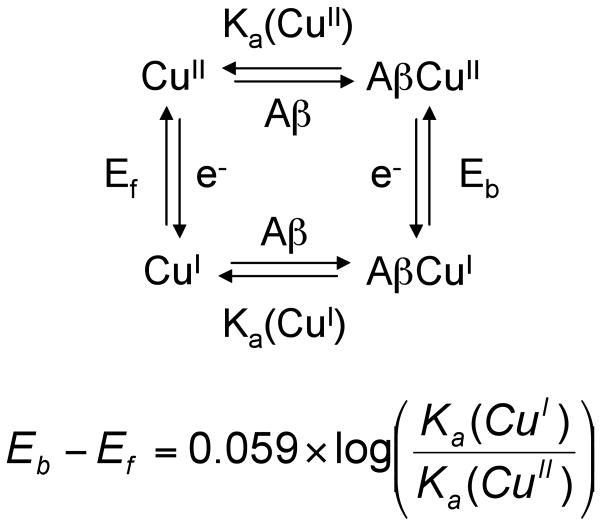Abstract
Oxidative stress has been suggested to contribute to neuronal apoptosis associated with Alzheimer's disease (AD). Copper may participate in oxidative stress through redox-cycling between its +2 and +1 oxidation states to generate reactive oxygen species (ROS). In vitro, copper binds to the amyloid-β peptide of AD and in vivo, copper is associated with amyloid plaques characteristic of AD. As a result, the AβCuI complex may be a critical reactant involved in ROS associated with AD etiology. To characterize the AβCuI complex, we have pursued X-ray absorption (XAS) and EPR spectroscopy of AβCuII and AβCuI (produced by ascorbate reduction of AβCuII). The AβCuII complex Cu K-edge X-ray absorption spectrum is indicative of a square-planar CuII center with mixed N/O ligation. Multiple scattering analysis of the extended X-ray absorption fine structure (EXAFS) data for AβCuII indicate that two of the ligands are imidazole groups of histidine ligands, indicating a (NIm)2(N/O)2 CuII ligation sphere for AβCuII. After reduction of the AβCuII complex with ascorbate, the edge region decreases by ∼4 eV in energy. The X-ray absorption near-edge spectrum (XANES) region of AβCuI displays an intense pre-edge feature at 8984.1(2) eV. EXAFS data fitting yielded a two coordinate geometry with two imidazole ligands coordinated to CuI at 1.877(2) Å in a linear geometry. Ascorbate reduction of AβCuII under inert atmosphere and subsequent air oxidation of AβCuI to regenerate AβCuII was monitored by low-temperature EPR spectroscopy. Slow re-appearance of the AβCuII EPR signal indicates that O2 oxidation of the AβCuI complex is kinetically sluggish, and Aβ damage is occurring following reoxidation of AβCuI by O2. Together, these results lead us to hypothesize that CuI is ligated by His13 and His14 in a linear coordination environment in Aβ, that Aβ may be playing a neuroprotective role, and that metal-mediated oxidative damage of Aβ occurs over multiple redox-cycles.
Introduction
Alzheimer's disease (AD) is a fatal neurodegenerative disorder that plagues approximately 26 million people worldwide.1 AD is characterized by a progressive cognitive decline that has been attributed to the deposition of extracellular protein plaques containing the amyloid-β (Aβ) peptide. A single determinant for AD etiology has not been established,2,3 but oxidative stress induced by reactive oxygen species (ROS) has been hypothesized to be a principal contributor.4 The redox-active metal ions iron and copper are found in AD plaques,5 suggesting that they might mediate ROS generation.6 Thus, answers to fundamental questions regarding the coordination environment and reactivity of these metal ions when bound to Aβ are essential to evaluating their potential role in AD etiology.
The majority of previous studies performed on copper-Aβ adducts were aimed at elucidating the coordination environment of CuII bound to the peptide.6-11 Although some work has focused on the ROS reactions catalyzed by the AβCu complex,10,12,13 information regarding the structure and reactivity of the AβCuI complex is scarce.14-16 Previous EXAFS measurements of AβCuII led to a model in which the CuII ion binds to Aβ in a 5-coordinate square-pyramidal ligand environment created by coordination to three His imidazoles, an oxygen atom donor (Tyr), and an axial water molecule. Results from other XAS measurements proposed a 6-coordinate CuII species bound to three His imidazoles, Glu/Asp residues, and a water molecule.17 These structural models are inconsistent with 4-coordinate CuII in the CuII–Aβ adduct, a geometry that has been suggested on the basis of other spectroscopic data and recent theoretical work.8,9,18,19
Recent XANES data indicated that the AβCuII complex can be reduced by ascorbate and 6-hydroxydopamine, but not by cholesterol or dopamine.15 In another study, Maiti et al. reduced the AβCuII complex with sodium borohydride, a non-physiological reductant, and followed the reduction reaction with EPR and fluorescence spectroscopies.14 Ascorbate radical and hydroxyl radical (either via fluorescent detection or spin-trapped adducts) have been detected upon redox-cycling of the AβCu system.13,20 None of these authors generated the AβCuI complex under inert atmosphere nor did they probe the O2 reactivity of the AβCuI complex over time.14 Furthermore, quantification of the amount of AβCuII produced following air-oxidation was never rigorously assessed.
Herein we present XAS data of both the AβCuII and AβCuI complexes along with EPR spectroscopic monitoring of the reduction and re-oxidation of the AβCu complex. These studies will demonstrate that the CuI ion forms a stable well defined adduct with Aβ. Furthermore, oxidation of CuI to CuII by O2 is kinetically sluggish, yet none the less affords Aβ damage. The biological implications of these new findings and their implications for AD will be discussed.
Materials and Methods
Materials
Aβ40 and Aβ16 peptides were purchased from Bachem (King of Prussia, PA) or rPeptide (Athens, GA) or synthesized on a Protein Technologies PS3 peptide synthesizer using standard Fmoc chemistries and subsequently purified by HPLC as previously described.11 The amino acid sequence for the Aβ40 peptide is DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV; the amino acid sequence of Aβ16 is the first 16 amino acids of Aβ40. Biological grade glycerol, sodium phosphate, 1,1,1,3,3,3-hexafluoroisopropanol (HFIP), and sodium chloride were purchased from Fisher Scientific (Pittsburgh, PA). Bovine serum albumin (BSA) standards were purchased from Sigma-Aldrich (St. Louis, MO). Quartz EPR tubes were purchased from Wilmad (Buena, NJ). Solutions were prepared in MilliQ™ water (resistivity > 18 mΩ, total organic content < 35 ppb). Sodium ascorbate was purchased from Spectrum Chemicals (Gardena, CA). Na2IrCl6 was from Alfa-Aesar (Ward Hill, MA).
Sample Preparation
Aβ peptides were monomerized with HFIP and stored at -80 °C in HFIP as previously reported.18,21 Peptide stock solution in HFIP was removed via a Hamilton gastight syringe that had been washed previously with multiple volumes of HFIP. An aliquot of the peptide stock in HFIP was removed for peptide concentration determination using a BSA calibration curve.18,21 Immediately prior to making peptide samples, HFIP was removed using a spin-vacuum system. This protocol produces homogeneous solutions of monomeric peptide.22
Samples of soluble Aβ16 or Aβ40 were prepared by resuspending dried peptide into buffer containing 50 mM NaPi, 75 mM NaCl, pH 7.2 with 50% glycerol (v/v). The Aβ16 peptide was used to model Cu(II) binding to soluble Aβ because it is well-established that this region contains the Aβ Cu-binding domain8,9,18,23 and because it does not contain the fibrillization domain (residues 17-21).23
CuII stock solutions were generated as previously reported.18,21,24 The CuII concentration in each EPR sample was determined on the basis of a calibration curve generated from CuII-imidazole standards in N-ethylmorpholine buffer at pH 7.22 containing 50% glycerol (v/v). The concentration of CuII in each of the EPR standards was assayed by chelation with bathocuproine disulfonic acid (BC) and reduction with ascorbate.25 The quantity of total copper as [Cu(BC)2]3- was quantified at 483 nm (ε483 = 12500 M-1 cm-1).25 The 0, 25, 50, and 100 μM CuII standards contained 2.0 ± 1.0, 21 ± 1.0, 61 ± 0.4, and 119 ± 1.2 μM CuII, respectively.
