Abstract
Brother of the Regulator of Imprinted Sites (BORIS/CTCFL) is an autosomal cancer germline (CG) or cancer-testis antigen gene and paralog of CTCF that has been proposed to function as an oncogene in human cancer via dysregulation of the cancer epigenome. Here we show that genetic disruption of DNA methylation in human cancer cells induces BORIS expression, coincident with DNA hypomethylation and an altered histone H3 modification pattern at the BORIS promoter. Rapid amplification of cDNA ends (RACE) mapping revealed that the transcriptional start site of BORIS in human testis, DNMT deficient human cancer cells, and human epithelial ovarian cancer (EOC) tissues, is similar and lies within the 5' CpG island. The BORIS promoter is repressed by CpG methylation in a dose-dependent fashion, indicating a direct role for DNA methylation in BORIS transcriptional regulation. In human ovarian cancer cell lines, 5-aza-2'-deoxycytidine treatment activates BORIS expression and reduces BORIS promoter DNA methylation. We quantitatively measured BORIS mRNA expression and promoter DNA methylation in normal ovary (NO; n = 10) and epithelial ovarian cancer (EOC; n = 77) and found that, compared to NO, EOC tumors show increased BORIS expression and decreased BORIS methylation. Importantly, BORIS promoter DNA methylation shows a significant inverse correlation with BORIS mRNA expression in EOC (Kendall's Tau = -0.235, P = 0.007, n = 63). These data establish promoter DNA hypomethylation as a mechanism leading to BORIS expression in human ovarian cancer.
Keywords: human, ovarian cancer, BORIS, gene expression, DNA methylation
Introduction
BORIS/CTCFL is a recently described paralog of CCCTC-binding factor (CTCF) (1). While CTCF is a key regulator of genomic imprinting, it is unknown whether BORIS is involved in this process. Unlike CTCF, which is ubiquitously expressed, BORIS expression appears to be restricted to testicular germ cells and human cancer cells (2, 3). The N and C termini of BORIS are distinct from CTCF, suggesting divergent or opposing regulatory roles for the two proteins (2). It has been proposed that BORIS expression in human cancer cells leads to epigenetic deregulation, possibly by disrupting methylation insulator boundaries in the genome normally imposed by CTCF (2, 4). This model implicates BORIS as an oncogene, which is in agreement with its localization to chromosome 20q13.2, a region frequently amplified in human cancer (2).
Based on its expression pattern, BORIS is classified as a cancer-germline (CG), or cancer-testis, antigen (1, 5). Approximately half of the known CG antigen genes are encoded on the X-chromosome (CG-X genes), while the other half are autosomal, including BORIS (5). It has been previously established that CG-X gene expression is regulated primarily at the transcriptional level by DNA methylation (6). We and others have demonstrated that genetic disruption of DNA methyltransferase (DNMT) enzymes by either recombination-based knockout or RNAi-mediated knockdown activates CG-X antigen gene expression in human cancer cells (7-9). DNA hypomethylation at CG-X promoters is associated with an altered histone H3 modification pattern, including increased levels of acetylated lysine 9 (H3K9ac) and dimethylated lysine 4 (H3K4me2), and decreased levels of dimethylated lysine 9 (H3K9me2) (9, 10).
Compared to CG-X genes, far less is known about the molecular mechanisms regulating the expression of autosomal CG antigen genes (5, 11). However, recent evidence suggests that BORIS is regulated by DNA methylation. First, BORIS is expressed in the spermatocytes of the testis, a cell population with reduced 5-methyl-deoxycytidine (5mdC) (1). Second, BORIS expression is induced in normal fibroblasts and tumor cell lines by treatment with the DNMT inhibitor 5-aza-2'-deoxycytidine (DAC) (3, 12, 13). In addition, two groups have reported that BORIS expression in lung cancer, sperm and testis tissues are inversely correlated with promoter methylation (12, 13). However, it should be noted that the interpretation of DAC treatment experiments is complicated by the pleiotropic effect of this agent on gene expression, including the activation of genes by methylation-independent mechanisms (14, 15). In addition, the tissue studies used to link methylation to BORIS expression status were either qualitative and limited to a few CpG sites (12), or utilized only a small number of samples, precluding significance testing (12, 13). Finally, the transcriptional start site of BORIS, which provides the critical context for epigenetic regulation, was not defined in the tissues under study (12, 13).
To resolve these issues, in the current study we have utilized human cancer cells with a genetically engineered deficiency in DNMT enzymes to examine the connection between DNA hypomethylation and BORIS expression. We have also defined the transcriptional start site of BORIS in normal and tumor tissues, as well as in cells with engineered DNA hypomethylation. In addition, we examined whether DNA methylation directly represses the BORIS promoter using a luciferase driven transgene approach. Most importantly, we have developed and utilized quantitative methods of BORIS expression and methylation analyses and applied these to a large set of human ovarian cancer tissue samples. Our data strongly support a role for promoter DNA hypomethylation in driving BORIS expression, particularly in the context of ovarian cancer.
Results
BORIS expression in DNMT deficient human cancer cells
BORIS expression was previously reported to be activated following DAC treatment of human cell lines (3, 12, 13). Because DAC treatment has pleiotropic effects on gene expression and can activate genes independently of DNA methylation changes (14, 16), we sought a genetic approach to mechanistically determine whether DNA hypomethylation is causally linked to BORIS expression. For this purpose, we utilized DNMT genetic knockout cell lines derived from HCT116 (17-19). The genotype of these cell lines is: DNMT1-/-, DNMT3b-/-, DNMT1-/-, 3b-/-, or DNMT3a-/-, 3b-/-. It should be noted that DNMT1-/- cells have recently been found to express an enzymatically competent aberrant DNMT1 splice variant, suggesting that these cells are not functionally null for DNMT1 (20, 21). However, the HCT116 DNMT genetic system still serves as a useful model to assess the relationship between DNMTs, DNA methylation, and gene expression in human cancer cells.
