Abstract
Importance of the field
Cardiac arrhythmias remain a major challenge for modern drug discovery. Clinical events are paroxysmal, often rare and may be asymptomatic until a highly morbid complication. Target selection is often based on limited information and though highly specific agents are identified in screening, the final efficacy is often compromised by unanticipated systemic responses, a narrow therapeutic index and substantial toxicities.
Areas covered in this review
Our understanding of complexity of arrhythmogenesis has grown dramatically over the last two decades, and the range of potential disease mechanisms now includes pathways previously thought only tangentially involved in arrhythmia. This review surveys the literature on arrhythmia mechanisms from 1965 to the present day, outlines the complex biology underlying potentially each and every rhythm disturbance, and highlights the problems for rational target identification. The rationale for in vivo screening is described and the utility of the zebrafish for this approach and for complementary work in functional genomics is discussed. Current limitations of the model in this setting and the need for careful validation in new disease areas are also described.
What the reader will gain
An overview of the complex mechanisms underlying most clinical arrhythmias, and insight into the limits of ion channel conductances as drug targets. An introduction to the zebrafish as a model organism, in particular for cardiovascular biology. Potential approaches to overcoming the hurdles to drug discovery in the face of complex biology including in vivo screening of zebrafish genetic disease models.
Take home message
In vivo screening in faithful disease models allows the effects of drugs on integrative physiology and disease biology to be captured during the screening process, in a manner agnostic to potential drug target or targets. This systematic strategy bypasses current gaps in our understanding of disease biology, but emphasizes the importance of the rigor of the disease model.
1. Introduction
The identification of novel anti-arrhythmic agents is a major challenge. Many of today’s anti-arrhythmic drugs are relatively ineffective or are plagued by substantial ‘on-target’ and ‘off-target’ toxicity[1–3]. The focus of the current anti-arrhythmic armamentarium is the modulation of myocardial automaticity, refractoriness and conduction: fundamental properties of myocardial tissue required for normal physiologic function. Our approaches to arrhythmias as disparate in risk as simple atrial premature beats and ventricular tachycardia are remarkably similar, suggesting that current targets are far downstream from the primary arrhythmic mechanisms and that existing anti-arrhythmic drugs are rather blunt instruments[4–6]. This inference is further bolstered by the observation that most effective anti-arrhythmic agents can also be highly pro-arrhythmic in particular contexts[7]. These lessons have been learned the hard way with large and expensive antiarrhythmic trials ending as a result of objective increases in mortality or morbidity associated with the drugs[3, 8]. In this article we will outline evidence that most existing anti-arrhythmic drug targets are poorly validated, review emerging data on the mechanisms of arrhythmogenesis in humans, and then discuss the implications of these findings for systematic approaches to the discovery of novel anti-arrhythmic agents in zebrafish.
2. Choosing anti-arrhythmic targets
2.1 The scope of the problem
Arrhythmias pose several unique problems for potential therapeutics[1, 9]. Many clinical arrhythmias are fatal at initial presentation or result in a catastrophic event such as resuscitated ‘sudden death’, yet the underlying diathesis to arrhythmia may be completely asymptomatic[10–12]. Arrhythmias are often paroxysmal, thus confounding strategies for detection and for the objective assessment of responses to therapy. For many arrhythmic disorders, while the risk associated with individual arrhythmic episodes is extremely high, the absolute lifetime risk of an episode may be quite low[13]. The fundamental mechanisms of most clinical arrhythmias remain poorly understood, and though exploration of the chronic myocardial abnormalities necessary for the initiation or maintenance of dysrhythmia has made substantial progress, the precise pathways that result in an actual arrhythmia after years of quiescence are largely unknown[14]. The prospect of long-term exposure to drugs with even small risks of toxicity in order to counteract a single morbid event is not attractive for physicians or for pharmaceutical companies[1]. However, agents that reduce the risk for symptomatic arrhythmias, and their downstream consequences such as stroke or heart failure, without increasing unpredictable catastrophic events such as sudden death will be of major importance as the global burden of heart disease grows[4].
Implicit in these arguments is a need to match the risks of the disease being treated with the risks of any new molecular entities under development. In recent years this has resonated with the emerging personalized medicine movement. However, the development of truly personalized medicine brings with it unvoiced requirements for much higher resolution diagnostics to define the pathophysiologic mechanisms in each patient, as well as for a paradigm shift in the scale of drug discovery. This degree of personalization to date only seems feasible in any meaningful way in clonal neoplastic disorders. In the anti-arrhythmic arena such tailoring of therapeutics will require the development of new insights into different arrhythmia substrates and their detection, as well as innovative approaches to the discovery of truly substrate-specific therapeutics[15].
2.2 Beyond transmembrane ionic fluxes
Over the last few decades, much of the effort to understand arrhythmia mechanisms has focused on transmembrane ionic fluxes and the channels or ion exchangers required to generate these[16]. This investigative direction was driven by the accessibility of single channel biophysics, as well as by the insights accumulating from pioneering work in the molecular genetics of inherited human arrhythmias[16]. Primary abnormalities in the genes encoding individual channel protein subunits are a now a well-established cause of inherited arrhythmic disorders such as the long QT (LQT) and Brugada syndromes [12]. As a result of our improved understanding of the molecular basis of membrane depolarization and repolarization, large discovery efforts were directed to identify inhibitors of specific channel conductances in the hope that it might be possible control the complex behavior of the membrane[17]; tuning automaticity, refractoriness and conduction through subtle orchestration of the various currents contributing to these phenomena.
The lack of success of this strategy is a result of many different factors. Fundamentally, the premise that arrhythmias result from abnormal conductance alone is difficult to substantiate. Attempts to model LQTS in the mouse strongly suggest that simply perturbing channel density or net ionic fluxes rarely leads to arrhythmia, though these efforts have also been complicated by the distinctive nature of rodent cardiac electrophysiology [18]. Powerful homeostatic responses appear to prevent major effects from a range of null alleles in several key ion channel genes. In stark contrast, targeted knock-in of human disease alleles in these same genes leads to spontaneous arrhythmias[19]. These data, combined with clinical observations, implicate very specific gains of channel functions rather than isolated effects on transmembrane conductance in arrhythmogenesis.
The importance of such confounders is only amplified by the failure of therapeutic targeting of single ion channels to discriminate adequately between health and disease. The very same channels implicated in LQT, Brugada or other arrhythmic syndromes are critical for the normal physiology of the heart. The distinctive roles of particular channels in different molecular contexts or in different functional states may explain some of the proarrhythmia seen when these channels are targeted for therapeutic effect[2, 16]. Similarly, the pharmacologic targeting of extra-cardiac isoforms (neuronal or smooth muscle for example) of these same channels may be another potential source of toxicity[20].
The very dynamic nature of myocardial electrophysiology is another major factor in the narrow therapeutic window observed with many existing anti-arrhythmic agents. In the setting of physiologic or pathologic changes in heart rate, contractility, metabolism and autonomic tone, drugs with little toxicity may become a liability. A well-known example is the rate related use-dependent toxicity of flecainide, but other examples exist[7]. The nature of normal cardiac rhythm dooms such agents not just to therapeutic failure but also to lethal toxicity.
Evidence from inherited arrhythmic syndromes also suggests that focused perturbation of single ionic currents is unlikely to prove successful as an antiarrhythmic strategy. Even in the original kindreds with familial LQTS, defining a simple correlation between baseline myocardial electrophysiology (as manifest on the surface electrocardiogram) and clinical arrhythmic events has proven remarkably difficult[21]. Many other factors including autonomic tone, acute changes in myocardial physiology, intercurrent illness and even specific sensory afferents have all been implicated in the initiation of lethal arrhythmias in LQTS family members[22–24]. A recent example of the limits of our current understanding of the mechanisms of arrhythmogenesis is the identification of mutations in the KCNQ1 gene that cause both prolongation of the QT interval and familial atrial fibrillation (where a longer action potential would be predicted to reduce the risk of the arrhythmia)[25, 26]. Subsequent mechanistic studies have highlighted the distinctive physiology of a single transmembrane protein in atrium and ventricle. Finally, given the very nature of arrhythmias, stochastic processes also are likely to play a major role. Together these same factors render genotype of limited predictive utility for clinical events in the LQT syndrome[27].
2.3 Ion channels as signal transducers
In inherited cardiac arrhythmias emerging evidence now implicates not only abnormalities of the ionic currents carried by the mutated channels, but also disruption of physical or functional interactions with adaptor proteins and downstream signaling pathways [12, 28, 29]. More recent genetic studies have identified channel accessory proteins and membrane scaffolding molecules in some forms of human LQT syndrome, while the murine modeling outlined above emphasizes the very distinctive pathways that may lead to final ‘common’, if low resolution, clinical phenotypes[3, 19, 30–32]. The lifecycles of ion channel subunits and those of many other membrane molecules (proteins, lipids, carbohydrates and a host of small molecules) are inextricably linked. Messenger RNA editing, splicing, multiple post-translational modifications, functional quality control, chaperoning, trafficking, complex assembly, and membrane insertion and turnover are all tightly regulated[16, 29, 33, 34]. Novel technologies are beginning to reveal parallel control of membrane lipid composition, the partitioning of membrane subdomains, and transitions in the plane of the membrane that determine protein endocytosis and turnover[35]. Relatively unbiased genetic screens have also implicated extracellular, cytoskeletal and nuclear pore proteins in the regulation of electrophysiologic events[36]. The interconnected nature of these pathways reflects the complex networks at play in cardiac electrical function.
