Abstract
Generating induced pluripotent stem cells (iPSCs) requires massive epigenome reorganization. It is unclear whether reprogramming of female human cells reactivates the inactive X chromosome (Xi), like in mouse. Here we establish that human (h)iPSCs derived from several female fibroblasts under standard culture conditions carry an Xi. Despite the lack of reactivation, the Xi undergoes defined chromatin changes, and expansion of hiPSCs can lead to partial loss of XIST RNA. These results indicate that hiPSCs are epigenetically dynamic and do not display a pristine state of X-inactivation with two active X’s as found in some female human embryonic stem cells. Furthermore, while fibroblasts are mosaic for the Xi, hiPSCs are clonal for the Xi. This non-random pattern of X chromosome inactivation in female hiPSCs, which is maintained upon differentiation, has critical implications for clinical applications and disease modeling, and could be exploited for a unique form of gene therapy for X-linked diseases.
INTRODUCTION
Induced pluripotent stem cells (iPSCs) have generated enormous interest as they provide a source of patient-specific cells for regenerative medicine. iPSCs are obtained from differentiated mouse or human cells by forced expression of a few transcription factors related to pluripotency, most commonly consisting of Oct4, Sox2, c-Myc, and Klf4 (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). It is recognized that iPSCs are functionally and molecularly very similar to embryonic stem cells (ESCs) (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007). Hence, mouse (m)iPSCs, like mESCs, can differentiate into all three germ layers in vitro or in the teratoma assay in vivo, and even give rise to animals entirely derived from these cells (Boland et al., 2009; Zhao et al., 2009). Due to ethical concerns, it is impossible to judge the extent of reprogramming of human (h)iPSCs by more stringent assays of pluripotency such as chimerism or tetraploid complementation, highlighting the importance of indirect methods to assay the character and quality of these cells. Here we focus on the epigenetic status of the somatically silenced X chromosome in female hiPSCs to better understand the equivalency of hESCs and hiPSCs and to compare the human and mouse reprogramming processes.
In female cells, one of the two X chromosomes is transcriptionally silenced through a process referred to as X-chromosome inactivation (XCI). XCI, best studied in the mouse system, is developmentally regulated and initiated when female mESCs or their in vivo equivalents of the blastocyst are induced to differentiate (Payer and Lee, 2008). In the embryo, XCI is random such that approximately half of the cells inactivate the maternally inherited X chromosome and the other half the paternal X. Initiation of XCI is absolutely dependent on the large non-coding RNA, Xist, encoded on the X-chromosome (Marahrens et al., 1997; Wutz and Jaenisch, 2000). Upon induction of differentiation, Xist is exclusively upregulated the future inactive X chromosome (Xi) and spreads along the chromosome in cis to initiate silencing. Silencing of the X is accompanied by exclusion of Polymerase II and activating chromatin marks followed by sequential accumulation of repressive chromatin marks (Payer and Lee, 2008). Coating of the Xi by Xist RNA and the heterochromatic state are then maintained for the lifetime of the organism. Intriguingly, upon reprogramming of somatic cells from female mice, the Xi is reactivated, such that miPSCs, just like mESCs, carry two active X chromosomes (Xa’s) that are competent for random XCI upon induction of differentiation (Maherali et al., 2007). Reactivation of the Xi is a late event in the reprogramming process, mirroring the activation kinetics of endogenous pluripotency genes such as Nanog and Oct4 (Stadtfeld et al., 2008), albeit the exact relationship between Xi-reactivation and establishment of the pluripotency transcription network remains unclear. Thus, two Xa’s appear to be a critical determinant of the pluripotent state of mESC/iPSCs.
Unlike mESCs, female hESC lines display a highly variable epigenetic status of the X chromosome, even differing for the same hESC line at different passages, under varying culture conditions, or among sub clones (Adewumi et al., 2007; Dhara and Benvenisty, 2004; Hall et al., 2008; Hoffman et al., 2005; Lengner et al., 2010; Shen et al., 2008; Silva et al., 2008). While a few female hESC lines can, at least partially, carry two Xa’s and undergo random XCI upon differentiation, many lines display complete XCI in the undifferentiated state with an XIST RNA- coated, heterochromatic Xi. Other hESC lines contain an Xi that lacks XIST expression and XIST RNA-dependent repressive chromatin marks and do not reactivate XIST upon differentiation (Silva et al., 2008). It remains unclear whether hiPSCs display the variable states of XCI described for hESCs, and whether reactivation of the Xi seen in mouse reprogramming is recapitulated during the generation of hiPSCs. Resolving these questions will be critical for our understanding of the epigenetic equivalency between hESCs and hiPSCs, and could impact on the use of hiPSCs in disease modeling and clinical applications. To address the XCI status upon human cell reprogramming, we analyzed XIST and X-linked gene expression as well as chromatin composition in a large number of female hiPSC lines derived from four independent female fibroblast populations with two viral reprogramming strategies.
RESULTS
Derivation of female hiPSCs using two different viral approaches
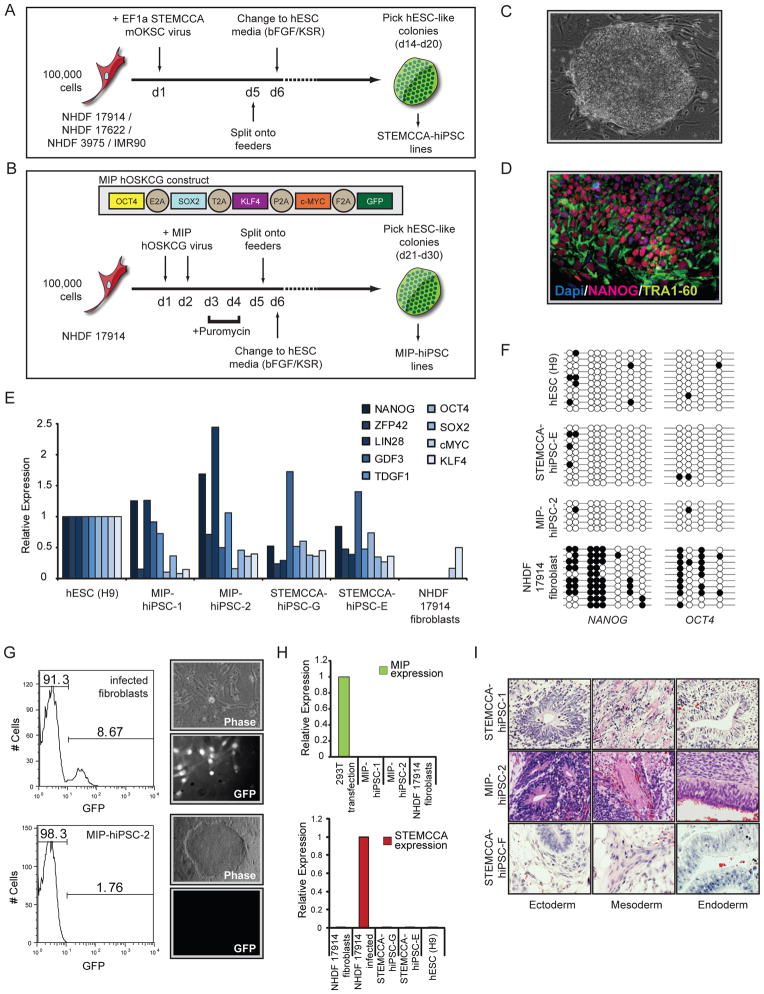
To gain insight into the epigenetic status of the X chromosomes upon reprogramming of female human somatic cells, we reprogrammed four normal fibroblast lines, including one from an embryonic source and three from females of varying ages (Table S1) using a single polycistronic lentiviral vector encoding Oct4, Klf4, Sox2, and c-Myc under control of the constitutive EF1α promoter (termed STEMCCA-vector) (Sommer et al., 2009). According to standard protocols, fibroblast cultures were split onto feeders five days following viral transduction and bFGF-containing hESC media was added on day six (Fig 1A). Dramatic changes in morphology were evident only a few days later and colonies with ES cell- like morphology were clonally expanded into stable cell lines as early as 14 days post infection and will subsequently be referred to as STEMCCA-hiPSC lines. In addition, we generated hiPSC lines from one of the adult female fibroblast populations using a new polycistronic retroviral vector encoding human OCT4, SOX2, KLF4, c-MYC, as well as GFP, separated by 2A sequences, and a puromycin resistance gene following an IRES element (called MIP-vector, Fig 1B). The only deviation from the prior reprogramming protocol was a brief addition of puromycin to the culture media to select successfully transduced fibroblasts. Colonies arose as early as 21 days post infection and lines are referred to as MIP-hiPSCs (Fig 1B).
