cells were infected with 200μl of supernatants. Cells were again maintained in Opti-MEM with Glutamax and TPCK-trypsin. Four to eight days post infection (p.i.) – as soon as a cytopathic effect became visible – supernatants were tested for haemagglutination (HA). HA tests were performed as previously reported (Regl et al, 1999) with 0.5% (v/v) of human erythrocytes obtained from the local blood bank. HA titres were expressed as the reciprocal of the highest virus dilution resulting in full agglutination of erythrocytes.
vRNA isolation, RT-PCR and sequencing of recombinant viruses
To confirm the rescued recombinant viruses, one-millilitre fractions of cell-culture supernatants with HA titres of 64 to 128 were purified by centrifugation through a 20% (w/v/) sucrose cushion for 90min at 116,000xg and 4°C. vRNA was isolated using the QiaAmp viral RNA mini kit (Qiagen) according to the manufacturer's recommendations and subsequently digested with DNase I to eliminate any plasmid contamination. For the recombinant wild-type C/JJ/50 virus, RT-PCR was performed with primers specific for segment P3 to amplify a 1287-nucleotide fragment containing the four silent mutations. For the recombinant virus with the NS segment from C/JHB/1/66, the first 560 nucleotides of NS were amplified. Finally, for virus r-C/JJ/50-PB1-5'-3-m, the 5'- and 3'-nc regions were amplified with primers complementary to the last 12 and 11 nucleotides, respectively, together with internal primers. The purified PCR products were sequenced by Eurofins MWG Operon (Germany). Primer sequences can be provided on request.
Nucleotide sequence accession numbers
The GenBank/EMBL/DDJ accession numbers for PB2, PB1, HEF, NP, M, NS, and vector pPMV are FR671420, FR671421, FR671422, FR671423, FR671424, FR671425, and FR669675, respectively.
RESULTS
Complete sequences of the 3' and 5'ends of all seven segments from influenza C/JJ/50 virus
First, we determined the 3' and 5'ends of the seven viral segments from strain C/JJ/50. In the 1980s, the complete cDNA sequences including the nc regions of segments PB2, PB1, P3, and M from influenza C/JJ/50 virus were published (Yamashita et al, 1988; 1989). Our analyses revealed some differences in comparison with these sequences. For the 3'-nc ends (see Table 1) we observed that the first eleven nucleotides and nucleotide 14 are identical in all seven segments, whereas nucleotides 12 and 13 show a slight variability: The two largest segments PB2 and PB1 have a U at position 12, while all other segments have a C at this position. Nucleotide 13 is a C in all segments except for NP, which has a U at this position. Regarding the viral 5'ends, the first twelve nucleotides as well as nucleotide 15' are conserved among the segments, with exception of the NS segment, where nucleotide 6' differs from the others (see Table 1). Concerning the two variable positions 13' and 14', PB2 and PB1 harbour an A at position 13', while the other segments have a G, and NP has an A at position 14', where all other segments have a G. Our results coincide with recent sequence analyses from strain C/JHB/1/66 (Crescenzo-Chaigne and van der Werf, 2007) and strain C/AnnArbor/1/50 (Muraki et al, 2007). In all segments, nucleotides 10-14 are complementary to nucleotides 11'-15', which supports the idea that influenza C virus RNAs form a double-stranded structure of five base pairs at this region (Crescenzo-Chaigne and van der Werf, 2001). Actually, segments PB2, PB1, P3, NP, and M of C/JJ/50 are even capable of forming six base pairs. If the poly(U) stretch was included, double-stranded regions of between five to ten base pairs would be possible. Additionally, in accordance with the corkscrew model (Flick et al, 1996) for influenza A virus RNAs, our results show that base pairing may occur between nucleotides 2 and 9, 3 and 8 as well as 2' and 9' and 3' and 8'.
Table 1.
