Abstract
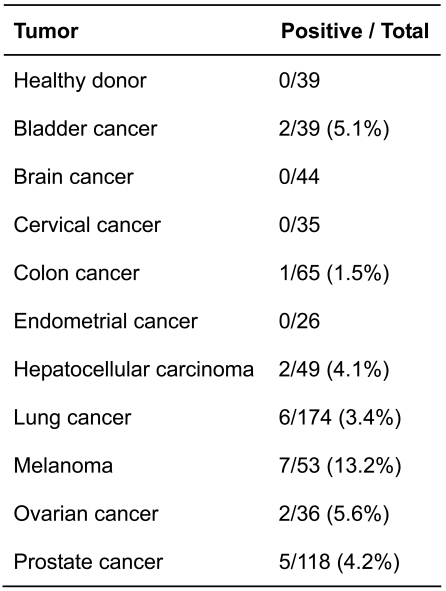
The prostate cancer HERV-K gag-related NGO-Pr-54 antigen was identified by SEREX analysis using autologous patient serum. NGO-Pr-54 mRNA was observed to be faintly expressed in normal prostate and strongly expressed in a variety of cancers, including ovarian cancer (5/8), prostate cancer (6/9), and leukemia (5/14). A phage plaque assay showed that a strong reaction was constantly observed with clone ZH042 in which the 5' end of NGO-Pr-54 is deleted, suggesting that it contained the sequence coding for the protein product. A TI-35 mAb was produced using a recombinant protein (438 aa) deduced from the sequence of ZH042. Transfection of clone ZH042 into 293T cells resulted in the production of an approximately 50-kDa molecule visualized by Western blotting. Natural production of the molecule was confirmed in a SK-MEL-23 melanoma cell line. An indirect immunofluorescence assay showed that NGO-Pr-54 protein was expressed on the cell surface as well as in the cytoplasm. Cell surface expression was confirmed by flow cytometry using the TI-35 mAb. The antibody response against NGO-Pr-54 was observed in patients with bladder (5.1%), liver (4.1%), lung (3.4%), ovarian (5.6%), and prostate (4.2%) cancer, as well as with malignant melanoma (13.2%).
Keywords: human, prostate cancer, SEREX, HERV-K, gag, tumor antigen
Introduction
Serological recombinant cDNA expression cloning (SEREX) has been utilized for the identification of tumor antigens (1, 2). More than 2,000 antigens including cancer/testis antigens, mutational antigens, over-expressed antigens, differentiation antigens, splice-variant antigens and viral antigens have been defined by SEREX (2). Some SEREX-defined antigens, such as NY-ESO-1 (3, 4, 5), Her2/neu (6), NY-BR-1 (7), etc., have also been identified as targets of cellular immune responses.
Human endogenous retroviruses (HERVs) are genomic sequences that result from ancient retroviral infections that became fixed in the germ line DNA (8, 9) and represent approximately 8% of the human genome (10). More than twenty HERV families have been identified (11). Most HERV families are defective; however, some families still contain open reading frames (ORFs) for retroviral genes (12). HERVs contain gag, pol, and env genes encoding polyproteins flanked by two long terminal repeats (LTRs) (9, 13). The HERV-K family is the most conserved family. It is present as 30-50 proviral copies in the human genome (14) and has intact ORFs for the gag, pol, or env genes (15, 16). No expression of HERVs has been observed in most normal tissues. However, HERVs have been shown to be expressed in normal placenta (17) and brain (18, 19) from patients with multiple sclerosis. In tumors, HERV-K was shown to be expressed in teratocarcinoma (20) and HERV-E in prostate cancer (21).
In this study, the NGO-Pr-54 antigen was identified by immunoscreening of cDNA expression libraries prepared from prostate cancer specimens obtained from a patient with autologous sera. NGO-Pr-54 is homologous to HERV-K. The mRNA expression was examined in various normal tissues and in a variety of tumors from different origins. The ORF was determined and mAb was produced. Its localization on the cell surface as well as in the cytoplasm was demonstrated. The immunogenicity of NGO-Pr-54, as evidenced by the production of antibody in cancer patients, was shown by ELISA using the recombinant protein.
Results
Identification of the NGO-Pr-54 gene in prostate cancer by SEREX using autologous serum
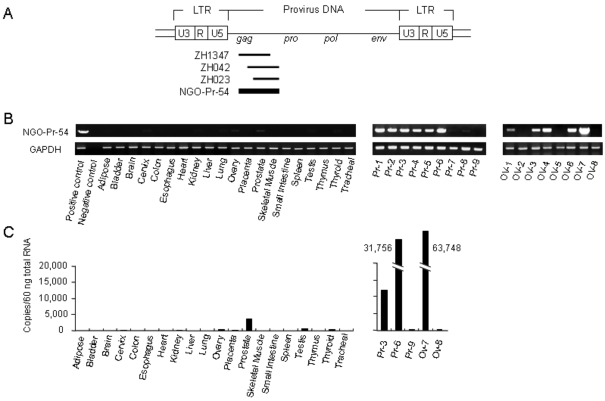
The prostate cancer specimens were obtained surgically from an 80 year-old patient and cDNA expression libraries were constructed from the mRNA. A total of 1.3 × 106 cDNA clones were prepared. Approximately 2.0 × 105 clones were screened with the autologous patient serum using SEREX methodology and 125 reactive clones were isolated. These clones correspond to 67 different genes, as determined by nucleotide sequencing analysis. As shown in Figure 1A, three clones (ZH1347, ZH042, and ZH023) represented the same gene which was named NGO-Pr-54 and which was found to be a part of the human endogenous retrovirus-K (HERV-K) element on chromosome 22q11.2 (GenBank accession number AP000346). The expression sequence tag (EST) database indicated a restricted expression pattern for NGO-Pr-54 in normal prostate tissue.