2.4 Cell-cell communication
Intercellular communication has also been directly implicated in arrhythmogenesis, in particular in the biology of one of the most malignant forms of inherited heart muscle disease; arrhythmogenic right ventricular cardiomyopathy (ARVC). ARVC is characterized fibro-adipocytic myocardial dystrophy and typically associated with arrhythmias and contractile dysfunction [37, 38]. Cardiac myocytes rely on the specialized function of intercalated discs for both mechanical and electrical coupling. Three distinct types of intercellular junction exist within the intercalated disc; gap junctions, adherens junctions and desmosomes. Gap junctions, composed of connexins, regulate the intercellular exchange of ions as well many physiologically important small molecules. Adherens junctions are thought to provide substantial mechanical coupling through the linkage of cadherin-catenin complexes to the actin cytoskeleton, while desmosomes offer mechanical support through the physical interaction of desmosomal cadherins with intermediate filaments. Adherens junctions and desmosomes also participate in a number of cell signaling pathways, apparently enabling sophisticated cross-talk between mechanical and biochemical stimuli. Human molecular genetic studies have identified mutations in several desmosomal proteins in ARVC [39–44]. Established links between the desmosome and canonical Wnt/beta-catenin signaling led to the exploration of these pathways in ARVC [45]. Although the role of Wnt signaling in early cardiovascular development and morphology has been described in detail, its role later in development, in adult tissue and after cardiac stress and repair is less well understood. Plakoglobin, a component of both desmosomes and adherens junctions, is closely related to beta-catenin, and there is evidence that in the context of a cardiac-restricted desmoplakin knockout, shuttling of plakoglobin from desmosomes to a cytoplasmic/nuclear pool may directly antagonize canonical Wnt/beta-catenin signaling in cardiomyocytes[45]. This hypothesis has been further strengthened by the observation that there is loss of membrane plakoglobin signal in tissue samples from ARVC irrespective of the causal desmosomal gene[46].
Together these data strongly implicate intercellular junction structure and function in arrhythmogenesis, but also imply that there is a sophisticated integration of mechanical and electrical information at the complex intercellular junctions in the heart. The differentiation of cardiomyocytes, and likely other intramyocardial cell types, is directly modified by cardiac functional activity, but these inputs appear to modulate cell polarity, junctional strength and stability, and the specificity of individual myocyte-myocyte connections. There is emerging evidence that these junctional complexes are also regulated by inflammatory mediators, unifying diverse proarrhythmic conditions such as heart failure and intercurrent infectious disease[6]. This work also suggests that cell-based screens suffer a major lack of representation.
2.5 Metabolism and other cellular mechanisms in arrhythmogenesis
The application of functional genomics technologies to myocardial samples from arrhythmia cases has identified several consistent features. Several studies, employing transcript profiling, proteomics or metabolomics, have detected abnormalities of myocardial metabolism in tissue from those with arrhythmias including atrial fibrillation and a broad range of cardiomyopathies[47]. These data suggest that, at least later in the natural history of these disorders, perturbed utilization of energy substrates may play a role in arrhythmogenesis. Importantly, these same defects have been observed in animal models after induction of arrhythmias. However, in all of the major atrial fibrillation models the arrhythmias and metabolic abnormalities spontaneously resolve in the absence of continued stimuli, suggesting that such perturbations may not represent the primary diathesis[48].
More common metabolic abnormalities have been proposed as contributors to the proarrhythmia observed in human cardiomyopathies[49]. In hypertrophic cardiomyopathy the metabolic stress of inefficient mutant sarcomeric contractile proteins has been suggested not only as a major driver of myocyte hypertrophy, but also as one of the factors underlying the elevated risk of malignant ventricular arrhythmias[50]. Molecular genetic work in the muscular dystrophies has identified a central role for the dystrophin-associated glycoprotein complex (DAGC) in organizing key membrane subdomains, coupling these specialized rafts and particular channels with specific cytoskeletal networks and their associated signaling compartments[51]. The DAGC has also been implicated in the orchestration of membrane turnover and the organization of post-synaptic receptor assembly in skeletal muscle. Similarly, several other proteins associated directly or indirectly with the DAGC result in comparable pathologies, most consistent with general dedifferentiation of the cardiomyocyte. This is a common feature of many cardiomyopathies, and undoubtedly contributes to the proarrhythmia seen in these disorders though the precise mechanisms remain obscure.
Notably, several rare primary metabolic abnormalities and mitochondrial dysfunction also may affect the electrical stability of the cardiomyocyte[52, 53]. Interestingly specific metabolic abnormalities will often cause extremely reproducible arrhythmias that are highly dependent on the precise defect[54]. These conditions are often free from any arrhythmias except when myocardial energetics are stressed by the lack of appropriate energy supplies or by intercurrent severe illness. These insights suggest that for many arrhythmias the minimal ‘target’ may be an organelle or a large macromolecular complex and difficult to represent in a reductionist in vitro system for traditional high-throughput screening.
2.6 Cellular heterogeneity in the heart
An additional level of complexity is introduced by the revelation of extensive heterogeneity among cardiomyocyte populations within the heart[55]. Specialized populations of pacemaking, nodal and conduction system cells long have been recognized, but their roles in arrhythmogenesis are only now beginning to be explored[56]. Perhaps the best-characterized source of myocardial heterogeneity is infarct-related scar[57], but elegant ex vivo models have demonstrated that there are physiologic heterogeneities in myocyte electrophysiology between endocardium, mid-myocardium, and epicardium[55]. Despite the syncytial nature of myocardium, the tendency for specific cardiomyopathies to lead to unique ECG abnormalities suggests that distinctive subpopulations of myocytes are afflicted[58, 59]. The normal and pathological roles for these and other myocardial subpopulations have yet to be fully understood.
Such tightly patterned heterogeneities within the cardiac syncytium are thought to play a role in the inherited cardiomyopathy and sudden death condition known as the Brugada syndrome[60]. Heterozygous loss of function mutations in the cardiac sodium channel, SCN5A, are known to cause the rare Brugada syndrome, characterized by abnormal repolarization in the right ventricle and a nocturnal pattern of sudden death[61]. Subsequent work has identified a physiologic endocardial to epicardial transmural gradient in the expression of SCN5A that is disrupted by mutations that can cause lethal arrhythmia[55, 62]. Recently, trophic influences emanating from neurons as they innervate the heart during development have been found to orchestrate such gradients of sodium and potassium channel expression[60, 62, 63]. These findings reveal the heart to be considerably more complex in terms of cardiomyocyte cell types and connectivity than had been appreciated. Indeed, many important analogies exist between the central nervous system and the myocardium. The concepts of neuronal pathways with discrete functional circuits, dynamic connectivity and functional modulation of coupling likely reflect the role that junctional plasticity plays in adult myocardial physiology more faithfully than our current framework of a relatively static homogeneous muscular syncytium.
More direct contributions by non-cardiomyocytes to arrhythmogenesis are also beginning to be explored. Coronary vascular cells, fibroblasts, histiocytes and infiltrating leukocytes, among other cell types, each may contribute to the arrhythmic substrate in different ways. Primary cellular defects in cardiomyocytes may also manifest in other cell types or the molecular machinery responsible for cross-talk between other cell types and cardiomyocytes may be deranged. For example perturbed coronary endothelial cell-cardiomyocyte interactions are thought to mediate some of the microvascular ischemia seen in cardiomyopathies, and are now known to be dependent on myocyte signals and not a result of primary abnormalities in the endothelium[64]. Inflammatory infiltration is usually a response to initial myocardial damage, either inherited or acquired, but occasionally may be primary[65]. Not only do lymphocytes and other inflammatory cell types invade the interstitium locally, but the associated cytokines also may lead to phenotypic changes in many aspects of cellular excitability and coupling throughout the myocardium. Similar effects are seen with systemic cytokines, and contribute to the proarrhythmic milieu reflected in myocarditis, Chagas disease, “VT storm” related to device infection or overt sepsis as well as to more subtle and chronic arrhythmia substrates[66, 67]. Fibroblasts migrating into healing myocardium will not only contribute collagen-based scar that results in fixed electrical barriers, but also signal through a host of paracrine effectors, many of which modulate cardiomyocyte automaticity, coupling and excitability[68–70].