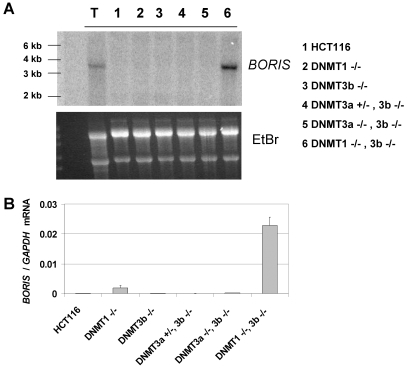
Northern blot analysis of BORIS expression in these various cell lines revealed strong expression of a single BORIS mRNA transcript in DNMT1-/-, 3b-/- cells, but not in the other cell lines (Figure 1A). The size of the BORIS transcript (approx. 3.5 kb) expressed in DNMT1-/-, 3b-/- cells matched the size of the BORIS mRNA detected in testis (Figure 1A). To confirm this finding, we utilized quantitative reverse transcriptase PCR (qRT-PCR) to measure BORIS mRNA expression in the DNMT deficient cell lines. Consistent with Northern blot analysis, qRT-PCR revealed a robust level of BORIS expression specifically in DNMT1-/-, 3b-/- cells (Figure 1B). We also attempted to measure the expression of endogenous human BORIS protein using commercial antibodies and were unable to obtain convincing repeatable results, although overexpressed BORIS protein was detected (data not shown). However, as the present studies are focused on relating DNA methylation to BORIS expression (a transcriptional control mechanism), this was not a significant concern.
Figure 1.
BORIS mRNA expression in DNMT deficient HCT116 cell lines. (A) Northern blot analysis of BORIS in the indicated cell lines. Total RNA from human adult testis tissue (Lane T) was run as a positive control. Ethidium bromide staining of the gel (lower panel) confirmed equivalent RNA loading. (B) Quantitative reverse transcriptase RT-PCR (qRT-PCR) analysis of BORIS in the indicated cell lines. BORIS expression was normalized to GAPDH. Error bars correspond to 1 SD.
BORIS transcriptional start site mapping in different tissue types
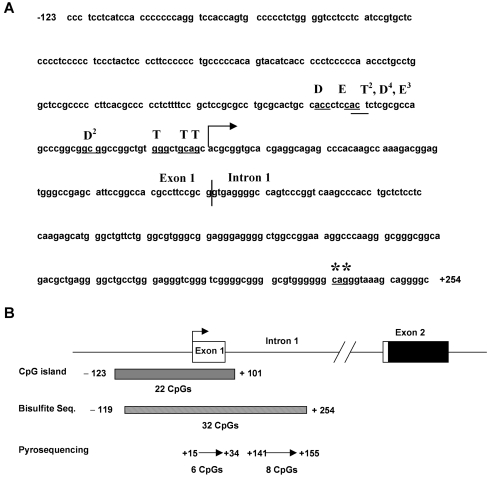
The data presented above suggest that DNMTs play an important role in regulating BORIS expression. It is well established that epigenetic modifications, including DNA methylation and histone code modifications, most crucially impact gene expression when located in the region adjacent to transcriptional start sites (22, 23). Thus, we mapped the 5' end of the BORIS mRNA in testis and DNMT1-/-, 3b-/- cells using the 5' RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) method (24, 25). In addition, we analyzed an epithelial ovarian cancer tissue (EOC#7) that expresses BORIS (see below). Our results, shown in Figure 2A, reveal two major findings. First, the transcriptional start site of BORIS mRNA was very similar in each tissue type examined and was positioned close to, but upstream of, the transcriptional start site predicted by the NCBI and UCSC Genome browsers (Figure 2A). This suggests that the transcriptional start site for BORIS may be controlled by similar mechanisms in normal tissues and during tumorigenesis. Second, the transcriptional start site was variable, consistent with other TATA box-less promoters (27). Notably, the RLM-RACE mapped start site is approximately 230 bp upstream of the originally reported transcriptional start site for BORIS (1) (Figure 2A, asterisks). This difference likely reflects the increased fidelity of RLM-RACE relative to the standard RACE method previously utilized (24, 25). In addition, the observed exon 1-intron 1 splice junction in each cell type examined was identical and coincided with the NCBI prediction (Figure 2A, vertical line). The intron 1-exon 2 boundary was also conserved and in agreement with the genome browser predictions (data not shown), confirming the presence of an 810 bp first intron (Figure 2B). Most importantly, the conserved transcriptional start site of the BORIS mRNA is contained within a CpG island (225 bp, 73% GC, Obs/Exp = 0.78) that overlaps the promoter region, exon 1, and intron 1 (Figure 2B).
Figure 2.
RLM-RACE mapping the 5' end of BORIS mRNA. (A) BORIS transcriptional start site in testis (T), DNMT1-/-, 3b-/- HCT116 cells (D), and epithelial ovarian cancer (EOC) sample #7 (E). Superscripted numbers indicate the number of sequenced clones displaying the indicated 5' end (first three nucleotides underlined). The right arrow shows the NCBI-predicted transcriptional start site. The vertical line indicates the exon 1-intron 1 boundary as mapped by RLM-RACE. Asterisks indicate the transcriptional start site of BORIS in testis reported in (1). (B) Diagram of the 5' region of BORIS. The right arrow displays the NCBI-predicted transcriptional start site, and the filled region in exon 2 shows the start of the open reading frame. EMBOSS CpG plot (26) identified a CpG island (225 bp, 22 CpGs, 73% GC, observed/expected CpG ratio = 0.73) overlapping the transcriptional start site. The regions analyzed by sodium bisulfite sequencing and pyrosequencing, along with the number of CpGs contained within each region, are shown.