Figure 1.
NGO-Pr-54 mRNA expression in normal and tumor tissues. (A) Genomic structure of the HERV-K provirus. The HERV-K provirus contains the gag, pro, pol, and env genes flanked by two long terminal repeats (LTRs). Three clones (ZH1347, ZH042, and ZH023) representing the same gene were recognized in prostate cancer cDNA libraries by SEREX using autologous sera; the gene was named NGO-Pr-54. (B) RT-PCR results for NGO-Pr-54 mRNA in a panel of normal tissues (left), prostate cancer (middle, Pr-1 to -9), and ovarian (right, OV-1 to -8) cancer specimens. (C) Quantitative real-time RT-PCR for a panel of normal tissues (left) and prostate and ovarian cancer specimens (right).
NGO-Pr-54 mRNA expression in normal and tumor tissues and in tumor cell lines
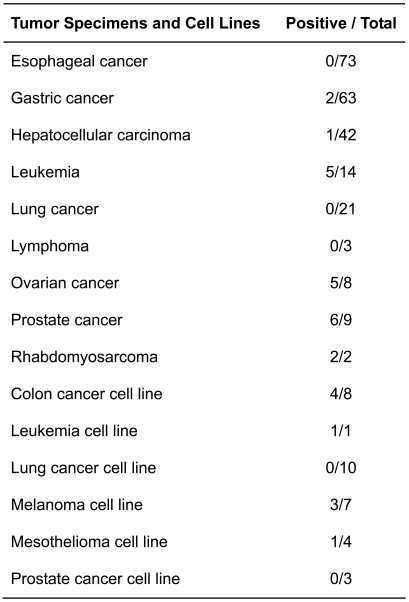
NGO-Pr-54 mRNA expression was investigated in a panel of normal tissues, tumors, and tumor cell lines by 35 cycle RT-PCR using specific primers. As NGO-Pr-54 contains no intron, the RNA was pretreated with DNase to remove genomic DNA before reverse transcription. As shown in Figure 1B, NGO-Pr-54 mRNA was faintly detectable in normal prostate. Quantitative real-time RT-PCR analysis confirmed the results (Figure 1C). In tumors, NGO-Pr-54 mRNA was observed to be strongly expressed in 6/9 prostate cancers, 5/8 ovarian cancers, and 5/14 leukemias (Figure 1B). Table 1 summarizes NGO-Pr-54 mRNA expression in various tumors and tumor cell lines as determined by RT-PCR analysis.
Table 1.
NGO-Pr-54 mRNA expression in tumors and tumor cell lines.
Production of monoclonal antibody (mAb) against NGO-Pr-54
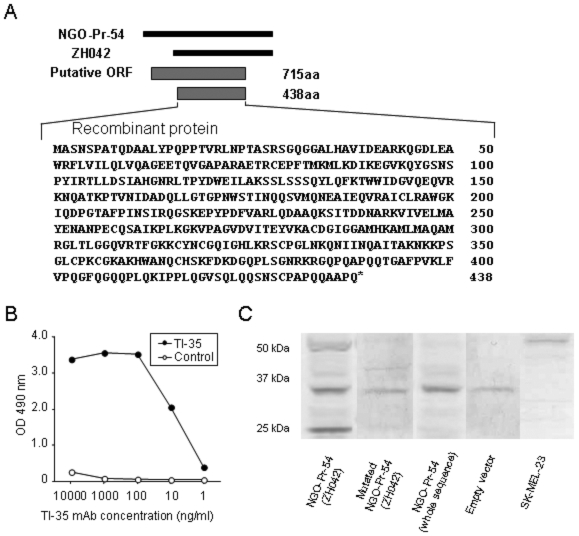
By phage plaque assay, 16/31 sera samples from prostate cancer patients reacted with NGO-Pr-54, but none of 30 control sera from healthy donors did. Within the three clones, ZH042 constantly gave a strong reaction despite lacking the N-terminal sequence of the putative ORF (715 amino acids) of NGO-Pr-54 (Figure 2A). Therefore, a recombinant protein consisting of the C-terminal 438 amino acids was produced and BALB/c mice were immunized with the protein to produce a mAb. Five clones were obtained: Three IgG1 and two IgG2. TI-35 mAb, which was IgG1, reacted strongly to the recombinant protein. Figure 2B shows the titration curve of the TI-35 mAb obtained by ELISA using the recombinant protein.