Comparison of the conserved 3'- and 5'-noncoding ends of the vRNA segments from influenza C/JJ/50 virus.
| Segment | 3' nc end (ultimate 20 nucleotides)a | 5' nc end (first 20 nucleotides)a |
|---|---|---|
| PB2b | UCCAAUCCUCUGCUUCUGCU | AGCAGUAGCAAGAGGAUUUU |
| PB1b | CAUAAUCCUCUGCUUCUGCU | AGCAGUAGCAAGAGGAUUUU |
| P3 | UCGGAUCCCCUGCUUCUGCU | AGCAGUAGCAAGGGGAUUUU |
| HEF | AUUAAACCCCUGCUUCUGCU | AGCAGUAGCAAGGGGAUUUU |
| NPb | CAAAAUCUCCUGCUUCUGCU | AGCAGUAGCAAGGAGAUUUU |
| M | AGAAAUCCCCUGCUUCUGCU | AGCAGUAGCAAGGGGAUUUU |
| NSb | AAAGUACCCCUGCUUCUGCU | AGCAGGAGCAAGGGGUUUUU |
Underlined: Conserved sequences.
In bold: Variations at positions 12, 13, 6', 13' and 14'.
Note that sequences are shown in 5'-3' orientation and numbering of nucleotides starts from both ends.
Cloning of the seven C/JJ/50 segments and the NS segment from C/JHB/1/66 into vector pPMV and sequencing of the whole C/JJ/50 genome
We constructed the bidirectional vector pPMV, which allows transcription of viral genomic RNA and mRNA from one template. The human RNA polymerase I promoter and the murine RNA polymerase I terminator ensure exact synthesis of viral RNA, while CMV promoter and SV40 poly(A) signal direct mRNA transcription. We amplified the seven full-length viral segments using primers complementary to the ultimate 12 nucleotides and cloned them into the two BsmBI restriction sites of pPMV (see Figure 1). To assure that rescued virus is recombinant and not the result of a contamination with wild-type virus, four silent mutations were introduced into the coding sequence of segment P3, the resulting plasmid was termed pPMV-P3m. At least three clones of every segment were sequenced, and the consensus sequence of the whole influenza C/JJ/50 virus genome was determined. The sequences of PB2, PB1, P3, and M were compared to the available sequences in the GenBank database. Our PB2 sequence differs from the published sequence M28061 by one nucleotide in the 3'-nc end. PB1 has one different nucleotide resulting in a silent mutation as well as one different nucleotide in each nc end compared to M28060. No difference was found for P3 in comparison with M28062. Segment M has one nucleotide difference leading to a different amino acid in the ORF of CM2 compared to the published sequence M22038. Not only sequencing of three different M clones but also direct cDNA end sequencing confirmed the nucleotide difference. Segments HEF, NP, and NS from C/JJ/50 had not been published before. The NP segment is known to be of variable length among different strains. We have determined that NP from strain C/JJ/50 is 1802 nucleotides long as NP from C/JHB/1/66 (AF170573). Compared to strains C/Cal/78 (M17700) and C/AnnArbor/1/50 (AB126195), it has a deletion of five nucleotides in the ORF near the 3'end (regarding positive-sense polarity) leading to an earlier stop codon, and is consequently nine amino acids shorter. NP from C/JJ/50 has another deletion of two nucleotides in the 5'-nc end (vRNA polarity) in comparison with NP from C/Cal/78. Most interestingly, the NS segment from strain C/JJ/50 is 21 nucleotides shorter than any to our knowledge published NS sequence. Sequence comparison revealed that nucleotides 239-259 (in cDNA polarity) are missing in C/JJ/50, leading to a deletion of amino acids 72-78 in the ORF of NS1. The NS segment from strain C/JJ/50 therefore encodes a 239-amino acid NS1 protein, in contrast to the 246-amino acid NS1 proteins from other influenza C virus strains (Alamgir et al, 2000). As this nucleotide deletion is within the region spliced from NEP/NS2 mRNA, the NEP/NS2 ORF from C/JJ/50 is not affected and codes for a 182-amino acid protein – as reported for other strains (Alamgir et al, 2000). We consequently decided to additionally clone the NS segment from strain C/JHB/1/66 (pPMV-NS-C/JHB/1/66). The sequences of the influenza C/JJ/50 virus genome with exception of P3 were deposited at the GenBank Database.