Altered epigenetic modifications at the BORIS promoter in DNMT deficient human cancer cells
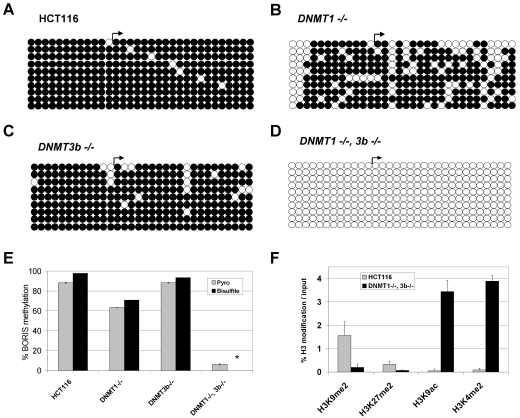
To determine the effect of DNMT gene disruption on epigenetic modifications at the BORIS 5' CpG island, we initially focused on DNA methylation and utilized sodium bisulfite sequencing to analyze a region containing a total of 32 CpG sites (Figure 2B). As expected, in wild-type HCT116 cells this region is heavily methylated (Figure 3A). In DNMT1-/- and DNMT3b-/- cells, methylation levels are reduced or unaffected, respectively (Figure 3, B and C). The low level hypomethylation of the BORIS promoter in DNMT1-/- cells is consistent with a low level of BORIS expression observed in this cell type (Figure 1B). Remarkably, in DNMT1-/-, 3b-/- cells, bisulfite sequencing indicated a complete loss of methylation at the BORIS promoter region (Figure 3D). To facilitate the analysis of BORIS promoter methylation in tissue samples (see below), as bisulfite sequencing is costly and time consuming, we next developed a quantitative pyrosequencing assay to determine the methylation level of the BORIS promoter region (Figure 2B). Pyrosequencing analysis of BORIS methylation in HCT116 cells and DNMT knockout cell lines showed close agreement with sodium bisulfite sequencing (Figure 3E), confirming the validity of our pyrosequencing assay.
Figure 3.
Epigenetic modifications at the BORIS promoter in DNMT deficient HCT116 cell lines. (A-D) The methylation status of the region diagrammed in Figure 2B was determined by sodium bisulfite DNA sequencing. Right arrows indicate the NCBI transcriptional start site. Rows show individually sequenced alleles and filled and open circles represent methylated and unmethylated CpG sites, respectively. (A) Wild-type HCT116 cells. (B) DNMT1-/- HCT116 cells. (C) DNMT3b-/- HCT116 cells. (D) DNMT1 -/-, 3b -/- HCT116 cells. (E) Comparison of BORIS promoter DNA methylation as determined by pyrosequencing and sodium bisulfite DNA sequencing. Pyrosequencing of the BORIS promoter region was performed as described in Materials and methods; the regions analyzed are shown in Figure 2b. The data shown represents the average methylation level of the 14 sequenced CpG sites. For bisulfite sequencing, the percentages indicate the average methylation level of the 32 sequenced CpG sites, taking into account all sequenced alleles. Error bars correspond to 1 SD and the asterisk (*) designates 0% methylation of this sample. (F) Histone H3 modification pattern at the BORIS promoter following DNA hypomethylation. Quantitative ChIP (qChIP) analysis of H3 modifications was performed as described in Materials and methods. Amplification values resulting from no antibody ChIP control were subtracted from the specifically immunoprecipitated DNA before normalizing to 2% input DNA. Mean values from triplicate data points are plotted, along with error bars corresponding to 1 SD.
DNA methylation and histone modifications are epigenetic mechanisms that act in concert to regulate gene expression (28). In particular, modifications of lysine residues on the N-terminal tails of histone H3 play a crucial role in this crosstalk (28, 29). At gene promoters, H3K9me2 and H3K27me2 have been linked to transcriptional repression, while H3K9ac and H3K4me2 have been linked to transcriptional activation (29). We used quantitative chromatin immunoprecipitation analyses (qChIP) to measure the level of each of these modifications at the BORIS promoter, in the context of cells showing DNA hypomethylation. Relative to wild-type HCT116 cells, H3K9me2 and H3K27me2 levels are reduced at the BORIS promoter in DNMT1-/-, 3b-/- cells, while H3K9ac and H3K4me2 levels are elevated (Figure 3F). These changes are consistent with the histone alterations seen at CG-X gene promoters in these cells (9), indicating that autosomal and X-linked CG antigen genes show similar histone modification changes in response to DNMT loss. In contrast, other genomic loci, including heterochromatic regions and constitutively active euchromatin, do not always show these histone modification changes in DNMT1-/-, 3b-/- cells (9), demonstrating a general specificity of this effect to transcriptionally inactive euchromatic regions.
Dose-dependent repression of BORIS promoter activity by DNA methylation
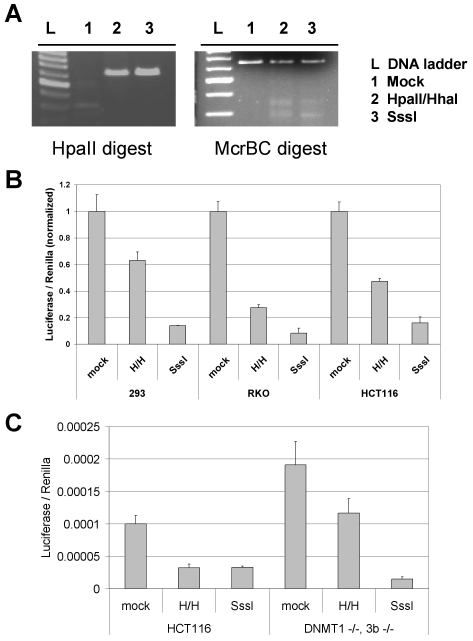
To examine whether DNA methylation directly represses BORIS promoter activity, we sub-cloned a 536 bp fragment of the 5' end of BORIS, which contains 33 CpG sites, into a luciferase reporter construct. We accomplished specific differential methylation of the cloned insert (but not the vector) using SssI methylase (methylation of 33 CpGs), HpaII and HhaI methylases (methylation of 8 CpGs), or no enzyme control (methylation of 0 CpGs) reactions, as described in Materials and methods. We confirmed the outcome of the methylation reactions by performing HpaII (methylation sensitive) and McrBC (methylation specific) enzymatic digests of the reaction products (Figure 4A). As shown in Figure 4B, DNA methylation elicits a dose-dependent inhibition of BORIS promoter activity in the three different mammalian cell lines, demonstrating that DNA methylation directly represses BORIS promoter activity.
Figure 4.