Figure 2.
Production of a monoclonal antibody, TI-35, against NGO-Pr-54. (A) Schematic representation of NGO-Pr-54 and its putative open reading frame (ORF). The recombinant protein was produced from the C-terminal 438 amino acids of the putative ORF. (B) Reactivity of monoclonal antibody TI-35 against recombinant NGO-Pr-54 protein. Control, isotype (IgG1) matched mouse mAb (anti-Lyt-2.1). (C) Western blot of the lysate of 293T cells transfected with NGO-Pr-54 (ZH042) plasmid, mutated NGO-Pr-54 (ZH042) plasmid with a point mutation in the start codon (ATG to TTG), NGO-Pr-54 (whole sequence) plasmid, and empty vector (only p3xFLAG-CMV-14), and SK-MEL-23 using TI-35 mAb.
NGO-Pr-54 protein expression in 293T transfectants and SK-MEL-23 by Western blot analysis
We examined NGO-Pr-54 protein expression in the transfectants by Western blot using TI-35 mAb. As shown in Figure 2C, TI-35 mAb recognized two bands of approximately 50 kDa and 20 kDa in the 293T lysate when transfected with the NGO-Pr-54 (ZH042) plasmid. The 50-kDa band size corresponds to the fusion protein of NGO-Pr-54 and FLAG-tag. Insertion of a point mutation in the start codon from ATG to TTG in NGO-Pr-54 (ZH042) (mutated NGO-Pr-54 (ZH042) plasmid) resulted in no bands being detectable in the 293T lysate. The 50-kDa band was also detected in the lysate of a melanoma cell line, SK-MEL-23, in which NGO-Pr-54 mRNA expression had been detected by RT-PCR.
On the other hand, TI-35 mAb detected no NGO-Pr-54 protein in the 293T lysate when transfected with NGO-Pr-54 (whole sequence) including the putative ORF.
Subcellular localization of NGO-Pr-54
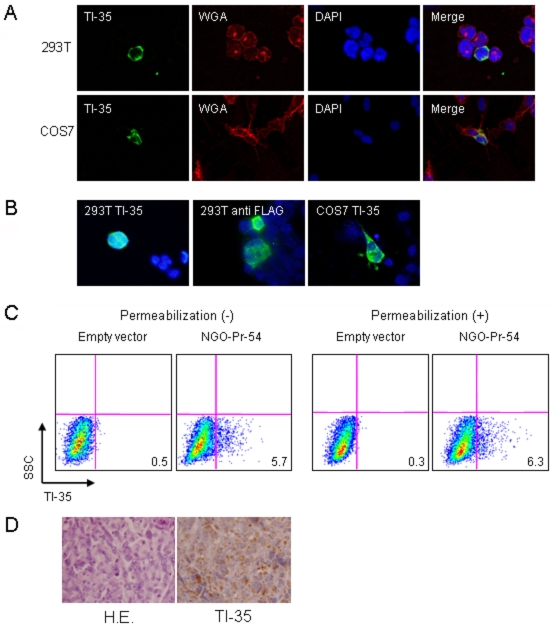
The subcellular localization of NGO-Pr-54 was investigated using 293T and COS7 cells transfected with NGO-Pr-54 (ZH042) by indirect immunofluorescence. As shown in Figure 3A, staining was observed in non-permeabilized 293T and COS7 transfectants by either TI-35 mAb or anti-FLAG mAb. Control staining of the membrane by rhodamine-labeled WGA gave similar results. In permeabilized cells, cytoplasmic staining was also observed (Figure 3B). As shown in Figure 3C, flow cytometry analysis confirmed the cell surface expression of NGO-Pr-54 protein.
Figure 3.
Subcellular localization of NGO-Pr-54 protein. (A) Immunofluorescence staining of 293T and COS7 transfected with NGO-Pr-54 (ZH042) plasmid without permeabilization. TI-35 and anti-FLAG antibody (not shown) detected NGO-Pr-54 protein (green) similarly. The cell membrane was marked with rhodamine-labeled WGA (red) and the nuclei were stained with DAPI. (B) 293T and COS7 cells were stained with TI-35 mAb and anti-FLAG antibody after permeabilization. (C) Flow cytometry analysis of 293T cells transfected with NGO-Pr-54 (ZH042) using TI-35 mAb with or without permeabilization. (D) Hematoxylin and eosin (H&E) and TI-35 mAb staining of a melanoma specimen obtained from a patient showing an antibody against NGO-Pr-54. Magnification, 400x.
A melanoma specimen obtained from a patient who showed antibody against the recombinant NGO-Pr-54 was examined further by immunohistochemistry. As shown in Figure 3D, diffuse staining possibly involving the cytoplasm and membrane was observed in the tumor cells.