Figure 1. Establishment of female hIPSCs using lenti- and retroviral polycistronic reprogramming cassettes.
(A) Schematic representation of reprogramming experiments performed with the STEMCCA lentiviral vector. This vector encodes the mouse reprogramming factors in the order of Oct4 (O), Klf4 (K), Sox2 (S), and cMyc (C). The four starting female human fibroblast lines are listed.
(B) As in (A) for the MIP- retroviral reprogramming approach with fibroblast line NHDF17914. The MIP-vector encodes the human reprogramming factors in the indicated order.
(C) Phase-contrast image of a representative STEMCCA-hiPSC colony.
(D) TRA1-60 (green) and NANOG (red) immunostaining of STEMCCA-hiPSC line E. Nuclei were detected with Dapi (blue).
(E) Graph summarizing real-time PCR data for transcript levels of endogenously expressed pluripotency genes in indicated cell lines. All values are relative to GAPDH expression.
(F) Bisulfite sequencing of promoter regions of NANOG and OCT4 in indicated cell lines.
(G) Analysis of MiP-based GFP expression at different steps of the reprogramming process. FACS analysis of GFP levels in MIP-retrovirally infected fibroblasts before puromycin selection demonstrating approximately 9% infection efficiency. Phase-contrast and GFP fluorescence images to the right indicate that almost all cells express GFP at day 5 of reprogramming upon puromycin selection. Resulting hiPSCs do not express GFP as shown in the GFP fluorescence image and the FACS analysis of MIP-hiPSC line 2 in the bottom row.
(H) Real-time PCR for ectopic expression from the MIP- (top) or STEMCCA- (bottom) reprogramming cassettes in indicated cell lines, relative to GAPDH expression.
(I) Teratoma formation with differentiation into all three germ layers in three representative hiPSC lines. See Fig S1 for additional data.
Multiple STEMCCA- and MIP-hiPSC lines (total of 30) were stably expanded and demonstrated typical characteristics of human pluripotent stem cells (Table S1). In addition to their hESC-like morphology (Fig 1C), they were strongly positive for NANOG and TRA1-60 in immunostainings (Fig 1D and Table S1) and re-expressed the endogenous copies of OCT4 and SOX2 and other key pluripotency genes (Fig 1E), indicating successful reactivation of crucial pluripotency markers, which was corroborated by bisulfite sequencing experiments that demonstrated the demethylation of the NANOG and OCT4 promoter regions (Fig 1F). As expected from these results, we observed silencing of the polycistronic MIP- and STEMCCA- expression cassettes in hiPSC clones compared to infected fibroblasts (Fig 1G/H) and residual ectopic expression progressively decreased upon continued passaging of the lines (Fig S1A). All analyzed hiPSC lines were karyotypically normal (Fig S1B) and had a genome-wide expression profile highly similar to that of hESCs as they clustered together closely with hESCs and apart from their fibroblast origin (Fig S1C). In directed in vitro differentiation assays, these cells generated neural progenitors (ectoderm), fibroblasts (mesoderm), and hepatocytes (endoderm), which were confirmed by immunostainings and genome-wide expression (Fig S1D/E and data not shown), and differentiated in vivo into teratomas with discernable elements of all embryonic germ layers (Fig 1I). Thus, our STEMCCA- and MIP-hiPSC lines met the most stringent criteria for pluripotency, an important prerequisite for the subsequent analysis of the XCI status.
XIST RNA coats one X chromosome in undifferentiated female hiPSC lines
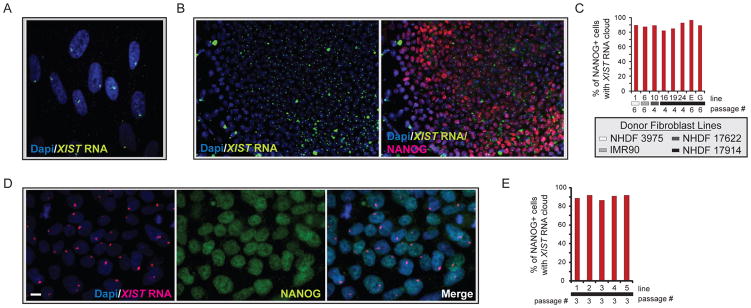
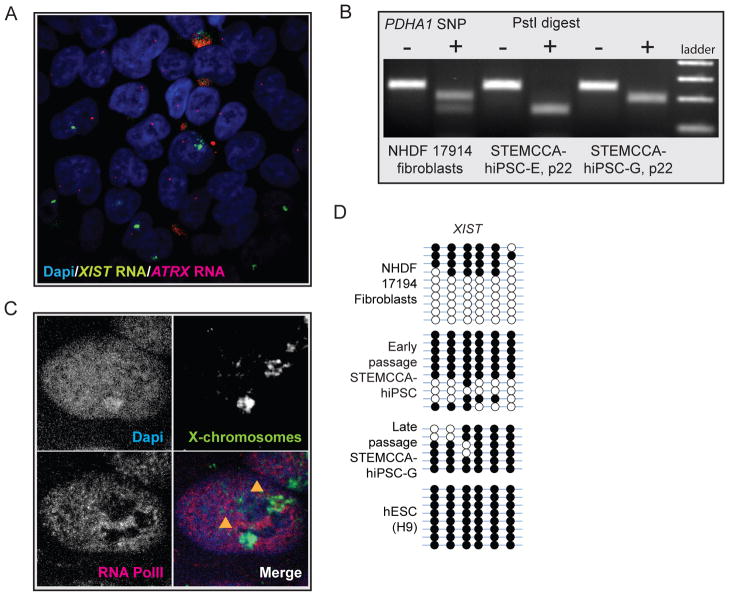
XIST RNA, the key molecule of XCI, is highly transcribed from the Xi and coats the chromosome in female fibroblasts, which can be visualized by RNA fluorescence in situ hybridization (FISH) as a large nuclear area enriched for the RNA signal (Fig 2A). To determine the XCI status upon successful reprogramming, we first characterized various STEMCCA-hiPSC lines for XIST RNA expression and localization. A single XIST RNA-coated chromosome occurred in around 88% of NANOG-positive cells in each hiPSC line (Fig 2B/C, and Table S1), closely resembling the pattern found in the starting female fibroblast populations. This result was confirmed with the MIP-hiPSC lines (Fig 2D/E). XIST RNA is released from the Xi during mitosis, potentially explaining the lack of RNA enrichment in some of the NANOG-positive cells (Clemson et al., 1996). High-level expression of XIST in all female lines was confirmed by expression arrays and, as expected, not found in male control cells (Fig S2). The presence of an XIST RNA- coated X chromosome suggested that female hiPSCs have an Xi independent of the fibroblast source or the type of virus used to overexpress the reprogramming factors.
Figure 2. Female hiPSCs carry a single XIST RNA- coated chromosome.
