Summary
Light plays a profound role in plant development, yet how photoreceptor excitation directs phenotypic plasticity remains elusive. One of the earliest effects of light is the regulated translocation of the red/far-red photoreceptors, phytochromes, from the cytoplasm to subnuclear foci called phytochrome nuclear bodies. The function of these nuclear bodies is unknown. We report the identification of hemera, a seedling lethal mutant of Arabidopsis with altered phytochrome nuclear body patterns. hemera mutants are impaired in all phytochrome responses examined, including proteolysis of phytochrome A and phytochrome-interacting transcription factors. HEMERA was identified previously as pTAC12, a component of a plastid complex associated with transcription. Here we show that HEMERA has a function in the nucleus, where it acts specifically in phytochrome signaling, is predicted to be structurally similar to the multiubiquitin-binding protein, RAD23, and can partially rescue yeast rad23mutants. Together, these results implicate phytochrome nuclear bodies as sites of proteolysis.
Plant development, physiology, and growth are impacted by environmental light cues. Light affects every major developmental transition of plants, from germination through flowering, and plays a particularly profound role during dicotyledonous seedling establishment. Upon germination, seedlings that do not perceive light undergo a mode of development that emphasizes stem elongation at the expense of leaf development, allowing the seedling to emerge from the soil and to transition from growth on seed reserves to photoautotrophic growth. In contrast, leaf development is initiated and the rate of hypocotyl elongation slows when a plant sees light after emerging from the ground and assumes a body plan that is suited for a photosynthetic lifestyle. Underlying the development of leaves are dramatic changes in gene expression and in plastid function.
The diverse responses that plants have to light, known collectively as photomorphogenesis, require sophisticated sensing of light's intensity, direction, duration, and wavelength. The action spectra of light responses provided assays to identify multiple photoreceptors absorbing in the red (R)/far-red (FR), blue/near-ultraviolet, and ultraviolet spectral ranges. Among these, the red/far-red absorbing photoreceptors, called phytochromes (PHYs), are the best characterized (Chen et al., 2004). PHYs have two long-lived spectral forms: a R-light absorbing inactive form (Pr) and a near FR-light absorbing active form (Pfr). Of the five phytochromes in Arabidopsis, PHYA and PHYB are the most prominent. PHYA is photolabile and plays a major role immediately after seedling emergence, followed by a dominant role of PHYB later in development.
During the dark to light transition, up to one third of Arabidopsis genes are differentially regulated (Ma et al., 2001). A central mechanism by which PHYs rapidly regulate gene expression is to control the abundance of key light signaling components, including a number of transcription factors. Photomorphogenesis requires the removal of negative regulators, in particular a number of PHY-interacting bHLH transcription factors called PIFs or PILs (PIF3-like), which are stable in the dark and are turned over within 30 minutes of exposure to light (Al-Sady et al., 2006; Lorrain et al., 2008; Shen et al., 2008). Most of the PIFs/PILs interact directly with the photo-activated Pfr form of PHY and act as negative regulators for various light-regulated responses, such as hypocotyl growth inhibition, seed germination, and chlorophyll accumulation (Leivar et al., 2008b). The degradation of PIFs requires a direct interaction with the Pfr form of PHY, followed by phosphorylation, which likely marks the PIFs for turnover (Al-Sady et al., 2006; Lorrain et al., 2008; Shen et al., 2008); however, proteins required for PIF degradation remain unknown. PHYA also accumulates in the dark and is rapidly degraded in the light, which is thought to be a desensitization mechanism (Seo et al., 2004). PHYA degradation is delayed, but not blocked, in cop1 mutants, suggesting that both COP1-dependent and -independent PHYA degradation pathways exist (Seo et al., 2004).
Light-regulated gene expression changes require PHY to be converted to its active Pfr form, which results in its translocation from the cytoplasm to the nucleus (Fankhauser and Chen, 2008). Upon import to the nucleus, both PHYA and PHYB become associated with nuclear foci (herein called nuclear bodies, NBs), and these NBs vary in size and number depending on the fluence rate of light, developmental stage, and phase of the diurnal cycle (Chen et al., 2003; Kircher et al., 2002; Yamaguchi et al., 1999). Functional PHY appears to be required for its association with NBs, as point mutations in PHYB and PHYA affect their signaling functions, as well as their association with nuclear bodies, but do not affect nuclear import (Chen et al., 2003; Chen et al., 2005; Kircher et al., 2002). Because both PHYA and PIF3 are localized to PHY NBs before their degradation, it has been proposed that PHY NBs are sites for protein degradation (Al-Sady et al., 2006; Bauer et al., 2004; Seo et al., 2004). However, direct evidence supporting this hypothesis has been lacking. Thus, the precise function of PHY NBs in light signaling is still unknown.
Here, we used a confocal microscopy-based screen to identify a new gene, HEMERA (HMR), required for the localization of PHYB::GFP to large NBs in high fluence rate R light. hmr mutants appear to define a unique class of Arabidopsis light signaling mutants that are albino, tall under continuous R and FR light, and die as seedlings. hmr seedlings are defective in all PHY responses examined, including high, low and very low light fluence response modes, indicating a role for HMR in both PHYA and PHYB signaling. hmr mutants are blocked in chloroplast development in response to light. Genetic analyses of hmr mutants demonstrate that HMR acts specifically between PHY and a downstream master repressor DET1. Moreover, PHYA, PIF1, and PIF3 are not degraded in hmr in the light, and HMR is structurally similar to RAD23. Expression of HMR partially rescues the yeast rad23Δ mutants, supporting a biochemical role for HMR and PHY NBs in light-mediated proteolysis. Surprisingly, HMR localizes to both the nucleus and chloroplasts, and localization to both compartments appears to be required for HMR function. We propose that the dual localization of HMR is responsible for the rapid adaptation of seedlings to light during emergence from soil, a crucial time when the expression of thousands of nuclear light-regulated genes must be coordinated with chloroplast gene expression and development.