Antibody response against NGO-Pr-54 in cancer patients
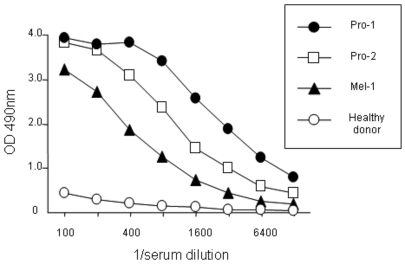
The antibody response against NGO-Pr-54 was investigated in cancer patients by ELISA using recombinant protein. Figure 4 shows the titration curves for positive sera from two prostate cancer patients, a melanoma patient, and serum from a healthy donor. The results are summarized in Table 2. Antibody against NGO-Pr-54 was found in sera from bladder, liver, lung, ovarian, and prostate cancer patients, as well as in melanoma patients. No antibody was detected in 39 sera samples from healthy donors.
Figure 4.
Antibody response against NGO-Pr-54 in cancer patients. ELISA for sera from prostate cancer patients (Pro-1 and -2) and a melanoma patient (Mel-1) using recombinant protein. Control, healthy donor.
Table 2.
Antibody response against NGO-Pr-54 in cancer patients.
Discussion
In this study, HERV-K gag-related NGO-Pr-54 antigen was identified by SEREX analysis in a prostate cancer using autologous patient serum. NGO-Pr-54 mRNA was observed to be faintly expressed in normal prostate and strongly expressed in a variety of cancers including ovarian cancers (5/8), prostate cancers (6/9), and leukemias (5/14). A phage plaque assay showed that a strong reaction was consistently observed with clone ZH042 having NGO-Pr-54 with a deleted 5' end, suggesting that it contained the sequence coding for the protein product. TI-35 mAb was produced using the recombinant protein (438 aa) deduced from the sequence of ZH042 as antigen for immunizing mice. Transfection of clone ZH042 into 293T cells resulted in the production of an approximately 50-kDa molecule visualized by Western blot. Natural production of the molecule was confirmed in the SK-MEL-23 melanoma cell line. An indirect immunofluorescence assay showed NGO-Pr-54 protein is expressed on the cell surface as well as in the cytoplasm. Cell surface expression was confirmed by flow cytometry using TI-35 mAb. The antibody response against NGO-Pr-54 was observed in patients with bladder (5.1%), liver (4.1%), lung (3.4%), ovarian (5.6%), and prostate (4.2%) cancer, as well as in patients with malignant melanoma (13.2%).
Most endogenous retroviruses are defective due to the presence of stop codons and frameshifts in the genes (9, 22). HERV-K is the most conserved family including intact ORFs (9). However, the presence of ORFs did not always result in translation of the protein (23). NGO-Pr-54 harbors an intact ORF spanning gag consisting of 715 aa. While the molecular size appeared to be consistent with that of the gag precursor determined previously (24), transfection of the full length sequence of NGO-Pr-54 failed to produce protein in 293T cells. On the other hand, transfection of ZH042 that lacked the 5' region from NGO-Pr-54 resulted in the production of a protein approximately 50 kDa in size that appeared to be the processed form of the gag protein. The exact reason why the transfection of the full length NGO-Pr-54 gag gene failed to produce a protein product, but transfection of the 5' region-deleted ZH042 construct resulted in the production of gag protein is unknown at present. The HERV-K genome has as a feature the nuclear retention of mRNAs, with their export being mediated by Rec proteins (25, 26). The lack of protein production by the full-length sequence could be due to the impairment of nuclear export of the HERV-K mRNA. An approximately 20-kDa protein was observed in ZH042-transfected 293T cells, which might be the mature gag processing product. However, the recombinant 438 aa protein used in this study contained no protease sequences adjacent to the gag protein. The absence of a 20-kDa molecule in the HERV-K-expressing melanoma cell line SK-MEL-23 might suggest that it is a degradation product due to cellular proteases in 293T cells.
Endogenous retroviral sequences have been shown to be expressed in human cancers. Increased expression of a HERV-K env transcript was shown in melanoma (27) and in breast (28) and ovarian (29) cancers as compared to the expression in normal tissues. HERV-E mRNA expression was shown in prostate cancer, but not in normal prostate (30). HERV-H expression was shown in gastrointestinal cancer (31). This study showed HERV-K gag expression in prostate cancer, ovarian cancer, and leukemia. Furthermore, cell surface expression of HERV-K gag was shown, something which could be useful for antibody therapy of cancer, as seen for HER2/neu (32) or NY-BR-1 (33). Localization of the HERV-K gag protein in the cell membrane has been shown by Western blotting using purified membrane and rabbit anti-HERV-K gag serum (34). Recently, HERV-K 22q11.23 was shown to be fused to ETV1, creating oncogenic fusions in prostate cancer (35). HERV-K 22q11.23 was expressed in normal prostate at higher levels than in other normal tissues. This gene was shown to be overexpressed in the LNCap prostate cancer cell line in response to synthetic androgen.
