Abstract
The human gut is populated by a microbiota composed of tens of trillions of organisms and millions of genes that together form a metabolic ‘organ’. Intra- and interpersonal variations in the structure and functions of this organ suggest that it likely contributes to our normal metabolic variations as well as the origins and treatment of metabolic disorders. This Commentary discusses experimental approaches for connecting microbial and host metabolism in the context of health and disease.
Introduction
Here is a striking and perhaps humbling perspective about ourselves: approximately 10 percent of our cells and <1 percent of our genes are human; the remainder are microbial. The vast majority of these microbes (tens of trillions), representing all three domains of life, live in our gastrointestinal tracts. The enormous genetic repository (microbiome) encoded in our gut microbial community (microbiota) differs from our human genome in terms of its origin (we are born in a pristine, essentially germ-free state), composition, plasticity, and extent of inter-personal diversity (Costello et al., 2009; Qin et al., 2010; Turnbaugh et al., 2010). Even genetically identical adult monozygotic co-twins harbor distinct collections of microbial species and genes in their guts (Turnbaugh et al., 2009a; Turnbaugh et al., 2010). This leads to the thought that we each house personal metabolic organs within our gastrointestinal tracts, composed of microbial cell lineages bound together in an elaborate co-evolved web of interspecies and interkingdom metabolic commerce that could have a profound impact on our metabolic phenotypes (metabotypes).
The potential to develop new microbial diagnostic biomarkers of our metabolic status, and new therapeutic strategies to maximize the contribution of our microbiota to our health has spawned many human microbiome projects (HMPs) worldwide. These HMPs reflect the rapid co-evolution of new experimental and computational tools that together form a field known as “metagenomics” which uses culture-independent, sequencing-based methods to characterize the composition and dynamic operations of microbial communities.
Much of what we consume, both beneficial and noxious, is first encountered and processed by our gut microbes. Our current understanding of the connections between metabolism, diet, and the microbiota has been the subject of several recent reviews (e.g. Backhed and Crawford, 2009; Musso et al., 2009). The goals of this Commentary are to (i) highlight experimental approaches that are likely to play important roles in forging connections between the microbiota and host metabolism, (ii) discuss some of the challenges facing the field and how they might be addressed, and (iii) envision therapeutic strategies that may emerge from these studies.
Experimental Strategies
Culture-independent approaches
The proportion and phylogenetic distribution of human gut microbial species that are recalcitrant to culture in vitro has not been clearly established. Nonetheless, culture-independent techniques have made important contributions to our current understanding of these microbial populations and how they may both induce and respond to metabolic changes in their hosts. Many of these culture-independent techniques are closely linked to advances in DNA sequencing technology: e.g. microbial community composition profiling via targeted sequencing of small subunit (SSU) ribosomal RNA genes; shotgun sequencing to predict encoded functions; and defining which functions are expressed by profiling mRNAs emanating from the microbiome. We can safely predict that the breadth and depth of microbial community sampling will change significantly as the throughput of highly parallel sequencing instruments continues to increase rapidly. For example, descriptions of human gut microbial communities that assimilate the results of detailed time-course studies of ever larger numbers of individuals living in distinct cultural contexts will be critical for determining how lifestyle (diet and other environmental exposures) impacts community structure and functions, and for defining healthy versus disease-relevant community configurations.
In addition to these sequencing-based approaches, culture-independent identification and quantification of proteins and metabolites by MS or NMR will be critical to connect the gut microbiota to metabolism of exogenously- as well as endogenously-derived molecules. Comparisons of blood metabolites in germ-free and conventionally-raised mice using non-targeted MS have revealed hundreds of compounds that are detectable in one but not the other type of animal, and even more whose concentrations are affected by the microbiota (Wikoff et al., 2009). For example, many compounds are conjugated to charged moieties such as sulfate, glycine, and glucoronide only in the presence of a microbiota: as these adducts are involved in the processing of xenobiotics, this finding suggests that the microbiota could determine individual responses to many drugs that are metabolized either inside or outside of the intestine. These results translate to humans: metabolism of the analagesic acetaminophen to either sulfonated or glucoronidated forms has been associated with pre-dose levels of p-cresol, a microbial tyrosine metabolite that competes with acetaminophen for its sulfonate donor and the relevant sulfotransferase enzyme. Higher levels of p-cresol correlate with decreased sulfonation and increased glucoronidation of acetaminophen (Clayton et al., 2009).