Results
Identification of hemera (hmr) using a PHYB∷GFP mislocalization screen
To identify factors required for PHYB NB localization, we took advantage of a transgenic Arabidopsis line, called PBG (PHYB∷GFP), which expresses constitutively a PHYB∷GFP fusion protein in a phyB null background (Yamaguchi et al., 1999). PBG seeds were mutagenized with ENU (N-ethyl-N-nitrosourea) and planted. Seeds from each M1 plant were collected individually. M2 seedlings were grown under 8 μmol m-2 sec-1 of R light for 4 days. In these conditions, PHYB∷GFP is localized exclusively to large NBs with a diameter of 1-2 μm (Figure 1 A). 30 M2 seedlings from each M1 plant were examined for PHYB∷GFP localization pattern by confocal microscopy. Putative mutants with abnormal PHYB∷GFP NB localization patterns were kept for further analyses.
Figure 1. Isolation and map-based cloning of hmr.

(A) Confocal images showing subnuclear localization of PHYB∷GFP in epidermal cells at the top of the hypocotyl of PBG and hmr-1 under 8 μmol m-2 sec-1 of R light. PHYB∷GFP was localized to large NBs with an average diameter of 1.6 μm in PBG, whereas PHYB∷GFP NBs in hmr-1 were smaller, with an average diameter of 0.4 μm. In a small fraction of hmr-1 hypocotyl cells, PHYB∷GFP was evenly dispersed in the nucleoplasm. (B) Protein levels of PHYB∷GFP remained the same in PBG and hmr-1 seedlings. Total protein extracts from 4-day R light grown hmr-1 and PBG seedlings were resolved by SDS-PAGE. Protein levels of PHYB∷GFP were detected by western blot using anti-GFP antibodies. Actin was used as a loading control. (C) Images of 4-day old PBG and hmr-1 seedlings grown under 8 μmol m-2 sec-1 of R light. The hmr-1 mutant was taller compared to PBG. (D) Map-based cloning of hmr. The hmr-1 mutation was mapped to chromosome II on BAC T31E10 between markers MC671672 and MC549550 based on an F2 mapping population of 1960 plants generated by crossing hmr-1 (Ler) and Col-0. The interval contains five predicted genes illustrated as arrows. The bold arrow represents the HMR gene, At2g34640. (E) Schematic illustration of the exon-intron structure of HMR with the shaded boxes representing exons. The mutations in hmr-1 and hmr-2 are indicated by red arrows. See also Figure S1.
After screening 24,000 M2 lines, one mutant was isolated and named hemera-1 (hmr-1, Hemera is the goddess of day in Greek mythology). Mutant hmr seedlings contain either no or smaller PHYB∷GFP NBs in epidermal cells along the top of the hypocotyl (Figure 1A). The PHYB∷GFP protein level in hmr-1 was the same as that in PBG (Figure 1B), suggesting that the hmr-1 mutation affected the localization of PHYB∷GFP, but not its stability.
hmr-1 seedlings were defective in multiple PHY-mediated responses, and had a unique phenotype. Under high irradiance of R light, hmr-1 seedlings had long hypocotyls, a striking albino phenotype, and died as seedlings (Figure 1C). Dark-grown hmr-1 seedlings were indistinguishable from PBG or wild-type seedlings. Backcrossing of hmr-1 to PBG demonstrated that hmr-1 is a recessive mutation. hmr was mapped to a 25 kb region between the markers MC671672 and MC549550 on BAC T31E10 on Chromosome 2 (Figure 1D). Sequencing of the five genes in the interval revealed a single G to A nucleotide change in the second exon of the gene At2g34640, resulting in a premature stop in codon 9 (Figure 1E). A 7 kb HMR genomic DNA was able to complement hmr-1 (Figure S1A), which confirmed that At2g34640 defines the HMR gene. A second hmr allele, hmr-2 (Salk_025099), was identified from the Salk T-DNA insertion mutant collection (Alonso et al., 2003). hmr-2 carries a T-DNA inserted in the last intron behind nucleotide 14589253 on chromosome II (Figure 1E). Additional characterization of the HMR gene and protein is described below.
The hmr mutant is impaired in both PHYB- and PHYA-mediated responses and multiple PHY response modes
PHY responses are categorized into four modes of action based on the fluence of light, including PHYB-mediated HIR-R (High Irradiance Response in R light) and LFR (Low Fluence Responses), and PHYA-mediated VLFR (Very Low Fluence Response) and HIR-FR (High Irradiance Response in FR light) (Chen et al., 2004). We examined all four response-modes in hmr-1 (in the PBG background) and hmr-2 (in the wild-type Col-0 background). Both alleles are likely to be null alleles based on western blots using an anti-HMR antibody (Figure S1B).
The PHYB HIR-R was examined in hmr seedlings by measuring hypocotyl lengths of 4-day-old seedlings grown under continuous R light (Rc). Both hmr-1 and hmr-2 seedlings had intermediate hypocotyl lengths compared either with their parental genotypes or phyB mutants under a range of R light intensities (Figure 2A-B), indicating that HMR is required for PHYB-mediated HIR-R. In addition, both hmr-1 and hmr-2 alleles were albino in Rc, suggesting HMR is required for the R-induced chloroplast biogenesis.
Figure 2. hmr mutants are defective in multiple PHYB- and PHYA-mediated responses.