The antigenicity of HERV elements has been shown serologically and by CD8 T cell recognition. Antibodies against HERV-K gag and env have been detected in patients with germ cell tumor (24, 36), ovarian cancer (29), leukemia (37), and malignant melanoma (27). Moreover, the association of antibody response to better prognosis was observed (38). On the other hand, Schiavetti et al. (39) identified the antigenic peptide presented by HLA-A2 molecules encoded by a very short open reading frame present in the env region of the gene belonging to HERV-K. The gene HERV-K-MEL was expressed in most malignant melanoma samples. It was also expressed in most naevi and as a part of carcinomas and sarcomas. However, in normal tissues, it is expressed only in testis and some skin samples. HERV-K-MEL is a pseudogene that does not code for a retroviral envelope protein. Because of the presence of many mutations, a main ORF can not be defined. Thus, the antigenic peptide can be encoded by a small ORF. Takahashi et al. (40) obtained donor-derived CD8 CTLs recognizing recipient renal cell cancer (RCC) in hematopoietic stem cell transplantation (HSCT) and identified the peptide epitope presented on HLA-A11. The peptide was derived from the gene belonging to HERV-E, which was expressed in renal cancer cell lines and tissues, but not in normal tissues including kidney.
The antibody response to HERV products and CTL recognition of HERV-derived peptides in cancer patients suggests that HERVs are immunogenic in cancer patients and can be utilized as target antigens for cancer immunotherapy.
Abbreviations
- HERV
human endogenous retrovirus
- ORF
open reading frame
- SEREX
serological recombinant cDNA expression cloning
Acknowledgements
We thank Ms. M. Isobe for excellent technical assistance and Ms. J. Mizuuchi for preparation of the manuscript. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by the Cancer Research Institute, New York.
References
- 1.Sahin U, Türeci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen YT. Identification of human tumor antigens by serological expression cloning: an online review on SEREX. Cancer Immun. 2004 http://www.cancerimmunity.org/SEREX/ [Google Scholar]
- 3.Jäger E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gnjatic S, Atanackovic D, Jäger E, Matsuo M, Selvakumar A, Altorki NK, Maki RG, Dupont B, Ritter G, Chen YT, Knuth A, Old LJ. Survey of naturally occurring CD4+ T cell responses against NY-ESO-1 in cancer patients: correlation with antibody responses. Proc Natl Acad Sci USA. 2003;100:8862–8867. doi: 10.1073/pnas.1133324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnjatic S, Nishikawa H, Jungbluth AA, Güre AO, Ritter G, Jäger E, Knuth A, Chen YT, Old LJ. NY-ESO-1: review of an immunogenic tumor antigen. Adv Cancer Res. 2006;95:1–30. doi: 10.1016/S0065-230X(06)95001-5. [DOI] [PubMed] [Google Scholar]
- 6.Fisk B, Blevins TL, Wharton JT, Ioannides CG. Identification of an immunodominant peptide of HER-2/neu protooncogene recognized by ovarian tumor-specific cytotoxic T lymphocyte lines. J Exp Med. 1995;181:2109–2117. doi: 10.1084/jem.181.6.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jäger D, Karbach J, Pauligk C, Seil I, Frei C, Chen YT, Old LJ, Knuth A, Jäger E. Humoral and cellular immune responses against the breast cancer antigen NY-BR-1: definition of two HLA-A2 restricted peptide epitopes. Cancer Immun. 2005;5:11. http://www.cancerimmunity.org/v5p11/051014.htm [PubMed] [Google Scholar]
- 8.Stauffer Y, Theiler G, Sperisen P, Lebedev Y, Jongeneel CV. Digital expression profiles of human endogenous retroviral families in normal and cancerous tissues. Cancer Immun. 2004;4:2. http://www.cancerimmunity.org/v4p2/040102.htm [PubMed] [Google Scholar]
- 9.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 11.Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths DJ. Endogenous retroviruses in the human genome sequence. Genome Biol. 2001;2:reviews1017.1–1017.5. doi: 10.1186/gb-2001-2-6-reviews1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Müller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tönjes RR, Löwer R, Boller K, Denner J, Hasenmaier B, Kirsch H, König H, Korbmacher C, Limbach C, Lugert R, Phelps RC, Scherer J, Thelen K, Löwer J, Kurth R. HERV-K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl 1):S261–S267. doi: 10.1097/00042560-199600001-00039. [DOI] [PubMed] [Google Scholar]
- 17.Blond JL, Beseme F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perron H, Garson JA, Bedin F, Beseme F, Paranhos-Baccala G, Komurian-Pradel F, Mallet F, Tuke PW, Voisset C, Blond JL, Lalande B, Seigneurin JM, Mandrand B. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc Natl Acad Sci USA. 1997;94:7583–7588. doi: 10.1073/pnas.94.14.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen T. Association of human endogenous retroviruses with multiple sclerosis and possible interactions with herpes viruses. Rev Med Virol. 2005;15:179–211. doi: 10.1002/rmv.465. [DOI] [PubMed] [Google Scholar]