Recent work in myocardial regeneration has highlighted potential roles of heterocellular coupling or even cell fusion in the biology of post-natal terminally differentiated cells[71]. These insights also bring into sharp relief our limited understanding of the electrical effects of cardiomyocyte cell division (even to the limited extent that this occurs) during development or in later life. Many of the responses of the adult heart appear to reflect blocked cell division, and the electrophysiologic consequences of myocardial remodeling may thus represent an intrinsic tension between cellular coupling and independent cell behaviors[72].
3. Drug discovery in the face of irreducible complexity
Drug discovery has traditionally involved the isolation of a specific target molecule, the design of a robust and scalable assay for this target and the completion of an empiric screen of large chemical libraries for entities with the desired activity in this assay. Clearly, the choice of target is a major decision in this process and often the source of downstream problems. Target choice is often based on a host of factors that may have little to do with the disease biology in humans, such as prior work in the area, perceived drugability of the target, previous successful drugs in the same field or most commonly data from mechanistically distinct animal models[73]. Rarely is the target chosen because it is known to be a specific cause of the underlying disease. Often the target will have been studied intensively in one particular arena, but little may be known of target function in other cell types or tissues, or target behavior in the context of commonly encountered stressors. Many preclinical animal models are expensive or are highly inbred, and it is not uncommon for drugs to reach the market having been tested on fewer than 1000 animals. It is perhaps not surprising therefore that many drugs suffer from unanticipated ‘on target’ effects as well as apparently idiosyncratic reactions in the face of rare genetic variants[2, 74].
Ironically, amiodarone, the most effective antiarrhythmic agent identified to date, was discovered serendipitously during its development as an antianginal drug[75]. Effects against multiple targets are now known to be important contributors to the final profile of any drug, with beneficial or adverse outcomes[76, 77]. As for any form of drug discovery, it is increasingly important to closely match any risk associated with the drug with the potential benefit: the acute termination of ventricular tachycardia is a very different setting than chronic suppression of a relatively benign atrial arrhythmia. A major impetus to the concept of in vivo drug discovery is the ability to screen a priori for such ‘dirty’ drugs using therapeutically relevant endpoints and counter-screens for toxicities. In many ways this approach is a systematized search for serendipity in the context of rigorous disease models.
An ideal drug discovery model would incorporate not just a single target but all conceivable targets in a mechanistically faithful native context, would allow parallel screening for ADME and toxicology and would also enable the study of drug-drug interactions. The ability to model interactions with other common disease processes and environmental exposures would also be beneficial. While such a detailed knowledge of biologic networks may be conceivable it is far from realization. The ideal animal model would recapitulate not just individual components of the causal chain leading to an arrhythmia, but each step along the way. Perhaps the closest we will come for some time is direct in vivo discovery in genetic disease models. True genetic modeling opens the possibility of recapitulation of most, if not all, of the events in the pathophysiology of these archetypal arrhythmias[73].
4. Integrative physiology and systems biology
In the last decade, a resurgence in quantitative biology has been stimulated by the generation of comprehensive, unbiased datasets from a range of functional genomics technologies[78, 79]. Innovative approaches to the analysis of the ‘networks’ of interacting genes, trancripts and proteins that underlie basic biological processes have begun to generate predictive computational models[80, 81]. Combining such ‘systems’ analyses with existing in silico physiologic models has begun, but for cardiac electrophysiology at present the links between genomic or proteomic data and traditional biophysical models remain limited [82]. Cardiac electrophysiology while long at a leading edge of rigorous in silico modeling, may prove one of the most challenging areas to integrate in this way. A major challenge for systems biology and systems pharmacology will be the integration of diverse datasets gathered from rapidly changing technologies, each often with very different temporal and spatial resolution.
Ultimately, if we are to rigorously model arrhythmias we must incorporate all the variables outlined above and in a context that allows for tremendous variation across timescales from milliseconds to years. The model must represent not only basal conditions but also informative perturbations. Superimposed on any arrhythmogenic substrate are a host of transient physiologic stimuli including autonomic activity, immune and environmental triggers each of which may contribute to the initiation of a clinical event[52, 83]. Any systems level analysis of cardiac function in health and disease must necessarily include the effects of diverse modifiers ranging from instantaneous loading conditions, through diurnal variation in cellular physiology, and ambient immunology all the way to factors as evanescent as emotion.
Perhaps the most important modifiable environmental contributors to clinical arrhythmias are drugs. Many cardioactive drugs, particularly antiarrhythmic agents themselves, are in some situations proarrhythmic[2]. To generate a framework for the treatment and prevention of rhythm disorders we must aim to describe systematically the relationships between complex functional networks and individual drugs. New anti-arrhythmics would be tested not only under basal physiologic conditions but also in the setting of disease network where the interacting components in the network and their connectivity may be very different. In a preview of what might be possible, Iyengar and colleagues recently used random-walk based distance algorithms exploiting existing LQT genes to identify modules of the human interactome associated with proarrhythmia, and were then able to predict other arrhythmia genes and drug targets associated with repolarization toxicity[84].
Even the most powerful computational strategies will require empiric validation and thus, implicitly, biologic models capable of a comparable scale of investigation. The precise models used will depend on the context, but ultimately the more comprehensive a model is the more likely it is to represent the relevant biology.
5. Next generation in vitro discovery
Heterologous expression systems are capable of spectacular resolution, and automation has been developed to allow this approach at tremendous scale[85]. These techniques have facilitated the screening of millions of compounds for specific effects on target membrane proteins. Many companies are screening for specific inhibitors or activators of a broad range of novel ion channels using these approaches. However, limited representation of the underlying biology remains a fundamental weakness of in vitro techniques in antiarrhythmic drug discovery. As already noted, recent insights from modeling the LQT syndrome strongly suggest that isolated conductance changes are rarely if ever the cause of arrhythmia[19, 86]. Rates of translation, trafficking, protein quality control, glycosylation and other modifications, as well as modified interactions with partners both at the membrane or in some intracellular compartment may all play a role in disease pathogenesis or in drug responses. Heterologous systems represent partner proteins, lipids and small molecules only fortuitously or by specific design, but for most traditional targets in antiarrhythmic discovery, the comprehensive repertoire of partner proteins is unknown[16, 87]. Much more representative molecular and cellular context will be necessary if drug discovery is ever to be personalized. Supplying this context will require in vivo modeling on a scale not previously encountered.
6. Cellular pathway profiling
Moving to relevant cell types may overcome some of the limitations of biologic representation in situations where the disease process is cell autonomous, but this is rarely the case in cardiac arrhythmias[12]. Nevertheless, many of the pathways implicated in arrhythmogenesis are also important in other biological contexts, and it is conceivable that testing cell lines to discern the ‘net effect’ of a drug on a specific pathway may play a role in matching drug to disease. The recent discovery of genetic and chemical manipulations that can induce differentiated cells from virtually any source (skin, muscle etc) to regain a pluripotent stem-cell like state has led to the promise of cell lines derived from individual patients for diagnostics, disease modeling and drug discovery[88, 89]. These induced pluripotent stem cells (iPS) cells can in principle be differentiated into a wide range of different somatic cell types, including multiple different cardiomyocyte subtypes. While such approaches may render considerably greater native context than simple cell culture, the cell networks, physiologic integration and remote cell types so central to arrhythmogenesis are unlikely to be completely accessible in this system[90].
7. In vivo discovery
In the face of intractable complexity in vivo drug discovery in mechanistically faithful animal models appears to be an ideal solution. In essence, this strategy allows one to directly interrogate each of the components integrated precisely in their native context. However, the feasibility and cost of screening in mammals has allowed this approach only in the later phases of drug discovery to discriminate among small numbers of compounds. The lack of comprehensive insight into cardiac physiology in more tractable models such yeast, C. elegans, or Drosophila has prevented their widespread use in anti-arrhythmic drug discovery, but these models have been successfully employed in other disease areas[91].
In the last decade, the emergence of the zebrafish as a screenable vertebrate model has revolutionized the scale of genetic study that is feasible for cardiovascular or other complex diseases[92–95]. Within 48 hours of fertilization, the larval fish has established complex physiology, yet can be sustained in large numbers for days in multi-well plates[96, 97], and is amenable to both genetic and chemical screening[98]. The zebrafish genome has been sequenced, and each gene in the genome is readily manipulated using morpholino antisense oligos[99]. In the last few months stable gene knockouts have become feasible using zinc finger nuclease technologies[100]. Saturation phenotype-driven screens for morphologic phenotypes have been successfully performed[101], and with the improvement in phenotyping technologies screens for complex physiologic and pharmacologic endpoints have begun[96]. Embryos can be readily arrayed in 96 or 384 well plates and the development of increasingly sophisticated automated assays has allowed initial in vivo screens for integrated physiologic phenotypes that previously were only accessible for cell-based assays[36, 102].