Rescue of infectious influenza C/JJ/50 virus
To rescue recombinant wild-type C/JJ/50 virus entirely from cloned cDNA, we transfected the seven bidirectional vectors encoding the seven segments (pPMV-PB2, -PB1, -P3m, -HEF, -NP, -M, and -NS) into Vero cells. Cells had to be 90% confluent to survive transfection and the following incubation in serum-free medium with trypsin added. Seven days after transfection, supernatants were collected but HA titration was not sensitive enough to determine virus yield. We therefore passed the supernatants to fresh Vero cells or MDCK cells, the latter of which highly promote growth of influenza C virus. Four to six days p.i. of MDCK cells with transfection supernatants, cells became cytopathic and supernatants were harvested. HA titres of 128 could be determined in five independent experiments (see Table 2). Passing the transfection supernatants to fresh Vero cells led seven to eight days p.i. to HA titres of 64. RNA of infectious supernatants was prepared, and a fragment of P3 covering the introduced silent mutations was reverse-transcribed and PCR-amplified. Sequence analysis verified the four silent mutations, confirming that the recovered virus was derived from plasmids.
Table 2.
Rescue of recombinant influenza C viruses by plasmid transfection of Vero cells and subsequent amplification on MDCK cells.
| Virusa | Expt 1 | Expt 2 | Expt 3 | Expt 4 | Expt 5 |
|---|---|---|---|---|---|
| rwt-C/JJ/50 | HA = 128 | HA = 128 | HA = 128 | HA = 128 | HA = 128 |
| r-C/JJ/50:NS-JHB | HA = 128 | HA = 64 | HA = 256 | ||
| r-C/JJ/50:PB1-5'-3'm | HA = 64 | HA = 16 |
Rescued viruses: rwt-C/JJ/50: recombinant wild-type C/JJ/50, r-C/JJ/50:NS-JHB: recombinant reassortant virus with the six large segments from C/JJ/50 and NS from C/JHB/1/66, r-C/JJ/50:PB1-5'-3'm: recombinant virus C/JJ/50 with one mutation in each nc region of PB1. HA titres are expressed as reciprocal of the highest virus dilution resulting in agglutination of red blood cells.
Replication of recombinant and wild-type viruses C/JJ/50 and C/JHB/1/66
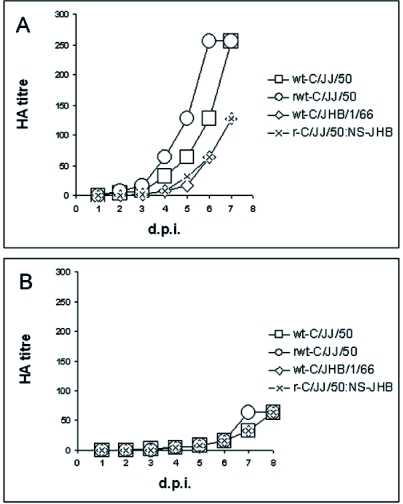
As mentioned before, the NS segment of strain C/JJ/50 has a deletion of 21 nucleotides in comparison with other strains, which results in a deletion of seven amino acid residues at positions 72–78 in the NS1 coding sequence. We therefore decided to generate an influenza C virus harbouring the six large segments (PB2, PB1, P3, HEF, NP, and M) from C/JJ/50 and the NS segment from strain C/JHB/1/66. HA titres of 128, 64 and 256 were measured in three independent experiments (see Table 2). The NS segment was sequenced to confirm the reassortant virus. In order to find out, whether there is any difference in replication of recombinant wild-type C/JJ/50 virus, wild-type C/JJ/50 virus and recombinant reassortant virus (NS: C/JHB/1/66), we determined the growth kinetics of these three viruses as well as of wild-type C/JHB/1/66 virus on MDCK and Vero cells. Both cell lines were infected with viruses of HA titres 128, which were diluted 1:10,000, and the amount of virus released to the medium was tested daily. Two days p.i. of MDCK cells, wild-type and recombinant wild-type C/JJ/50 viruses were measurable (HA titres of 4 and 8, respectively, see Fig. 2A). Virus yield doubled approximately each day, reaching an HA titre of 256 six to seven days p.i. No higher titres could be gained, as cells were already cytopathic. Wild-type C/JHB/1/66 and recombinant reassortant virus with the NS segment from C/JHB/1/66 exhibited a slightly delayed growth (Figure 2A). Three days p.i., titres of 2 were determined. Seven days p.i., when cells were cytopathic, titres of 128 were measured.