Dose-dependent repression of BORIS promoter activity by DNA methylation. The BORIS 5' promoter region insert was methylated in vitro with no enzyme (mock), HpaII and HhaI, or SssI methylases, then was re-ligated into pGL3-Basic and transfected into cell lines. (A) Confirmation of the methylation status of the BORIS promoter. Following in vitro methylation reactions, BORIS promoter fragments were digested with either HpaII (a methylation-sensitive restriction enzyme) or McrBC (a methylation-specific restriction enzyme). (B) Dual luciferase activity assay measurement of differentially methylated BORIS promoter constructs following transfection into 293, RKO, and HCT116 cell lines. The mean values from triplicate data points are plotted, along with error bars corresponding to 1 SD. Data from each cell line are normalized to the mock-methylated control of the same cell line. (C) Activity of BORIS promoter constructs following transfection into wild-type and DNMT1-/-, 3b-/- HCT116 cells. To illustrate differences in basal activity of the mock methylated constructs in the two cell lines, luciferase values were not normalized to mock-methylated control samples in this instance.
The fact that endogenous BORIS is robustly activated in DNMT1-/-, 3b-/- cells suggests that its expression is controlled by promoter DNA methylation, but does not exclude the possibility that BORIS activation in these cells results from DNA methylation-independent effects. If true, this alternative model predicts that DNA methylation may not block BORIS promoter activity in DNMT1-/-, 3b-/- cells. To address this question, we measured the activity of differently methylated BORIS promoter constructs in DNMT1-/-, 3b-/- cells. DNA methylation led to a dose-dependent repression of BORIS promoter activity in these cells (Figure 4C), supporting the idea that endogenous BORIS activation in these cells is a direct consequence of promoter hypomethylation. Notably, the activity of the mock and partially methylated BORIS promoter constructs were increased two-fold in DNMT1-/-, 3b-/- cells as compared to control HCT116 cells (Figure 4C). This may reflect activation of the BORIS promoter transgene by endogenous BORIS expressed in this cell type (Figure 1), as BORIS appears to activate its own expression (3). Further studies are required to rigorously test this model.
BORIS expression and methylation in ovarian cancer
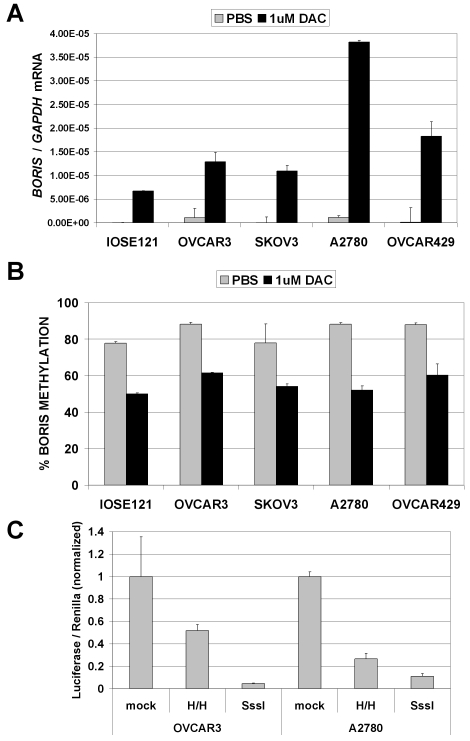
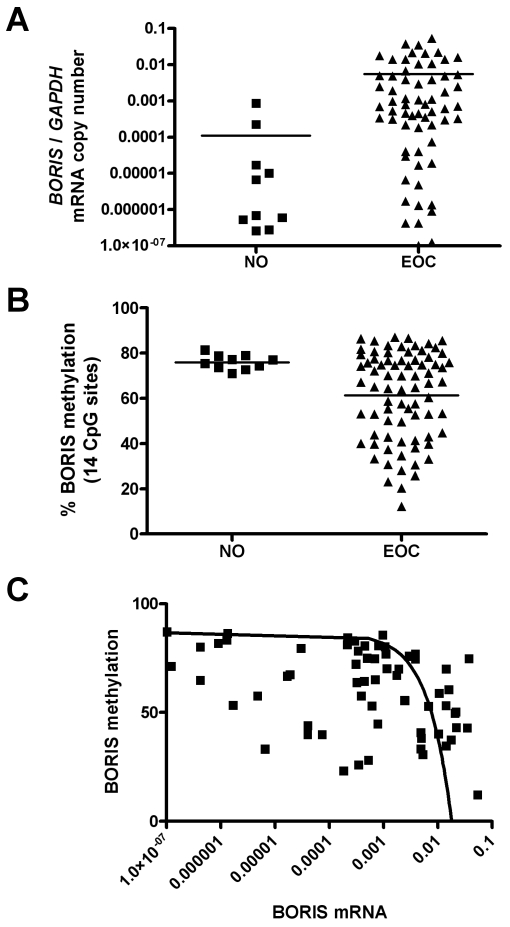
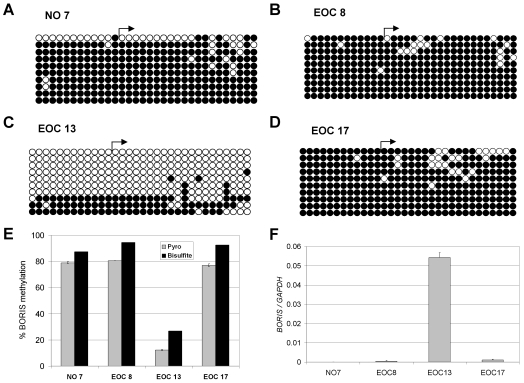
The above data suggest promoter DNA hypomethylation as a potential mechanism leading to BORIS expression in human cancer. To test this hypothesis, we focused our studies on ovarian cancer. Initially, we determined the level of BORIS expression in human ovarian cancer cell lines and immortalized ovarian epithelial cells before and after treatment with DAC. DAC treatment induced BORIS expression in each of these cell lines and, as expected, DAC-mediated induction of BORIS expression coincided with reduced BORIS promoter methylation in each cell type (Figure 5, A and B). As in other mammalian cell lines (see above), methylation of a BORIS promoter construct elicited dose-dependent repression of BORIS promoter activity in ovarian cancer cell lines (Figure 5C), further suggesting a role for DNA methylation in regulating BORIS expression in ovarian tissues. Therefore we next sought to analyze the relationship between BORIS expression and promoter methylation in primary ovarian tissues. For this task, we prepared RNA and genomic DNA samples from normal ovary (NO) (n = 10) and epithelial ovarian cancer (EOC) (n = 77) tissues. Figure 6A shows BORIS mRNA expression in NO and EOC samples as determined by qRT-PCR. BORIS expression is extremely low or undetectable in the NO samples, while it is highly expressed in many EOC tumors (Figure 6A). This difference showed a trend towards, but did not reach statistical significance, likely because of the small number of NO samples available (Figure 6A). We next utilized quantitative pyrosequencing to determine BORIS promoter methylation levels in NO and EOC (Figure 6B). As shown, BORIS promoter methylation levels are uniformly high in NO, but become greatly reduced in numerous EOC tumors; the difference in methylation between the two groups was statistically significant (Figure 6B). To verify the BORIS pyrosequencing methylation data, we conducted sodium bisulfite sequencing analysis on one normal ovary and three EOC tumor samples (Figure 7A-D). These data confirmed close agreement between the two assays, supporting the use of pyrosequencing in the larger data set (Figure 7E). Furthermore, BORIS expression in the analyzed tissues confirmed a close agreement between promoter DNA hypomethylation and BORIS expression (Figure 7F). Finally, we utilized statistical correlation analysis to determine the relationship between BORIS mRNA expression and BORIS promoter methylation in EOC. This analysis revealed a highly significant inverse correlation between the two parameters (Figure 6C), providing clear evidence that BORIS expression is associated with promoter hypomethylation in EOC.