- 20.Bieda K, Hoffmann A, Boller K. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J Gen Virol. 2001;82:591–596. doi: 10.1099/0022-1317-82-3-591. [DOI] [PubMed] [Google Scholar]
- 21.Wang-Johanning F, Frost AR, Jian B, Azerou R, Lu DW, Chen DT, Johanning GL. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 22.Leib-Mösch C, Brack-Werner R, Werner T, Bachmann M, Faff O, Erfle V, Hehlmann R. Endogenous retroviral elements in human DNA. Cancer Res. 1990;50:5636S–5642S. [PubMed] [Google Scholar]
- 23.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauter M, Schommer S, Kremmer E, Remberger K, Dölken G, Lemm I, Buck M, Best B, Neumann-Haefelin D, Müller-Lantzsch N. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J Virol. 1995;69:414–421. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Löwer R, Tönjes RR, Korbmacher C, Kurth R, Löwer J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Bogerd HP, Peng S, Wiegand H, Truant R, Cullen BR. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc Natl Acad Sci USA. 1999;96:13404–13408. doi: 10.1073/pnas.96.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 28.Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, Strong TV. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553–1560. [PubMed] [Google Scholar]
- 29.Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2006;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 30.Wang-Johanning F, Frost AR, Jian B, Azerou R, Lu DW, Chen DT, Johanning GL. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 31.Wentzensen N, Coy JF, Knaebel HP, Linnebacher M, Wilz B, Gebert J, von Knebel Doeberitz M. Expression of an endogenous retroviral sequence from the HERV-H group in gastrointestinal cancers. Int J Cancer. 2007;121:1417–1423. doi: 10.1002/ijc.22826. [DOI] [PubMed] [Google Scholar]
- 32.Coussens L, Yang-Feng TL, Liao YC, Chen E, Gray A, McGrath J, Seeburg PH, Libermann TA, Schlessinger J, Francke U, Levinson A, Ullrich A. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science. 1985;230:1132–1139. doi: 10.1126/science.2999974. [DOI] [PubMed] [Google Scholar]
- 33.Seil I, Frei C, Sültmann H, Knauer SK, Engels K, Jäger E, Zatloukal K, Pfreundschuh M, Knuth A, Tseng-Chen Y, Jungbluth AA, Stauber RH, Jäger D. The differentiation antigen NY-BR-1 is a potential target for antibody-based therapies in breast cancer. Int J Cancer. 2007;120:2635–2642. doi: 10.1002/ijc.22620. [DOI] [PubMed] [Google Scholar]
- 34.Boller K, König H, Sauter M, Mueller-Lantzsch N, Löwer R, Löwer J, Kurth R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 35.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 36.Sauter M, Roemer K, Best B, Afting M, Schommer S, Seitz G, Hartmann M, Mueller-Lantzsch N. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 1996;56:4362–4365. [PubMed] [Google Scholar]
- 37.Denner J, Phelps RC, Löwer J, Löwer R, Kurth R. Expression of the human endogenous retrovirus HERV-K in tumor and normal tissues and antibody response of pregnant women, tumor and AIDS patients against HERV-K Gag and Env peptides. AIDS Res Hum Retroviruses. 1995;11:103. [Google Scholar]
- 38.Kleiman A, Senyuta N, Tryakin A, Sauter M, Karseladze A, Tjulandin S, Gurtsevitch V, Mueller-Lantzsch N. HERV-K(HML-2) GAG/ENV antibodies as indicator for therapy effect in patients with germ cell tumors. Int J Cancer. 2004;110:459–461. doi: 10.1002/ijc.11649. [DOI] [PubMed] [Google Scholar]
- 39.Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62:5510–5516. [PubMed] [Google Scholar]
- 40.Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, Hanada K, Mena O, Kurlander R, Tawab A, Srinivasan R, Lundqvist A, Malinzak E, Geller N, Lerman MI, Childs RW. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J Clin Invest. 2008;118:1099–1109. doi: 10.1172/JCI34409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köhler G, Howe SC, Milstein C. Fusion between immunoglobulin-secreting and nonsecreting myeloma cell lines. Eur J Immunol. 1976;6:292–295. doi: 10.1002/eji.1830060411. [DOI] [PubMed] [Google Scholar]
- 43.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 44.Takaki T, Hiraki A, Uenaka A, Gomi S, Itoh K, Udono H, Shibuya A, Tsuji T, Sekiguchi S, Nakayama E. Variable expression on lung cancer cell lines of HLA-A2-binding MAGE-3 peptide recognized by cytotoxic T lymphocytes. Int J Oncol. 1998;12:1103–1109. doi: 10.3892/ijo.12.5.1103. [DOI] [PubMed] [Google Scholar]