Assessment of Aβ16 Damage Following Multiple Redox Cycles by HPLC
A 1.0 mL 0.10 mM solution of Aβ16CuII was prepared as outlined above using a 1:1 CuII:Aβ16 stochiometry. Throughout the experiment aliquots were sampled from the main reaction vessel, and were analyzed by HPLC on a Waters DeltaPrep 60 equipped with an X-Terra C-18 reverse phase column (5 μm; 4.6 × 100 mm) using a gradient of 10-45% MeCN (0.1 % trifluoroacetic acid) in H2O (0.1% trifluoroacetic acid) over 25 min. Following the initial analysis of the Aβ16CuII solution, the sample was then reduced with 1.0 equivalent of ascorbate. After 19 hours the sample was reanalyzed by HPLC, rereduced with 1.0 equivalent of ascorbate, and then reanalyzed after 19 hours. The sample was then rereduced with an additional 1.0 equivalent of ascorbate and analyzed a final time after an additional 19 hours. In each case 50 μL of sample was added to 50 μL of a stock solution containing 0.100 mM of the peptide H2N-GAATDAQ-COOH as an internal standard and then analyzed by HPLC. Samples were not stirred throughout the course of the experiment, but were agitated by wrist for ∼15 sec. upon addition of ascorbate, and remained exposed to air throughout the experiment. All chromatograms were quantified using the software program PeakFit (SeaSolve Software, Inc.; Framingham, MA). The chromatograms displayed are baseline corrected.
EPR Spectroscopy
EPR spectra were collected on a Bruker EMX 6/1 spectrometer equipped with a microwave frequency meter and an Oxford Instruments ESR900 liquid He cryostat system. All spectra were collected with the following experimental parameters: microwave frequency = 9.38 GHz, microwave power = 0.5 mW, modulation amplitude = 10 G, time constant = 40.96 ms, conversion time = 40.96 ms, gain = 5 × 104, eight scans, temperature 20 K. Spectra of calibration standards were collected every run and a calibration curve to determine the CuII concentration was generated. The error associated with CuII concentrations determined using the calibration curve is estimated to be a maximum of 5-10 μM (about 10%) on the basis of comparison of spectra of the calibration standards collected on different days.
EPR samples for experiments with ascorbate were prepared in quartz EPR tubes capped with a tip-off manifold from Wilmad. Samples were degassed under inert atmosphere (dinitrogen or argon) using 3 freeze-pump-thaw cycles on a vacuum line. Stock solutions of sodium ascorbate for EPR experiments were prepared by adding solid sodium ascorbate to MQ water that had been thoroughly degassed on the vacuum line prior to addition of the solid. Aliquots of sodium ascorbate were removed from the stock solution under a blanket of inert gas and injected into the EPR sample, which was maintained under an inert atmosphere during the process. The addition and mixing was complete in less than 2 min typically.
Air was introduced into the EPR samples by freezing them to 77K, evacuating the sample headspace on the vacuum line, and admitting air into the headspace. Subsequently, the sample was thawed and mixed by inversion of the tube and its contents several times.
XAS Data Measurement and Analysis
The copper-containing peptide solutions were injected into aluminum sample holders in between two windows made of Kapton tape (3M, cat. #1205; Minneapolis, MN) and quickly frozen in liquid nitrogen. The Aβ40CuII sample was prepared and handled at 0 °C to maintain the monomeric peptide state.26 Data were then collected at the National Synchrotron Light Source (Brookhaven National Laboratories; Upton, NY) on beamlines X9b and X3b (ring operating conditions: 2.8 GeV; 200 – 305 mA). A focused Si(111) double monochrometer was used for energy selection along with a low-angle (4.5 mrad) Ni mirror for harmonic rejection. Energy calibrations were performed by recording a reference spectrum of Cu foil (first inflection point assigned to 8980.3 eV) simultaneously with the samples. All samples were maintained at 20 K throughout the data collection using a helium Displex cryostat. The spectra are reported as fluorescence data, which were recorded utilizing a 13-element Ge solid-state fluorescence detector (Canberra). Total count rates were maintained under 20 kHz per channel, and a deadtime correction of 3 μs was utilized (this had a negligible influence on the data). For XANES spectra the primary hutch aperture height was set to 0.4 mm to obtain the maximum resolution (theoretical maximum is ∼0.9 eV), while the hutch aperture was set to 1 × 2 mm, and data were obtained in 10 eV steps in the pre-edge region (8779 – 8958 eV), 0.3 eV steps in the edge region (8959 – 9023 eV), and 2.0 eV steps in the near-edge region. For EXAFS spectra the primary hutch aperture was set to 0.8 mm, the hutch aperture was set to 1 × 8 mm, and data were obtained in 5.0 eV steps in the pre-edge region (8779 – 8958 eV), 0.5 eV steps in the edge region (8959 – 9023 eV), 2.0 eV steps in the near-edge region (9024 – 9278 eV), and 5.0 eV steps in the far-edge region (9279 eV – 14.0 k). The EXAFS spectra represent the averaged sum of 20 spectra, while the XANES spectra represent the averaged sum of 3 spectra. After every other scan the beam was moved to a different position on the sample to avoid potential radiation damage. All spectra were individually inspected prior to data averaging to insure that sample decomposition in the beam was not occurring.
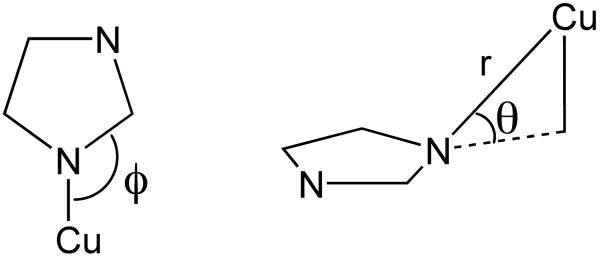
Data analysis was performed as previously described using the XAS analysis package EXAFS123.27,28 The only deviation is that the number of scatterers in the individual shells were initially left as free parameters, and then restrained to the nearest whole number. Single scatterer functions for Cu-N, Cu-O, and Cu-C interactions were constructed as previously described.27,29 Multiple scattering (MS) pathways for the Cu-Im moiety were constructed by removal of the Cu-N single scattering interaction from 48 simulated reference spectra. These reference spectra were then used to construct the MS pathways in an analogous manner as before, where three refinable parameters are considered: the Cu-“N” distance (r(Im)), the in-plane angle (ϕ), and the out of place angle (θ; see Chart 1). For utilization of these MS pathways in the EXAFS analysis we restrained r(Im) to be equal to that of the single scatterer Cu-N distance, and then allowed the two angles to refine freely. Although data were collected to 14 k, data refinements were only performed out to k = 12.0 Å−1 due to noise at higher values of k. Best fits to the experimental data were determined by selecting the model that gave both chemically reasonable refinement parameters and the lowest value for the goodness of fit parameter:
Chart 1.
| (1) |
where ni is the number of independent data points and np is the number of parameters used in the data simulations.
The bond valence sum (BVS) analysis30 on all refined EXAFS models was applied according to:
| (2) |
| (3) |
where r is the experimentally derived (EXAFS) bond length for ligand i and ro is the reference bond-length. The values used for ro include: rCu(II)N = 1.751 Å; rCu(II)O = 1.679 Å; and rCu(I)N = 1.595 Å.
Results and Discussion
Aβ16CuII Cu K-edge XAS
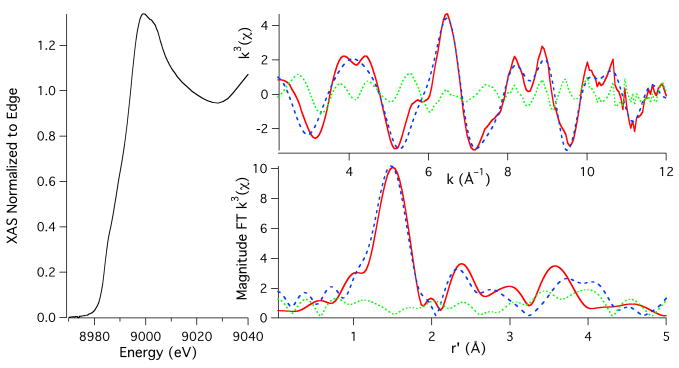
The Cu K-edge XAS obtained for the CuII adduct of Aβ16 is depicted in Figure 1. The edge region is characterized by a weak transition at 8979.7(1) eV that corresponds to the parity forbidden Cu(1s → 3d) transition and a shoulder in the edge region at 8986.8(1) eV that corresponds to a Cu(1s → 4p + shakedown) transition.31,32 The corresponding normal Cu(1s → 4p) transition is not resolvable from the edge.