Figure 5.
Regulation of BORIS expression by DNA methylation in ovarian cancer cell lines. (A) The indicated cell lines were treated with 1 µM DAC for 48 hours and RNA was harvested and used to measure BORIS expression by qRT-PCR. BORIS expression was normalized to GAPDH. Triplicate data points are plotted, and error bars correspond to 1 SD. Data are representative of three independent experiments. (B) The indicated cell lines were treated with DAC as in A and BORIS promoter methylation was measured by quantitative pyrosequencing. The mean of quadruplicate data points from two independent experiments are plotted and error bars correspond to 1 SD. (C) Dual luciferase activity assay measurement of differentially methylated BORIS promoter constructs following transfection into OVCAR3 and A2780 ovarian cancer cell lines. The mean values from triplicate data points are plotted, along with error bars that correspond to 1 SD. Data from each cell line are normalized to the mock-methylated control of the same cell line.
Figure 6.
BORIS mRNA expression and promoter methylation in normal ovary and ovarian cancer. (A) BORIS mRNA expression in normal ovary (NO) and epithelial ovarian cancer (EOC) tissues. BORIS expression was measured by qRT-PCR and was normalized to GAPDH. The difference between the two groups was not statistically significant (unpaired, two-tailed t-test; P = 0.1056), although a trend towards increased expression in EOC is apparent. (B) BORIS promoter DNA methylation in NO and EOC tissues. Methylation was measured by quantitative pyrosequencing. The difference between the two groups was statistically significant (unpaired, two-tailed t-test; P = 0.02). (C) Correlation between BORIS mRNA expression and promoter methylation in EOC. There is a highly significant inverse correlation between the two parameters (Kendall's Tau = -0.235, P = 0.007).
Figure 7.
BORIS methylation and expression in normal ovary (NO) and ovarian tumors (EOC). (A-D) The methylation status of the region diagrammed in Figure 2B was determined by sodium bisulfite DNA sequencing for samples (A) NO #7, (B) EOC #8, (C) EOC #13 and (D) EOC #17. Right arrows indicate the NCBI transcriptional start site. Rows show individually sequenced alleles and filled and open circles represent methylated and unmethylated CpG sites, respectively. (E) Comparison of BORIS promoter DNA methylation as determined by pyrosequencing and sodium bisulfite DNA sequencing of the samples shown in A-D. Pyrosequencing of the BORIS promoter region was performed as described in Materials and methods; the regions analyzed are shown in Figure 2B. The data shown represents the average methylation level of the 14 sequenced CpG sites. For bisulfite sequencing, the percentages indicate the average methylation level of the 32 sequenced CpG sites, taking into account all sequenced alleles. Error bars correspond to 1 SD. (F) BORIS expression was measured in the indicated samples using qRT-PCR. BORIS expression was normalized to GAPDH. Triplicate data points are plotted, and error bars correspond to 1 SD.
Discussion
BORIS has been proposed to mediate oncogenic changes to the cancer epigenome (2, 4). Given the key role of epigenetic deregulation in oncogenesis (30), achieving a better understanding of the mechanisms controlling BORIS expression in human cancer is imperative. Here, we present novel information that solidifies the mechanistic link between DNA hypomethylation and BORIS expression in human cancer. Specifically, our studies reveal that genetic disruption of DNA methylation, which bypasses the pleiotropic effects of DAC and similar agents, leads to robust BORIS expression. Additionally, BORIS expression in DNMT deficient cells directly correlates with promoter DNA hypomethylation and an altered histone H3 modification pattern, in a region encompassing the transcriptional start site. These data provide the first characterization of the histone modification pattern at the BORIS promoter. Using a luciferase reporter strategy, we show that DNA methylation directly represses BORIS promoter activity in mammalian cell lines in a dose-dependent manner. Notably, this repression is retained in DNMT deficient cells, implying that endogenous BORIS expression in DNMT deficient cells is a direct consequence of BORIS promoter hypomethylation. Finally, the similar transcriptional start site of BORIS in normal and tumor tissues and deliberately hypomethylated cells suggests that DNA methylation controls BORIS transcriptional activity in multiple tissue types.
BORIS is a paralog of the well-characterized chromatin insulator protein CTCF, which plays a key role in the regulation of genomic imprinting (2). While CTCF is expressed both in cancer and normal somatic tissues, BORIS is only expressed in human cancers and germ cells (2, 3). Aberrant expression of BORIS in human cancer has been proposed to lead to displacement of CTCF from its normal target sites, re-patterning of chromatin insulator boundaries, and widespread epigenetic deregulation (2, 4). This model is supported by the observation that overexpression of BORIS in mammalian cells induces the expression of CG antigens, including BORIS itself and MAGE-A1 (3). Relevant to BORIS auto-regulation, we observed that the unmethylated BORIS promoter construct shows higher activity in DNMT1-/-, 3b-/- cells than wild-type HCT116 cells. These data are consistent with auto-regulation of BORIS, but only when the BORIS promoter is hypomethylated, as the methylated BORIS promoter reporter construct is equivalently silenced in DNMT knockout and wild-type HCT116 cells. Overall, these observations suggest that DNA methylation plays a dominant role in BORIS promoter repression, even in the presence of high levels of endogenous BORIS protein. Thus, in human cancers, hypomethylation of the BORIS promoter may be the critical initiating event leading to BORIS expression.