Figure 2.
Comparison of the growth behaviour of wild-type C/JJ/50 and C/JHB/1/66 viruses with recombinant wild-type virus and recombinant reassortant virus on (A) MDCK cells and (B) Vero cells. Confluent cells were infected with the respective virus (amplified on MDCK cells to HA titres of 128, diluted 1:10,000) in the presence of 1 μg/ml TPCK-trypsin. Samples of supernatants were collected every day and virus titres were determined by HA test. Viruses: wild-type C/JJ/50, recombinant wild-type C/JJ/50, wild-type C/JHB/1/66, r-C/JJ/50:NS-JHB: recombinant reassortant virus with the six large segments from C/JJ/50 and NS from C/JHB/1/66.
In Vero cells, the four strains replicated identically fast (see Figure 2B). An HA titre of 4 was measured four days p.i. Seven to eight days p.i., virus titres of 64 were reached. At that time, Vero cells exhibited a cytopathic effect.
Generation of recombinant virus with mutations in the 3' and 5' nc ends of PB1 and growth kinetics
We exploited our established reverse-genetics system to study the effect of mutations in the double-stranded region of the proposed panhandle/corkscrew-structured vRNA ends (see Figure 3A). To this end we constructed two bidirectional plasmid vectors for PB1. Vector pPMV-PB1-5'm has a single mutation at position 13' of the viral 5'-nc region: The wild-type A was replaced by a G (A→G[13']), which is noticeably the wild-type nucleotide 13' of segments P3, HEF, NP, M and NS. But this single nucleotide exchange disrupts the base-paired duplex region. Vector pPMV-PB1-5'-3'm has a base-pair mutation: It harbours the above described nucleotide exchange at the 5'end as well as a nucleotide exchange at position 12 of the 3'end. Here, the wild-type U was replaced by a C (A–U→G–C[13'–12]), which again represents the wild-type nucleotide 12 of segments P3, HEF, NP, M, and NS. (Figure 3B). Transfection of Vero replaced cells with the wild-type vectors for PB2, P3, HEF, NP, M, and NS along with pPMV-PB1-5'm and passage of the supernatant to MDCK cells for further viral growth did not result in the generation of a recombinant virus in two independent experiments. On the other hand, when transfecting the six wild-type segments along with pPMV-PB1-5'-3'm, which has one nucleotide exchange in each nc end restoring complementarity (Figure 3), recombinant viruses with HA titres of 64 and 16 were gained after six days of growth on MDCK cells (see Table 2, this virus was named r-C/JJ/50:PB1-5'-3'm). The presence of the two mutations was confirmed by sequencing the viral ends. As the yield was lower than that of recombinant wild-type virus, we compared the growth kinetics of recombinant wild-type C/JJ/50 virus and virus r-C/JJ/50-PB1-5'-3'm. We infected MDCK cells with viruses of HA titres 64, diluted 1:5,000, and tested the HA titres daily. Recombinant wild-type virus again reached an HA titre of 256 six days p.i. (Figure 4), while virus r-C/JJ/50-PB1-5'-3'm showed a delayed growth kinetic reaching a maximum HA titre of 64 five days p.i. In conclusion, the base pair-mutated virus exhibited a 4-fold reduction in growth.
Figure 3.