Siuzdak and colleagues have used non-targeted MS to identify a metabolite, indole-3-propionic acid (IPA), in the serum of conventionally-raised but not germ-free mice. This compound is an antioxidant and a potential therapeutic agent for Alzheimer’s disease. They subsequently screened cultured human gut symbionts and found a species, Clostridium sporogenes, capable of producing IPA in vitro. When introduced into the guts of germ-free mice, C. sporogenes is capable of restoring serum IPA to levels encountered in their conventionally-raised counterparts (Wikoff et al., 2009). In another example, non-targeted MS-based analyses of age-matched and genetically identical germ-free versus conventionally-raised mice (Velagapudi et al., 2009) have indicated that there is increased trafficking of triglycerides between the circulation and adipose storage depots in animals with a microbiota. Concurrent increases in serum and liver levels of phosphatidylcholine (16:0/18:1), an agonist of the lipoprotein lipase (LPL) transcriptional activator PPAR-α, are consistent with other observations that PPAR-α gene expression can be activated by the gut microbiota (Crawford et al., 2009). Microbiota-dependent suppression of intestinal epithelial expression of Fiaf (Angptl4), a circulating LPL inhibitor, also contributes to increased triglyceride storage in adipocytes (Velagapudi et al., 2009).
Engraftment of complete microbiotas into germ-free mice
Several difficult-to-control variables complicate studies of the metabolic properties of the gut microbiota in humans, including variation in host genotypes, incomplete histories of food consumption and prior environmental exposures, the absence of replicate individuals with identical gut microbial communities, and limitations in sampling more remote regions of the gastrointestinal tract. Gnotobiotic (“known life”) animals, in which microbes are either entirely absent (germ-free) or intentionally introduced at selected times, provide an approach for connecting microbial communities and their genes to host metabolism (Figure 1A). Encouragingly, culture-independent SSU (16S) rRNA-based community composition analysis suggests that nearly all of the bacterial diversity present in a human fecal microbiota (even samples archived at −80°C for more than a year) can be stably engrafted into germ-free mice (Turnbaugh et al., 2009b). Inter-generational transfer of the human gut microbiota from these humanized mice to their offspring produces animals that inherit a foreign microbiota “naturally” and thus allows experiments to be performed and hypotheses to be generated about how human gut microbial community assembly may shape and even imprint a host metabotype in animals where the innate and adaptive arms of the immune system co-evolve with the microbiota from birth. Transplantation of gut communities from human donors with different metabotypes to germ-free animals with or without (host) genetic mutations postulated to contribute to donor phenotypes, and who do or do not consume diets analogous to those of the donors, could allow proof-of-principle, proof-of-mechanism studies to be performed. These studies could directly address the question of how much of a host’s physiologic characteristics are a reflection of traits encoded in their microbiome versus their human genome, and how environmental factors, such as diet, contribute to eliciting a metabotype.