Figure 1.
Cu K-edge XAS of Aβ16CuII. The left portion of the figure depicts the XANES region of the XAS. The bottom and top right spectra depict the magnitude FT k3(χ) and k3(χ), respectively. The experimental data are given as the red spectra, the simulations to the data are given as the dashed blue spectra, and the difference between the experimental and simulations are given as the dotted green spectra.
The EXAFS region was best modeled with CuII contained in a square-planar coordination environment with N/O ligation (Table 1; Figure 1). These refinements yielded three N/O scatterers at 1.938(12) Å and a fourth N/O scatterer at 2.07(2) Å. As can be seen, the magnitude Fourier Transformed (FT) EXAFS shows strong outersphere scattering between r = 2 to 4 Å, which is characteristic of multiple scattering (MS) interactions between the Cu-center and rigid ligands (i.e. imidazole rings from histidine residues). Using a MS analysis we were able to determine that two of the three shorter N/O scatterers are imidazole scatterers. In addition to the distance parameter, the phase and amplitude functions utilized to refine the MS pathways also yielded the in (ϕ) and out (θ) of plane Cu-His angles (see Chart 1). The resulting angles are best refined to a θ = 8(3)° and a ϕ = 129(8)°. These are both consistent and reasonable for Cu-His ligation. We note that this represents the average of the two imidazole scatterers, and an analysis where the two imidazole scatterers were separated into different shells gave identical results. Addition of more or fewer imidazole functions to these refinements yielded either poorer fits to the data, unrealistic angles for the Cu-Im moiety, or instability in the data refinements. Attempts were made to identify a fifth longer ligand without success.
Table 1.
Parameters used for the refinements to the Cu K-edge X-ray Absorption data for Aβ16CuII and Aβ16CuI.
| Aβ16CuII | Aβ16CuI | |
|---|---|---|
| Pre-edge Peak #1 (eV) | 8979.3(7) | 8984.1(2) |
| Pre-edge Peak #2 (eV) | 8984.0(6) | ---- |
| Eo (eV)a | 8988.7 | 8982.2 |
| N/O-shell | ||
| nb | 3 | 2 |
| r (Å) | 1.938(12) | 1.877(2) |
| σ2 (Å2) | 0.0030(17) | 0.0038(1) |
| N/O-shell | ||
| n | 1 | ---- |
| r (Å) | 2.07(2) | ---- |
| σ2 (Å2) | 0.001(1) | ---- |
| Im-shell | ||
| n | 2 | 2 |
| r (Å)C | 1.938 | 1.877 |
| σ2 (Å2) | 0.003(2) | 0.0057(1) |
| θ | 8(3)° | 2(2)° |
| ϕ | 129(8)° | 130(11)° |
| GOF | 0.54 | 0.22 |
| BVS | 2.05 | 0.94 |
For both refinements Eo was initially refined for the N-shell and then restrained.
Because of the way in which the MS pathways for the Im-shell were constructed, the first N/O shell contains the SS pathway for the Im-shell. Therefore, the number of non-imidazole N/O scatterers is equal to the number of N/O scatterers in the first N/O shell minus the number of imidazole scatterers.
The distance of the imidazole shell was restrained to the distance of the first N/O scatterer.
A bond valence sum (BVS) analysis was utilized to determine if these refinements make chemical sense.30 In this empirical analysis the metal-ligand bond length derived from the EXAFS data is compared to a reference bond length, and a bond valence (si) is obtained. Summing up the bond valences for all of the metal-ligand distances in the complex gives the BVS. The strength of the BVS analysis is that it yields the same value for all transition metal complexes of the same metal type and oxidation state (e.g. CuII) irrespective of coordination number and ligand type. In the most widely applied variant of the BVS analysis, the reference bond lengths are chosen such that the resulting BVS will equal the transition metal ion's oxidation state. Therefore, for CuII the BVS should ideally be equal to 2. A BVS analysis is extremely helpful in evaluating EXAFS data, where the number of scatterers is the least reliable parameter obtained, because models that only differ by the number of scatterers in the same shells can be readily compared and contrasted on the basis of the BVS to determine which is most likely.
When a BVS analysis is applied to the above EXAFS data (CuII ligated by two N-imidazole scatterers and two N or O ligands) we calculate a BVS ranging from: 2.05 for a CuII(NIm)2(O)2 coordination model, either 2.13 or 2.16 for a CuII(NIm)2(N)(O) coordination model (depending on the long vs. short ligand identity), and 2.23 for the CuII(NIm)2(N)2 coordination model. The three possible models above with both N and O ligands in the primary coordination sphere gives BVS values that are most consistent with CuII, while the coordination model with four nitrogen based ligands gives a BVS that is likely too large to be consistent with CuII. Increasing the coordination number to five causes the resulting BVS to range between 2.40 to 2.84 depending on the number of N or O ligands, which are far too high to be consistent with CuII. Therefore our XAS data are fully consistent with a four coordinate CuII center ligated by two N-imidazole scatterers and two N or O ligands.
On the basis of these data alone it is impossible to identify if the two non-imidazole ligands are nitrogen or oxygen donors. It is also impossible to determine the identity of the ligands. There is good evidence, however, that one of these ligands is an N-atom donor derived from the N-terminal amine nitrogen.8,9,18 One consideration is whether the N-terminal amine group coordinates inter- or intramolecularly. In an intermolecular coordination mode, a CuII ion could be anchored to histidine residues in one peptide and bind the amino terminus from a second peptide. If each peptide binds one CuII ion and provides a ligand to another CuII ion via the amino terminus, a 1:1 Cu:peptide stoichiometry would result, but the molecular mass of the species would be twice that of a simple intramolecular AβCu complex. We performed gel filtration chromatography of AβCuII to rule out this possibility (Figure S2) and find that the molecular weight of the AβCuII species corresponds to a monomeric complex. In addition, the mass spectrometry results of Jiang et al. show that only a 1:1 complex is detected for Aβ40CuII.10 Although neither of these results rule out a small population (< ∼1%) of dimer resulting from intermolecular copper-ligation, the vast majority is due exclusively to intramolecular copper-ligation.
We speculate that the other N/O atom donor is likely an O-atom of a carbonyl group, perhaps from a carbonyl group of the peptide backbone. We suggest this possibility because one of our CuII-N/O atom distances determined from the EXAFS refinements (2.07 Å) is close to the CuII-O atom carbonyl distances measured in crystallographically-characterized CuII-peptide complexes. CuII bound to a prion octarepeat peptide fragment has a CuII O-atom distance of 2.067 Å.33 In that structure, CuII is coordinated to two backbone amides, one histidine, and a backbone carbonyl in the equatorial plane.33 In addition, a 2.04 Å distance was reported for CuII coordinated to a carbonyl O-atom in the equatorial plane in a cylic bis-histidine peptide.34 It should be noted that tyrosine has been ruled out as the O-atom donor ligand to CuII by numerous spectroscopic experiments.9,11,14,18
Our model for Aβ16CuII can be contrasted with a recently published model for Aβ40CuII by Morante and coworkers based on Cu K-edge XAS data.35 Because the CuII coordinating residues are all found within the first 16 residues of the N-terminus of Aβ, Aβ16 is a good model of CuII coordination to Aβ40.8,9,36 In the previous EXAFS study the CuII ion was modeled in a five coordinate square pyramidal geometry ligated by: two His scatterers at 1.94 Å, one shorter His scatterer at 1.85 Å, a Tyr oxygen scatterer at 2.00 Å, and a water at 1.91 Å. In our view the data presented by Morante are of insufficient quality to reliably separate similar scatterers at those distances; because the quality of the data presented by Morante are sufficient for analysis only between k = 3.5 – 10.5, the spectral resolution of their EXAFS data is only Δr > 0.22 Å at best (calculated from Δr = π/(2Δk)). Even with a sophisticated MS analysis the reliable separation of the two different imidazole shells and the phenol shell would be both extremely difficult (if not impossible) and still yield statistically meaningless structural parameters given the quality of the data.