(A) Images of 4-day old Col-0, phyB-9, hmr-2, PBG, and hmr-1 seedlings grown in 8 μmol m-2 sec-1 R light. (B) Fluence response curves for Rc. Relative hypocotyl length of 4-day grown Col-0 (◆), phyB-9 ( ), hmr-2 (●), PBG (
), hmr-2 (●), PBG ( ), hmr-1 (
), hmr-1 ( ), and phyB-5 (
), and phyB-5 ( ) seedlings under different fluence of Rc and dark conditions. (C) EOD-FR responses of Col-0, hmr-2, phyB-9, PBG, and hmr-1. Filled columns represent hypocotyl lengths of 4-day old seedlings under 8-hour day/16-hour night; open columns represent hypocotyl lengths of 4-day old seedlings under the same short day conditions with an additional 15 min FR treatment at the end of the day. Red columns represent the percentage of increase in hypocotyl length of the treated seedlings compared to untreated seedlings. (D) Cotyledon opening responses for VLFR measurement. Cotyledon images of Col-0, phyA-211, hmr-2, PBG, and hmr-1 seedlings grown under hourly 3 min 1 μmol m-2 sec-1 FR pulse for 4 days. (E) Images of 4-day old Col-0, phyA-211, hmr-2, PBG, and hmr-1 seedlings grown in 1 μmol m-2 sec-1 FR light for 4 days. (F) Fluence response curves for FRc. Relative hypocotyl length of 4-day old Col-0 (◆), phyA-211 (■), hmr-2 (❄), PBG (
) seedlings under different fluence of Rc and dark conditions. (C) EOD-FR responses of Col-0, hmr-2, phyB-9, PBG, and hmr-1. Filled columns represent hypocotyl lengths of 4-day old seedlings under 8-hour day/16-hour night; open columns represent hypocotyl lengths of 4-day old seedlings under the same short day conditions with an additional 15 min FR treatment at the end of the day. Red columns represent the percentage of increase in hypocotyl length of the treated seedlings compared to untreated seedlings. (D) Cotyledon opening responses for VLFR measurement. Cotyledon images of Col-0, phyA-211, hmr-2, PBG, and hmr-1 seedlings grown under hourly 3 min 1 μmol m-2 sec-1 FR pulse for 4 days. (E) Images of 4-day old Col-0, phyA-211, hmr-2, PBG, and hmr-1 seedlings grown in 1 μmol m-2 sec-1 FR light for 4 days. (F) Fluence response curves for FRc. Relative hypocotyl length of 4-day old Col-0 (◆), phyA-211 (■), hmr-2 (❄), PBG ( ), and hmr-1 (
), and hmr-1 ( ) seedlings grown under different fluence of FRc and dark conditions. Error bars represent standard error.
) seedlings grown under different fluence of FRc and dark conditions. Error bars represent standard error.
To assess the PHYB-mediated LFR in hmr mutants, we measured the length of hypocotyls after an end-of-day FR (EOD-FR) treatment (Elich and Chory, 1997). As shown in Figure 2C, both Col-0 and PBG seedlings had a dramatic hypocotyl elongation response after an EOD-FR treatment. This response was more pronounced in the PBG background, perhaps due to the overexpression of PHYB∷GFP. In contrast, phyB-9 seedlings had no response, while both hmr-1 and hmr-2 had dramatically reduced responses to EOD-FR treatment (Figure 2C), suggesting that HMR is required for the LFR.
Next, we asked whether PHYA-mediated VLFR and HIR-FR were affected in hmr-1 and hmr-2 mutants. For VLFR, we examined the cotyledon-opening response under very low fluence of FR light. Both hmr-1 and hmr-2 were lacking this response, having closed cotyledons and mimicking a phyA null allele, phyA-211 (Figure 2D). HIR-FR was examined by measuring hypocotyls of 4-day old seedlings grown under continuous FR light (FRc). Figure 2E and 2F show that both hmr-1 and hmr-2 had intermediate hypocotyl lengths between their parental genotypes and phyA-211. Taken together, these results demonstrate that HMR is required for both PHYB- and PHYA-mediated response modes.
HMR activity is required downstream of PHYs and upstream of the global negative regulator, DET1
Although defective in both R and FR responses, both hmr-1 and hmr-2 had a normal hypocotyl response under blue light (Figure S2A). This suggests that HMR is involved specifically in PHY-mediated hypocotyl inhibition in R and FR but not required for crytochrome and PHYA mediated hypocotyl responses in blue light. Surprisingly, both hmr-1 and hmr-2 alleles also had a normal hypocotyl response to white light (Figure S2B and S2C). This distinguishes hmr from phyB and chromophore-deficient mutants, such as hy1 and hy2, because phyB, hy1 and hy2 have elongated hypocotyls in R, FR, and white light conditions (Parks and Quail, 1991).
To further demonstrate that HMR plays a specific role in PHY signaling, we performed double mutant analyses between hmr-2 and phyA-211 or phyB-9 mutants. Whereas hmr-2 was taller than Col-0 in R light, a hmr-2/phyB-9 double mutant was not taller than phyB-9 (Figure 3A-B). Likewise, a hmr-2/phyA-211 double mutant was not taller than phyA-211 in FR light (Figure 3C-D). These results suggest that the hypocotyl phenotype of hmr-2 is PHY-dependent. We also generated a double mutant between hmr-1 and a constitutively active PHYB allele, PHYBYH. Dark-grown PHYBYH seedlings are de-etiolated with PHYBYH proteins constitutively localizing to large NBs (Su and Lagarias, 2007). As shown in Figure 3E-F, hmr-1 was able to suppress both the cotyledon opening and hypocotyl inhibition phenotypes of PHYBYH in the dark. More interestingly, PHYBYH failed to localize to large NBs, instead it localized to many small NBs in hmr-1/PHYBYH double mutants (Figure 3G). The PHYBYH localization pattern resembles that of PHYB∷GFP in hmr-1 in the light. These results demonstrate that HMR is a downstream component for PHY signaling. In addition, because PHYBYH could still be observed based on the fluorescence of its chromophore in hmr-1 (Figure 3G), it suggests that neither PHY chromophore biosynthesis nor its incorporation to PHY is blocked in hmr-1 mutants.
Figure 3. HMR acts specifically in phytochrome signaling pathways between PHY and DET1.
(A) Images of 4-day old Col-0, hmr-2, phyB-9, hmr-2/phyB-9 double mutant seedlings grown under 8 μmol m-2 sec-1 R light. (B) Hypocotyl length measurements of seedlings in (A). (C) Images of 4-day old Col-0, hmr-2, phyA-211, hmr-2/phyA-211 double mutant seedlings grown under 3.6 μmol m-2 sec-1 FR light. (D) Quantitative hypocotyl length measurements of seedlings in (C). (E) Images of 4-day old dark-grown PHYBYH, hmr-1/PHYBYH, and Ler seedlings. (F) Hypocotyl measurements of 4-day old dark-grown PHYBYH, hmr-1/PHYBYH, and Ler seedlings. (G) Confocal images of PHYBYH subnuclear localization patterns in 4-old dark-grown PHYBYH and hmr-1/PHYBYH seedlings. DIC and merge images show the location of the nucleus. NB, nuclear body; P, plastid; N, nucleus. (H) Images of 4-day old det1-1 and hmr-1/det1-1 seedlings grown in the dark. Error bars represent standard error. See also Figure S2.