Representation of the nc terminal sequences of PB1 vRNA from influenza C virus in the corkscrew model in A (Flick et al, 1996; Crescenzo-Chaigne and van der Werf, 2001), and single/base-pair mutations at positions 13' and 12 within the proposed six base pairs long double-stranded region in B. Numbering of residues starts from the 3'end and the 5' end; 5' residues are marked with a prime, and base pairs are joined by lines in (A).
Figure 4.
Growth kinetics of recombinant virus with base-pair mutation in the noncoding ends of PB1. Confluent monolayers of MDCK cells were infected with recombinant wild-type C/JJ/50 virus (rwt-C/JJ/50) and with recombinant virus harbouring the base-pair mutation A–U→G–C(13'–12) in the nc terminal sequences of PB1 (r-C/JJ/50:PB1-5'-3'm). Both viruses were grown on MDCK cells to an HA titre of 64 and were diluted 1:5,000 prior to infection. Samples of the supernatants were tested for haemagglutination daily.
DISCUSSION
In the present study, we report the successful generation of recombinant influenza C virus entirely from seven bidirectional plasmids. While the employment of bidirectional vectors has already been described for influenza A and B viruses (Dauber et al, 2004; de Wit et al, 2004; Hoffmann et al, 2000; 2002), this is the first report for influenza C viruses.
As a prerequisite to produce recombinant influenza C viruses, we determined the consensus sequence of the C/JJ/50 genome. Surprisingly, we found that the NS segment from C/JJ/50 is 21 nucleotides shorter than the respective of other strains. This deletion results in an NS1 protein missing amino acids 72–78 in comparison with other strains. As a consequence, we included the NS segment from strain C/JHB/1/66 in our experimental system.
We designed a bidirectional vector for vRNA replication and mRNA transcription from one template (see Fig. 1), as initially established by Hoffmann et al (2000). We decided to transfect our plasmids into Vero cells instead of 293T cells, which were used to produce recombinant influenza C viruses by two other groups (Crescenzo-Chaigne and van der Werf, 2007; Muraki et al, 2007), as we observed that Vero cells promote growth of influenza C viruses and are well transfectable. Although they cannot compete with 293T cells in transfection efficiency, 293T cells on the other hand are very sensitive to trypsin and therefore hard to handle. We transferred the Vero cell-supernatants seven days post transfection to MDCK cells, which highly support influenza C virus replication, and could consequently abstain from using embryonated chicken eggs for further growth. It should be mentioned that Vero as well as MDCK cells are permitted for vaccine production. Infection of MDCK cells resulted in average HA titres of 128 five days p.i. for recombinant wild-type viruses (see Table 2). Plaque assays previously performed in our lab with wild-type C/JJ/50 virus let us conclude that an HA titre of 128 approximately equals 109 plaque forming units (p.f.u.)/ml. In comparison, Muraki et al (2007) recovered recombinant influenza C viruses with maximum titres of 2,5 X 103 EID50/ml by transfection of 11 to 16 plasmids into 293T cells and inoculation of transfection supernatants after 48 to 60 hours in chicken eggs. Crescenzo-Chaigne and van der Werf (2007) transfected 11 plasmids into 293T cells and measured 103 to 104 p.f.u./ml eight to ten days post transfection. Transferring the supernatants into chicken eggs led to titres of 109 p.f.u./ml, whereas further growth on SK 93/2 cells yielded 105 to 106 p.f.u./ml. We conclude that our system is competitive in comparison with the above described ones regarding virus yield referred to time expended. Moreover, our seven-plasmid system is less time-consuming. Seven plasmids are faster constructed, and transfection experiments are easier to handle. Using this system, mutations can be easily introduced into the viral genome.