To discount changes in the microenvironment about the Cu-center induced by the differences in the length of the Aβ fragment in this vs. Morante's study, a reanalysis of the EXAFS data as presented in the manuscript of Stellato et al. was undertaken. This reanalysis yields structural parameters consistent with our refinements from above (see Supporting Information; Table S2, Figure S3). In fact, we find that because of the “penalty” imposed by the inclusion of additional refinement parameters in our error analysis, our 4-coordinate (2 imidazole/2 N or O scatterers) model actually affords a slight statistical improvement over their 5-coordinate model using Morante's data.
Our re-analysis of the Stellato et al. EXAFS data in-and-of itself obviously does not preclude a 5-coordinate model, but a BVS analysis of the structural models does. Using their original refinement35 (and our 5-coordinate re-refinement of their data), we arrive at a BVS between 2.87 – 2.92, while 4-coordinate refinements of these data yield a BVS that ranges between 2.02 – 2.31 depending on the CuN2O2 or CuN4 models. This result means that a five coordinate formulation for Aβ16CuII or Aβ40CuII is most likely incorrect. The issues with the original refinement of the low resolution data collected by Stellato et al.,35 our re-refinement of their data indicating that a 5-coordinate geometry is untenable, and refinement of our own higher resolution data, makes us strongly favor a 4-coordinate CuII model with two imidazole scatterers over a 5-coordinate model for the Aβ-CuII adduct.
A very recent combined DFT/Cu K-edge XAS study by Streltsov et. al.16 on AβCuII and several AβCuII derivatives was performed at a similar resolution to our study (in fact our data are virtually superimposable), and at a considerably higher resolution than the study of Stellato et. al.35 That study arrived at a different Cu-coordination environment than either Stellato or us; Streltsov et. al. propose a six coordinate structure with CuII ligation provided by three His imidazole nitrogens, both carboxylate oxygens from either Glu11 or Asp1, and another oxygen from a water molecule. Although the fit to the EXAFS data appears to be statistically valid there are two reasons why the chemical interpretation of Streltsov et. al. is questionable. There is general agreement from other spectroscopic data and recent theoretical work that CuII binds in a 4-coordinate square planar coordination environment within Aβ.8,9,18,19 Several of the spectroscopic methods applied to the AβCuII complex cannot directly detect axial ligands, which means that a 5 or 6-coordinate CuII coordination geometry cannot be definitively ruled out, but the BVS produced by the Streltsov model is 2.64, which is considerably too high for CuII. Our BVS analysis is therefore key to affirming the general consensus from other spectroscopic and theoretical work that CuII is bound to Aβ in a 4-coordinate geometry.
Aβ16CuI Cu K-edge XAS
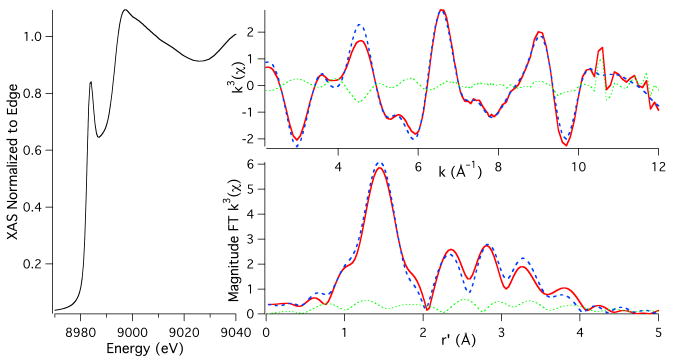
The Cu K-edge XAS of ascorbate reduced Aβ16CuI is presented in Figure 2; similar data were obtained for Aβ40CuI (Supporting Information; Figure S4) indicating identical coordination modes for Aβ40CuI and Aβ16CuI. A comparison of the edge regions of Aβ16CuII (Figure 1) vs. Aβ16CuI shows that upon reduction the edge energy decreases by 4.2(6) eV, which is consistent with the reduction of CuII to CuI. Consistent with our results, Streltsov et al. report a >5 eV shift in edge energy upon reduction of Aβ16CuII with ascorbate.15 The XANES region of the XAS for Aβ16CuI displays an intense pre-edge feature at 8984.1(2) eV, which is assigned as the Cu(1s → 4p) transition.31 For CuI this feature is highly dependent on coordination geometry about the CuI center. The position and intensity of the Cu(1s → 4p) transition observed for Aβ(16)CuI is consistent with either a two-coordinate linear geometry about CuI, or possibly a three coordinate T-shaped geometry.31,37 However, as will be shown, the reactivity of AβCuI is most consistent with a two-coordinate linear geometry.
Figure 2.
Cu K-edge XAS of Aβ16CuI. The left portion of the figure depicts the XANES region of the XAS. The bottom and top right spectra depict the magnitude FT k3(χ) and k3(χ), respectively. The experimental data are given as the red spectra, the simulations to the data are given as the dashed blue spectra, and the difference between the experimental and simulations are given as the dotted green spectra.
A best fit to the EXAFS yielded a two coordinate geometry with two imidazole ligands coordinated to CuI at 1.877(2) Å (θ = 2(2)°; ϕ = 130(11)°; Scheme 1). Refinement of the data splitting the two imidazole scatters into different shells resulted in similar bond angles for the imidazole ligands (Figure S5). Attempts were made to solve for a three coordinate ligation environment for Aβ16CuI.16 In all cases a T-shaped geometry could not be refined: the shell containing a third imidazole scatterer was always placed at distance nearly identical to the shell containing the other two imidazole scatterers. A BVS analysis of the two refinements yields a BVS = 0.94 for the two coordinate complex and 1.42 for the three coordinate complex. We therefore favor a linear two coordinate CuI coordination environment for Aβ16CuI on the basis of the Cu K-edge XAS coupled with the BVS analysis.
Scheme 1.
AβCuII/AβCuI EPR spectroscopy
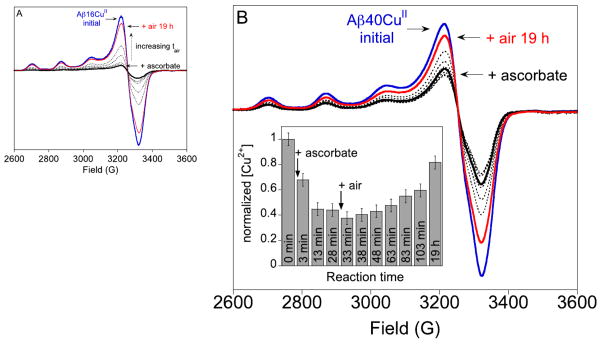
Spectra for both Aβ16CuII and Aβ40CuII samples before and after the addition of ascorbate are shown in Figure 3. After the addition of ascorbate, the EPR signal decreases due to the disappearance of the AβCuII complex, indicating the formation of the d10, EPR-silent AβCuI complex. Maiti et al. were the first to report that the reduction of Aβ16CuII and O2-reoxidation of Aβ16CuI are completely reversible as observed by fluorescence spectroscopy.14 This observation is inconsistent with our EPR spectroscopic data.
Figure 3.
20 K EPR spectra of the reduction of AβCuII by ascorbate and subsequent re-oxidation in air. (A) 250 μM Aβ16CuII (blue) + 1.2 equiv ascorbate (solid black line) followed by addition of air (dotted lines, red line). (B) 100 μM Aβ40CuII (blue) + approximately 0.5 equiv ascorbate (solid black line) followed by addition of air (dotted lines, red line). Panel (B) inset, normalized Aβ40CuII signal area as a function of total reaction time. EPR conditions are given in the Materials and Methods Section.