The permeability of the larval zebrafish to small molecules has popularized chemical screens for modifiers of specific pathways or for the suppression of disease traits that have been modeled in the zebrafish[103, 104]. The zebrafish is also being developed as a tool in toxicology for both pharmacologic and environmental contexts. Counter-screens for toxicities and secondary screens of derivatives are immediately possible to optimize lead compounds[97, 105]. In addition, it should be possible to screen large numbers of other drugs for drug-drug interactions or genetically diverse pools of individuals to identify rare gene-drug interactions[96, 102]. For many diseases the fish can also be used for subsequent higher resolution studies to explore drug mechanisms at lower throughput, but still on a scale and at a cost that is difficult to replicate with other vertebrates. For example, in cardiac electrophysiology it is possible to evaluate the effects of drugs or genes on a host of electrophysiologic parameters including conduction, excitability and automaticity at a resolution similar to that in mouse or man. Similarly, high-fidelity calcium imaging can be used for the investigation of arrhythmic mechanisms. Staged use of these assays can enable the balance between sensitivity and specificity to be optimized[36, 106].
7.1 The importance of the model
At the core of this approach is the assumption that screening all conceivable targets together is worthwhile even though the target or targets are unknown. A foundation for this premise is once again the primary fidelity of the model. A uniquely balanced molecular entity that perfectly ‘tunes’ a disease pathway through activities on multiple targets is just as flawed as any other agent if the primary screening model does not share the underlying mechanisms of the relevant human disease.
While a shared primary etiology, genetic or otherwise, is helpful, it is also critical to validate the fidelity of the biologic response to this initial insult to ensure that there is relevance across a broad spectrum. Here functional genomics can be extremely useful, offering robust assessments of molecular responses across a wide dynamic range. It is also vital to ensure that the basic physiology and pharmacology of the relevant system are recapitulated. This type of rigorous validation often yields surprising results. For example, the 48 hour old zebrafish exhibits cardiac electrophysiology that is considerably more representative of human electrophysiology than the mouse or other small mammals[36, 107]. While this advantage partly reflects the very high heart rates and the apparent dependence of rodent repolarization on Ito, the zebrafish exhibits remarkably rapid maturation of cardiac electrophysiology, and multiple individual currents, whole heart electrophysiology and drug responses that closely mirror the adult human[36, 97, 107–109]. Similar fidelity has been observed for cardiac contractility at baseline and in the setting of a range of pharmacologic manipulations[110]. This fidelity has formed the basis for zebrafish work in predictive toxicology and cardiovascular drug discovery.
7.2 Zebrafish and arrhythmias
Techniques have been developed for measuring heart rate, contractility, and blood flow at high throughput in the zebrafish, as well as a repertoire of secondary assays including optical voltage mapping, Ca2+ imaging, and specific transgenic reporters for subcellular Ca2+ compartments[36, 96, 97, 109]. These tools enable efficient large-scale screens for genetic or chemical modifiers of known arrhythmia disease pathways in a completely native context[36].
It was possible to show that over 90% of drugs that cause repolarization toxicity in humans, result in cognate electrophysiologic effects in the zebrafish even as early as 48 hours post fertilization (hpf)[97]. Initial assays for heart rate using image analysis to explore cardiotoxicity were based on the dominant frequency component of the heart rate, but traded specificity for both sensitivity and throughput[97]. The complexity of arrhythmogenesis suggested that more sophisticated modeling was necessary to fully understand the underlying biology. To enable mechanistic evaluation of genetic or chemical modifiers methods were developed to directly measure cardiac action potentials in zebrafish embryos using optical mapping with voltage sensitive dyes at a stage when the fish are amenable to morpholino gene knockdown (Figure 1). Normal embryos display subtle differences in atrial and ventricular action potential profiles as anticipated[36].
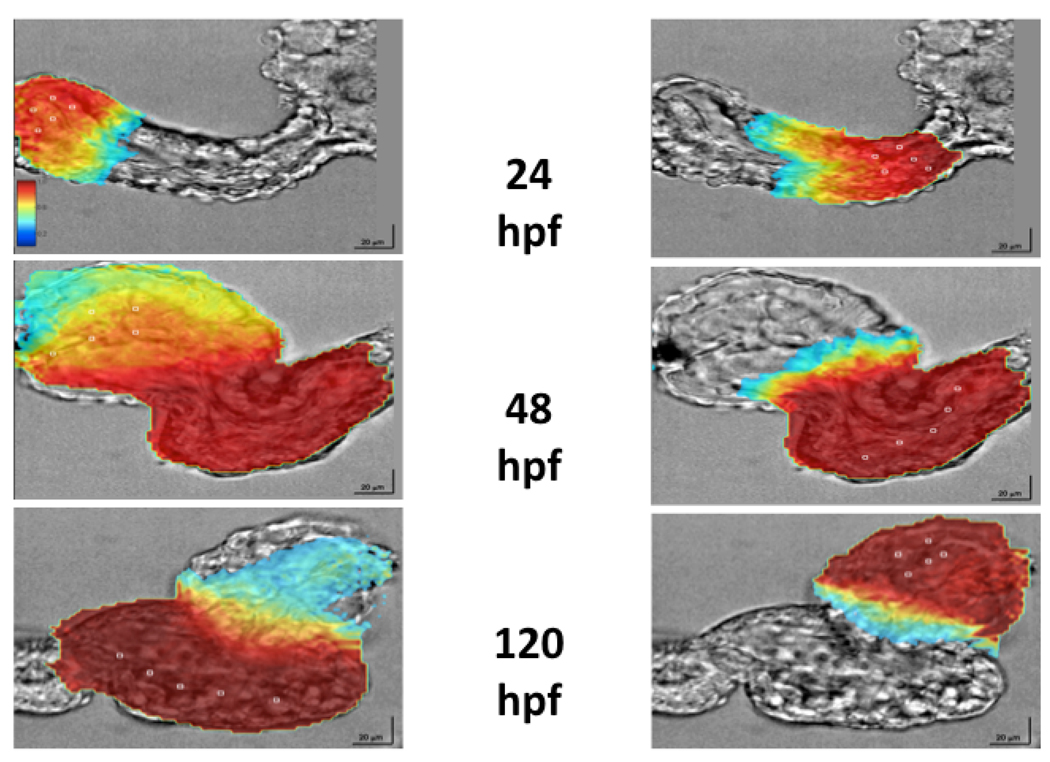
Figure 1. Ex vivo optical voltage mapping in zebrafish heart.
Using voltage sensitive dyes it is possible to characterize the electrical activity of the developing heart at virtually cellular resolution. Within 24 hours of fertilization of the oocyte, the fish heart is beating rhythmically (upper panel). By 48 hours the heart is representative of adult human cardiac electrophysiology, even though it consists of fewer than 400 cells and is less than 200 microns in size (middle panel). At 120 hours the heart has completed looping, the ventricle is growing rapidly and there is further subtle physiologic maturation (lower panel).
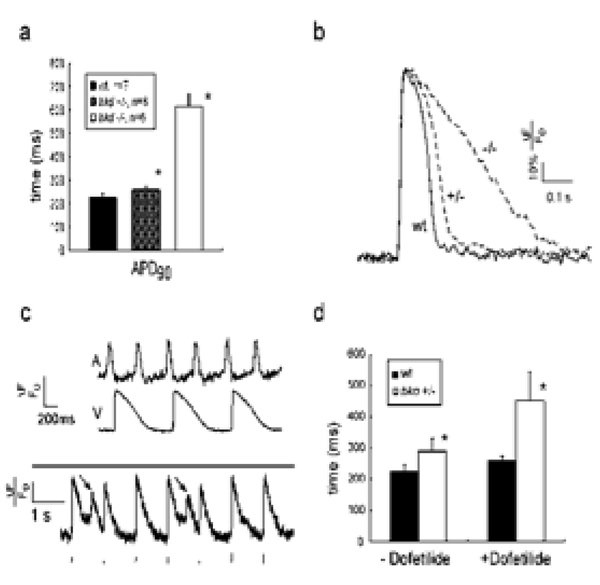
These techniques were validated in a comprehensive study of one form of human arrhythmia, inherited repolarization perturbation, using a zebrafish mutant breakdance that carries a missense mutation in the cardiac KCNH2 gene, the major subunit of the potassium channel responsible for IKr[36]. In breakdance homozygote action potentials there was evidence of significant “triangulation”, or prolongation of the APD25–75, a phenomenon observed in human repolarization disorders with high arrhythmic risk (Figure 2a and 2b[111]. The mechanism of 2:1 AV block was evident from recordings that demonstrated alternate atrial impulses encountering refractory ventricular myocardium (Figure 2c)[112]. Importantly, breakdance homozygotes also exhibited spontaneous early afterdepolarizations, the postulated triggers of fatal arrhythmias in both inherited and acquired repolarization disorders (Figure 2c). Treatment with doses of dofetilide as low as 10nM caused subtle prolongation of wild type action potentials (64ms ±45, 28% increase) while the same concentration of dofetilide resulted in marked prolongation of heterozygote action potentials (194 ms ±92, 75% increase) (Figure 2d). A final confirmation of the fidelity of the model was the extension of these observations to novel repolarization genes such as NOS1AP, first identified in large human genetic studies[36].