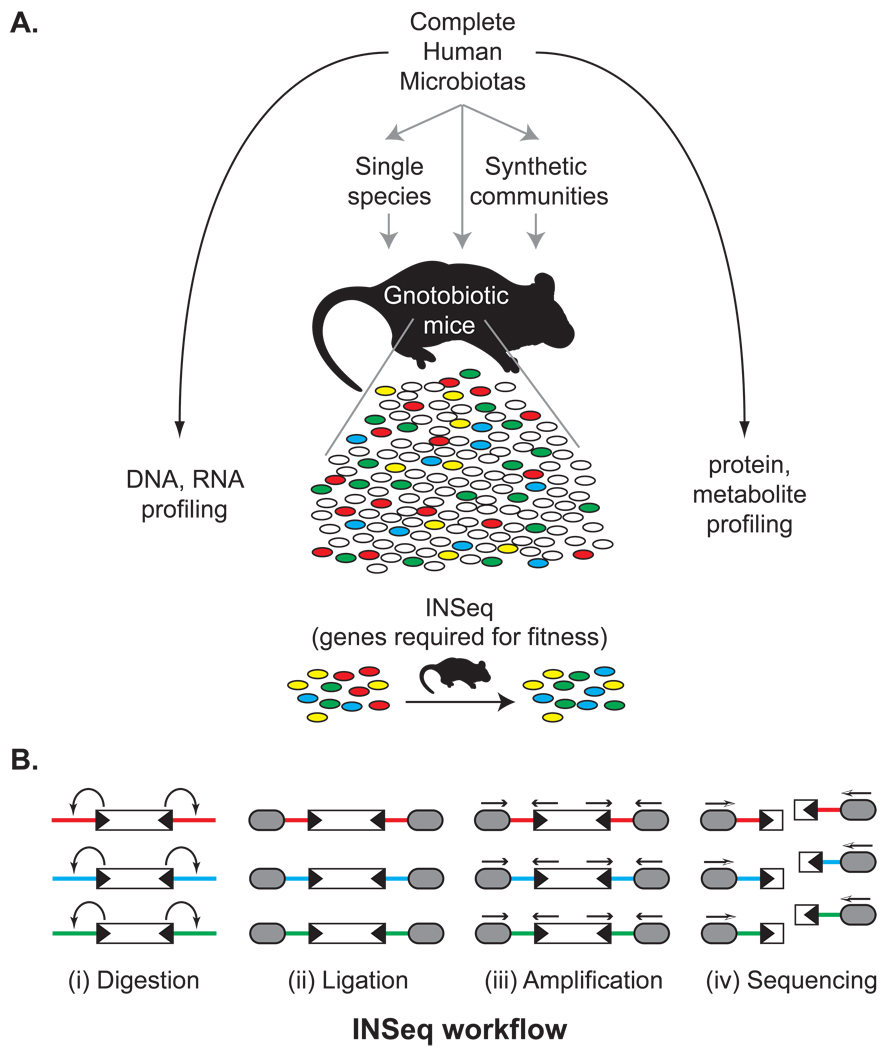
Figure 1. Connecting microbiota and metabolism through gnotobiotics.
(A) A complete human gut (fecal) microbiota, obtained from individuals varying in their diet or physiology, can be surveyed by highly parallel DNA-, RNA-, protein-, and metabolite-directed methods. These microbial communities also serve as source material for transplantation into germ-free mice. They can also be fractionated into component species and assembled into “synthetic communities” of defined composition in order to directly test the roles of specific phylotypes in shaping host physiology: these phylotypes can be intentionally added, removed, or substituted with mutagenized populations prior to colonization of germ-free animals to identify genes required for fitness in specific host/dietary contexts using a method known as insertion sequencing (INSeq). (B) Quantifying the relative abundance of tens of thousands of transposon mutants of a given microbial species by INSeq. (i) The INSeq transposon (depicted as an open rectangle) contains recognition sites for the Type IIs restriction enzyme MmeI in its inverted repeats (black triangles); MmeI digests adjacent chromosomal DNA 16bp outside of the transposon. (ii) Following digestion, sequencing adapters (grey ovals) are appended by ligation. These adapters can contain sample-specific barcodes to allow sample pooling. (iii) A limited number of cycles of PCR amplification of these uniformly-sized molecules are performed. (iv) The resulting amplicons are sequenced using a massively parallel sequencing instrument. The sequence of each read indicates the genomic location of the source transposon, and the relative abundance of each sequence mirrors the relative abundance of the corresponding transposon mutant in the population. Comparison of these relative abundances in input versus output microbial populations identifies genes whose functions are required for fitness in vivo.