For the Aβ16CuII sample, spectral integration indicates that approximately 90% of the initial AβCuII content is recovered in a sample treated with 1.2 equivalents of ascorbate under inert atmosphere followed by 19 h of air exposure. We note that if an ascorbate-treated sample is immediately treated with excess Ir(IV), the initial AβCuII signal hyperfine intensity recovers entirely (Figure S6). In this case, spectral integration to quantify the AβCuII concentration is not reliable because of a contribution from the broad EPR signal of unreacted IrIV. For Aβ40CuII, a similar pattern of sluggish O2 reactivity was observed even under conditions where only 0.5 equiv of ascorbate was added (Figure 3B). The CuII concentrations for the 13 and 28 min time points for the AβCuII + ascorbate reaction are the same within error (Figure 3B, inset), which indicates that the reduction reaction is complete after this time. After 19 h in air, the CuII concentration recovers to approximately 80% of its initial value. A matched sample treated with a 2-fold excess of ascorbate recovered only 60% of the initial CuII signal after 3 d in air (Figure S7). Finally, Aβ40CuII was reduced with a 4-fold excess of ascorbate in air-saturated solution (Figure S8). The EPR signal from Aβ40CuII decreased to about 15% of the initial signal. Like the sample treated with excess ascorbate under Ar, only about 60% of the initial CuII intensity is recovered in this sample after extended incubation (5 d) at room temperature.
In all of the samples prepared anaerobically and then exposed to oxygen, the CuII spectral shapes are the same pre- and post-ascorbate addition, suggesting that CuII is bound in its original coordination site after its reduction and reoxidation. The experiment performed in air, however, has slight differences in the spectral shape pre and post-ascorbate addition (Figure S8, inset). The post-ascorbate addition spectrum appears to contain contributions from two different CuII species. The appearance of a new CuII signal could be due to protein modification from carbonate radicals that are generated upon redox cycling of Cu.38,39 Under conditions where the sample was degassed, all dissolved gases were removed prior to admitting air. Thus, samples made in air-saturated buffer contain more CO2 than samples that were degassed. Addition of carbonate radicals to proteins does not fragment the protein,39 but instead introduces modifications. Such modifications might change the protein structure and/or metal binding sites leading to our observation of two types of CuII EPR signals in aerobic samples treated with ascorbate.
The fluorescence data of Miati et al. indicate that the Aβ16CuII complex is completely regenerated upon exposure of the AβCuI complex to O2.14 In contrast, our EPR spectroscopic results indicate that the initial full concentration of AβCuII complex is not regenerated upon oxidation of AβCuI. As EPR spectroscopy is a direct indicator of the CuII coordination environment, whereas tyrosine fluorescence quenching is an indirect measure of Aβ–CuII coordination, our more sensitive data strongly suggest the loss of CuII binding affinity toward Aβ.
There are several possible explanations for why an EPR signal from AβCuII does not reappear entirely upon air exposure of anaerobic samples treated with ascorbate. One possibility is that some fraction of CuII is released from the peptide because of oxidative decomposition of the peptide (vide infra). This released CuII will bind to phosphate buffer to make an antiferromagnetically-coupled CuII polymer that is EPR silent. Another possibility is that a stable CuI compound forms or some other type of copper cluster that is EPR silent. Our HPLC results (vide infra) are consistent with oxidative damage of the peptide.
The reaction of AβCuII with a large excess of reductant has been reported to produce hydrogen peroxide, hydroxyl radicals, and tyrosine radicals.3,13,40-42 Tyrosyl radical, in particular, has been proposed as an intermediate in ROS generation by the AβCu complex.40,42,43 Under our conditions, AβCuII with ascorbate does not produce a detectable amount of tyrosine radical, a species that is easily identifiable by low-temperature EPR spectroscopy.44 This result indicates that this radical does not reach a steady-state concentration above our detection limit. Barnham et al. proposed catalytic tyrosine radical formation as part of the ROS-generating mechanism for AβCu,42 which contradicts peptide oxidation detected by mass spectrometry showing that histidine oxidation appears prior to tyrosine oxidation and crosslinking reactions.45,46 Thus, it remains unclear whether tyrosine radical formation precedes, is concerted with, or follows hydrogen peroxide/hydroxyl radical production.
Assessment of Oxidative Damage of Aβ16 Following Multiple Cu Redox Cycles
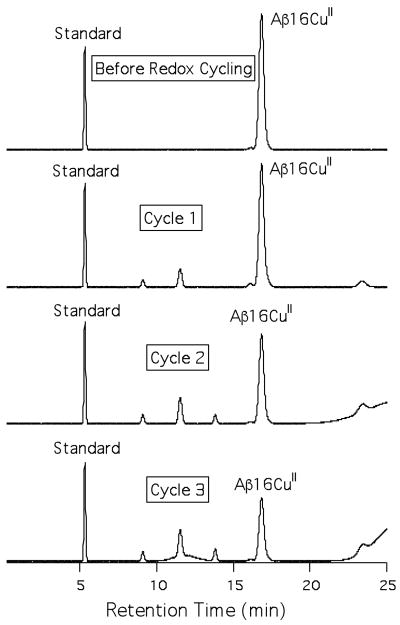
Our EPR results clearly show that regeneration of AβCuII is not quantitative following redox-cycling, which may be indicative of peptide damage to the copper ligating residues of Aβ or other processes that change the physiochemical nature of Aβ16, as has previously been observed.47 We probed the supposition that peptide damage may be responsible for the non-quantitative regeneration of AβCuII by following multiple redox cycles using HPLC (Figure 4). Prior to reduction of Aβ16CuII by ascorbate the metallopeptide shows one distinct peak in the chromatogram. Following reduction of Aβ16CuII with one equivalent of ascorbate and 19 hours of air oxidation, three new peaks have appeared in the chromatogram, and the amount of Aβ16CuII has decreased by 33% of its original concentration compared to a peptide standard. After a second redox cycling under identical conditions more Aβ16CuII has been lost (56% of its original concentration), the additional peaks have increased in intensity, a fourth peak has grown in, and there is a significant amount of material that comes off the column only towards the end of the HPLC run (and in the column wash), which is indicative of peptide oligomerization. After a third and final redox cycling, the amount of Aβ16CuII remaining in solution is 39% of its original concentration, while the amount of material coming off at the end of the run has increased a significant amount indicating increased peptide aggregate formation, and a fifth broad peak between 11-13 min, has appeared in the chromatogram.
Figure 4.
HPLC chromatograms of Aβ16CuII following multiple redox cycling using ascorbate in air.
The HPLC redox cycling results confirm the EPR results from above: damage to Aβ16CuII is occurring following multiple redox cycles. Control HPLC and GPC studies demonstrate that Aβ16CuII does not undergo significant damage or oligimerization if a reductant is not added to solution (Figures S2 and S9). This strongly suggests that redox cycling is accelerating the deterioration of monomeric Aβ16CuII. It is likely that similar oxidative damage to Aβ16 is occurring here as was previously observed by Schöneich and Williams, who observed predominant oxidation of His13 and His14.47 We are currently in the process of identifying the Aβ modification sites and identity of the oxidized products to confirm this supposition.
These HPLC results indicate that slightly more oxidative damage to Aβ16CuII is occurring during redox cycling than indicated by EPR, as there is a significantly larger decrease in soluble monomeric Aβ16CuII observed by HPLC than EPR. There are at least three possible explanations for this observation. The first is that one (or more) of the modified Aβ16 products is capable of coordinating CuII in a ligand environment nearly identical to that of unmodified Aβ16. Another explanation is that redox cycling produces soluble, crosslinked Aβ16CuII oligomers, which are spectroscopically similar by EPR, but distinguishable by HPLC. The third explanation is that the EPR samples contained less oxygen than the HPLC samples. The buffer was the same for both sets of samples so the O2 solubility in the solvent system is the same. However, following air exposure, EPR samples were sealed and not reopened, which means that no additional air (and, therefore, O2) was introduced. We calculate that there are 14.7 μmol of O2 in the headspace of the EPR tube compared to 0.01 μmol of Cu in the sample, which represents a 1500-fold excess of O2 over Cu. Nevertheless, the HPLC samples were prepared and handled in air so that the amount of air in the samples could be higher than in the EPR samples.