(A) FISH for XIST RNA (green) in the Dapi-stained fibroblast line NHDF 17430. The accumulation of XIST RNA on the Xi can be seen as focal nuclear enrichment of the RNA signal.
(B) FISH for XIST RNA (green) in the STEMCCA-hiPSC line E, merged with Dapi in the left image and overlaid with an immunostaining for NANOG (red) in the right image.
(C) The graph depicts the percentage of NANOG-positive cells with a single Xi-like accumulation of XIST RNA for various STEMCCA-hiPSC lines. Fibroblast origin is marked with gray scale indicators and passage of hiPSCs is indicated.
(D) As in (B) with NANOG in green and XIST RNA in red for MIP-hiPSC line 1. Scale bar denotes 10um.
(E) As in (C) for various MIP-hiPSC lines. See Fig S2 for additional data.
The XIST RNA- coated chromosome is silent in female hiPSCs
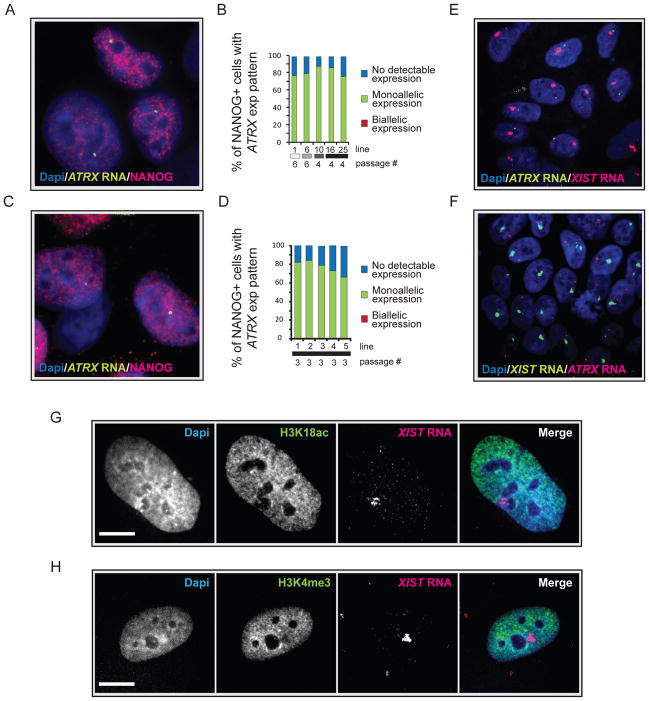
It is possible that XIST RNA expression and its coating of the chromosome are uncoupled from XCI in hiPSCs. To determine the expression state of the XIST RNA- associated chromosome, we carried out RNA FISH for the X-linked gene ATRX in both MIP- and STEMCCA-hiPSC lines, which allows the detection of nascent transcripts at the site of transcription. The majority of cells displayed a single nuclear spot of ATRX transcripts in NANOG-positive cells, which was never associated with XIST RNA-coated X chromosome, and biallelic expression of ATRX was consistently not observed (Fig 3A–F). The exclusive expression of XIST and ATRX indicated that the XIST RNA-coated chromosome is transcriptionally inactive in hiPSCs. However, to ensure that expression of the ATRX gene, which is located close to XIST on the X, reflects that of the entire X chromosome, we performed XIST RNA FISH in combination with immunostainings for two active histone modifications, acetylation of lysine (K) 18 and methylation of K4 of histone H3, both of which should be excluded from the entire X chromosome area, when it is inactive. Both histone marks were depleted from the area occupied by XIST RNA (Fig 3G/H) and similar experiments demonstrated exclusion of Polymerase II from the XIST RNA coated region (data not shown) confirming that the XIST RNA-coated chromosome is transcriptionally inactive in faithfully reprogrammed hiPSCs.
Figure 3. The XIST RNA- coated chromosome is inactive in female hiPSCs.
(A) NANOG immunostaining (red) combined with RNA FISH for ATRX nascent transcripts (green) in the Dapi-stained STEMCCA-hiPSC line 19. A pinpoint of ATRX marks the site of nascent transcription and occurs from only one X chromosome.
(B) The graph depicts the quantification of NANOG-positive cells with monoallelic, biallelic, or no ATRX expression for various STEMCCA-hiPSC lines at the indicated passage. Fibroblast origin is given as in Fig 2C.
(C) As in (A) for MIP-hiPSC line 1.
(D) As in (B) for various MIP-hiPSC lines.
(E) FISH for XIST RNA (red) and ATRX transcripts (green) in STEMCCA-hiPSC line G at passage 8 with Dapi staining the nucleus. The XIST RNA cloud signal does not overlap with the single ATRX signal.
(F) As in (E) for MIP-hiPSC line 5 at passage 3 with XIST RNA in green and ATRX transcripts in red.
(G) Immunostaining for H3K18ac (green) and FISH for XIST RNA (red) in STEMCCA-hiPSC line E at passage 8. Exclusion of the histone mark can be seen at the site of XIST RNA accumulation and was found in 96% of cells in the population indicating silencing of the entire XIST RNA-coated X in hiPSCs. Scale bar = 5um.
(H) As in (G), except that Xi exclusion of H3K4 me3 was analyzed. Exclusion of this histone mark was found in 92% of hiPSCs. See Fig S3 for additional data.
Previously, it had been found that the single Xa in female and male somatic cells is dosage compensated, doubling its transcription to achieve an X to autosome expression ratio of 1 for both sexes (Gupta et al., 2006; Nguyen and Disteche, 2006). In female mESCs, the X:A expression ratio is enhanced to 1.5 due to the presence of two Xa’s (Lin et al., 2007). Based on our analysis, the calculated X:A ratio in human fibroblasts and female hiPSC lines is close to 1 (Fig S3), consistent with the presence of the Xi in our female hiPSCs. However, it still needs to be determined if hESCs with two Xa’s would have an increased X:A ratio.