Other examples have also highlighted the utility of transplanting foreign microbiotas into germ-free mice. In a recent study, mice lacking toll-like receptor 5 (TLR5) exhibited several features of metabolic syndrome: increases in adiposity, serum triglycerides and cholesterol, hyperinsulinemia, as well as reduced response to insulin challenge (Vijay-Kumar et al., 2010). TLR5, which recognizes bacterial flagellin, is highly expressed in the intestine. Antibiotic treatment, coupled with 16S rRNA pyrosequencing, suggested that changes in distal gut microbiota community composition correlate with these metabolic features. Importantly, transplantation of the distal gut microbiota from TLR5 knockout animals to wildtype germ-free recipients transmitted many of these phenotypic characteristics to the recipients.
Synthetic human gut communities
Transplantation of a complete microbiota into germ-free mice allows host and environmental factors to be controlled, but the microbial component of these experiments remains difficult to completely describe or manipulate. Gnotobiotic mice provide an opportunity for defining the metabolic properties of synthetic human gut microbial communities composed of selected sets of cultured representatives of a human (gut) microbiota; these representatives can be chosen from private or public culture collections based on a number of criteria including their consistent association with specific human metabolic states and their representation in a fecal microbiota that when transferred en masse confers a metabotype to recipient germ-free mice. They can also be selected based on their phylogenetic features as defined by SSU rRNA gene sequencing, and/or by the results of in silico predictions of their metabolic potential based on their genome sequences (Martin et al., 2007).
Members of these assembled model human microbial communities can be systematically removed or replaced prior to colonization of germ-free recipient mice to test the connections between microbiota composition and the metabolic consequences for a host. These synthetic microbial communities also provide an opportunity to directly identify genes that human gut symbionts require in specific in vivo contexts. Such genes can (i) highlight microbial metabolic pathways that are critical to competitive fitness in certain host or microbial contexts; (ii) provide a mechanistic basis for observed changes in microbial community structure in response to various perturbations; and (iii) include potential targets for therapeutic manipulation of the gut microbiota by small-molecule or other approaches. Insertion-sequencing (INSeq), a method that combines genome-wide transposon mutagenesis with massively parallel sequencing, represents one approach for identifying such genes (Goodman et al., 2009). In this approach, complex populations of tens of thousands of transposon mutants are simultaneously introduced into wild-type or genetically manipulated germ-free mice in the presence or absence of other microbes. The representation of each of these mutants in the input community is determined by targeted, massively parallel sequencing of transposon-adjacent chromosomal DNA (Figure 1B) and compared to their representation in the output community recovered from the gnotobiotic mouse host. Differences in mutant representation in the input versus output communities can be used to infer which microbial genes confer a fitness advantage as a function of whatever selective pressure is intentionally applied to the system (e.g., manipulations of host genotype, host diet, or which other wild-type microbial species are introduced together with the collection of mutagenized strains of the indicator species, etc). In one example, INSeq highlighted the importance of a largely unappreciated family of small molecules (vitamin B12-like compounds called corrinoids) as a microbial currency produced by some species and required by others in the mammalian gut, and identified the broadly conserved, but previously unannotated genes that likely mediate the interspecies exchange of these corrinoids (Goodman et al., 2009). Related transposon mutagenesis strategies have also been successfully applied to microbial pathogens (Gawronski et al., 2009; Langridge et al., 2009; van Opijnen et al., 2009).
Taken together, genomic, transcriptomic and INSeq datasets emanating from these synthetic human gut communities provide the substrate for training and evaluating algorithms for mapping metabolic networks. One such method is based on predictions of molecules that function as ‘seeds’ (acquired exogenously from the environment), ‘intermediates’, or ‘products’ (end products of cellular metabolism) in an organism’s metabolic reactions (Borenstein et al., 2008). The extent of syntrophy (cross-feeding) for any two organisms can thus be viewed as the proportion of seeds for one that are intermediates or products in another, while competition reflects the number of seeds that are shared. Similar community metabolic reconstructions that also integrate host diet plus host seed/intermediate/product metabolites should provide a powerful tool for dissecting microbiota and host co-metabolism.