Previous studies have established a genetic pathway for PHY signaling (Ang and Deng, 1994; Chory, 1993). Downstream of both PHYs and cryptochromes, there are a group of genes referred to as global repressors of photomorphogenesis, including DET1, COP1, and proteins in the COP9 signalosome (Chen et al., 2006). To pinpoint where HMR acts in this genetic pathway, we crossed hmr-1 with det1-1. Dark-grown det1-1 seedlings, similar to PHYBYH, show characteristics of wild-type seedlings in the light, such as short hypocotyls and opened cotyledons. However, hmr-1 failed to suppress the photomorphogenetic phenotypes of det1-1 in the dark (Figure 3H), suggesting that HMR acts genetically between PHY and DET1. Interestingly, det1-1 cannot suppress the albino phenotype of hmr-1, as the double mutant was still albino in the light, suggesting that chloroplast defects in hmr are not entirely dependent on DET1.
HEMERA gene and protein
HMR encodes a predicted 527 amino acid protein and is a single copy gene in Arabidopsis. A PSI-BLAST failed to reveal any significant similarity between HMR and known protein classes. Using a pattern and profile search software InterPro Scan (http://www.ebi.ac.uk/Tools/InterProScan/), a few putative domains were identified in HMR, including a glutamate rich region, two bipartite nuclear localization signals (NLS), and a PEST domain, which is a signature for short-lived proteins (Rogers et al., 1986) (Figure 4A). HMR orthologs can be found in all plant genomes sequenced. Protein sequences similar to the Arabidopsis HMR gene can be traced back to two HMR-like genes in Physcomitrella patens (GI:168063606 and GI:168038429), suggesting that HMR is highly conserved in land plants.
Figure 4. Spatial and temporal expression of HMR RNA and protein.
(A) Predicted domain structure of HMR, including a glutamate (GLU) rich region, two bipartite nuclear localization signals (NLSa and NLSb), and a PEST domain. (B) Steady-state HMR mRNA levels in 3-day old Col-0 seedlings grown under D, R, FR, B, and WL measured by qRT-PCR. Error bars represent standard deviation. (C-E) GUS staining of 2-day or 4-day old transgenic lines carrying the HMRp∷GUS construct. The seedlings were grown in the dark (B), red light (C), or far-red light (D). (F) Western blot using anti-HMR antibodies showing HMR proteins in 3-day old seedlings grown under D, R, FR, B, and WL. Levels of RPN6 were used as loading controls.
HMR expression is temporally and spatially regulated
To examine whether the expression of HMR is regulated, we first compared the steady-state mRNA levels of HMR in 3-day old Col-0 seedlings grown under dark, R, FR, blue, or white light by quantitative real-time PCR (qRT-PCR). As shown in Figure 4B, the mRNA level of HMR remains relatively unchanged (within 2-fold range) under different light conditions, suggesting that the expression of HMR is not regulated by light at the transcript level. To determine the spatial expression pattern of HMR, we fused a 4 kb HMR promoter region (HMRp) with a β-glucuronidase (GUS) reporter gene and transformed it to Col-0 plants. The GUS staining pattern is not changed under different light conditions, but surprisingly, it was regulated by developmental stage (Figure 4C-E). In 2-day old seedlings, HMR was expressed predominantly in cotyledons, at the top of the hypocotyl, and in the root tip regardless of light conditions. In contrast, in 4-day old seedlings, HMR was expressed only in unexpanded cotyledons of dark-grown seedlings but not in fully expanded cotyledons of seedlings grown in either R or FR light. These results suggest that HMR is expressed right before tissue expansion. Lastly, we examined the steady-state protein level of HMR. In contrast to HMR mRNA level, HMR protein level was higher in the light compared to that in the dark (Figure 4F), suggesting that HMR might be regulated by light at the posttranslational level.
HMR is required for PHYA, PIF1, and PIF3 degradation
In addition to the translocation of PHYs to PHY NBs, another early light-mediated event is the degradation of PHYA and PIFs (Al-Sady et al., 2006; Clough and Vierstra, 1997; Lorrain et al., 2008; Shen et al., 2008). Because PHYA and PIF3 colocalize to PHY NBs prior to their degradation (Al-Sady et al., 2006; Bauer et al., 2004; Seo et al., 2004), we asked whether the proteolysis of PHYA and PIFs were affected in hmr mutants. We were unable to measure PHYA and PIF degradation during the dark-to-light transition, because we do not have hymozygous hmr seeds and dark-grown hmr mutants were indistinguishable from the wild type. Rather, we directly examined whether PHYA and PIFs were depleted in light-grown hmr seedlings. To our surprise, PHYA accumulated in both hmr alleles in R light (Figure 5A), suggesting hmr is defective in PHYA degradation in the light. It is worth noting that despite an increase in PHYA protein levels, hmr mutants were defective in PHYA responses. This suggests that PHYA degradation is required for PHYA function as proposed by Maloof et al. (Maloof et al., 2001), rather than acting as a desensitization mechanism for light signaling (Seo et al., 2004)
Figure 5. HMR is required for light-dependent PHYA, PIF1, and PIF3 proteolysis and partially rescues the yeast rad23Δrpn10Δ mutant.