Figure 2. Parallels between zebrafish and human physiology and pharmacology.
(1a) Ventricular action potential (AP) durations in wildtype (wt) and breakdance heterozygotes (+/−) and homozygotes (−/−) at 6 days post fertilization. * denotes p<0.05. (1b) Typical ventricular APs are displayed for wildtype, breakdance heterozygote and homozygote embryos. The heterozygote AP is subtly prolonged, while the homozygote shows marked AP prolongation. Vertical calibration bar denotes 20% ΔF/F0, horizontal bar denotes 100ms. (1b) Upper panel: simultaneous atrial and ventricular voltage recordings from breakdance (−/−) heart showing the mechanism of 2:1 atrioventricular block: APs are so prolonged in the ventricle that alternate atrial impulses encroach on the refractory plateau of the previous ventricular repolarization. Lower panel: Early afterdepolarizations (arrows) in breakdance (−/−) embryos during ventricular pacing; the pacing train is shown below the AP recording. (1d) Heterozygote breakdance embryos display increased sensitivity to 10nM dofetilide.
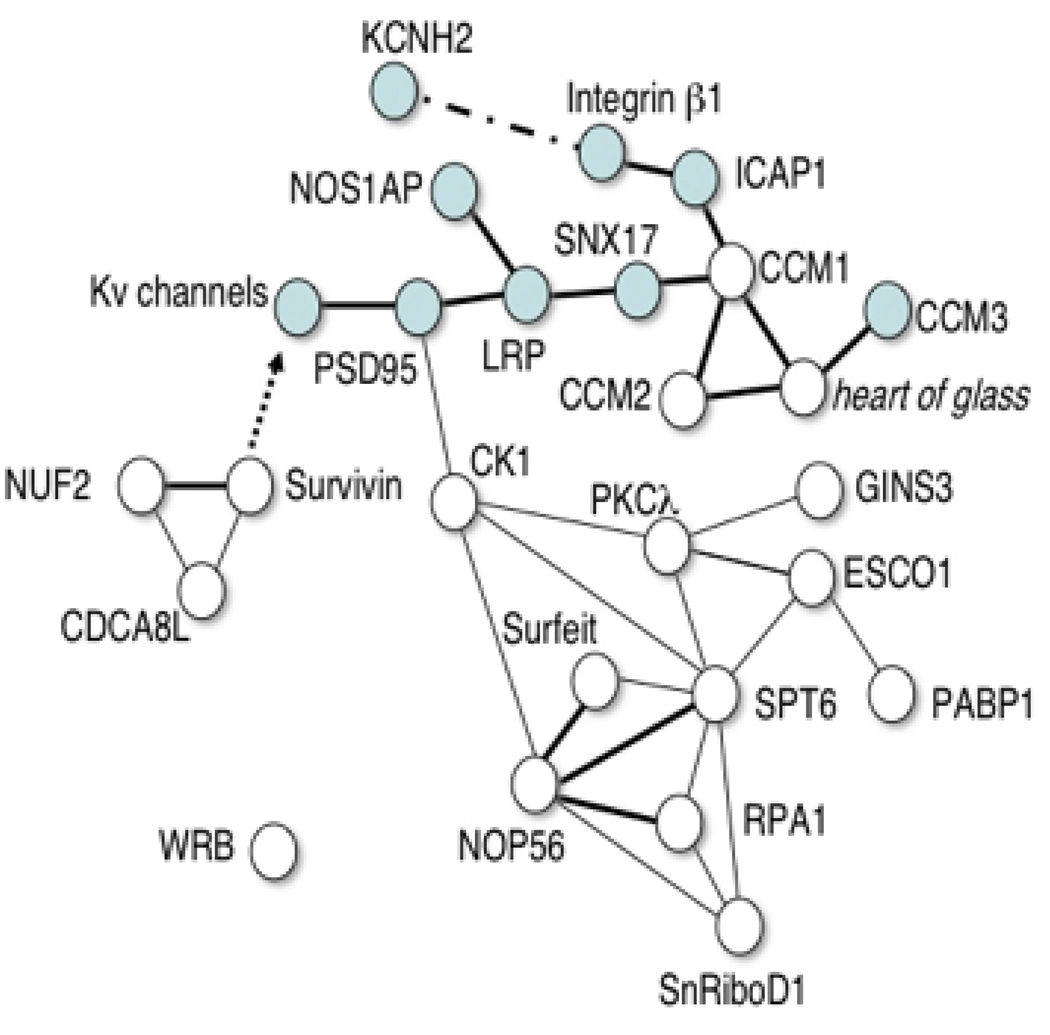
Having established the fidelity of the model in physiologic and disease states, it was then possible to exploit the throughput of the zebrafish model system to undertake a pharmacogenetic screen of a library of insertional zebrafish mutants[36]. This was designed to identify, in an unbiased manner, new genes that modify the cardiac response to IKr blockade. Despite intense efforts, to date there have been few biologically relevant repolarization or drug response modifier loci identified. The robust parallels between zebrafish and human cardiac repolarization suggested that formal genetic analysis of this clinically important complex trait might be feasible. To optimize sensitivity, specificity and throughput an initial high-throughput screen for abnormal heart rate response to dofetilide was combined with a second high-resolution assay in which confirmed mutants are studied using optical mapping. Subsequent testing in the absence of dofetilide allowed discrimination between pure drug response phenotypes and intrinsic heart rate defects. In the initial shelf screen of 340 insertional mutants 15 genes with major effects on repolarization were identified, none of which had been implicated previously in this process. Interestingly, the majority of these genes appear to belong to an integrin-associated network modulating channels and their adaptor proteins (Figure 3). These findings suggest potential links between mechanical loading conditions, inflammation and repolarization that may shed light on arrhythmogenesis in a number of conditions. Subsequently, some of these genes have been shown to modify human repolarization, confirming the utility of zebrafish screens for the discovery of genetic modifiers in physiologic or pharmacologic pathways[36].
Figure 3. A simple interaction diagram depicts interactions between known repolarization genes (blue symbols) and the genes identified in a zebrafish pharmacogenetic screen.
Single lines indicate genetic interactions supported by data from multiple model organisms. Bold lines show direct physical interactions. A dashed line represents a physical interaction that may not be direct. The arrow represents a downstream regulatory effect, the mechanism of which is unknown. Details of data used to construct diagram in reference 36.
7.3 Scalable in vivo models complement other emerging technologies
Validated in vivo models of physiology or disease that can be efficiently scaled also complement the broad range of ‘omics technologies that are emerging from academia and industry. Functional genomics have identified a host of new ion channels, many of which are now being studied as potential drug targets. In many instances relatively little is known of the integrated physiology of these channels, and their roles at different stages or in different disease settings may be difficult to define on the scale necessary for prioritization in drug discovery. Morpholino and transgenesis technologies have brought the zebrafish to the forefront in the initial modeling of data from genome-wide association studies, expression profiling, metabolomic and other ‘omics experiments. While the precise germline manipulations feasible in the mouse are not yet feasible in the zebrafish, the speed and cost of the zebrafish have favored its use in situations where the phenotypic parallels have been validated carefully[113, 114]. The ease of modeling extends to many aspects of the pathohysiology of disease and using specific zebrafish strains it is possible to test interactions with heart failure, hypertrophy and ischemia to name but a few[93, 115].
7.4 Translation to other models and limitations of the zebrafish
Once small molecules of interest have been identified in the zebrafish, it is straightforward to translate these findings to higher animals. While there may be some publication bias, where this has been attempted the results have proven reassuring, and reflect the remarkable conservation of the majority of vertebrate pathways[103, 104, 116]. Given the efficiency of the zebrafish as a model it is envisioned that a mammalian validation step might be included in a screen as a final definitive test-if an appropriate mammalian model exists for the phenotype in question. Thus, in a screen for suppressors of genetic forms of atrial fibrillation one might be circumspect if the same compounds were not effective to some extent in large animal atrial pacing models, but aware that these large animal models might not be recapitulate the primary substrate for the arrhythmia[73].
The zebrafish has some intrinsic limitations that must be considered in the overall design of in vivo screening strategies. Only a limited number of zebrafish disease models have been rigorously validated, and, like all other models, empiric assessment of the representation of a given physiology or disease state is a critical initial step. While the fish genome is remarkably tractable, homologous recombination for precise manipulations is not yet feasible. Drug penetration is often difficult to measure, and in many instances compounds with no activity in the assay may not reach the embryo in significant concentration. This may indirectly reflect the utility of the molecule as a drug, and can be at least partially anticipated on the basis of simple physicochemical parameters[97]. Pharmacokinetics and pharmacodynamics are only feasible in lower throughput, but the technological hurdles have been slowly overcome. Unbiased profiling technologies such as mass spectrometry have been used to measure drug absorption and might also be applied to assess drug distribution and metabolism[117]. Perhaps the single most restrictive limitation is the fact that no drug identified using zebrafish as the primary screening platform has yet made it to market. Several candidates in multiple disease areas are under development, but the success of even one new molecular entity identified using this approach may open the door to the wider incorporation of zebrafish technology into drug discovery[116].