Summary and Biological Implications
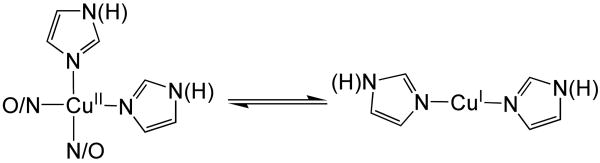
In this study we have shown that CuII adducts of Aβ can readily undergo reduction by the biologically relevant reductant ascorbate.48 By XAS we have shown that upon reduction of Aβ16CuII and Aβ40CuII with ascorbate linear His2-CuI adducts are formed (Scheme 1). Our XAS analysis is therefore consistent with the observed O2 reactivity displayed by Aβ16CuI and Aβ40CuI. Following reduction of AβCuII with stoichiometric amounts of ascorbate, solutions of Aβ16CuI or Aβ40CuI are not highly sensitive to O2, requiring hours to undergo conversion from CuI to CuII. Similarly, solutions of two coordinate CuI complexes are not susceptible to oxidation by dioxygen.37,49 In contrast, three coordinate CuI centers are highly reactive towards dioxygen, typically undergoing oxidation to CuII instantaneously upon exposure to dioxygen.37,50 Therefore, the dioxygen reactivity of Aβ16CuI is consistent with the our XAS analysis.
A recent study by Karlin and coworkers is also consistent with our formulation of a linear diimidazole CuI ligation scheme for Aβ16CuI.37 In that study, a series of His-His dipeptides coordinated to CuI formed linear two coordinate copper centers with Cu-imidazole bond lengths ranging from 1.863 – 1.876 Å. These two-coordinate CuI-dipeptides were not prone to oxidation by O2. However, upon the addition of a third imidazole-based ligand to the CuI-dipeptides the rapid oxidation of CuI to CuII was noted in that study. Therefore, the O2 reactivity of Aβ16CuI and Aβ40CuII are both most consistent with a linear diimidazole-CuI ligation scheme.
In addition to providing experimental precedence in support of our coordination mode for Aβ16CuI and Aβ40CuI, Karlin's study hints at the identity of the two His residues ligated to CuI in these metallopeptides. The copper-binding domain of Aβ contains two adjacent His residues at positions 13 and 14, making it reasonable to speculate that His13 and His14 are the residues involved in CuI coordination in Aβ. Computational studies by both us and Karlin indicate that the linear CuI bis-His motif is not strained, and we in-fact arrive at nearly identical structural parameters for the [CuI(His)2]+ metallopeptide using high-level double hybrid DFT methods as we determined by EXAFS (Figure S10, Table S3). Indeed, mass spectrometry of oxidative damage products generated upon reduction of AβCuII with ascorbate indicates that His13 and His14 are preferentially oxidized at short reaction times,46 not His6, suggesting His13 and His14 are the sidechains bound to CuI. Although it is possible that one of these two histidines is damaged because of proximity to copper and not direct ligation, it is difficult to explain why His6 is not damaged if it is one of the two histidine ligands to copper.
Another finding of this study that has broad implications for our understanding of the etiology of the AβCu complex is the oxidative damage of the reduced metallopeptides following oxidation by O2. It has been shown that AβCuII in the presence of reductants generates ROS.13,51 We ascribe incomplete recovery of the AβCuII EPR signal to ROS-induced peptide damage, in particular histidine oxidation,12,40,47,52 that precludes CuII binding in the native coordination site. Our HPLC results also indicate that multiple redox cycling produces significantly more Aβ damage following each cycle; there is a non-linear increase in Aβ damage following each redox cycle. This leads us to predict that when aerobic solutions of AβCuII are redox-cycled via addition of excess ascorbate, an evolving and heterogeneous mixture of Cu species is created: copper ions bound to modified and unmodified peptide and copper ions not bound to the peptide are all present.
It has been shown that addition of an exogenous chelator to solutions of AβCuII treated with ascorbate retards hydrogen peroxide production.53 This result is entirely consistent with the fact that chelated CuII is reduced more slowly by ascorbate than “free” CuII ion.54 Such heterogeneity of the Aβ/CuII/reductant reaction mixture and details of the rate constant/mechanism differences between bound and unbound copper have not been considered carefully previously. In our view, these critical issues call into question measurements of the rates of hydrogen peroxide and/or hydroxyl radical production by AβCuII, which have assumed a single species (Cu-ligated Aβ) is responsible for ROS formation.10,13 Both CuII that is not bound to peptide and copper-ligated Aβ may be producing ROS, but we believe CuII released from oxidatively-modified Aβ is the principal ROS generator in this system.
Together, these findings suggest several hypotheses concerning Aβ and its role in neurodegeneration. First, under redox-cycling conditions, oxidative damage to Aβ inhibits its ability to bind Cu, and thus will promote oxidative stress in neuronal tissue. Also, these data point towards the supposition that copper-induced ROS generation mediates Aβ aggregation, as has been previously suggested.40,55 Upon multiple rounds of redox cycling of Aβ16CuII, it appears that significant Aβ16 aggregation, perhaps via covalent crosslinking, occurs. Oxidative modification of Aβ increases its propensity to aggregate, which has been hypothesized to increase Aβ oligomer formation and/or plaque deposition in AD.40,42,56
Finally, we propose that AβCu may be neuroprotective57 if it forms a CuI complex rather than a CuII complex in vivo. We come to the conclusion that AβCu might be neuroprotective based on our EXAFS results, which are consistent with the O2-stable linear CuI coordination geometry, and a thermodynamic square scheme (Scheme 2). This scheme predicts a higher affinity of Aβ for CuI than for CuII because the midpoint potential of AβCuII/AβCuI is higher (0.28 V10 vs. NHE) than that of free CuII/CuI (0.158 V vs. NHE).58 Note that the magnitude of the affinity of Aβ for CuII has no effect on the relative affinities of Aβ for CuI vs. CuII. Our assumption that the “free” CuII/CuI potential is lower than that of the AβCuII/AβCuI complex is supported by electrochemistry experiments that showed that CuII in buffer is reduced at a lower potential than AβCuII/AβCuI in the same buffer.10 If Aβ has a higher affinity for CuI than CuII and the AβCuI complex is sluggish to react with O2, damage from Cu ions will be abrogated in the presence of Aβ compared to CuII/CuI that is not bound to peptide. The redox-cycling of AβCu depends on the concentrations of ascorbate and O2 in vivo. Extracellular fluid that surrounds mammalian neurons, and is in equilibrium with cerebral spinal fluid (CSF), contains 200 – 400 μM ascorbate.48 The pO2 of CSF has been measured to be 31-51 mm Hg by invasive methods.59 More recently, values obtained using fluid-attenuated inversion recovery (FLAIR) magnetic resonance imaging on humans are 54 ± 18 (lateral ventricles), 67 ± 1 (cisterna magna), 106 ± 42 (cortical sulci), and 130 ± 49 mm Hg (third ventrical), which gives an average of about 90 mm Hg in CSF.60 For comparison, the pO2 in the trachea is about 150 mm Hg. Thus, the concentration of O2 in CSF is about 2-fold lower than in air-saturated solutions at 37 °C. Although our experiments were performed at room temperature instead of 37 °C and, therefore, the rates are different from those at 37 °C, we find that the Aβ40CuII species can be reduced to Aβ40CuI in an environment containing substantially more O2 (air-saturated buffer) than is present in CSF. Although speculative, our idea that AβCuI is the dominant species in vivo is consistent with our findings and represents a neuroprotective mechanism for AβCu that has not been proposed previously.
Scheme 2.
Supplementary Material
Acknowledgments
Funded by Alzheimer's Association Grant IIRG-07-5821 (V.A.S.) and NIH grant number P20 RR-016464 from the INBRE Program of the National Center for Research Resources, (J.S.) Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
Footnotes
Supporting Information Available: XAS spectra of Aβ40CuI, additional refinements to the EXAFS region of Aβ16CuII and Aβ16CuI, EPR spectra of the reoxidation of Aβ16CuI by Ir(IV), reduction/air reoxidation of Aβ40CuII with excess ascorbate in anaerobic solution and in air-saturated buffer, digitized XAS data from Stellato et al.,35 our re-refinement of EXAFS data from Stellato et al.,35 geometry optimized structure for (CuI(His)2)+, control HPLC and GPC chromatograms of air exposed AβCuII, and complete reference 51 are available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Jason Shearer, Email: shearer@unr.edu.
Veronika A. Szalai, Email: vszalai@umbc.edu.