A role for DNA methylation in regulating BORIS expression was initially suggested by studies of its expression in the murine testis, where BORIS was found to be primarily expressed in pre-meiotic spermatocytes, while CTCF was expressed exclusively in post-meiotic spermatids (1). BORIS expressing spermatocytes were negative for staining with an antibody directed against 5-methylcytosine, implying that BORIS may be expressed as a consequence of global DNA hypomethylation (1). Our data are consistent with this idea, in that we observe that BORIS is robustly induced only in DNMT1-/-, 3b-/- cells, which alone show global genomic DNA hypomethylation (18, 31). However, a recent study of urogenital malignancies found no association between BORIS expression and hypomethylation of the LINE-1 repetitive element (13). It will be relevant to determine whether this lack of association is related to the differences between LINE-1 hypomethylation status and genomic 5mdC levels, or the malignancy under study.
Here we demonstrate that promoter hypomethylation drives BORIS expression in vivo, in the context of ovarian cancer. First, BORIS expression in ovarian cancer cell lines is induced by DAC treatment, coincident with BORIS promoter methylation. Second, primary EOC specimens display increased BORIS expression and decreased BORIS promoter methylation as compared to NO. Most importantly, quantitative expression and methylation studies reveal that BORIS methylation shows a strong inverse correlation with BORIS expression in EOC. These findings define promoter hypomethylation as a key mechanism leading to BORIS expression in human ovarian cancer. Furthermore, these data set the stage for studies investigating the relationship between BORIS expression and methylation in EOC with other critical epigenetic parameters, including CG-X antigen gene expression, global genomic methylation, and tumor suppressor gene hypermethylation.
Abbreviations
- BORIS
Brother of the Regulator of Imprinted Sites
- CG
cancer germline (antigen)
- CG-X
CG (antigen) gene located on the X-chromosome
- DAC
5-aza-2'-deoxycytidine
- DNMT
DNA methyltransferase
- EOC
epithelial ovarian cancer
- NO
normal ovary
- qRT-PCR
quantitative reverse transcriptase PCR
- RLM-RACE
5' RNA ligase-mediated rapid amplification of cDNA ends
Acknowledgements
We thank Drs. Bert Vogelstein and Kurt Bachman (Johns Hopkins University School of Medicine) for providing the DNMT deficient HCT116 cell lines. We thank Dr. Nelly Auersperg and the Canadian Ovarian Tissue Bank for the IOSE121 cells. We thank Dr. Ivan Still of Roswell Park Cancer Institute for providing ovarian cancer cell lines. We thank Dr. Sriharsa Pradhan and Rick Feehery of New England Biolabs for providing ChIP protocols and reagents. This work was supported by grants from the NCI (RO1CA11674) and the Roswell Park Alliance Foundation (to A.R.K.), a Cancer Vaccine Collaborative Grant from the Cancer Research Institute/Ludwig Institute for Cancer Research (to K.O.), and NCI Center grant CA16056 (to Roswell Park Cancer Institute).
References
- 1.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, Cui H, Niemitz EL, Rasko JE, Docquier FM, Kistler M, Breen JJ, Zhuang Z, Quitschke WW, Renkawitz R, Klenova EM, Feinberg AP, Ohlsson R, Morse HC 3rd, Lobanenkov VV. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci U S A. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klenova EM, Morse HC 3rd, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 3.Vatolin S, Abdullaev Z, Pack SD, Flanagan PT, Custer M, Loukinov DI, Pugacheva E, Hong JA, Morse H 3rd, Schrump DS, Risinger JI, Barrett JC, Lobanenkov VV. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 2005;65:7751–7762. doi: 10.1158/0008-5472.CAN-05-0858. [DOI] [PubMed] [Google Scholar]
- 4.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 5.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 6.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koslowski M, Bell C, Seitz G, Lehr HA, Roemer K, Muntefering H, Huber C, Sahin U, Tureci O. Frequent nonrandom activation of germ-line genes in human cancer. Cancer Res. 2004;64:5988–5993. doi: 10.1158/0008-5472.CAN-04-1187. [DOI] [PubMed] [Google Scholar]
- 8.Loriot A, De Plaen E, Boon T, De Smet C. Transient down-regulation of DNMT1 methyltransferase leads to activation and stable hypomethylation of MAGE-A1 in melanoma cells. J Biol Chem. 2006;281:10118–10126. doi: 10.1074/jbc.M510469200. [DOI] [PubMed] [Google Scholar]
- 9.James SR, Link PA, Karpf AR. Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene. 2006;25:6975–6985. doi: 10.1038/sj.onc.1209678. [DOI] [PubMed] [Google Scholar]
- 10.Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res. 2006;4:339–349. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 11.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. http://www.cancerimmunity.org/v4p1/031220.htm [PubMed] [Google Scholar]
- 12.Hong JA, Kang Y, Abdullaev Z, Flanagan PT, Pack SD, Fischette MR, Adnani MT, Loukinov DI, Vatolin S, Risinger JI, Custer M, Chen GA, Zhao M, Nguyen DM, Barrett JC, Lobanenkov VV, Schrump DS. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763–7774. doi: 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann MJ, Muller M, Engers R, Schulz WA. Epigenetic control of CTCFL/BORIS and OCT4 expression in urogenital malignancies. Biochem Pharmacol. 2006;72:1577–1588. doi: 10.1016/j.bcp.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Karpf AR, Moore BC, Ririe TO, Jones DA. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2'-deoxycytidine. Mol Pharmacol. 2001;59:751–757. [PubMed] [Google Scholar]
- 15.Karpf AR, Jones DA. Reactivating the expression of methylation silenced genes in human cancer. Oncogene. 2002;21:5496–5503. doi: 10.1038/sj.onc.1205602. [DOI] [PubMed] [Google Scholar]
- 16.Karpf AR, Lasek AW, Ririe TO, Hanks AN, Grossman D, Jones DA. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine. Mol Pharmacol. 2004;65:18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- 17.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 18.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, Baylin SB, Vogelstein B. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 19.Jair KW, Bachman KE, Suzuki H, Ting AH, Rhee I, Yen RW, Baylin SB, Schuebel KE. De novo CpG island methylation in human cancer cells. Cancer Res. 2006;66:682–692. doi: 10.1158/0008-5472.CAN-05-1980. [DOI] [PubMed] [Google Scholar]