- 45.Niiya M, Niiya K, Kiguchi T, Shibakura M, Asaumi N, Shinagawa K, Ishimaru F, Kiura K, Ikeda K, Ueoka H, Tanimoto M. Induction of TNF-alpha, uPA, IL-8 and MCP-1 by doxorubicin in human lung carcinoma cells. Cancer Chemother Pharmacol. 2003;52:391–398. doi: 10.1007/s00280-003-0665-1. [DOI] [PubMed] [Google Scholar]
- 46.Shimono M, Uenaka A, Noguchi Y, Sato S, Okumura H, Nakagawa K, Kiura K, Tanimoto M, Nakayama E. Identification of DR9-restricted XAGE antigen on lung adenocarcinoma recognized by autologous CD4 T-cells. Int J Oncol. 2007;30:835–840. [PubMed] [Google Scholar]
- 47.Usami N, Fukui T, Kondo M, Taniguchi T, Yokoyama T, Mori S, Yokoi K, Horio Y, Shimokata K, Sekido Y, Hida T. Establishment and characterization of four malignant pleural mesothelioma cell lines from Japanese patients. Cancer Sci. 2006;97:387–394. doi: 10.1111/j.1349-7006.2006.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ono T, Sato S, Kimura N, Tanaka M, Shibuya A, Old LJ, Nakayama E. Serological analysis of BALB/c methylcholanthrene sarcoma Meth A by SEREX: identification of a cancer/testis antigen. Int J Cancer. 2000;88:845–851. doi: 10.1002/1097-0215(20001215)88:6<845::aid-ijc1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
Materials and methods
Mice
BALB/c and SCID mice were purchased from Japan SLC (Shizuoka, Japan). The mice were bred at the Laboratory Animal Center in Okayama University. The experiments were conducted according to the Guidelines for Animal Experiments at Okayama University, the Japanese Government Animal Protection and Management Law (No. 105) and the Japanese Government Notification on Feeding and Safekeeping of Animals (No. 6).
Tissues and sera
Prostate cancer specimens used for SEREX were obtained surgically from a patient at Aichi Cancer Center (Nagoya, Japan). Tumor specimens used for reverse transcription (RT)-PCR analysis were obtained surgically from patients at Okayama University Hospital (Okayama, Japan) and Osaka University Hospital (Osaka, Japan). Sera were obtained from 39 healthy donors and cancer patients at Okayama University Hospital. Informed consent was obtained from each healthy donor and each patient for the use of specimens and sera.
Cell lines
293T is human embryonic kidney cell line 293 transfected with the SV40 T gene (41). NS-1 is a myeloma cell line derived from BALB/c MOPC-21 cells (42). COS7 is a cell line derived from SV40-transformed African green monkey kidney cells (43). Colon cancer cell lines SK-CO-1, WiDr, LS 174T, SW480, COLO 201, COLO 320DM, and LoVo were obtained from the American Type Culture Collection (ATCC, Rockville, MD). DLD-1 was obtained from the RIKEN BioResource Center (Ibaraki, Japan). Lung cancer cell lines OU-LC-MS, OU-LU-5, 1-87, 11-18, PC-9, LK-87, QG-56, EBC-2, and LC-1sq were described previously (44, 45, 46). OU-LU-17 was provided by Dr. M. Shimono (Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan). Prostate cancer cell lines LNCap, DU 145, and PC-3 were obtained from the American Type Culture Collection. Mesothelioma cell lines C-13 and YM were established in our laboratory. ACC-MESO-1 and ACC-MESO-4 were obtained from the RIKEN BioResource Center (47). Melanoma cell lines SK-MEL-19, SK-MEL-23, SK-MEL-27, SK-MEL-28, SK-MEL-37, SK-MEL-64, CLL-MEL were provided Dr. R. Ueda (Nagoya City University Graduate School of Medical Sciences, Nagoya, Japan). The acute T cell leukemia cell line Jurkat was obtained from the American Type Culture Collection. These cell lines were maintained in RPMI 1640 supplemented with 10% FCS.
Total RNA isolation and cDNA synthesis
Total RNA was isolated from frozen tumor specimens and pellets of washed cell lines using an RNeasy Mini Kit (QIAGEN, Hilden, Germany). Isolated RNA was treated with DNase (TURBO DNA-free, Ambion, Austin, TX, USA) to remove genomic DNA contamination. Treated RNA (2 µg) was reverse transcribed into single-stranded cDNA using Moloney murine leukemia virus reverse transcriptase (Ready-To-Go You-Prime First-Strand Beads, Amersham Biosciences, Piscataway, NJ, USA) and oligo (dT)15 as a primer. Complementary DNA was tested for integrity by amplifying the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript for over 30 cycles. An RNA panel from normal tissues was purchased from Ambion and treated with DNase as described above.
Reverse transcription (RT)-PCR analysis
To amplify the NGO-Pr-54 cDNA segment, primers specific for NGO-Pr-54 were designed. Primers for RT-PCR were: 5'-CGTCTAATTCACCAGCAACAC-3' (forward), 5'-TAGACTTTTGAGCAGCATCTTG-3' (reverse). The amplification program for NGO-Pr-54 was 10 s at 98˚C, 30 s at 55˚C, and 1 min at 72˚C for 35 cycles after denaturing at 98˚C for 2 min. These cycles were followed by a 10-min elongation step at 72˚C. The PCR products (702 bp) were analyzed on a 0.8% agarose gel.