(A) Western blot showing PHYA protein levels in 4-day old R light grown Col-0, hmr-2, PBG, hmr-1, and phyB-9. (B) Western blots showing PIF1 and PIF3 protein levels in 4-day R light grown Col-0, hmr-2, PBG, and hmr-1 seedlings. Tubulin was used as a loading control. (C) A growth assay showing serial dilutions of rad23Δrpn10Δ, RAD23rpn10Δ, and HMRrpn10Δ grown in 30°C either with Gal (Galactose) in the upper panel or with Glc (Glucose). The growth defect of rad23Δrpn10Δ was partially rescued only in the presence of Gal, which induces HMR expression in yeast. (D) Western blot showing multiubiquitylated proteins detected by anti-ubiquitin (anti-ubi) antibodies. The SDS-PAGE gel (lower panel) was used as a loading control. (E) UV survival assay using rad23Δ (rad23Δrpn10Δ), RAD23 (RAD23rpn10Δ), and HMR (HMRrpn10Δ). Error bars represent standard error from three independent replica. See also Figure S3.
Because PIF1 and PIF3 are involved in both chloroplast differentiation and hypocotyl growth during the dark-to-light transition and are known to undergo rapid light-dependent degradation (Al-Sady et al., 2006; Shen et al., 2008), we raised antibodies specifically against PIF1 or PIF3 (Figure S3), and compared PIF1 and PIF3 protein levels in hmr-1 and hmr-2 to their parental types. As shown in Figure 5B, both PIF1 and PIF3 accumulate in R light grown hmr-1 and hmr-2 seedlings, but not in PBG or Col-0. These experiments demonstrate that HMR is required for PIF1 and PIF3 degradation in the light, and provide genetic evidence supporting the involvement of PHY NBs in protein degradation.
HMR partially complements the yeast rad23rpn10 mutant
The primary amino acid sequence of HMR does not provide any clue for its biochemical function. However, the predicted secondary structure of HMR is highly similar to the secondary structure of a human protein hHR23A with a very significant E value of 0.00884 by 3D-PSSM (Kelley et al., 2000). Human hHR23A is an ortholog of the yeast RAD23 (Raasi et al., 2004). RAD23 contains an ubiquitin-like domain (UbL), which interacts with the proteasome, and ubiquitin-associated domains (UBAs), which interact with multiubiquitin chains (Chen and Madura, 2002; Lambertson et al., 1999). RAD23 and RPN10, which is a subunit of the proteasome and another multiubiquitin-binding protein, play synergistic roles in the recognition of ubiquitylated proteins by the proteasome (Chen and Madura, 2002). The loss of both RAD23 and RPN10 causes reduced growth and defects in proteolysis (Lambertson et al., 1999). RAD23 is also required for nucleotide-excision-repair, which does not overlap with RPN10 (Lambertson et al., 1999). We tested whether HMR can complement the rad23Δrpn10Δ mutant in yeast. We expressed HMR under the GAL1 promoter in the rad23Δrpn10Δ double mutant and used a rad23Δrpn10Δ mutant strain expressing RAD23 or GFP as a positive or a negative control. HMR partially rescued both the growth defect and the DNA damage repair defect of rad23Δrpn10Δ (Figure 5C-E). Of even greater interest, expression of HMR reduced the global level of multi-ubiquitylated proteins in rad23Δrpn10Δ (Figure 5D), suggesting that HMR shares some biochemical properties with RAD23 and likely functions in protein degradation.
HMR localizes to both the nucleus and chloroplasts
Our biochemical studies suggest that HEMERA functions in the nucleus, which is in agreement with its predicted nuclear localization by PSORT (http://psort.ims.u-tokyo.ac.jp/form.html). However, a proteomics experiment identified HMR (At2g34640) as a plastid-localized protein, pTAC12, which is associated with plastid transcriptionally active chromosomes and was suggested to be involved in transcription in chloroplasts (Pfalz et al., 2006). pTAC12's subcellular localization has not been verified by other criteria.
To solve this dilemma, we made HMR constructs with either a C-terminal CFP fusion (HMR∷CFP) or an N-terminal YFP fusion (YFP∷HMR) and expressed them under the CaMV 35S constitutive promoter in the Col-0 background. Surprisingly, HMR∷CFP was localized exclusively in chloroplasts (Figure S4A), however, YFP∷HMR was localized to the nucleus and cytoplasm but not chloroplasts (Figure S4B). To determine which localization was required for HMR's function, we transformed each of these constructs into the hmr-2 background and looked for rescue of the mutant. To our surprise, we did not obtain any rescued lines after examining more than 40 transgenic lines for each construct. It is possible that the YFP or CFP tag interferes with the function of HMR. Alternatively, HMR is localized to both the nucleus and chloroplasts, and both pools of HMR are essential for its function.
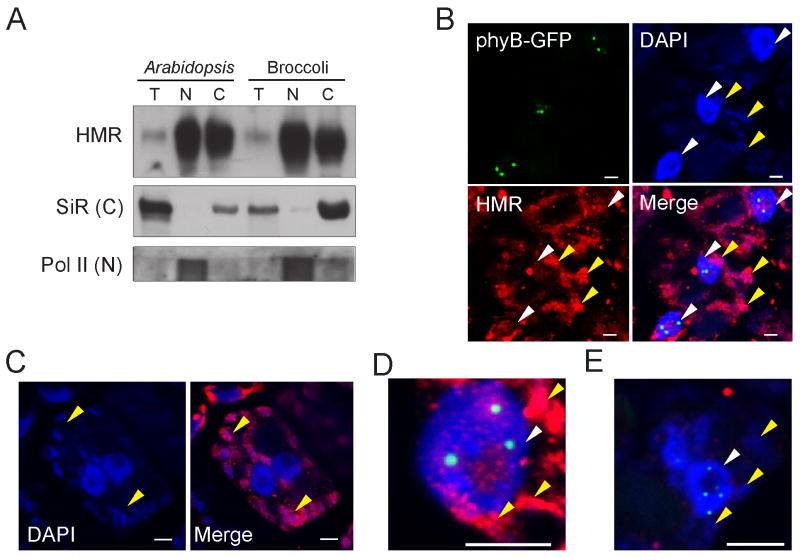
To test the latter possibility, we fractionated nuclei and chloroplasts from 2-day old wildtype seedlings grown under R light, and looked for HMR in these fractions by western blots. As shown in Figure 6A, HMR was present in both the nuclear and chloroplast fractions, suggesting that HMR is dual localized to both of these compartments. As the anti-HMR antibody can recognize the HMR ortholog in Broccoli, we performed the same fractionation experiments using Broccoli and observed similar results (Figure 6A), suggesting that the dual nuclear/chloroplast localization pattern of HMR is conserved in different plant species.