- 20.Egger G, Jeong S, Escobar SG, Cortez CC, Li TW, Saito Y, Yoo CB, Jones PA, Liang G. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci U S A. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Langst G, Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 23.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, Jones PA. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Gorovsky MA. Mapping the 5' and 3' ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res. 1993;21:4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 26.EBI Tools: EMBOSS CpGPlot/CpGReport/Isochore. http://www.ebi.ac.uk/emboss/cpgplot/
- 27.Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 28.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 29.Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 30.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–510. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- 32.Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, Jones DA. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci U S A. 1999;96:14007–14012. doi: 10.1073/pnas.96.24.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
Materials and methods
Cell lines and drug treatments
Wild-type, DNMT1-/-, DNMT3b-/-, and DNMT1-/-, 3b-/- HCT116 colorectal cancer cell lines were generously provided by Dr. Bert Vogelstein (Johns Hopkins University School of Medicine), and were cultured as described previously (31). DNMT3a+/-, DNMT3b-/- and DNMT3a-/-, 3b-/- cells were generously provided by Dr. Kurt Bachman (Johns Hopkins University School of Medicine) and were cultured as described previously (19). The colon adenocarcinoma cell line RKO and the human embryonic kidney cell line 293 were obtained from ATCC (Rockville, MD). RKO cells were cultured as described previously (9), and 293 cells were cultured in DMEM supplemented with 10% FBS, 2 mM L-glutamate, and 0.5% penicillin-streptomycin (Pen-Strep). The ovarian cancer cell lines, A2780, OVCAR3 and OVCAR429, were obtained from Dr. Ivan Still (Roswell Park Cancer Institute) and were grown in DMEM supplemented with 10% FBS, 0.5% Pen-Strep, and 2 mM L-glutamine. The ovarian cancer cell line SKOV3 was obtained from ATCC and was grown in McCoy's media supplemented with 10% FBS, 0.5% Pen-Strep, 2 mM L-glutamine, and 1 mM sodium pyruvate. IOSE121 cells (SV40 immortalized normal human ovarian surface epithelium cells) were a generous gift from the Canadian Ovarian Tissue Bank and Dr. Nelly Auersperg (University of British Columbia). IOSE121 cells were grown in a 1:1 mix of 199 and MCDB105 media containing 5% FBS and 50 µg/ml gentamicin. Ovarian cancer cell lines were treated with 1 µM 5-aza-2'-deoxycytidine (DAC) (Sigma Chemical Company, St. Louis, MO) once, and RNA and DNA samples were harvested 48 hours post-treatment. Total RNA was purified using TRIzol® (Invitrogen, Carlsbad, CA), and genomic DNA was isolated using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN).
Human tissue samples
Normal ovary (NO) and epithelial ovarian cancer (EOC) tissue samples were obtained from patients undergoing surgical resections at Roswell Park Cancer Institute. All samples were collected under an IRB-approved protocol, with appropriate patient consent. Twenty milligrams of tissue were used for RNA extractions. Flash-frozen tissues were homogenized using an electric homogenizer with disposable microtube pestles. Following tissue homogenization, RNA was extracted using TRIzol® reagent (Invitrogen) and 2 µg of each RNA sample was converted to cDNA using random oligo-dT primer (Fermentas, Hanover, MD), and M-MuLV reverse transcriptase enzyme (Fermentas, Hanover, MD). Thirty milligrams of tissue were used for genomic DNA extractions. Flash-frozen tissue samples were crushed using liquid nitrogen pre-chilled mortar and pestles. Upon addition of lysis buffer (Gentra Systems, Minneapolis, MN), tissues were further homogenized with an electric homogenizer. Genomic DNAs were isolated using the Puregene DNA isolation kit (Gentra Systems).
Northern blot analysis
Northern blotting was performed as previously described (32). The BORIS probe was produced by end-point RT-PCR amplification of a region corresponding to the 5' end of BORIS mRNA (not found in CTCF). The primers used for amplification of the BORIS probe were: +67 F: 5'-CCGCGGCCAAGTCATTAT-3' and +633 R: 5'-TCCTCGAGCTTGATCAGTCC-3'.
Quantitative reverse transcriptase PCR (qRT-PCR)
Total RNA and cDNA were prepared and qRT-PCR was performed as described previously (9). Primers for BORIS were as follows: +1012 F: 5'-CAGGCCCTACAAGTGTAACGACTGCAA-3' and +1282 R: 5'-GCATTCGTAAGGCTTCTCACCTGAGTG-3'. GAPDH-specific primers were as reported previously (9).
5' RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) mapping
The transcription start site of BORIS in different tissues was determined using the FirstChoice RLM-RACE kit (Ambion, Austin, TX), according to the manufacturer's instructions. RNA for RLM-RACE analysis was obtained from three sources: human normal testis (Biochain Institute, Inc., Hayward, CA), DNMT1-/-, 3b-/- HCT116 cells, and EOC specimen #7, using TRIzol® reagent (Invitrogen). RLM-RACE allows for the specific amplification of 5' capped RNA, which is found only on full-length mRNA transcripts (25). Briefly, the specific primers used for BORIS amplification (in combination with adaptor-specific primers) were: BORIS outer primer: 5'-TTTGAATTTGAAACTGTGAGAACAA-3' and BORIS inner (nested) primer: 5'-CCTCTAGAGCGTGGAACTGC-3'. As negative controls, non-TAP (tobacco alkaline phosphatase) treated aliquots from the three RNA sources were utilized; in all cases, this did not yield any specific product amplification. In contrast, BORIS-specific PCR products from TAP-containing reactions yielded products of the expected size. PCR products were separated on 2% agarose gels, excised, and subsequently purified using the QIAquick gel extraction kit (Qiagen, Valencia, CA). Gel-purified PCR products were cloned using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA), and individual clones were sequenced using standard methods.