XCI is non-random in female hiPSCs
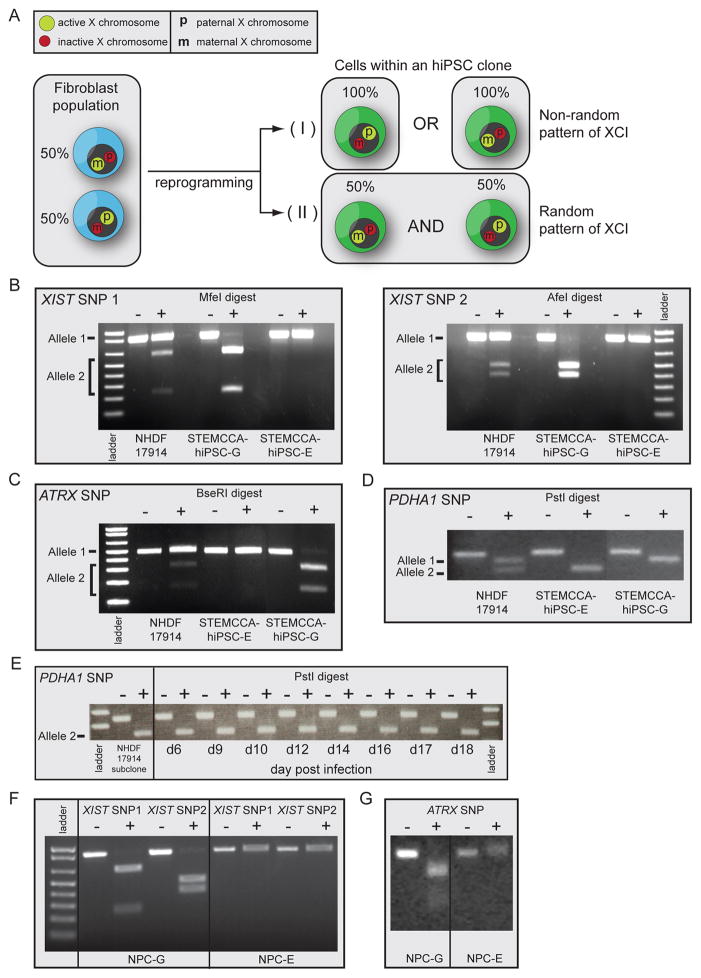
The presence of an Xi in female hiPSCs raised the question of which of the two X chromosomes is inactive in these cell lines. During mouse and human embryonic development, XCI results in silencing of one X chromosome in every female somatic cell, but the selection of paternal versus maternal X chromosome for inactivation is an independent and random event in each cell (Payer and Lee, 2008). Therefore, cells in the fibroblast population are thought to be mosaic for the Xi (Fig 4A, left). To test whether hiPSCs also have a random pattern of XCI (Fig 4A–II) or are clonal, with one of the two X chromosomes exclusively being inactive (Fig 4A–I), we identified single nucleotide polymorphisms (SNPs) that led to differential restriction enzyme digest patterns between the two X chromosomes, by sequencing of relevant genomic regions in one female fibroblast line (NHDF17914). SNPs were selected within exons of XIST and ATRX, and the 3’UTR of PDHA1, encoded on the distal tip of the X chromosome about 53Mb away from XIST, to address the allele-specific expression status along the X (Fig S4A). As expected, random XCI was found in the starting fibroblast population by detection of transcripts from both alleles of XIST, PDHA1, and ATRX (Fig 4B/C/D, Fig S4B). In contrast, only transcripts from one allele of XIST, PDHA1, and ATRX were detected in each NHDF17914-derived hiPSC line (Fig 4B/C/D, Fig S4B) indicating that all cells in a given hiPSC line exclusively express one of the two X chromosomes. Interestingly, different hiPSC lines derived from the same fibroblast population can differ in which X chromosome displays inactivation as they express opposite XIST, ATRX, and PDHA1 alleles (Fig 4B/C/D - compare STEMCCA-hiPSC lines G and E; Fig S4B for different MIP-hiPSC lines). This mutually exclusive, non-random pattern of XCI in hiPSCs obtained from one fibroblast population (Fig 4A–I) suggests that each hiPSC line is derived from a single fibroblast in the donor cell population without intermittent reactivation of the Xi, and that the XCI status in iPSCs reflects that of the starting fibroblast.
Figure 4. Non-random XCI in female hiPSC lines.
(A) Schematic representation of random XCI in fibroblasts and potential XCI states in female hiPSC lines.
(B) Analysis of allele-specific expression of XIST using restriction enzyme-sensitive SNPs. RNA from NHDF17914 fibroblasts and STEMCCA-hiPSC lines E and G (passages 8–9) derived from this fibroblast population was reverse transcribed and two regions of XIST surrounding different SNPs were amplified by PCR and analyzed with and without the respective restriction digest as indicated. SNPs were chosen such that the PCR product derived from the two X alleles are differentially cleaved as indicated. Note that fibroblasts express both XIST alleles, while hiPSC line G exclusively expresses one and line E the other allele.
(C) As in (B) except for allele-specific expression of ATRX.
(D) As in (B) except for allele-specific expression of PDHA1.
(E) A subclone of NHDF17914 fibroblasts obtained upon delivery of a lentiviral shRNA targeting p53 was reprogrammed with the STEMCCA approach and allele-specific expression of PDHA1 determined at different times of the reprogramming process using the same assay as in (D). This fibroblast subclone only expresses allele 2 and does not reactivate allele 1 during reprogramming.
(F) As in (B) but, allele-specific expression of XIST was analyzed upon differentiation of STEMCCA hiPSC lines E and G into neural precursors (NPCs), indicating that non-random XCI is preserved upon differentiation.
(G) As in (F) except for allele-specific expression of ATRX in hiPSC-derived NPCs. See Fig S4 for additional information.
To further substantiate that the Xi does not reactivate during human cell reprogramming, we immortalized NHDF17914 fibroblasts using a constitutive knockdown of p53 and isolated a clonal fibroblast line that only expressed the X carrying allele 2 of PDHA1. We then asked whether allele 1 of PDHA1, located on the Xi, can become expressed during the reprogramming process (Fig 4E). Albeit the sensitivity of the SNP-based expression assay may be limited, allele 1 transcripts were not detected, further supporting the notion that the Xi does not reactivate during the reprogramming process.
Non-random XCI is preserved upon differentiation of iPS cell lines
Since XCI, at least in the mouse system, is tightly linked to differentiation, we next tested whether the non-random pattern of XCI found in undifferentiated hiPSCs is maintained upon induction of differentiation. Two female STEMCCA-hiPSC lines, differing in which X chromosome they express, were differentiated into neural precursors as confirmed by immunostaining (Fig S1D/E). Using our SNP-based method of detection for allelic expression, we found that hiPSC- derived neural precursors displayed the same non-random pattern of XCI that is found in the undifferentiated state (Fig 4F/G). Similarly, we observed conservation of the non-random pattern of XCI when hiPSCs were differentiated into fibroblasts (Fig S4C). Thus, differentiation of hiPSCs does not alter the XCI pattern present in the undifferentiated state.
Changes in Xi chromatin structure during human cell reprogramming
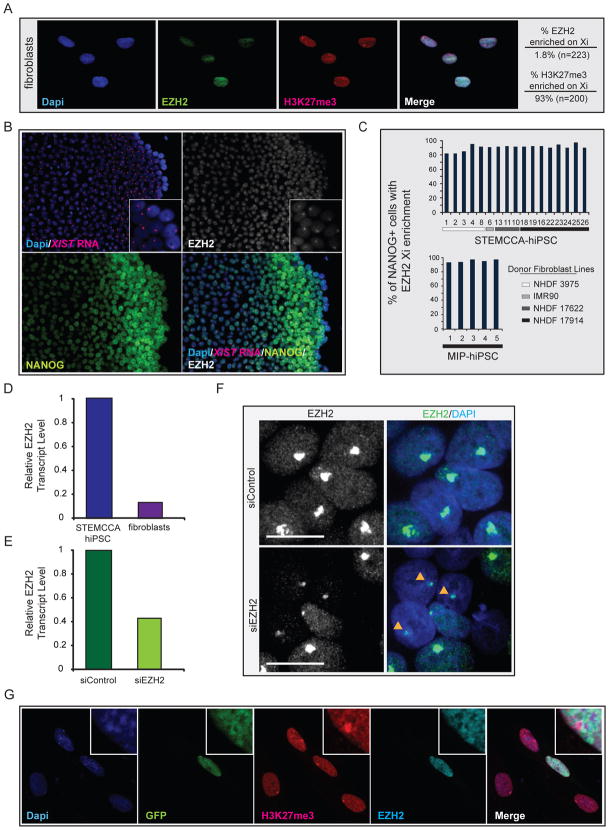
In mouse, XCI during development is associated with a loss of active chromatin modifications and the sequential accumulation of repressive chromatin marks (Payer and Lee, 2008). Accordingly, the chromatin state of the Xi differs at various stages of the XCI process. For example, Polycomb Group (PcG) proteins of the Polycomb Repressive Complex 2 (PRC2) complex accumulate immediately upon initiation of XCI to mediate the enrichment of histone H3 K27 trimethylation on the Xi (Plath et al., 2003; Silva et al., 2003). The accumulation of PRC2 on the Xi is lost upon establishment of the inactive state when analyzed by immunostaining, even though high-level H3K27 trimethylation persists on the Xi for the life of the organism. Here, we assayed the status of the PRC2 subunit EZH2 on the Xi in fibroblasts and hiPSCs by immunostaining to determine whether this mark of the Xi changes during reprogramming. As expected, 93% of fibroblasts carried an Xi enrichment of H3K27 trimethylation, but only around 2% of these Xi’s accumulated EZH2 (Fig 5A). Intriguingly, various MIP- and STEMCCA- hiPSC lines consistently displayed an Xi enrichment of EZH2 in an average of 88% of NANOG positive cells (Fig 5B/C). A similar proportion of cells displayed XIST RNA coating in Fig 1 in agreement with the notion that EZH2 recruitment to the Xi is XIST RNA-dependent. Interestingly, the enrichment of EZH2 on the Xi only occurs after NANOG is upregulated during the reprogramming process (data not shown). Therefore, reprogramming appears to return the Xi chromatin to a state resembling the initiation phase of XCI.