Challenges
What is the range of normal variation in microbiome structure and function?
Independent studies suggest that our gut microbiota may be a deeply personal part of our biology (Costello et al., 2009; Turnbaugh et al., 2010; Qin et al., 2010). As emphasized above, the environmental, genetic, and/or dietary factors that drive this variation are ill-defined and thus identifying pathologic configurations of the microbiota (dysbiosis) is difficult. Core (shared) gene families present in the gut microbiomes of sampled populations have been identified from searches of databases of hierarchically organized annotations of gene function (e.g. COG/STRING, KEGG). However, several caveats apply: the largest study to date (Qin et al., 2010) found that only one-third of orthologous gene clusters conserved across all 124 individuals in their survey could be associated with even a broad functional assignment. Notably, functional classification schemes are based on varying degrees of experimental validation, little of which has been carried out in bacterial phyla that are prominently represented in the gut.
The importance of considering the dynamic nature of the gut ecosystem in defining a normal configuration of the microbiota is illustrated by the following observations: controlled diet manipulation in gnotobiotic mice colonized with a complete human gut (fecal) microbiota revealed that the composition of their human gut microbial communities changed dramatically within a single day after the animals were switched from a plant polysaccharide-rich chow to a high-fat, high-sugar “Western” diet (Turnbaugh et al., 2009b). The microbiota of RELMβ knockout mice also changes its configuration in response to a high-fat diet, although the animals themselves are resistant to diet-induced obesity (Hildebrandt et al., 2009). Thus, it will be very important to develop ways to “correct for” or normalize the effects of long- and short-term dietary choices when connecting microbiota composition to host metabotype. One approach could be to develop a standard meal that is administered over a period of time prior to sampling of the microbiota, much like a standard meal of glucose is consumed prior to oral glucose tolerance tests.
Deciphering the contributions of microbial metabolism, host metabolism, and microbial-host co-metabolism
Dietary preferences may also influence gut microbial community composition and function in ways that produce persistent and transmissible changes. A β-porphyranase encoded by a gene originally cloned and characterized from the seaweed-associated bacterium Zobellia glactanivorans, was recently identified in the genome of the human gut symbiont Bacteroides plebeius. The presence of this glycoside hydrolase confers the ability to process otherwise indigestible marine plant polysaccharides present in nori and other edible seaweeds. To date, B. plebius has only been isolated from individuals living in Japan, and inspection of gut microbiome datasets from North American and Japanese individuals revealed the presence of this β-prophyranase in the latter but not the former. These remarkable observations are consistent with dietary selection of a microbiome-associated metabolic characteristic within a human population - a characteristic that apparently was acquired by horizontal gene transfer from marine microbes associated with non-sterile food (e.g. sushi) to an entrenched member of Japanese gut microbial communities (Hehemann et al., 2010). This finding provides a microbiome counterpart to diet selection of H. sapiens genotypes, such as amylase gene copy number (Perry et al., 2007), that affect our metabolic capabilities.