8. Expert opinion
A fundamental problem with modern drug discovery is not the screening strategies that have been used, but rather the complexity of the underlying disease biology. Selection of a single target is simply not feasible, even where the specific molecular cause has been identified, because we do not fully understand the downstream biology. In arrhythmias a focus on transmembrane conductance has obscured the structural, signaling and other roles of ion channels in the cell, and led to the choice of specific targets that may not offer the therapeutic margin necessary for successful drug discovery as a result of their physiologic importance. The human genome project has re-taught us that integration across the entire organism is vital to capture the full picture in a biologic system. Understanding how biologic systems vary in disease and between individuals will be vital for truly personalized medicine.
It will be increasingly important to know precise disease mechanisms, ultimately at the level of the individual patient. This will lead to better, more mechanistically faithful, disease models that recapitulate not just one aspect of the disease process but all of its nuances. Screening for drugs in vivo in validated vertebrate model organisms, such as the zebrafish, offers a complementary strategy for drug discovery where the perturbed disease networks can be ‘tuned’ toward a better outcome. This empiric approach in a truly native biological context offers unique possibilities. It allows for the systematic discovery of ‘dirty’ drugs with effects on multiple targets, or agents with more subtle gain or loss of function effects at individual targets that are only apparent in the integrated context. Counter-screens for toxicology or in vivo optimization in the setting of other drugs or other disease contexts are also feasible. In vivo discovery also offers unique opportunities for parallel diagnostics as well as for rigorous pharmacogenetics and genomics. Widespread use of the zebrafish as an integral component of drug discovery platforms will probably not occur until the approach has been validated through successful commercial application, but this is likely to occur in the near future.
Article highlights
Most arrhythmia models in use fail to recapitulate large parts of the disease pathophysiology
Simple abnormalities of ion channel conductance do not explain the majority of arrhythmic risk
Unbiased approaches to understanding arrhythmia pathogenesis are identifying novel mechanisms
Parallel unbiased approaches to drug discovery are required
In vivo drug discovery in high throughput is feasible in the zebrafish
Ongoing work will place this approach in the appropriate stage of the discovery pipeline
Acknowledgments
Declaration of Interest
The author’s work was supported by the NHLBI as well as NIH/NIGMS and NIH/NINDS grants GM075946 and NS063563 respectively. The author also received funding from the British Heart Foundation and March of Dimes.
References
- 1.Wellens HJ. Cardiac arrhythmias: the quest for a cure: a historical perspective. J Am Coll Cardiol. 2004;44(6):1155–1163. doi: 10.1016/j.jacc.2004.05.080. [DOI] [PubMed] [Google Scholar]
- 2.Roden DM. Drug-induced prolongation of the QT interval. N Engl J Med. 2004;350(10):1013–1022. doi: 10.1056/NEJMra032426. [DOI] [PubMed] [Google Scholar]
- 3.The cardiac arrhythmia suppression trial. N Engl J Med. 1989;321(25):1754–1756. doi: 10.1056/NEJM198912213212510. [DOI] [PubMed] [Google Scholar]
- 4.Waldo AL. A perspective on antiarrhythmic drug therapy to treat atrial fibrillation: there remains an unmet need. Am Heart J. 2006;151(4):771–778. doi: 10.1016/j.ahj.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Waldo AL. Mechanisms of atrial fibrillation. J Cardiovasc Electrophysiol. 2003;14(12 Suppl):S267–S274. doi: 10.1046/j.1540-8167.2003.90401.x. [DOI] [PubMed] [Google Scholar]
- 6.Eckardt L, Haverkamp W, Johna R, et al. Arrhythmias in heart failure: current concepts of mechanisms and therapy. J Cardiovasc Electrophysiol. 2000;11(1):106–117. doi: 10.1111/j.1540-8167.2000.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 7.Roden DM. Mechanisms and management of proarrhythmia. Am J Cardiol. 1998;82(4A):49I–57I. doi: 10.1016/s0002-9149(98)00472-x. [DOI] [PubMed] [Google Scholar]
- 8.Ruskin JN. The cardiac arrhythmia suppression trial (CAST) N Engl J Med. 1989;321(6):386–388. doi: 10.1056/NEJM198908103210608. [DOI] [PubMed] [Google Scholar]
- 9.Spooner PM, Albert C, Benjamin EJ, et al. Sudden cardiac death, genes, and arrhythmogenesis : consideration of new population and mechanistic approaches from a national heart, lung, and blood institute workshop, part I. Circulation. 2001;103(19):2361–2364. doi: 10.1161/01.cir.103.19.2361. [DOI] [PubMed] [Google Scholar]
- 10.MacRae CA, Ellinor PT. Genetic screening and risk assessment in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44(12):2326–2328. doi: 10.1016/j.jacc.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 11.Vatta M, Dumaine R, Varghese G, et al. Genetic and biophysical basis of sudden unexplained nocturnal death syndrome (SUNDS), a disease allelic to Brugada syndrome. Hum Mol Genet. 2002;11(3):337–345. doi: 10.1093/hmg/11.3.337. [DOI] [PubMed] [Google Scholar]
- 12.Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104(4):569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd-Jones DM, Wang TJ, Leip EP, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110(9):1042–1046. doi: 10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 14.Shah M, Akar FG, Tomaselli GF. Molecular basis of arrhythmias. Circulation. 2005;112(16):2517–2529. doi: 10.1161/CIRCULATIONAHA.104.494476. [DOI] [PubMed] [Google Scholar]
- 15.Ho CY, Sweitzer NK, McDonough B, et al. Assessment of diastolic function with Doppler tissue imaging to predict genotype in preclinical hypertrophic cardiomyopathy. Circulation. 2002;105(25):2992–2997. doi: 10.1161/01.cir.0000019070.70491.6d. [DOI] [PubMed] [Google Scholar]
- 16.Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev. 2005;85(4):1205–1253. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 17.Roden DM, Balser JR, George AL, Jr, et al. Cardiac ion channels. Annu Rev Physiol. 2002;64:431–475. doi: 10.1146/annurev.physiol.64.083101.145105. [DOI] [PubMed] [Google Scholar]
- 18.Salama G, London B. Mouse models of long QT syndrome. J Physiol. 2007;578(Pt 1):43–53. doi: 10.1113/jphysiol.2006.118745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casimiro MC, Knollmann BC, Yamoah EN, et al. Targeted point mutagenesis of mouse Kcnq1: phenotypic analysis of mice with point mutations that cause Romano-Ward syndrome in humans. Genomics. 2004;84(3):555–564. doi: 10.1016/j.ygeno.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Emmi A, Wenzel HJ, Schwartzkroin PA, et al. Do glia have heart? Expression and functional role for ether-a-go-go currents in hippocampal astrocytes. J Neurosci. 2000;20(10):3915–3925. doi: 10.1523/JNEUROSCI.20-10-03915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priori SG, Napolitano C, Schwartz PJ. Low penetrance in the long-QT syndrome: clinical impact. Circulation. 1999;99(4):529–533. doi: 10.1161/01.cir.99.4.529. [DOI] [PubMed] [Google Scholar]
- 22.Koren G. Electrical remodeling and arrhythmias in long-QT syndrome: lessons from genetic models in mice. Ann Med. 2004;36 Suppl 1:22–27. doi: 10.1080/17431380410032643. [DOI] [PubMed] [Google Scholar]
- 23.Ali RH, Zareba W, Moss AJ, et al. Clinical and genetic variables associated with acute arousal and nonarousal-related cardiac events among subjects with long QT syndrome. Am J Cardiol. 2000;85(4):457–461. doi: 10.1016/s0002-9149(99)90772-5. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz PJ, Priori SG, Spazzolini C, et al. Genotype-phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation. 2001;103(1):89–95. doi: 10.1161/01.cir.103.1.89. [DOI] [PubMed] [Google Scholar]
- 25.Chen YH, Xu SJ, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299(5604):251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 26.Otway R, Vandenberg JI, Guo G, et al. Stretch-sensitive KCNQ1 mutation A link between genetic and environmental factors in the pathogenesis of atrial fibrillation? J Am Coll Cardiol. 2007;49(5):578–586. doi: 10.1016/j.jacc.2006.09.044. [DOI] [PubMed] [Google Scholar]
- 27.MacRae CA. Closer look at genetic testing in long-QT syndrome: will DNA diagnostics ever be enough? Circulation. 2009;120(18):1745–1748. doi: 10.1161/CIRCULATIONAHA.109.900415. [DOI] [PubMed] [Google Scholar]
- 28.Jentsch TJ, Hubner CA, Fuhrmann JC. Ion channels: function unravelled by dysfunction. Nat Cell Biol. 2004;6(11):1039–1047. doi: 10.1038/ncb1104-1039. [DOI] [PubMed] [Google Scholar]