References
- 1.Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, Hall K, Hasegawa K, Hendrie H, Huang Y, Jorm A, Mathers C, Menezes PR, Rimmer E, Scazufca M. Lancet. 2005;366:2112. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castellani RJ, Lee HG, Zhu X, Perry G, Smith MA. J Neuropathol Exp Neurol. 2008;67:523. doi: 10.1097/NEN.0b013e318177eaf4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sergeant N, Bretteville A, Hamdane M, Caillet-Boudin ML, Grognet P, Bombois S, Blum D, Delacourte A, Pasquier F, Vanmechelen E, Schraen-Maschke S, Buee L. Expert Rev Proteomics. 2008;5:207. doi: 10.1586/14789450.5.2.207. [DOI] [PubMed] [Google Scholar]; Cechetto DF, Hachinski V, Whitehead SN. Expert Rev Neurother. 2008;8:743. doi: 10.1586/14737175.8.5.743. [DOI] [PubMed] [Google Scholar]; Xia W. Curr Alzheimer Res. 2008;5:172. doi: 10.2174/156720508783954712. [DOI] [PubMed] [Google Scholar]; Windisch M, Wolf H, Hutter-Paier B, Wronski R. Neurodegener Dis. 2008;5:218. doi: 10.1159/000113707. [DOI] [PubMed] [Google Scholar]; Luheshi LM, Crowther DC, Dobson CM. Curr Opin Chem Biol. 2008;12:25. doi: 10.1016/j.cbpa.2008.02.011. [DOI] [PubMed] [Google Scholar]; Shi Q, Gibson GE. Alzheimer Dis Assoc Disord. 2007;21:276. doi: 10.1097/WAD.0b013e31815721c3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Skovronsky DM, Lee VM, Trojanowski JQ. Annu Rev Pathol. 2006;1:151. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]; Irvine GB, El-Agnaf OM, Shankar GM, Walsh DM. Mol Med. 2008;14:451. doi: 10.2119/2007-00100.Irvine. [DOI] [PMC free article] [PubMed] [Google Scholar]; Selkoe DJ. Behav Brain Res. 2008;192:106. doi: 10.1016/j.bbr.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green KN, Smith IF, Laferla FM. Subcell Biochem. 2007;45:507. doi: 10.1007/978-1-4020-6191-2_19. [DOI] [PubMed] [Google Scholar]
- 4.Zhu X, Su B, Wang X, Smith MA, Perry G. Cell Mol Life Sci. 2007;64:2202. doi: 10.1007/s00018-007-7218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong J, Atwood CG, Anderson VE, Siedlak SL, Smith MA, Perry G, Carey PR. Biochemistry. 2003;42:2768. doi: 10.1021/bi0272151. [DOI] [PubMed] [Google Scholar]; Beauchemin D, Kisilevsky R. Anal Chem. 1998;70:1026. doi: 10.1021/ac970783f. [DOI] [PubMed] [Google Scholar]; Miller LM, Wang Q, Telivala TP, Smith RJ, Lanzirotti A, Miklossy J. J Struct Biol. 2006;155:30. doi: 10.1016/j.jsb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Adlard PA, Bush AI, Adman ET. J Alzheimers Dis. 2006;10:145. doi: 10.3233/jad-2006-102-303. [DOI] [PubMed] [Google Scholar]
- 7.Karr JW, Szalai VA. Biochemistry. 2008;47:5006. doi: 10.1021/bi702423h. [DOI] [PubMed] [Google Scholar]; Kowalik-Jankowska T, Dolejsz-Ruta M, Wisniewska K, Lankiewicz L. J Inorg Biochem. 2001;86:535. doi: 10.1016/s0162-0134(01)00226-4. [DOI] [PubMed] [Google Scholar]; Ma QF, Hu J, Wu WH, Liu HD, Du JT, Fu Y, Wu YW, Lei P, Zhao YF, Li YM. Biopolymers. 2006;83:20. doi: 10.1002/bip.20523. [DOI] [PubMed] [Google Scholar]; Danielsson J, Pierattelli R, Banci L, Graslund A. FEBS Journal. 2007;274:46. doi: 10.1111/j.1742-4658.2006.05563.x. [DOI] [PubMed] [Google Scholar]; Lim KH, Kim YK, Chang YT. Biochemistry. 2007;46:13523. doi: 10.1021/bi701112z. [DOI] [PubMed] [Google Scholar]; Hou L, Zagorski MG. J Am Chem Soc. 2006;128:9260. doi: 10.1021/ja046032u. [DOI] [PubMed] [Google Scholar]
- 8.Syme CD, Nadal RC, Rigby SEJ, Viles JH. J Biol Chem. 2004;279:18169. doi: 10.1074/jbc.M313572200. [DOI] [PubMed] [Google Scholar]
- 9.Kowalik-Jankowska T, Ruta M, Wisniewska K, Lankiewicz L. J Inorg Biochem. 2003;95:270. doi: 10.1016/s0162-0134(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 10.Jiang D, Men L, Wang J, Zhang Y, Chickenyen S, Wang Y, Zhou F. Biochemistry. 2007;46:9270. doi: 10.1021/bi700508n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilloreau L, Damian L, Coppel Y, Mazarguil H, Winterhalter M, Faller P. J Biol Inorg Chem. 2006;11:1024. doi: 10.1007/s00775-006-0154-1. [DOI] [PubMed] [Google Scholar]
- 12.Kowalik-Jankowska T, Ruta M, Wisniewska K, Lankiewicz L, Dyba M. J Inorg Biochem. 2004;98:940. doi: 10.1016/j.jinorgbio.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Guilloreau L, Combalbert S, Sournia-Saquet A, Mazarguil H, Faller P. Chembiochem. 2007;8:1317. doi: 10.1002/cbic.200700111. [DOI] [PubMed] [Google Scholar]
- 14.Maiti NC, Jiang D, Wain AJ, Patel S, Dinh KL, Zhou F. J Phys Chem B. 2008;112:8406. doi: 10.1021/jp802038p. [DOI] [PubMed] [Google Scholar]
- 15.Streltsov VA, Varghese JN. ChemComm. 2008;27:3169. doi: 10.1039/b803911a. [DOI] [PubMed] [Google Scholar]
- 16.Raffa DF, Rickard GA, Rauk A. J Biol Inorg Chem. 2007;12:147. doi: 10.1007/s00775-006-0175-9. [DOI] [PubMed] [Google Scholar]
- 17.Streltsov VA, SJ JT, Epa VC, Barnham KJ, Masters CL, Varghese JN. Biophys J. 2008 doi: 10.1529/biophysj.108.134429. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karr JW, Akintoye H, Kaupp LJ, Szalai VA. Biochemistry. 2005;44:5478. doi: 10.1021/bi047611e. [DOI] [PubMed] [Google Scholar]
- 19.Mantri Y, Fioroni M, Baik MH. J Biol Inorg Chem. 2008 doi: 10.1007/s00775-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura M, Shishido N, Nunomura A, Smith MA, Perry G, Hayashi Y, Nakayama K, Hayashi T. Biochemistry. 2007;46:12737. doi: 10.1021/bi701079z. [DOI] [PubMed] [Google Scholar]
- 21.Karr JW, Kaupp LJ, Szalai VA. J Am Chem Soc. 2004;126:13534. doi: 10.1021/ja0488028. [DOI] [PubMed] [Google Scholar]
- 22.Stine WB, Jr, Dahlgren KN, Krafft GA, LaDu MJ. J Biol Chem. 2003;278:11612. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 23.Miura T, Suzuki K, Kohata N, Takeuchi H. Biochemistry. 2000;39:7024. doi: 10.1021/bi0002479. [DOI] [PubMed] [Google Scholar]
- 24.Karr JW, Szalai VA. J Am Chem Soc. 2007 doi: 10.1021/ja068952d. [DOI] [PubMed] [Google Scholar]
- 25.Moffett J, Zika RG, Petasne RG. Anal Chim Acta. 1985;175:171. [Google Scholar]