Figure 6. HMR is localized to both the nucleus and chloroplasts.
(A) HMR protein is enriched in both the nuclear and chloroplast protein fractions. Protein extracts of whole plant (T), nuclear (N), or chloroplast (C) fractions from either 2-day old Col-0 seedlings or Broccoli flower buds were separated by SDS-PAGE, and HMR protein was detected by the anti-HMR antibody. Ferrodoxin: Sulfite reductase (SiR) and RNA Pol II were used as controls for the chloroplast and nuclear fractions respectively. (B) Confocal images showing the subcellular localization of HMR in 2-day old R light grown PBG seedlings by immunofluorescent labeling. PHYB∷GFP (green) remains intact under the fixation condition. Both the nuclei (marked by white arrows) and plastid chromosome (marked by yellow arrows) were labeled by DAPI. HMR (red), labeled by anti-HMR antibodies and Alexa 555 conjugated anti-Rabbit secondary antibodies, was detected both in the plastids and the nuclei. (C) Confocal images showing SiR subcellular localization using immunofluorescent labeling. (D) Confocal image showing subnuclear localization of HMR. HMR (red) localizes to foci within the nucleolus and the nucleoplasm. The HMR foci are often adjacent to or sometimes partially overlapping with the PHYB∷GFP (green) NBs. (E) Immunofluorescent labeling using pre-immune serum (red) for anti-HMR antibodies. See also Figure S4.
We further explored HMR's dual localization by immunofluorescent labeling, which confirmed that HMR localizes to both the nucleus and chloroplasts (Figure 6B-E). Within the nucleus, HMR is not evenly dispersed, but rather localizes to subnuclear domains in the nucleolus and in the nucleoplasm (Figure 6D). HMR subnuclear domains are often at the periphery of PHYB∷GFP NBs, even though they are not completely overlapping (Figure 6D), suggesting a rather intimate relationship between HMR domains and PHYB∷GFP NBs.
Discussion
The regulated movement of PHYs from the cytoplasm to PHY NBs is one of the earliest effects of light in plants and, as such, provides an excellent assay to unravel the initial events in PHY signaling. Using a new mutant screen, we identified HMR as a regulator of PHYB's location within the nucleus. Characterization of hmr mutants, localization of HMR protein within cells, and analysis of biochemical function indicate that HMR is a specific and early PHY signal transduction component that is required for light-dependent proteolysis of PHYA, PIF1 and PIF3. Moreover, expression of Arabidopsis HMR can partially rescue a yeast rad23Δrpn10Δ mutant. These latter results implicate a direct role for HMR in proteolysis. HMR's localization to the periphery of PHYB NBs further suggests that NBs are sites of protein degradation in the nucleus. HMR also localizes to plastids. Pfalz et al. showed that HMR/pTAC12 is associated with plastid transcriptionally active chromosomes and may be involved in the transcription of photosynthesis related genes in chloroplasts (Pfalz et al., 2006), although its function is not known.
hmr defines a new class of photomorphogenetic mutant
Why have the HMR gene and its role in PHY signaling eluded detection until now? Traditional screens for light signaling mutants have used hypocotyl length as a read-out of photoreceptor action pathways. Unlike our screening strategy, which allowed us to recover seedling lethal mutants, most early screens relied on the homozygous recessive mutant to be both viable and fertile. Therefore, early signaling components encoded by essential genes were missed. It is interesting to note that null alleles of the global repressors for light signaling, such as det1, cop1, etc., are also seedling lethal (Reed and Chory, 1994; Wei and Deng, 1996).
In addition to arguments of lethality, hypocotyl length is an endpoint response after prolonged exposure to a particular light environment and may not be a sensitive reporter of early PHY signaling effects (Figure S5). Recently, several studies have demonstrated clear distinctions between mechanisms involved in prolonged response to light vs. those in early light signaling events that occur within a few hours after the dark-to-light transition (Al-Sady et al., 2008; Khanna et al., 2006; Leivar et al., 2008a). Based on this notion, it is not surprising that genetic screens based only on an endpoint measurement after a prolonged exposure to light may miss factors involved in early light responses (Figure S5).
hmr mutants are albino and tall in R and FR, which distinguishes it from other known mutants. The chromophore of PHYs is synthesized in chloroplasts, making it reasonable to suspect that hmr's pleiotropic phenotype is caused by chromophore deficiency by impaired plastids. However, hmr-1 and hmr-2 have normal hypocotyl responses in white light (Figure S2B and S2C) and PHYBYH is still fluorescent in the hmr-1 background (Figure 3G), arguing against this possibility. In addition, the hmr mutation does not affect cryptochrome and PHYA-mediated hypocotyl inhibition in blue light (Figure S2A). As such, HMR appears to be a unique and very early acting component of PHY signaling. hmr thus defines a new subset of albino mutants whose unique characteristics were previously missed.
HMR links phytochrome NBs to sites of proteolysis
Perhaps the most compelling phenotype of hmr mutants is their inability to degrade PHYA, PIF1, and PIF3 in the light. This observation links their light-regulated turnover to the function of PHY NBs. Light-dependent PHYA degradation has been known for 35 years (Pratt et al., 1974); however, the mechanism and significance of PHYA degradation remain unclear. Although, COP1 was proposed to be an E3-ubiquitin ligase for PHYA degradation, PHYA degradation is not completely dependent on COP1, as PHYA turnover is delayed, but still occurs, in cop1 mutants (Clough and Vierstra, 1997; Seo et al., 2004). Progress to a better understanding of PHYA degradation has been hindered mainly because of the lack of mutants defective in this process (Clough and Vierstra, 1997).