- 29.Ficker E, Dennis AT, Wang L, et al. Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circ Res. 2003;92(12):e87–e100. doi: 10.1161/01.RES.0000079028.31393.15. [DOI] [PubMed] [Google Scholar]
- 30.Mohler PJ, Schott JJ, Gramolini AO, et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421(6923):634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 31.Mohler PJ, Splawski I, Napolitano C, et al. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci U S A. 2004;101(24):9137–9142. doi: 10.1073/pnas.0402546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vatta M, Ackerman MJ, Ye B, et al. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114(20):2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 33.Petrecca K, Atanasiu R, Akhavan A, et al. N-linked glycosylation sites determine HERG channel surface membrane expression. J Physiol. 1999;515(Pt 1):41–48. doi: 10.1111/j.1469-7793.1999.041ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furutani M, Trudeau MC, Hagiwara N, et al. Novel mechanism associated with an inherited cardiac arrhythmia: defective protein trafficking by the mutant HERG (G601S) potassium channel. Circulation. 1999;99(17):2290–2294. doi: 10.1161/01.cir.99.17.2290. [DOI] [PubMed] [Google Scholar]
- 35.Kiss E, Nagy P, Balogh A, et al. Cytometry of raft and caveola membrane microdomains: from flow and imaging techniques to high throughput screening assays. Cytometry A. 2008;73(7):599–614. doi: 10.1002/cyto.a.20572. [DOI] [PubMed] [Google Scholar]
- 36.Milan DJ, Kim AM, Winterfield JR, et al. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120(7):553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corrado D, Basso C, Thiene G, et al. Spectrum of clinicopathologic manifestations of arrhythmogenic right ventricular cardiomyopathy/dysplasia: a multicenter study. J Am Coll Cardiol. 1997;30(6):1512–1520. doi: 10.1016/s0735-1097(97)00332-x. [DOI] [PubMed] [Google Scholar]
- 38.Sen-Chowdhry S, Syrris P, McKenna WJ. Desmoplakin disease in arrhythmogenic right ventricular cardiomyopathy: early genotype-phenotype studies. Eur Heart J. 2005;26(16):1582–1584. doi: 10.1093/eurheartj/ehi343. [DOI] [PubMed] [Google Scholar]
- 39.Coonar AS, Protonotarios N, Tsatsopoulou A, et al. Gene for arrhythmogenic right ventricular cardiomyopathy with diffuse nonepidermolytic palmoplantar keratoderma and woolly hair (Naxos disease) maps to 17q21. Circulation. 1998;97(20):2049–2058. doi: 10.1161/01.cir.97.20.2049. [DOI] [PubMed] [Google Scholar]
- 40.Rampazzo A, Nava A, Malacrida S, et al. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71(5):1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerull B, Heuser A, Wichter T, et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36(11):1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 42.Syrris P, Ward D, Evans A, et al. Arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in the desmosomal gene desmocollin-2. Am J Hum Genet. 2006;79(5):978–984. doi: 10.1086/509122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heuser A, Plovie ER, Ellinor PT, et al. Mutant desmocollin-2 causes arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2006;79(6):1081–1088. doi: 10.1086/509044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asimaki A, Syrris P, Wichter T, et al. A novel dominant mutation in plakoglobin causes arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2007;81(5):964–973. doi: 10.1086/521633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Gras E, Lombardi R, Giocondo MJ, et al. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116(7):2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asimaki A, Tandri H, Huang H, et al. A new diagnostic test for arrhythmogenic right ventricular cardiomyopathy. N Engl J Med. 2009;360(11):1075–1084. doi: 10.1056/NEJMoa0808138. [DOI] [PubMed] [Google Scholar]
- 47.Nattel S, Shiroshita-Takeshita A, Cardin S, et al. Mechanisms of atrial remodeling and clinical relevance. Curr Opin Cardiol. 2005;20(1):21–25. [PubMed] [Google Scholar]
- 48.Brundel BJ, Henning RH, Kampinga HH, et al. Molecular mechanisms of remodeling in human atrial fibrillation. Cardiovasc Res. 2002;54(2):315–324. doi: 10.1016/s0008-6363(02)00222-5. [DOI] [PubMed] [Google Scholar]
- 49.Sack MN, Kelly DP. The energy substrate switch during development of heart failure: gene regulatory mechanisms (Review) Int J Mol Med. 1998;1(1):17–24. doi: 10.3892/ijmm.1.1.17. [DOI] [PubMed] [Google Scholar]
- 50.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7(8):589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 51.McNally EM, Pytel P. Muscle diseases: the muscular dystrophies. Annu Rev Pathol. 2007;2:87–109. doi: 10.1146/annurev.pathol.2.010506.091936. [DOI] [PubMed] [Google Scholar]
- 52.Kass RS, Kurokawa J, Marx SO, et al. Leucine/isoleucine zipper coordination of ion channel macromolecular signaling complexes in the heart. Roles in inherited arrhythmias. Trends Cardiovasc Med. 2003;13(2):52–56. doi: 10.1016/s1050-1738(02)00211-6. [DOI] [PubMed] [Google Scholar]
- 53.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4(7):517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 54.Feillet F, Steinmann G, Vianey-Saban C, et al. Adult presentation of MCAD deficiency revealed by coma and severe arrythmias. Intensive Care Med. 2003;29(9):1594–1597. doi: 10.1007/s00134-003-1871-3. [DOI] [PubMed] [Google Scholar]
- 55.Antzelevitch C, Fish J. Electrical heterogeneity within the ventricular wall. Basic Res Cardiol. 2001;96(6):517–527. doi: 10.1007/s003950170002. [DOI] [PubMed] [Google Scholar]
- 56.Christoffels VM, Burch JB, Moorman AF. Architectural Plan for the Heart: Early Patterning and Delineation of the Chambers and the Nodes. Trends Cardiovasc Med. 2004;14(8):301–307. doi: 10.1016/j.tcm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 57.de Bakker JM, van Capelle FJ, Janse MJ, et al. Reentry as a cause of ventricular tachycardia in patients with chronic ischemic heart disease: electrophysiologic and anatomic correlation. Circulation. 1988;77(3):589–606. doi: 10.1161/01.cir.77.3.589. [DOI] [PubMed] [Google Scholar]
- 58.van der Merwe PL, Rose AG, van der Walt JJ, et al. Progressive familial heart block type I. Clinical and pathological observations. S Afr Med J. 1991;80(1):34–38. [PubMed] [Google Scholar]
- 59.Brink PA, Ferreira A, Moolman JC, et al. Gene for progressive familial heart block type I maps to chromosome 19q13. Circulation. 1995;91(6):1633–1640. doi: 10.1161/01.cir.91.6.1633. [DOI] [PubMed] [Google Scholar]
- 60.Brugada R, Brugada J, Antzelevitch C, et al. Sodium channel blockers identify risk for sudden death in patients with ST-segment elevation and right bundle branch block but structurally normal hearts. Circulation. 2000;101(5):510–515. doi: 10.1161/01.cir.101.5.510. [DOI] [PubMed] [Google Scholar]
- 61.Weiss R, Barmada MM, Nguyen T, et al. Clinical and molecular heterogeneity in the Brugada syndrome: a novel gene locus on chromosome 3. Circulation. 2002;105(6):707–713. doi: 10.1161/hc0602.103618. [DOI] [PubMed] [Google Scholar]
- 62.Antzelevitch C, Yan GX, Shimizu W. Transmural dispersion of repolarization and arrhythmogenicity: the Brugada syndrome versus the long QT syndrome. J Electrocardiol. 1999;32 Suppl:158–165. doi: 10.1016/s0022-0736(99)90074-2. [DOI] [PubMed] [Google Scholar]
- 63.Costantini DL, Arruda EP, Agarwal P, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123(2):347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heydemann A, Huber JM, Kakkar R, et al. Functional nitric oxide synthase mislocalization in cardiomyopathy. J Mol Cell Cardiol. 2004;36(2):213–223. doi: 10.1016/j.yjmcc.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 65.Frustaci A, Chimenti C, Bellocci F, et al. Histological substrate of atrial biopsies in patients with lone atrial fibrillation. Circulation. 1997;96(4):1180–1184. doi: 10.1161/01.cir.96.4.1180. [DOI] [PubMed] [Google Scholar]
- 66.Issac TT, Dokainish H, Lakkis NM. Role of inflammation in initiation and perpetuation of atrial fibrillation: a systematic review of the published data. J Am Coll Cardiol. 2007;50(21):2021–2028. doi: 10.1016/j.jacc.2007.06.054. [DOI] [PubMed] [Google Scholar]
- 67.Ramos-Mondragon R, Galindo CA, Avila G. Role of TGF-beta on cardiac structural and electrical remodeling. Vasc Health Risk Manag. 2008;4(6):1289–1300. doi: 10.2147/vhrm.s3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin CS, Pan CH. Regulatory mechanisms of atrial fibrotic remodeling in atrial fibrillation. Cell Mol Life Sci. 2008;65(10):1489–1508. doi: 10.1007/s00018-008-7408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellinor PT, Sasse-Klaassen S, Probst S, et al. A novel locus for dilated cardiomyopathy, diffuse myocardial fibrosis, and sudden death on chromosome 10q25–26. J Am Coll Cardiol. 2006;48(1):106–111. doi: 10.1016/j.jacc.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 70.Weber KT. Fibrosis and hypertensive heart disease. Curr Opin Cardiol. 2000;15(4):264–272. doi: 10.1097/00001573-200007000-00010. [DOI] [PubMed] [Google Scholar]