- 26.Hou L, Shao H, Zhang Y, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon IJ, Ray DG, Vitek MP, Iwashita T, Makula RA, Przybyla AB, Zagorski MG. J Am Chem Soc. 2004;126:1992. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 27.Shearer J, Soh P. Inorg Chem. 2007;46:710. doi: 10.1021/ic061236s. [DOI] [PubMed] [Google Scholar]
- 28.Scarrow RC. Haverford College; Haverford, PA: 2005. www.Haverford.edu/chem./Scarrow/EXAFS123/ ed. [Google Scholar]
- 29.Scarrow RC, S SB, Ellison JJ, Shoner SC, Kovacs JA, Cummings JG, Nelson MJ. J Am Chem Soc. 1998;120:9237. [Google Scholar]; Scarrow RC, Brennan BA, Cummings JG, Jin H, Duong DJ, Kindt JT, Nelson MJ. Biochemistry. 1996;35:1078. doi: 10.1021/bi960164l. [DOI] [PubMed] [Google Scholar]
- 30.Brown ID, Altermatt D. Acta Crystallogr, Sect B. 1985;41:244. [Google Scholar]; Thorp HH. Inorg Chem. 1992;31:1585. [Google Scholar]
- 31.Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. J Am Chem Soc. 1987;109:6433. [Google Scholar]
- 32.DuBois JL, Mukherjee P, Stack TDP, Hedman B, Solomon EI, Hodgson KO. J Am Chem Soc. 2000;122:5775. [Google Scholar]
- 33.Burns CS, Aronoff-Spencer E, Dunham CM, Lario P, Avdievich NI, Antholine WE, Olmstead MM, Vrielink A, Gerfen GJ, Peisach J, Scott WG, Millhauser GL. Biochemistry. 2002;41:3991. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kojima Y, Hirotsu K, Matsumoto K. Bull Chem Soc Japan. 1977;50:3222. [Google Scholar]
- 35.Stellato F, Menestrina G, Serra MD, Potrich C, Tomazzolli R, Meyer-Klaucke W, Morante S. Eur Biophys J. 2006:340. doi: 10.1007/s00249-005-0041-7. [DOI] [PubMed] [Google Scholar]
- 36.Curtain CC, Ali F, Volitakis I, Cherny RA, Norton RS, Beyreuther K, Barrow CJ, Masters CL, Bush AI, Barnham KJ. J Biol Chem. 2001;276:20466. doi: 10.1074/jbc.M100175200. [DOI] [PubMed] [Google Scholar]
- 37.Himes RA, Park GY, Barry AN, Blackburn NJ, Karlin KD. J Am Chem Soc. 2007;129:5352. doi: 10.1021/ja0708013. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez DC, Mejiba SE, Mason RP. J Biol Chem. 2005;280:27402. doi: 10.1074/jbc.M504241200. [DOI] [PubMed] [Google Scholar]
- 39.Ramirez DC, Gomez-Mejiba SE, Corbett JT, Deterding LJ, Tomer KB, Mason R. Biochem J. 2008 doi: 10.1042/BJ20070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atwood CS, Perry G, Zeng H, Kato Y, Jones WD, Ling KQ, Huang X, Moir RD, Wang D, Sayre LM, Smith MA, Chen SG, Bush AI. Biochemistry. 2004;43:560. doi: 10.1021/bi0358824. [DOI] [PubMed] [Google Scholar]
- 41.Haeffner F, Smith D, Barnham K, Bush A. J Inorg Biochem. 2005;99:2403. doi: 10.1016/j.jinorgbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Barnham KJ, Haeffner F, Ciccotosto GD, Curtain CC, Tew D, Mavros C, Beyreuther K, Carrington D, Masters CL, Cherny RA, Cappai R, Bush AI. Faseb J. 2004;18:1427. doi: 10.1096/fj.04-1890fje. [DOI] [PubMed] [Google Scholar]
- 43.Tickler AK, Smith DG, Ciccotosto GD, Tew DJ, Curtain CC, Carrington D, Masters CL, Bush AI, Cherny RA, Cappai R, Wade JD, Barnham KJ. J Biol Chem. 2005;280:13355. doi: 10.1074/jbc.M414178200. [DOI] [PubMed] [Google Scholar]
- 44.Szalai VA, Brudvig GW. Biochemistry. 1996;35:15080. doi: 10.1021/bi961117w. [DOI] [PubMed] [Google Scholar]; Szalai VA, Brudvig GW. Biochemistry. 1996;35:1946. doi: 10.1021/bi952378t. [DOI] [PubMed] [Google Scholar]; Szalai VA, Kühne H, Lakshmi KV, Brudvig GW. Biochemistry. 1998;37:13594. doi: 10.1021/bi9813025. [DOI] [PubMed] [Google Scholar]; Stubbe J, van der Donk WA. Chem Rev. 1998;98:705. doi: 10.1021/cr980059c. [DOI] [PubMed] [Google Scholar]
- 45.Schoneich C. Ann N Y Acad Sci. 2004;1012:164. doi: 10.1196/annals.1306.013. [DOI] [PubMed] [Google Scholar]
- 46.Pogocki D, Schoneich C. Chem Res Toxicol. 2002;15:408. doi: 10.1021/tx0101550. [DOI] [PubMed] [Google Scholar]
- 47.Schoneich C, Williams TD. Chem Res Toxicol. 2002;15:717. doi: 10.1021/tx025504k. [DOI] [PubMed] [Google Scholar]
- 48.Rice ME. Trends Neurosci. 2000;23:209. doi: 10.1016/s0166-2236(99)01543-x. [DOI] [PubMed] [Google Scholar]
- 49.Sorrell TN, Jameson DL. J Am Chem Soc. 1983;105:6013. [Google Scholar]; Sanyal I, Strange RW, Blackburn NJ, Karlin KD. J Am Chem Soc. 1991;113:4692. [Google Scholar]; Sanyal I, Strange RW, Blackburn NJ, Karlin KD. J Am Chem Soc. 1993;115:11259. [Google Scholar]
- 50.Lewis EA, Tolman WB. Chem Rev. 2004;104:1047. doi: 10.1021/cr020633r. [DOI] [PubMed] [Google Scholar]
- 51.Huang X, et al. J Biol Chem. 1999;274:37111. doi: 10.1074/jbc.274.52.37111. [DOI] [PubMed] [Google Scholar]
- 52.Lim J, Vachet RW. Anal Chem. 2003;75:1164. doi: 10.1021/ac026206v. [DOI] [PubMed] [Google Scholar]
- 53.Deraeve C, Pitie M, Meunier B. J Inorg Biochem. 2006;100:2117. doi: 10.1016/j.jinorgbio.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Taqui Khan MM, Martell AE. J Am Chem Soc. 1967;89:7104. doi: 10.1021/ja01002a046. [DOI] [PubMed] [Google Scholar]; Taqui Khan MM, Martell AE. J Am Chem Soc. 1967;89:4176. doi: 10.1021/ja00992a036. [DOI] [PubMed] [Google Scholar]
- 55.Smith DG, Cappai R, Barnham KJ. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Yoburn JC, Tian W, Brower JO, Nowick JS, Glabe CG, Van Vranken DL. Chem Res Toxicol. 2003;16:531. doi: 10.1021/tx025666g. [DOI] [PubMed] [Google Scholar]; Smith DP, Ciccotosto GD, Tew DJ, Fodero-Tavoletti MT, Johanssen T, Masters CL, Barnham KJ, Cappai R. Biochemistry. 2007;46:2881. doi: 10.1021/bi0620961. [DOI] [PubMed] [Google Scholar]
- 57.Zou K, Gong JS, Yanagisawa K, Michikawa M. J Neurosci. 2002;22:4833. doi: 10.1523/JNEUROSCI.22-12-04833.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weast RC, editor. Handbook of Chemistry & Physics. 51st. The Chemical Rubber Company; Cleveland, OH: 1970. [Google Scholar]
- 59.Jarnum S, Lorenzen I, Skinhoj E. Neurology. 1964;14:703. doi: 10.1212/wnl.14.8_part_1.703. [DOI] [PubMed] [Google Scholar]; Ganshirt H. Wien Med Wochenschr. 1966;116:953. [PubMed] [Google Scholar]
- 60.Zaharchuk G, Martin AJ, Rosenthal G, Manley GT, Dillon WP. Magn Reson Med. 2005;54:113. doi: 10.1002/mrm.20546. [DOI] [PubMed] [Google Scholar]; Zaharchuk G, Busse RF, Rosenthal G, Manley GT, Dillon WP. Proc Intl Soc Mag Reson Med. 2005;13:66. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.