Sodium bisulfite DNA sequencing
Sodium bisulfite sequencing was accomplished as previously described (9). Briefly, DNAs were purified using the Puregene DNA isolation kit (Gentra Systems) and were bisulfite converted using the EZ DNA Methylation Kit (Zymo Research, Orange, CA). Primers for amplification of the BORIS 5' CpG island were: -119 F: 5'-TTTTTTTTAATTTTGTTTGGTTT-3' and +254 R: 5'-TAACAAAACCCCTACTTTACCCTAC-3'.
Pyrosequencing
Genomic DNAs were isolated and bisulfite converted as described above. The methylation status of two regions of the BORIS promoter were analyzed by quantitative pyrosequencing (33). For the first region, the primers used were: (PCR F): -22 5'-GGAGGTAGGTTGTGGGTTGTAGTA-3', (PCR R): +173 5'-biotin-CCCCCCCCTTAAACCTTT-3', (sequencing): +15 5'-TAGAGTTTATAAGTTAAAGA-3'. For the second region, the primers used were: (PCR F) +108 5'-TTTTAAGAGTATGGGTTGTTTTGG-3', (PCR R): +256 5'-biotin-CCTAACAAAACCCCTACTTTACC-3', (sequencing): +141 5'-GAGGGAGGGGTTGGT-3'. PCR cycling conditions were 95˚C for 30 seconds, 60˚C (for region 1) and 61˚C (for region 2) for 30 seconds, and 72˚C for 1 minute, for 45 cycles. The resulting biotinylated PCR product was bound to Streptavidin Sepharose High Performance beads (Amersham Biosciences, Uppsala, Sweden), and the immobilized PCR product was purified using Pyrosequencing Vacuum Prep Tool (Biotage AB, Uppsala, Sweden), denatured with 0.2 M NaOH, and washed using Tris, pH 7.6. Pyrosequencing of the purified single-stranded PCR product was accomplished using the PSQ HS96 Pyrosequencing System (Biotage AB). Non-CpG cytosines served as internal controls to verify efficient sodium bisulfite DNA conversion, and unmethylated and methylated DNAs were also run as controls. Pyrosequencing was performed on duplicate samples, and pyrosequencing assays were performed a minimum of two times.
Quantitative chromatin immunoprecipitation-PCR (qChIP)
The ChIP method utilized was provided courtesy of Dr. Sriharsa Pradhan at New England Biolabs (NEB Simple ChIP Kit, beta test version). Briefly, approximately 2 x 107 cells were fixed to crosslink histones to DNA with 1% formaldehyde at 37˚C for 10 minutes. Nuclease S7 (New England Biolabs, Ipswich, MA) was used to digest DNA to approximately 500-1000 bp fragments, followed by brief sonication (Power 2, single 30 second pulse) with a Branson Sonifier 150 (VWR International, Bridgeport, NJ) to break open the nucleus. The lysate was pre-cleared with Protein G magnetic beads (New England Biolabs) at 4˚C for 30 minutes. The beads were separated using a magnetic separation rack (New England Biolabs), and a fraction of the cleared supernatant served as the input. The remaining supernatant was divided equally to perform six different immunoprecipitations, which included a no antibody control and normal rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) control. Four histone H3 lysine tail modifications were measured: H3K9ac, H3K4me2, H3K9me2, and H3K27me2. H3K9ac, H3K4me2 and H3K27me2 antibodies for ChIP were obtained from Upstate (Charlottesville, VA) and H3K9me2 was obtained from New England Biolabs. DNAs were recovered after immunoprecipitation and crosslinks reversed by phenol-chloroform extraction. qChIP PCR primers for the BORIS promoter were: -55 F: 5'-ACTGCCACCCTCCACTCTC-3' and +33 R: 5'-TTTGGCTTGTGGGCTCTG-3'. The probe, 5'-FAM-GGCGGCGG-quencher-3' (Universal Probe library, probe # 70), was purchased from Roche (Indianapolis, IN). qChIP PCR was performed using Faststart TaqMan Probe Master Mix (Roche). For quantification, standard curves of BORIS amplicons were generated and an absolute quantification method was utilized. In all experiments, the following cycling parameters were used: 95˚C for 10 min, 40 cycles of 95˚C for 15 s, and 60˚C for 1 min. Samples were run in triplicate, and data were normalized to 2% input DNA amplifications, after subtraction of signals obtained from no antibody control immunoprecipitations. All qChIP experiments were performed a minimum of three times.
BORIS promoter luciferase assays
The BORIS promoter region, including the 5' CpG island, was amplified from genomic DNA according to standard methods using the following primers: -252 F: 5'-CCTGTATGGGACCCTCCTC-3' and +284 R: 5'-GGCCAGACCAAGCACACT-3'. The amplified promoter fragment was gel purified and cloned into the pCR2.1 TOPO vector (Invitrogen). The insert was sub-cloned into pGL3-Basic vector (Promega, Madison, WI), using XhoI and HindIII restriction enzymes and T4 DNA ligase (Fermentas). To accomplish specific methylation of the insert, the cloned fragment was removed from pGL3-Basic and methylated in vitro with SssI methylase (New England Biolabs, Ipswich, MA), HpaII and HhaI methylases (New England Biolabs), or no enzyme (mock), all in the presence of the cofactor S-adenosyl methionine (New England Biolabs). Methylation efficiency was confirmed using HpaII and McrBC digests (New England Biolabs). Methylated or mock-methylated fragments were purified by phenol-chloroform extraction and ligated back into pGL3-Basic using standard methods. Individual cell lines were transfected with 150 ng of the differentially methylated pGL3-Basic constructs along with 50 ng of a Renilla luciferase control construct (Promega), to normalize for transfection efficiency. All transfections were carried out in triplicate wells of 24 well plates. Cells were harvested between 24 and 48 hours post-transfection, and firefly and Renilla luciferase activity were measured using the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer's instructions.