- 71.Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132(4):537–543. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87(2):521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milan DJ, MacRae CA. Animal models for arrhythmias. Cardiovasc Res. 2005;67(3):426–437. doi: 10.1016/j.cardiores.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 74.Roden DM, Altman RB, Benowitz NL, et al. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145(10):749–757. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rutitzky B, Girotti AL, Rosenbaum MB. Efficacy of chronic amiodarone therapy in patients with variant angina pectoris and inhibition of ergonovine coronary constriction. Am Heart J. 1982;103(1):38–43. doi: 10.1016/0002-8703(82)90526-9. [DOI] [PubMed] [Google Scholar]
- 76.Zhang S, Zhou Z, Gong Q, et al. Mechanism of block and identification of the verapamil binding domain to HERG potassium channels. Circ Res. 1999;84(9):989–998. doi: 10.1161/01.res.84.9.989. [DOI] [PubMed] [Google Scholar]
- 77.Kaski JC, Girotti LA, Elizari MV, et al. Efficacy of amiodarone during long-term treatment of potentially dangerous ventricular arrhythmias in patients with chronic stable ischemic heart disease. Am Heart J. 1984;107(4):648–655. doi: 10.1016/0002-8703(84)90310-7. [DOI] [PubMed] [Google Scholar]
- 78.Strange K. The end of "naive reductionism": rise of systems biology or renaissance of physiology? Am J Physiol Cell Physiol. 2005;288(5):C968–C974. doi: 10.1152/ajpcell.00598.2004. [DOI] [PubMed] [Google Scholar]
- 79.Ge H, Walhout AJ, Vidal M. Integrating 'omic' information: a bridge between genomics and systems biology. Trends Genet. 2003;19(10):551–560. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 80.Giallourakis C, Henson C, Reich M, et al. Disease gene discovery through integrative genomics. Annu Rev Genomics Hum Genet. 2005;6:381–406. doi: 10.1146/annurev.genom.6.080604.162234. [DOI] [PubMed] [Google Scholar]
- 81.Li S, Armstrong CM, Bertin N, et al. A map of the interactome network of the metazoan C elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rudy Y. From genome to physiome: integrative models of cardiac excitation. Ann Biomed Eng. 2000;28(8):945–950. doi: 10.1114/1.1308484. [DOI] [PubMed] [Google Scholar]
- 83.Verrier RL, Antzelevitch C. Autonomic aspects of arrhythmogenesis: the enduring and the new. Curr Opin Cardiol. 2004;19(1):2–11. doi: 10.1097/00001573-200401000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berger SI, Ma'ayan A, Iyengar R. Systems pharmacology of arrhythmias. Sci Signal. 2010;3(118):ra30. doi: 10.1126/scisignal.2000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng CS, Alderman D, Kwash J, et al. A high-throughput HERG potassium channel function assay: an old assay with a new look. Drug Dev Ind Pharm. 2002;28(2):177–191. doi: 10.1081/ddc-120002451. [DOI] [PubMed] [Google Scholar]
- 86.Casimiro MC, Knollmann BC, Ebert SN, et al. Targeted disruption of the Kcnq1 gene produces a mouse model of Jervell and Lange-Nielsen Syndrome. Proc Natl Acad Sci U S A. 2001;98(5):2526–2531. doi: 10.1073/pnas.041398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nerbonne JM. Studying cardiac arrhythmias in the mouse--a reasonable model for probing mechanisms? Trends Cardiovasc Med. 2004;14(3):83–93. doi: 10.1016/j.tcm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 88.Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008;41 Suppl 1:51–56. doi: 10.1111/j.1365-2184.2008.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 90.Domian IJ, Chiravuri M, van der Meer P, et al. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326(5951):426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Potet F, Petersen CI, Boutaud O, et al. Genetic screening in C. elegans identifies rho-GTPase activating protein 6 as novel HERG regulator. J Mol Cell Cardiol. 2009;46(2):257–267. doi: 10.1016/j.yjmcc.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Langheinrich U. Zebrafish: a new model on the pharmaceutical catwalk. Bioessays. 2003;25(9):904–912. doi: 10.1002/bies.10326. [DOI] [PubMed] [Google Scholar]
- 93.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8(5):353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 94.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4(1):35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 95.MacRae CA, Peterson RT. Zebrafish-based small molecule discovery. Chem Biol. 2003;10(10):901–908. doi: 10.1016/j.chembiol.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 96.Burns CG, Milan DJ, Grande EJ, et al. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1(5):263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- 97.Milan DJ, Peterson TA, Ruskin JN, et al. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation. 2003;107(10):1355–1358. doi: 10.1161/01.cir.0000061912.88753.87. [DOI] [PubMed] [Google Scholar]
- 98.Fishman MC. Genomics. Zebrafish--the canonical vertebrate. Science. 2001;294(5545):1290–1291. doi: 10.1126/science.1066652. [DOI] [PubMed] [Google Scholar]
- 99.Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26(2):216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 100.Meng X, Noyes MB, Zhu LJ, et al. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26(6):695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Driever W, Solnica-Krezel L, Schier AF, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 102.Kokel D, Bryan J, Laggner C, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6(3):231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yeh JR, Munson KM, Chao YL, et al. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development. 2008;135(2):401–410. doi: 10.1242/dev.008904. [DOI] [PubMed] [Google Scholar]
- 104.Peterson RT, Shaw SY, Peterson TA, et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22(5):595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- 105.Cuny GD, Yu PB, Laha JK, et al. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett. 2008;18(15):4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee JS, Yu Q, Shin JT, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11(6):845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 107.Langheinrich U, Vacun G, Wagner T. Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia. Toxicol Appl Pharmacol. 2003;193(3):370–382. doi: 10.1016/j.taap.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 108.Baker K, Warren KS, Yellen G, et al. Defective "pacemaker" current (Ih) in a zebrafish mutant with a slow heart rate. Proc Natl Acad Sci U S A. 1997;94(9):4554–4559. doi: 10.1073/pnas.94.9.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Milan DJ, Jones IL, Ellinor PT, et al. In vivo recording of adult zebrafish electrocardiogram and assessment of drug-induced QT prolongation. Am J Physiol Heart Circ Physiol. 2006;291(1):H269–H273. doi: 10.1152/ajpheart.00960.2005. [DOI] [PubMed] [Google Scholar]
- 110.Shin JT, Pomerantsev EV, Mably JD, et al. High-resolution cardiovascular function confirms functional orthology of myocardial contractility pathways in zebrafish. Physiol Genomics. 2010 doi: 10.1152/physiolgenomics.00206.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shah RR, Hondeghem LM. Refining detection of drug-induced proarrhythmia: QT interval and TRIaD. Heart Rhythm. 2005;2(7):758–772. doi: 10.1016/j.hrthm.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 112.Lupoglazoff JM, Cheav T, Baroudi G, et al. Homozygous SCN5A mutation in long-QT syndrome with functional two-to-one atrioventricular block. Circ Res. 2001;89(2):E16–E21. doi: 10.1161/hh1401.095087. [DOI] [PubMed] [Google Scholar]
- 113.Ho SY, Thorpe JL, Deng Y, et al. Lipid metabolism in zebrafish. Methods Cell Biol. 2004;76:87–108. doi: 10.1016/s0091-679x(04)76006-9. [DOI] [PubMed] [Google Scholar]
- 114.Weinstein BM. What guides early embryonic blood vessel formation? Dev Dyn. 1999;215(1):2–11. doi: 10.1002/(SICI)1097-0177(199905)215:1<2::AID-DVDY2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 115.Shin JT, Fishman MC. From Zebrafish to human: modular medical models. Annu Rev Genomics Hum Genet. 2002;3:311–340. doi: 10.1146/annurev.genom.3.031402.131506. [DOI] [PubMed] [Google Scholar]
- 116.Yu PB, Deng DY, Lai CS, et al. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat Med. 2008;14(12):1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Berghmans S, Butler P, Goldsmith P, et al. Zebrafish based assays for the assessment of cardiac, visual and gut function--potential safety screens for early drug discovery. J Pharmacol Toxicol Methods. 2008;58(1):59–68. doi: 10.1016/j.vascn.2008.05.130. [DOI] [PubMed] [Google Scholar]