Figure 5. Characterization of Xi chromatin in female hiPSCs.
(A) Immunostaining for EZH2 (green) and H3K27me3 (red) in NHDF17914 fibroblasts, nuclei were detected with Dapi. The proportion of cells with an Xi-like accumulation for H3K27me3 and EZH2 are given.
(B) Immunostaining for EZH2 (grey, from far-red channel), NANOG (green) and FISH for XIST RNA (red) in STEMCCA-hiPSC line G at passage 8.
(C) Proportion of NANOG-positive cells with an Xi-like accumulation of EZH2 for various MIP and STEMCCA-hiPSC lines at around passages 4–8. See Fig S5 for additional data.
(D) Real-time PCR data for EZH2 transcript levels in given cell types, normalized to GAPDH.
(E) Real-time PCR data for EZH2 transcript levels in early passage STEMCCA-hiPSCs upon treatment with scrambled siRNA controls (siControl) or siRNAs targeting EZH2 (siEZH2), normalized to GAPDH.
(F) Immunostaining for EZH2 in an early passage STEMCCA-hiPSC line upon transfection of control or EZH2 siRNAs. Cells are mosaic for depletion of EZH2 and arrowheads point to EZH2 Xi enrichment in cells with clear EZH2 knockdown.
(G) Immunostaining for histone H3K27me3 (red) and EZH2 (far-red, pseudocolored in cyan) in fibroblasts two days after transfection of a GFP-EZH2 overexpression plasmid. The zoom in suggests that higher global levels of EZH2 in fibroblasts do not lead to Xi-enrichment of EZH2.
Other chromatin marks were also different between fibroblasts and hiPSCs but were much less dramatically affected, with 43% versus 89% of fibroblasts and hiPSCs, respectively, displaying an Xi-like accumulation of macroH2A1 (Fig S5A), and 3% versus 10% cells carrying strong H4K20 monomethylation (Fig S5B). In contrast, the exclusion of activating histone marks occurred to the same degree between fibroblasts and hiPSC lines (Fig 3G/H and data not shown). Collectively these data indicate that certain aspects of the Xi chromatin state are changed during reprogramming from a pattern typically seen in the maintenance phase of XCI to one that is only found when XCI is initiated during development.
Enrichment of EZH2 was the most pronounced change in Xi chromatin structure during reprogramming and can likely be extended to other PRC2 components. To begin to address the underlying mechanism for this transition in chromatin composition, we first determined the expression level of EZH2 and, in accordance with data in the mouse system, found higher transcript levels of EZH2 in hiPSCs than fibroblasts (Fig 5D), which is also reflected in higher protein levels (data not shown). To test whether differential global EZH2 levels represent a possible explanation for the Xi enrichment of this protein in hiPSCs, we altered its expression levels in fibroblasts and hiPSCs, respectively. Knockdown of EZH2 by RNAi in hiPSCs (Fig 5E) led to a visible reduction of EZH2 protein levels in around 50% of the cells under maintenance of the Xi-like enrichment as determined by immunostaining (Fig 5F). Conversely, raising EZH2 levels in fibroblast by overexpression of an EZH2-GFP fusion protein did not lead to Xi-like accumulation of the protein (Fig 5G). Together, these data suggest that differential EZH2 protein levels between the different developmental stages are not a simple explanation for the divergence in EZH2 Xi enrichment between fibroblasts and hiPSCs.
Prolonged passaging affects Xi characteristics
Our results indicated that female hiPSCs, when first analyzed at around passages 4–10, consistently carried an XIST RNA- coated Xi. We reasoned that that the Xi could be maintained but lose XIST expression, as has been described for some female hESCs upon extended culturing, or still reactivate at later passage or under altered culture conditions. To test this idea, we expanded several hiPSC lines in bFGF- containing media on mouse fibroblast support cells to passage 22 and reanalyzed Xi markers. In some hiPSC lines, XIST RNA expression and EZH2 enrichment on the Xi were maintained in the majority of cells (data not shown). However, we also had lines in which extended culturing led to changes in the XCI status with a loss of the nuclear XIST RNA enrichment in most cells (Fig 6A) and dramatic reduction in XIST transcript levels (Fig S2, see STEMCCA-hiPSCs E and G at passage 22). RNA FISH demonstrated that ATRX continued to be expressed from only one X chromosome in XIST-negative hiPSC colonies (Fig 6A) and our SNP-based expression assay indicated the maintenance of monoallelic expression for PDHA1 (Fig 6B), suggesting that the inactive state of the X is preserved even in the absence of XIST RNA coating. Cytogenetic analysis demonstrated the presence of two X chromosomes allowing us to exclude the loss of one of the two X chromosomes in these cultures as a possible explanation for XIST loss (Fig S1B). In agreement with this finding, X chromosome painting in combination with immunostaining for RNA polymerase II confirmed that one of the two X chromosomes is depleted for the basic transcriptional machinery (Fig 6C). Even though our data indicate the presence of an Xi, it remains possible that loss of XIST may result in partial derepression of the silent chromosomes as previously suggested for hESCs with an Xi that lacks XIST coating (Lengner et al., 2010; Shen et al., 2008).
Figure 6. hiPSC lines are prone to XIST loss upon extended passaging but maintain the inactive state.
(A) RNA FISH for XIST (green) and ATRX (red) transcripts in STEMCCA-iPS line G at passage 16. At this passage, many cells without XIST RNA clouds were found. Note that ATRX continues to be expressed from only one chromosome. At around passage 22, this hiPSC line lost XIST RNA expression and coating in almost all cells (data not shown).
(B) Analysis of allele-specific expression of PDHA1 in STEMCCA-hiPSC lines G and E at passage 22 (upon loss of XIST) using our SNP-based assay. NHDF17914 fibroblast analysis was added as reference for a cell line expressing both alleles of PDHA1. Upon XIST loss, both hiPSC lines continue to express only the PHDA1 allele that was active at earlier passage (compare with Fig 4D) and do not reactivate the other allele.
(C) X chromosome paint (green) in STEMCCA-hiPSC lines G at passage 22 to detect the presence of two X chromosomes. The combination of the X-paint with RNA polymerase II immunostaining (red) illustrated that one of the two X chromosomes labeled with arrowheads is completely excluded from RNA polymerase II staining indicative of an Xi.
(D) Bisulfite sequencing of promoter region of XIST in indicated cell lines. Half of the clones were completely methylated in female fibroblasts and early passage STEMCCA-hiPSC in agreement with the notion that XIST is silenced on the Xa and actively expressed from the Xi. Complete methylation was found in STEMCCA-hiPSC line G at passage 22, which has lost XIST expression, and our H9 hESC culture, which also lacks XIST expression but contains an Xi (data not shown).
Bisulfite sequencing of the XIST promoter region implicated DNA methylation in the silencing of XIST as cells lacking XIST expression from the Xi exhibited complete methylation, while cells expressing XIST from the Xi, like female fibroblasts or early passage hiPSCs, were characterized by demethylation of 50% of the clones (Fig 6D). In agreement with the XIST-dependence of certain chromatin marks, we found that loss of XIST RNA from the Xi in hiPSCs was accompanied by the loss of the Xi accumulation of EZH2, macroH2A, and H4K20me1 (data not shown), suggesting that XIST-independent chromatin marks must keep the X silent in cells that lack XIST expression. Collectively, these data suggest that adaptation to culture conditions can affect the epigenetic state of the Xi in hiPSCs.