As studies increase our understanding of normal variation of the human gut microbiota and of the metabolic potential of differing microbial community configurations, we will be challenged to decode how host metabolism reacts to this enormous microbial population (e.g., Claus et al., 2008). Nontargeted MS surveys will likely represent the beginnings of this line of research, but obstacles in metabolite annotation (assigning MS peaks) will be even more significant than the difficulties in gene annotation. In addition to quantifying the direct impact of microbial metabolites, another important aspect of this challenge will be to identify host receptors that translate microbial activity in the lumen to the epithelium and beyond. For example, a diverse family of proteins, the G-protein coupled receptors, populate members of the enteroendocrine cell lineage present in the gut epithelium and have been shown to respond to short-chain fatty acids (SCFAs) and other microbial metabolites, suggesting that microbial SCFAs could play an important signaling role in addition to making a direct contribution to energy harvest (Samuel et al., 2008). Similarly, microbial de-conjugation and modification of bile acids influences both their detergent activity and their signaling ability (as activators of farnesoid X receptor and other nuclear receptors that influence host metabolism) (Hylemon et al., 2009). Non-targeted MS studies have clearly demonstrated that these are but a small fraction of the metabolically important molecules under microbial control (Martin et al., 2007, Wikoff et al., 2009). As emphasized above, strategies that integrate (i) gnotobiotic mouse models, colonized with carefully chosen synthetic human gut microbiotas of increasing complexity, or complete communities harvested from humans with well characterized metabotypes, with (ii) metagenomic and metabolomic pipelines, should allow the connections between community composition and metabolic function to be ascertained.
Diagnostic and Therapeutic Avenues
An understanding of the relationships between diet, gut microbes, and host biology promises to create another dimension of personalized medicine, where the flow of human and microbial genes between members of a family imprint metabotypes during postnatal and subsequent periods of the human lifecycle, and where intrapersonal (temporal) and interpersonal variations in host metabolism are found to have a microbial underpinning.
This axis of personalized medicine, guided by knowledge gleaned from metagenomic studies of a person’s gut community, may evolve to include a number of strategies. Microbial species, genes, and gene products could provide a new class of biomarkers for defining various host physiologic and pathophysiologic states and for evaluating therapeutic interventions. New ways will likely be found for highly selective manipulation of the representation and/or metabolic activities of organisms, or groups of organisms whose niches (professions) optimize the operations of the gut microbiota in various host contexts. This latter approach could involve use of new generations of ‘narrow spectrum’, taxa-specific antimicrobials whose targets are identified with tools such as INSeq, administration of seed compounds for metabolic networks (‘designer prebiotics’), or consumption of metabolically engineered probiotic strains or groups of microbes harvested from donors with desired traits. Products normally generated by a healthy microbiota or their synthetic derivatives could serve as new generation therapeutic agents delivered through a variety of routes, including gut microbes themselves. Identifying host signaling and metabolic pathways that are targeted by the microbiota opens the door to new therapeutic targets. In principle, each of these approaches could be applied to children at risk for microbiome-associated metabolic disorders, since this is a period when the microbiota may be particularly amenable to manipulation and reconfiguration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Backhed F, Crawford PA. Coordinated regulation of the metabolome and lipidome at the host-microbial interface. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbalip.2009.09.009. DOI 10.1016/j.bbalip.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein E, Kupiec M, Feldman MW, Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci USA. 2008;105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, Rezzi S, Ross A, Kochhar S, Holmes SE, Nicholson JK. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219–232. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host and Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann J, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffman C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High Fat Diet Determines the Composition of the Murine Gut Microbiome Independently of Obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, Sprenger N, Fay LB, Kochhar S, van Bladeren P, Holmes E, Nicholson JK. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G, Gambino R, Cassader M. Gut microbiota as a regulator of energy homeostasis and ectopic fat deposition: mechanisms and implications for metabolic disorders. Curr Opin Lipidol. 2009 doi: 10.1097/MOL.0b013e3283347ebb. DOI: 10.1097/MOL.0b013e3283347ebb. [DOI] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, Carter NP, Lee C, Stone AC. Diet and the evolution of human amylase gene copy number variation. Nature Genetics. 2007;39:1188–1190. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium. Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI. Effects of the gut microbiota on host adiposity are modulated by the short-chain fattyacid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Science Translational Medicine. 2009b;1:6–14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, Gordon JI. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1002355107. DOI 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu PV, Yetukuri L, Islam S, Felin J, Perkins R, Boren J, Oresic M, Backhed F. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2009 doi: 10.1194/jlr.M002774. DOI 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-Like Receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]