The intracellular site of PHYA degradation remains a mystery. It was suggested that PHYA degradation occurs on cytoplasmic foci, because PHYA localizes to cytoplasmic foci rapidly after R light exposure (Mackenzie et al., 1975). Recently, it was reported that PHYA degradation is normal when PHYA nuclear import is blocked, supporting the notion that PHYA degradation can occur in the cytoplasm (Rosler et al., 2007). PHY NBs were also proposed as sites for degradation, as PHYA, PIF3 and FHY1 localize to PHY NBs prior to their degradation (Chen, 2008). In addition, PHYA co-localizes with COP1 on NBs (Bauer et al., 2004; Seo et al., 2004). Here, we showed that a mutant with defective PHY NBs is also defective in PHYA, PIF1 and PIF3 degradation, thereby providing genetic evidence that supports a role of PHY NBs in protein turnover (Figure 7).
Figure 7.
Schematic illustration of a model for HMR functions in the nucleus and chloroplasts. In the nucleus, HMR is essential for PHY NB formation, which is required for the proteolysis of PHYA, PIF1 and PIF3. By controlling PIF1 and PIF3 stability, HMR could indirectly regulate the expression of PIF1/PIF3-controlled genes encoding chloroplast proteins. In chloroplasts, HMR/pTAC12 directly regulates the expression of photosynthetic genes as a transcriptionally active chromosome protein. See also Figure S5.
It is intriguing that HMR localizes to the periphery of PHYB NBs. As HMR could act functionally like RAD23 linking multi-ubiquitylated substrates and the proteasome, this result suggests that the proteolysis of PHYA, PIF1, and PIF3 could occur at the periphery of PHY NBs. Consistent with this notion, PHYB partially co-localized with CRY2 in tobacco NBs (Mas et al., 2000), and NBs were suggested to be involved in the degradation of CRY2 (Yu et al., 2009). In mammalian cells, components of the ubiquitin-proteasome pathway have been shown to localize to subnuclear foci called clastosomes (Lafarga et al., 2002). PHYB NBs could be plant clastosomes. Alternatively, PHY NBs could be sites of protein modifications. For example, PIF phosphorylation may occur on PHY NBs (Castillon et al., 2007) before PIF degradation occurs elsewhere. This latter model is consistent with the observation that PIF7 is localized to PHY NBs but is stable in the light (Leivar et al., 2008a). Future experiments to examine the localization of the proteasome relative to the PHY NBs, as well as characterization of the precise function of HMR in protein degradation will further test these models.
Dual localization of HMR
HMR's dual nuclear and chloroplast localization is an unexpected and intriguing feature. Two other plant proteins have been reported to localize to both the nuclear and plastid compartments: MFP1, a DNA-binding protein (Jeong et al., 2003; Meier et al., 1996), and WHY1, a nuclear DNA- and RNA-binding protein involved in salicylic acid-mediated disease resistance, telomere homeostasis, and chloroplast biogenesis (Desveaux et al., 2004; Prikryl et al., 2008; Yoo et al., 2007). Interestingly, WHY1, also known as pTAC1, was co-purified with HMR as a plastid transcriptionally active chromosome protein (Pfalz et al., 2006). In animal systems, several proteins, including p53 and estrogen receptors, have been localized to both the nucleus and mitochondria (Lee et al., 2008). The significance of HMR's dual localization, how partitioning to both compartments is regulated, and how dual localization has evolved are important questions for the future.
The basis for hmr's albino phenotype is currently not known. HMR may regulate chloroplast transcription directly (Pfalz et al., 2006), in addition to its role in nuclear gene expression. It is also possible that HMR regulates chloroplast biogenesis indirectly through its control of PIF1 and PIF3 accumulation. Because PIF1 and PIF3 are negative regulators of chloroplast development (Bauer et al., 2004; Leivar et al., 2009; Shen et al., 2008), it is conceivable that accumulation of PIF1 and PIF3 proteins might explain the albino phenotype of hmr mutants by interfering with chloroplast biogenesis (Figure 7).
In thinking of the incredible amount of stress that a seedling experiences when it first encounters light, we prefer a model in which HMR influences gene expression directly in both the nucleus and chloroplasts (Figure 7). In this way, a plant could mount a very rapid response to high light stress and then adjust to the ambient light environment as it acclimates. A similar model has been proposed for the coordinated action of both nuclear and mitochondrial-localized tumor suppressor gene product p53 in the initiation of apoptosis in mammalian cells. In this model, nuclear p53 regulates the transcription of pro-apoptotic genes, such as BAX and PUMA, while mitochrondrial p53 is directly involved in permeabilization of the outer mitochondrial membrane allowing cytochrome c release (Moll et al., 2006).
HEMERA is another example of the possible shared history of phytochrome signaling pathway components with proteins involved in the surveillance pathway associated with global nucleotide excision repair. DET1, a negative regulator of light-regulated gene expression is found in complex with COP10, DDB1 and DDB2, and is thought to play a role in chromatin remodeling (Schroeder et al., 2002; Zhang et al., 2008). Like RAD23, DDB proteins play an essential role in scanning the genome for UV-induced DNA damage. Light causes a global reorganization of gene expression with up to 1/3 of the Arabidopsis genome showing changes in gene expression. Thus, these proteins may have been co-opted to regulate transcription globally in response to light. Since Arabidopsis has a canonical RAD23 gene, which has not been implicated in light sensing (Farmer et al., 2010), HEMERA may be an example of convergent evolution.
Experimental Procedures
Standard Protocols
The protocols used for protein extraction, western blot analysis, qRT-PCR, GUS staining, cell fractionation, and immunofluorescent labeling are outlined in the Extended Experimental Procedures.
Plant materials
The PHYB∷GFP (PBG) (Ler) line was described in (Yamaguchi et al., 1999). The PHYBYH line was described in (Su and Lagarias, 2007). Wild-type Col-0, phyB-9 (Col-0) and the phyB-5 (Ler) mutants were used as controls for physiological studies. The hmr-1 (PBG) mutant line was isolated from a PHYB∷GFP mislocalization screen and backcrossed to PBG twice. The hmr-2 (Col-0) mutant line was Salk T-DNA insertion line, Salk_025099. Plant growth conditions and hypocotyl measurements are described in Extended Experimental Procedures.