Development of female Duchenne Muscular Dystrophy (DMD)-hiPSC lines
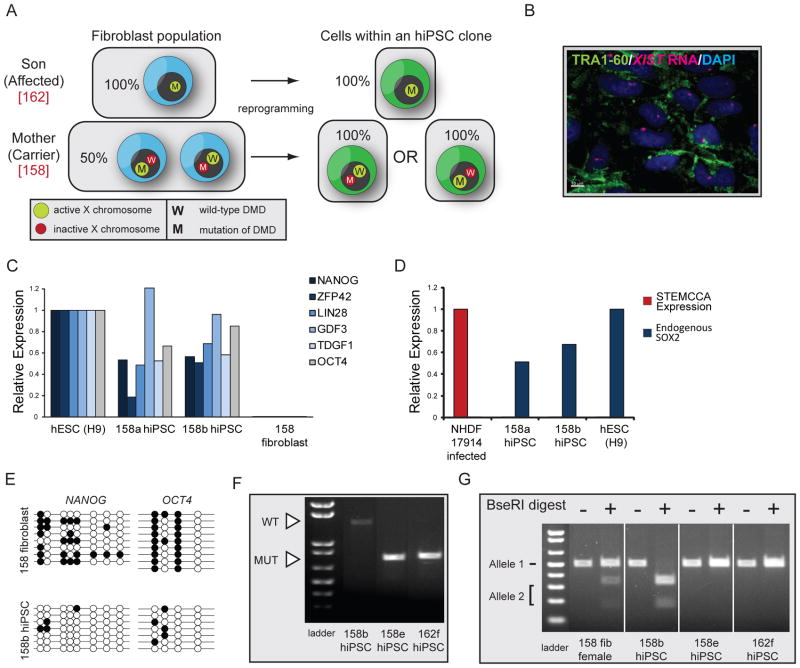
Because the finding of nonrandom XCI in female hiPSCs could be exploited for studies of X-linked diseases (see Discussion), we finally generated hiPSC lines from heterozygous females that are carriers of an X-linked disease. Duchenne Muscular Dystrophy (DMD) is an X-linked recessive disease that arises from mutations in the DYSTROPHIN gene (Hoffman et al., 1987). DMD occurs at an incidence of 1 in 3,500 live male births and is the most common and severe muscular disorder in childhood. To test if we can generate hiPSCs expressing either the wild-type or mutant form of DYSTROPHIN from a female carrier of the disease (Fig 7A), fibroblasts from a 47-year-old female that is clinically unaffected but heterozygous for a deletion of exons 46–50 of the DYSTROPHIN gene (EX46-50DEL; DMD fibroblast line 158), were reprogrammed using the STEMCCA approach. For control purposes, we also generated hiPSCs from fibroblasts from the 13-year-old son that is clinically affected with DMD and carries the same deletion (DMD fibroblast line 162). An initial characterization of stably expanded male and female DMD-hiPSC clones, referred to as 162 and 158, respectively (6 female and 4 male lines; Table S1), demonstrated typical characteristics of pluripotent stem cells: morphology similar to hESCs (data not shown); expression of TRA1-60 and other pluripotency genes (Fig 7B/C); silencing of the ectopic STEMCCA reprogramming cassette (Fig 7D); demethylation of the OCT4 and NANOG promoter regions (Fig 7E); suggestive of complete reprogramming. As expected, all female DMD-hiPSC lines also carried an XIST-RNA coated X chromosome confirming our original results (Fig 7B).
Figure 7. Female DMD hiPSCs exclusively express the mutant or wildtype allele of DYSTROPHIN.
(A) Schematic representation of the expected result in the reprogramming experiment with male and female DMD fibroblasts using in the STEMCCA approach.
(B) TRA1-60 (green) immunostaining and FISH for XIST RNA (red) in a representative female DMD-hiPSC line at passage 3. Nuclei were detected with Dapi (blue).
(C) Graph summarizing real-time PCR data for transcript levels of endogenously expressed pluripotency genes in indicated cell lines, relative to GAPDH expression.
(D) As in (C) for ectopic expression from the STEMCCA cassette and endogenous levels of SOX2.
(E) Bisulfite sequencing of promoter regions of NANOG and OCT4 in female DMD fibroblasts and the descendant hiPSC clone 158b.
(F) PCR analysis of DMD expression in female and male DMD-hiPSC clones. The wild-type and mutant alleles of DYSTROPHIN can easily be distinguished by size (1411 bp versus 716 bp). As expected, the male line (162) expressed only the mutant allele and only one of the two DYSTROPHIN alleles was transcribed in each female hiPSC line.
(G) SNP-based analysis of allele-specific expression of ATRX in female DMD fibroblasts, two descendant hiPSC clones (158b and 158e), and one male DMD hiPSC line (162f). Both ATRX alleles are present in fibroblasts, but only one can be detected in each female hiPSC line.
To test whether female hiPSCs expressed either the wild-type or mutant form of DYSTROPHIN, we took advantage of the two facts that the gene is expressed at very low levels in human pluripotent cells and that the two forms can be distinguished by PCR fragment length polymorphisms. We found that the female DMD-hiPSC line 158b expressed the wild-type but not the mutant form of DYSTROPHIN, while only the mutant allele could be detected in another line (158e), which, as expected, was also present in the disease-derived male hiPSC line 162f (Fig 7F). These data suggested that hiPSC lines 158b and 158e express different X chromosomes, a result that could be extended to the other female DMD-hiPSC lines (data not shown). This conclusion was further tested with our SNP-based assay for allele-specific expression of ATRX, which showed that female DMD-hiPSC lines expressed only one of the two ATRX alleles present in fibroblasts (Fig 7G and data not shown for additional lines). Importantly, female DMD-hiPSC lines that expressed different DYSTROPHIN forms also expressed opposite alleles of ATRX confirming that mutually exclusive X chromosomes are expressed among various DMD-hiPSC lines. We conclude that female hiPSCs expressing only the mutant allele of an X‐linked gene can be generated from a female heterozygous carrier of an X‐linked mutation as a result of the clonality of the reprogramming process and lack of Xi reactivation.
DISCUSSION
In this study, we provide the first extensive characterization of the XCI status upon reprogramming of female human somatic cells. We discovered that completely reprogrammed hiPSCs, derived under standard bFGF culture conditions, carry an Xi with XIST RNA coating and classic markers of Xi-heterochromatin. This outcome is consistent among all hiPSC lines generated here, irrespective of the age of the person from which the starting fibroblast population was obtained or the particular viral reprogramming approach applied. Female hiPSC lines display non-random XCI and different lines obtained from the same fibroblast culture differ in which of the two X chromosomes they express. This mutually exclusive, non-random pattern of XCI among hiPSC lines is consistent with the notion that the Xi does not reactivate during reprogramming of human somatic cells and that a single cell from the mosaic fibroblast population that has silenced either the maternally or the paternally inherited X chromosome gives rise to the reprogramming event (model proposed in Fig 4A–I). Upon expansion in culture, female hiPSC cells are prone to lose XIST expression, likely through methylation of its promoter, similar to what has been observed with female hESCs (Hall et al., 2008; Lengner et al., 2010; Shen et al., 2008; Silva et al., 2008). The absence of XIST RNA coating leads to the loss of XIST-dependent chromatin marks but not to the reactivation of the Xi, albeit we can currently not exclude partial de-repression of the X chromosome accompanying XIST loss. A better understanding of the cause of this dynamic epigenetic behavior of hiPSCs will be important, as the XCI status may reflect the global epigenetic status of human pluripotent cells.