After having established this system, we created a reassortant virus bearing the NS segment of strain C/JHB/1/66 to assess whether the seven amino acids smaller NS1 from C/JJ/50 has any effect on growth behaviour. Viral titres comparable to those gained from transfection of all seven C/JJ/50 segments were measured. Growth curves indicated that wild-type C/JHB/1/66 virus and reassortant virus replicated more slowly than wild-type and recombinant wild-type C/JJ/50 viruses on MDCK cells, while growth on Vero cells was equal (Figure 2, A and B). In summary, the recombinant wild-type C/JJ/50 virus replicated as fast as the mother strain on MDCK and Vero cells. There was no remarkable difference in virus titres between wild-type C/JJ/50 and C/JHB/1/66 viruses and the two recombinant strains. The missing seven amino acids of NS1-C/JJ/50 do not appear to play an important role for growth on MDCK and Vero cells.
We next tried to recover recombinant viruses with mutations in the nc ends of PB1. The nc ultimate nucleotides of the viral segments are described as partially double-stranded panhandle/RNA fork/corkscrew structures for influenza A viruses (Flick et al, 1996; Fodor et al, 1994; Hsu et al, 1987). We introduced a single mutation at position 13' of the 5' nc end of segment PB1 (A→G[13']), which abolishes the postulated base pairing between nucleotides 10 to 15 of the 3' end and 11' to 16' of the 5' end. Indeed, this mismatch prevented the rescue of a recombinant virus. When this single mutation at the 5' end was supplemented by a second mutation at the putative base-pairing partner at the 3' end (A–U→G–C[13'–12]), a recombinant virus was gained (see Fig. 3), but growth kinetics were slower in this strain (see Fig. 4). While the wild-type recombinant virus grew to an HA titre of 256, the recombinant mutant virus replicated to an HA titre of 64. According to previously performed plaque assays with wild-type virus, an HA titre of 256 corresponds to 2.5 X 109 p.f.u./ml, and an HA titre of 64 to 4 X 108 p.f.u./ml. Importantly, only segments PB2 and PB1 have A and U at positions 13' and 12, respectively. All other segments have the G–C base pair. Fodor et al (1998) suggested that impaired growth of such base-pair mutants in influenza A viruses is due to impaired polyadenylation properties. They observed that neither vRNA nor cRNA levels were declined but mRNA and protein levels. Initiation and elongation of transcription equalled wild type, but the mutations interfered with polyadenylation of the mRNA transcripts. We have not determined the exact reason for the lower viral titres of our base-pair mutant, but find that not only the base-pairing ability, but also a particular nucleotide sequence at the panhandle/corkscrew region is important.
In the future, our seven plasmid-based system may be useful to study transcription, replication and packaging events and to elucidate the function of proteins from influenza C virus. The influenza C virus NS1 protein for example would be a candidate antagonist of the cellular interferon system, which could be assessed by generating NS1-deficient influenza C viruses. Several melanoma cell lines have a defective interferon signalling and express the cellular receptor for influenza C viruses, which is 9-O-acetyl sialic acid (reviewed in: Kohla et al, 2002; Varki and Varki, 2007). Thus an attenuated recombinant influenza C virus could be employed as gene delivery vector and/or tumour-specific oncolytic virus.
CONCLUSIONS
The generation of recombinant influenza C viruses from seven bidirectional plasmids is an efficient system with regards to virus yield and time spent. Employing certified Vero and MDCK cells, this system is independent of the use of embryonated chicken eggs for virus amplification. Studies on the generation of influenza C viruses with mutations in the nc ends of PB1 indicate that base pairing between the viral ends is indispensable for virus rescue, and the exact nature of each nucleotide in the nc regions strongly influences viral titres.
ACKNOWLEDGEMENTS
This work was supported by the Austrian Science Fund (FWF project number P20080), Salzburg Research Fellowship P147200-06, and Salzburg University Fellowship SF0706V to KP. We are grateful to Dr. Markus Wolschek and Dr. Christian Kittl (Avir Green Hills Biotechnology AG, Vienna) for the protocol to sequence viral nc ends and for helpful discussions. We also thank the Universitätsklinik für Blutgruppenserologie und Transfusionsmedizin der Paracelsus Medizinischen Privatuniversität Salzburg for human erythrocytes.