ENU (N-ethyl-N-nitrosourea) mutagenesis
0.2g of freshly collected PBG seeds were hydrated in 45 ml of ddH2O with 0.005% Tween-20 and left on a tube rotator for 4 h. The seeds were washed with ddH2O twice, and then soaked in 1 mM ENU solution for 15 hrs.
Positional cloning
hmr-1 (Ler) was crossed to Col-0 to generate an F2 mapping population. Detailed mapping procedures are described in Extended Experimental Procedures.
rad23Δrpn10Δ complementation
Growth and UV survival assays were performed as described in (Romero-Perez et al., 2007). The detailed protocol is described in Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Dr. Akira Nagatani for providing the PBG line, Dr. Clark Lagarias for the PHYBYH line, Drs Peter Quail and Sabine Heinhorst for anti-PHYA and anti-SiR antibodies, Dr. Kiran Madura for the yeast rad23Δrpn10Δ double mutant, and Dr. Enamul Huq for pif1-2 seeds. Benjamin Cole and Kazumasa Nito made critical comments and suggestions regarding this manuscript. The work was supported by a grant from the National Institutes of Health (R01GM52413) to J.C., the Howard Hughes Medical Institute (J.C.), a startup fund (4314268) to M.C. from Duke University, and a grant from the National Science Foundation to M.C. (IOS-0923722).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Sady B, Kikis EA, Monte E, Quail PH. Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci (United States of America) 2008;105:2232–2237. doi: 10.1073/pnas.0711675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Ang LH, Deng XW. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adam E, Fejes E, Schafer E, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD Complex in mediating light control of development. Plant Cell. 2006;18:1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. Phytochrome nuclear body: an emerging model to study interphase nuclear dynamics and signaling. Current Op Plant Biol. 2008;11:503–508. doi: 10.1016/j.pbi.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Ann Review Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Chen M, Schwab R, Chory J. Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci (USA) 2003;100:14493–14498. doi: 10.1073/pnas.1935989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tao Y, Lim J, Shaw A, Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol. 2005;15:637–642. doi: 10.1016/j.cub.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Chory J. Out of darkness: mutants reveal pathways controlling light-regulated development in plants. Trends Genet. 1993;9:167–172. doi: 10.1016/0168-9525(93)90163-c. [DOI] [PubMed] [Google Scholar]
- Clough R, Vierstra R. Phytochrome degradation. Plant Cell Environ. 1997;20:713–721. [Google Scholar]
- Desveaux D, Subramaniam R, Despres C, Mess JN, Levesque C, Fobert PR, Dangl JL, Brisson N. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell. 2004;6:229–240. doi: 10.1016/s1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- Elich TD, Chory J. Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell. 1997;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. Transposing phytochrome into the nucleus. Trends Plant Sci. 2008;13:596–601. doi: 10.1016/j.tplants.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Farmer LM, Book AJ, Lee KH, Lin YL, Fu H, Vierstra RD. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell. 2010;22:124–142. doi: 10.1105/tpc.109.072660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Meier I. MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucl Acids Res. 2003;31:5175–5185. doi: 10.1093/nar/gkg693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Toledo-Ortiz G, Kikis EA, Johannesson H, Hwang YS, Quail PH. Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell. 2006;18:2157–2171. doi: 10.1105/tpc.106.042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognar L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adam E, Schafer E, Nagy F. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell. 2002;14:1541–1555. doi: 10.1105/tpc.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M, Berciano MT, Pena E, Mayo I, Castano JG, Bohmann D, Rodrigues JP, Tavanez JP, Carmo-Fonseca M. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol Biology Cell. 2002;13:2771–2782. doi: 10.1091/mbc.E02-03-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sharma S, Kim J, Ferrante RJ, Ryu H. Mitochondrial nuclear receptors and transcription factors: who's minding the cell? J Neuro Res. 2008;86:961–971. doi: 10.1002/jnr.21564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008a;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008b;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Jr, Coleman RA, Briggs WR, Pratt LH. Reversible redistribution of phytochrome within the cell upon conversion to its physiologically active form. Proc Natl Sci (USA) 1975;72:799–803. doi: 10.1073/pnas.72.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al. Natural variation in light sensitivity of Arabidopsis. Nat Gen. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- Mas P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- Meier I, Phelan T, Gruissem W, Spiker S, Schneider D. MFP1, a novel plant filament-like protein with affinity for matrix attachment region DNA. Plant Cell. 1996;8:2105–2115. doi: 10.1105/tpc.8.11.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LH, Kidd GH, Coleman RA. An immunochemical characterization of the phytochrome destruction reaction. Biochem Biophys Acids. 1974;365:93–107. doi: 10.1016/0005-2795(74)90253-0. [DOI] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucl Acids Res. 2008;36:5152–5165. doi: 10.1093/nar/gkn492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Bio. 2004;341:1367–1379. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Reed JW, Chory J. Mutational analyses of light-controlled seedling development in Arabidopsis. Sem Cell Biol. 1994;5:327–334. doi: 10.1006/scel.1994.1039. [DOI] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Romero-Perez L, Chen L, Lambertson D, Madura K. Sts1 can overcome the loss of Rad23 and Rpn10 and represents a novel regulator of the ubiquitin/proteasome pathway. J Biol Chem. 2007;282:35574–35582. doi: 10.1074/jbc.M704857200. [DOI] [PubMed] [Google Scholar]
- Rosler J, Klein I, Zeidler M. Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc Natl Acad Sci (USA) 2007;04:10737–10742. doi: 10.1073/pnas.0703855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol. 2002;12:1462–1472. doi: 10.1016/s0960-9822(02)01106-5. [DOI] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell. 2008;20:1586–1602. doi: 10.1105/tpc.108.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YS, Lagarias JC. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HH, Kwon C, Lee MM, Chung IK. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 2007;49:442–451. doi: 10.1111/j.1365-313X.2006.02974.x. [DOI] [PubMed] [Google Scholar]
- Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C. Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell. 2009;21:118–130. doi: 10.1105/tpc.108.061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, Xiong Y, Deng XW. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell. 2008;20:1437–1455. doi: 10.1105/tpc.108.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.