In contrast to our findings with mouse reprogramming, which showed that female miPSCs consistently carry two Xa’s, our current analysis did not reveal any sign of complete reactivation of the somatically silenced X chromosome in hiPSCs, indicating that the reversal of XCI is not required for human cell reprogramming. Even though mouse reprogramming leads to the reactivation of the Xi in a late step of the reprogramming process, it remains to be determined how the heterochromatin of the Xi is transformed into euchromatin, and whether this process is essential for the establishment of the pluripotent state in mouse. Addressing these questions will help to reveal why human cell reprogramming does not include the reactivation of the Xi.
However, one possible explanation for the XCI differences of mouse and human reprogramming could be that the epiblast progenitor cells of the human blastocyst, in contrast to those of the mouse blastocyst, do not carry two Xa’s. The X chromosome activation status in human blastocysts has not been firmly established and direct evidence that, as in mouse, both X chromosomes are transcriptionally active in epiblast cells of the blastocyst is still lacking (van den Berg et al., 2009). Thus, it is possible that the Xi/Xa pattern found in hiPSCs and most hESCs reflects the state of the X chromosomes found at this developmental stage in vivo, which would suggest a dramatic difference in the developmental regulation of X chromosome activity between mouse and human embryos. However, a recent report described the derivation of hESCs with two Xa’s from blastocysts under physiological oxygen conditions (Lengner et al., 2010). The same study nicely demonstrated that hESCs with two Xa’s initiate XCI in response to different kinds of cellular stress, including exposure to atmospheric oxygen levels, possibly explaining why most hESC lines cultured in labs to date carry an Xi. Thus, it appears likely that the epiblast progenitor cells of the human blastocyst have two Xa’s and that cellular stress during derivation and maintenance of hESCs leads to progressive perturbations of this epigenetic state and precocious XCI in the undifferentiated state (Lengner et al., 2010; Silva et al., 2008). Importantly, once XCI is initiated in hESCs, hypoxic conditions or antioxidant treatments do not appear to enable reactivation of the Xi in hESCs (Lengner et al., 2010). Thus, we currently favor the idea that the inability to reproduce the XaXa pattern in hiPSCs may be explained by the irreversibility of existing XCI in standard hESC culture conditions. In agreement with this conclusion, the addition of several antioxidants to established hiPSC lines or during the reprogramming process or reprogramming experiments under hypoxic conditions did not result in reactivation the Xi in our preliminary experiments (data not shown). However, theoretically the failure to completely reprogram our hiPSCs to the hESC state with two Xa’s remains possible. It also remains possible that the presence of the reprogramming cassette could affect the XCI status seen in hiPSCs, even though ectopic expression is strongly downregulated in our hiPSC lines.
Another explanation for XCI differences between the human and mouse reprogramming may be that hESC/iPSCs represent a developmentally more advanced pluripotent state than mESC/iPSCs. Accordingly, hESC/iPSCs are thought to be in a primed pluripotent state, while mESCs represent the naïve pluripotent state (Nichols and Smith, 2009). Differences in culture conditions for these two pluripotent stages with hESC/iPSCs requiring bFGF signaling and mESC/iPSCs depending on the LIF/STAT pathway, may be tightly associated with their different XCI states. Intriguingly, a more hESC-like stem cell population has been derived from the mouse post-implantation epiblast (EpiSCs) and displays an Xi/Xa pattern with Xist RNA coating (Guo et al., 2009; Nichols and Smith, 2009). Conversely, it has also been suggested recently that a more naive pluripotent state with two Xa’s can be established in hESCs by histone deacetylase inhibition (Ware et al., 2009) or constant overexpression of KLF4 in combination with small molecule cocktail that supports mESC growth (Hanna et al., 2010). Additional studies are needed to reveal the origin of the epigenetic differences found between mouse and human ESC/iPSCs and the epigenetic state of the X chromosome may serve a valuable tool for elucidating these differences.
hiPSCs have garnered great interest in part because disease-specific hiPSCs, which theoretically can form any cell type of the human body, offer an unprecedented opportunity to examine disease states and may form the basis for drug development approaches. To date, hiPSCs have been generated from patients with a variety of genetic diseases including ALS, SMA, Parkinson disease, Huntington disease, juvenile-onset, type 1 diabetes mellitus, and Down syndrome (Dimos et al., 2008; Ebert et al., 2009; Park et al., 2008), and should also be invaluable for studies of X-linked diseases (Agarwal et al., 2010; Urbach et al., 2010). Our finding that female hiPSCs exhibit non-random XCI should have implications for the use of female hiPSCs with X-linked mutations in clinical applications or disease studies. Devastating X-linked genetic diseases include for example fragile X syndrome (mutation in FMR1), a-thalassemia (ATRX), Rett Syndrome (MECP2), Coffin-Lowry Syndrome (RSK2), DMD, Lesch-Nyhan syndrome (HPRT), and Wiskott-Aldrich Syndrome (WASP). Furthermore, approximately 11% of X-linked genes are implicated in mental retardation (Gecz et al., 2009). It has long been recognized that XCI has important consequences for the clinical phenotype of X-linked diseases between the two sexes. The cellular mosaicism with respect to XCI is often advantageous for womens’ health, ameliorating the deleterious effects of X-linked mutations and leading to less severe manifestations of the disease compared to males. However, there are X-linked mutations that are often lethal in males in utero and severely affect females (for example Rett Syndrome and Incontinentia Pigmenti), or that manifest in females even in the heterozygous case (X-linked Adrenoleukodystrophy). Our finding that each female hiPSC line, in contrast to the starting fibroblast population, is clonal for X-linked gene expression, leads to the prediction that hiPSC lines derived from females with an X-linked disorder will express either the X carrying the mutant or the normal allele, which we were able to confirm with our female DMD-hiPSC derived from a heterozygous carrier. For studies of X-linked diseases with female hiPSCs, one therefore needs to be careful about which X chromosome is expressed, and one could argue that hiPSCs from females with X-linked disorders thereby represent the perfect pair of control (expressing the normal allele) and experimental (expressing the mutant allele) cell types for investigation of the disease phenotype. More importantly, we propose that one could be able to exploit the non-random XCI feature of female hiPSCs for a form of gene therapy, as a mosaic population of cells expressing the mutant and wildtype allele from the female patient, can be turned into a pluripotent, isogenic cell population that carries the mutant allele on the Xi in all cells and thus solely expresses the normal form of the gene.
Experimental Procedures
Reprogramming methods, hiPSC culture, and differentiation
NHDF and IMR90 fibroblast cell lines were obtained from Lonza and ATCC, respectively. DMD-fibroblasts GM05158 and GM05162 were obtained from the NIGMS Human Genetic Cell Repository from Coriell. For STEMCCA reprogramming, approximately 100,000 fibroblast cells were infected overnight with 2–20 ul of concentrated STEMCCA lentivirus (~5×10e8 TU/ml) in 1 mL of standard fibroblast media (DMEM supplemented with 10% FBS, L-glutamine, non-essential amino acids, and penicillin–streptomycin) with 5ug/ml polybrene. Cells were trypsinized and replated onto 10 cm dishes containing mitomycin-treated male CF1 mouse embryonic fibroblasts on day 5 post infection (p.i.) and media was replaced with standard hESC media (DMEM/F12 supplemented with 20% knockout serum replacement, L-glutamine, nonessential amino acids, penicillin-streptomycin, 2-mercaptoethanol, and 20 ng/ml bFGF) the following day and changed every day thereafter. hESC-like colonies were picked between weeks 2–3 p.i. and enzymatically passaged with collagenase IV. Retroviral reprogramming with the MIP vectors (details on the construct can be found in Supplemental Experimental Procedures) was performed similarly, except that cells were double infected with 1 mL of unconcentrated pMIP retrovirus overnight. Media was supplemented with 1 μg/mL puromycin on days 3 and 4 p.i. and hESC-like colonies picked between weeks 3–5 p.i. MIP and STEMCCA viruses were produced using standard procedures. Cytogenetic analysis was performed by Cell Line Genetics (Madison, WI). hiPSCs were specified towards neural progenitors as described (Karumbayaram et al., 2009) and teratoma assays were performed by testis injection following standard procedures.