LIST OF ABBREVIATIONS
- d.p.i.
days post infection
- HA
haemagglutination
- nc
noncoding
- NP
nucleoprotein
- p.f.u.
plaque forming units
- p.i.
post infection
- RNP
ribonucleoprotein
COMPETING INTERESTS
None declared.
REFERENCES
- Alamgir ASM, Matsuzaki Y, Hongo S, et al. Phylogenetic analysis of influenza C virus nonstructural (NS) protein genes and identification of the NS2 protein. J Gen Virol. 2000;81:1933–1940. doi: 10.1099/0022-1317-81-8-1933. [DOI] [PubMed] [Google Scholar]
- Crescenzo-Chaigne B, van der Werf S. Nucleotides at the extremities of the viral RNA of influenza C virus are involved in type-specific interactions with the polymerase complex. J Gen Virol. 2001;82:1075–1083. doi: 10.1099/0022-1317-82-5-1075. [DOI] [PubMed] [Google Scholar]
- Crescenzo-Chaigne B, van der Werf S. Rescue of influenza C virus from recombinant DNA. J Virol. 2007;81:11282–11289. doi: 10.1128/JVI.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber B, Heins G, Wolff T. The influenza B virus nonstructural NS1 protein is essential for efficient viral growth and antagonizes beta interferon induction. J Virol. 2004;78:1865–1872. doi: 10.1128/JVI.78.4.1865-1872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Spronken MI, Bestebroer TM, et al. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 2004;103:155–161. doi: 10.1016/j.virusres.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Flick R, Neumann G, Hoffmann E, Neumeier E, Hobom G. Promoter elements in the influenza vRNA terminal structure. RNA. 1996;2:1046–1057. [PMC free article] [PubMed] [Google Scholar]
- Flick R, Hobom G. Interaction of influenza virus polymerase with viral RNA in the 'corkscrew' conformation. J Gen Virol. 1999;80:2565–2572. doi: 10.1099/0022-1317-80-10-2565. [DOI] [PubMed] [Google Scholar]
- Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E, Palese P, Brownlee GG, Garcia-Sastre A. Attenuation of influenza A virus mRNA levels by promoter mutations. J Virol. 1998;72:6283–6290. doi: 10.1128/jvi.72.8.6283-6290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Mahmood K, Yang CF, et al. Rescue of influenza B virus from eight plasmids. Proc Natl Acad Sci U S A. 2002;99:11411–11416. doi: 10.1073/pnas.172393399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci U S A. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohla G, Stockfleth E, Schauer R. Gangliosides with O-acetylated sialic acids in tumors of neuroectodermal origin. Neurochem Res. 2002;27:583–592. doi: 10.1023/a:1020211714104. [DOI] [PubMed] [Google Scholar]
- Matz MV. Amplification of representative cDNA pools from microscopic amounts of animal tissue. Methods Mol Biol. 2003;221:103–116. doi: 10.1385/1-59259-359-3:103. [DOI] [PubMed] [Google Scholar]
- Muraki Y, Murata T, Takashita E, et al. A mutation on influenza C virus M1 protein affects virion morphology by altering the membrane affinity of the protein. J Virol. 2007;81:8766–8773. doi: 10.1128/JVI.00075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Shaw ML. Orthomyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Philadelphia, USA: Lippincott, Williams and Wilkins, a Wolters Kluwer Business; 2007. pp. 1647–1689. [Google Scholar]
- Regl G, Kaser A, Iwersen M, et al. The hemagglutinin-esterase of mouse hepatitis virus strain S is a sialate-4-O-acetylesterase. J Virol. 1999;73:4721–4727. doi: 10.1128/jvi.73.6.4721-4727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest. 2007;87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Krystal M, Palese P. Evidence that the matrix protein of influenza C virus is coded for by a spliced mRNA. J Virol. 1988;62:3348–3355. doi: 10.1128/jvi.62.9.3348-3355.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Krystal M, Palese P. Comparison of the three large polymerase proteins of influenza A, B, and C viruses. Virology. 1989;171:458–466. doi: 10.1016/0042-6822(89)90615-6. [DOI] [PubMed] [Google Scholar]