Immunostaining and FISH
Antibodies used for immunostaining are: H3K27me3 (Upstate 07-449), EZH2 (BD 612666), macroH2A1 (rabbit polyclonal), NANOG (Abcam ab21624), TRA1-60 (Millipore MAB4360), H4K18ac (kindly provided by the Grunstein laboratory, UCLA), H3K4me3 (Abcam ab8580), H4K20me1 (Abcam ab9051). For immunostaining, cells grown on coverslips were washed in PBS and fixed for 10 min at room temperature (RT) in 1× PBS containing 4% PFA. Cells were then permeabilized by incubation with 1× PBS containing 0.5% Triton X-100 for 5 min at RT, transferred into 1× PBS with 0.2% Tween-20 (PBS/Tween), and then incubated for 30 min in blocking buffer (5% goat serum, 0.2% fish skin gelatin, 0.2% Tween in 1× PBS). Primary antibody incubations were performed for 2hrs at RT in blocking solution, cells were washed in three times in PBS/Tween, and incubated with Alexa 488, Alexa 546, or Cy5 labeled secondary antibodies in blocking buffer for 30min. Cells were then washed with PBS/Tween, stained with DAPI, and mounted in Aquapolymount (Polysciences Inc.). When combining FISH and immunostaining, immunostaining was performed as described above with the addition of tRNA (Invitrogen) and RNaseOUT (Invitrogen) to blocking buffer. After immunostaining cells were fixed again with 1×PBS/4% PFA and XIST and ATRX RNA FISH was performed with double strand DNA probes labeled with FITC or Cy3 generated with a Bioprime kit (Invitrogen) from a full-length XIST cDNA construct or BACs for ATRX (RP11-1145J4 and RP11-42M11) following standard procedures. X chromosome paint was performed following standard procedures using probes from Cambio. To quantify cells with Xi enrichment of chromatin marks, XIST RNA, or allelic expression of ATRX, at least 200 single cells were inspected under the microscope and the pattern of enrichment determined. In some quantifications, only NANOG or TRA1-60 positive cells were considered and cases in which fewer cells were counted are indicated.
Expression analysis
In order to confirm expression of the deleted or wild type DMD allele, we used PCR fragment length polymorphisms with primer sequences specific to detect the DMD region between exons 43 to 52 (sequences are available from Dr. van Deutekom; Aartsma-Rus et al., 2003). To identify X-linked SNPs in NHDF17914 and DMD fibroblasts, several regions of XIST, ATRX and PDHA1 were amplified from genomic DNA by PCR, TOPO XL (Invitrogen) cloned and sequenced. Four regions, each carrying a single SNP that confers allele-specific differences in restriction enzyme digests, were found within the coding regions of these three genes (Fig S4). To allow allele-specific expression analysis, cDNA was generated from RNA of different cell lines using the First Strand cDNA synthesis (Invitrogen), SNP-containing regions amplified by PCR with primers given in Table S2, and analyzed by restriction digest with Mfe1, Afe1, BseRI, and Pst1, respectively (Fig S4). Similar cDNA preparations were used in real time PCR experiments (primers given in Table S2) to determine transcript levels of pluripotency or virally encoded genes in triplicate reactions. Global gene expression analysis was performed as described previously (Chin et al., 2009) and data are deposited at NCBI’s Gene Expression Omnibus under accession number GSE22246.
Bisulfite Sequencing
Bisulfite conversion of genomic DNA was performed with the EpiTect Bisulfite Kit (QIAGEN) followed by PCR using HotStarTaq DNA polymerase. Primer sequences are given in Table S2.
EZH2 RNAi and overexpression experiments
siRNAs targeting EZH2 were obtained from Dharmacon. 50nM of siRNA were transfected into hiPSCs with Lipofectamine RNAiMax (Invitrogen) as described by the manufacturer. Cells were fixed and analyzed 72 hours after transfection. For overexpression, EZH2 was cloned into pEGFP-C3 (Clontech), transfected into primary fibroblasts, and analyzed 48 hrs later.
Highlights.
Reactivation of the somatically silent X chromosome is not seen in female human iPSCs.
The chromatin state of the inactive X (Xi) differs between human fibroblasts and iPSCs.
Human iPSCs are prone to lose XIST expression upon extended culture but maintain an Xi.
Human iPSC lines have non-random X-chromosome inactivation (XCI).
Non-random XCI in female human iPSCs will affect studies of X-linked diseases.
Supplementary Material
Acknowledgments
We are grateful to Alissa Minkovsky for critical reading of the manuscript; Dr. Gustavo Mostoslavsky (Boston University) for providing the STEMCCA reprogramming vector; Dr. Donald Kohn (UCLA) for generating concentrated lentiviral STEMCCA preparations; Dr. Michael Grunstein (UCLA) for providing antibodies; Dr. Nissim Benvenisty (Hebrew University) for expression profiling and karyotyping of various hiPSC lines; Dr. Silvia Diaz-Perez (UCLA) for consultation on X chromosome painting; Genevieve Kendall, Rebecca Anderson, Drs. Stan Nelson and Carrie Miceli (ULCA) for providing assistance with DMD-hiPSC RNA isolation and PCR experiments. JT is supported by a training grant of the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, MHC by a Ruth L. Kirschstein Institutional Training Grant (NRSA # T32CA009056), SPS by CIRM TG2-01169, KP by the NIH Director’s Young Innovator Award (DP2OD001686) and a CIRM Young Investigator Award (RN1-00564), and AP by the NIH P30 Supplement-# 3P30AR057230-01S1 and Department of Defense (DOD) funds # 08109003. WEL holds the Maria Rowena Ross Term Chair in Cell Biology and Biochemistry and is supported by CIRM #RS1-00259-1 and the Basil O’Connor Starter Scholar Award of The March of Dimes. This work was also supported by the CIRM New Cell Line grant to JZ, WEL and KP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, van Ommen GJ, van Deutekom JC. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. The Journal of Cell Biology. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhara SK, Benvenisty N. Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 2004;32:3995–4002. doi: 10.1093/nar/gkh746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gecz J, Shoubridge C, Corbett M. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 2009;25:308–316. doi: 10.1016/j.tig.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Butler J, Becker KA, Nelson A, Amit M, Itskovitz-Eldor J, Stein J, Stein G, Ware C, et al. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol. 2008;216:445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Hoffman LM, Hall L, Batten JL, Young H, Pardasani D, Baetge EE, Lawrence J, Carpenter MK. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- Karumbayaram S, Novitch BG, Patterson M, Umbach JA, Richter L, Lindgren A, Conway AE, Clark AT, Goldman SA, Plath K, et al. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells. 2009;27:806–811. doi: 10.1002/stem.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of Pre-X Inactivation Human Embryonic Stem Cells under Physiological Oxygen Concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O'Neill LP, Turner BM. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS biology. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold S, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes & Development. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nature Genetics. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annual Review of Genetics. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Developmental Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining Molecular Cornerstones during Fibroblast to iPS Cell Reprogramming in Mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg IM, Laven JS, Stevens M, Jonkers I, Galjaard RJ, Gribnau J, van Doorninck JH. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet. 2009;84:771–779. doi: 10.1016/j.ajhg.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, Askari B, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Molecular Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, Wang L, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.