Quantitative real-time RT-PCR
Two-step real-time RT-PCR was run on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The gene-specific primers and TaqMan probe for NGO-Pr-54 were designed using Primer Express software (version 1.5) (Applied Biosystems). The forward primer was 5'-CAGCGATGGCGTCTAATTCA-3' and the reverse primer was 5'-AGTGGGCGGCTGAGGATAC-3'. The TaqMan probe was 5'-FAM-AGCAACACAGGACGCGGCGC-TAMRA-3'. PCR was performed with TaqMan Universal PCR Master Mix (Applied Biosystems), the primer pair, TaqMan probe, and cDNA solution (corresponding to 60 ng total RNA). The thermal cycling conditions comprised an initial denaturation step at 95˚C for 10 min, followed by 40 cycles of 95˚C for 15 s and 60˚C for 1 min.
Construction of cDNA libraries
Messenger RNA was isolated from prostate cancer specimens using a Fast Track mRNA purification kit (Invitrogen, Life Technologies, Carlsbad, CA, USA). Complementary DNA expression libraries were prepared in a λZAP expression vector using a cDNA library kit (Stratagene, La Jolla, CA, USA).
Immunoscreening of cDNA expression libraries
Complementary DNA expression libraries were screened with autologous patient serum as described previously (48). Briefly, the serum was diluted 1:10 and preabsorbed with phage-transfected Escherichia coli lysate. Nitrocellulose membranes containing the phage plaques at a density of 4,000-5,000 pfu per 130 mm plate were incubated overnight at room temperature with the serum diluted 1:200. After washing, the filters were incubated with alkaline phosphatase-conjugated goat anti-human Fcγ. The reactive clones were visualized with 5-bromo-4-chloro-3-indolyl-phosphatase and nitroblue tetrazolium.
Sequence analysis of reactive clones
pBK-CMV phagemids were excised in vivo from positive-staining phage. cDNA inserts were subjected to DNA sequencing using an ABI PRISM Model 377 automated sequencer (Perkin Elmer, Norwalk, CT, USA). The sequence alignments of the clones were analyzed for similarity using BLAST software, Genbank, EST, and SEREX databases.
Preparation of plasmid vectors
The NGO-Pr-54 cDNA was excised from pBK-CMV phagemids and ligated into a pcDNA3.1(+) vector (Invitrogen) and a p3xFLAG-CMV-14 expression vector (Sigma-Aldrich, Munich, Germany). TOP10 Escherichia coli cells were transformed with the recombinant vector. Plasmid DNA was purified using a QIAprep Spin Miniprep Kit (QIAGEN). Insertion of cDNA was confirmed by DNA sequencing.
Production of the recombinant NGO-Pr-54 protein
The NGO-Pr-54 cDNA corresponding to the C-terminal 438 amino acids of the putative ORF was amplified by PCR. The amplified DNA was ligated into the histidine-tag-containing vector pQE30 (QIAGEN). NGO-Pr-54/pQE30 was introduced into M13. His-tagged recombinant NGO-Pr-54 protein was purified by nickel ion affinity chromatography (HisTrap HP, GE Healthcare, Uppsala, Sweden).
Generation of a monoclonal anti-NGO-Pr-54 antibody, TI-35
BALB/c mice were immunized intramuscularly with NGO-Pr-54/pcDNA3.1 (100 µg) four times at 2-week intervals using an electric pulse generator (CUY-21, BEX, Tokyo, Japan). Mice were boosted intraperitoneally with recombinant NGO-Pr-54 protein (100 µg) twice at a 2-week interval. Spleen cells from immunized mice were fused with NS-1 myeloma cells. Hybridoma cells were cultured in ClonaCell-HY Medium D (StemCell Technologies, Vancouver, BC, Canada). The production of NGO-Pr-54 specific monoclonal antibody from the hybridoma was assessed by ELISA using recombinant NGO-Pr-54 protein. SCID mice were administered intraperitoneally with hybridoma cells. The monoclonal antibody was purified from ascites of the mice via protein G affinity chromatography (Amersham Biosciences).
ELISA
Recombinant NGO-Pr-54 protein (1 µg/ml) in 0.05 mol/l carbonate buffer (pH 9.6) was adsorbed onto 96-well plates (Nunc, Rochester, NY, USA) at 4˚C overnight. Plates were washed with 0.05% Tween-20/PBS and blocked with 200 µl/well of 5% FCS/PBS for 1 h at room temperature. After washing, sera serially diluted with 5% FCS/PBS were added to each well and incubated for 2 h at room temperature. After washing, diluted goat anti-human Fcγ or goat anti-mouse IgG, labeled with horseradish peroxidase (Jackson ImmunoResearch, Baltimore, PA, USA) was added and incubated for 1 h at room temperature. After washing, the substrate solution [50 mmol/l citric acid, 100 mmol/l Na2HPO4, 0.03% ortho-phenylenediamine, and 0.1% H2O2 in distilled water (pH 5.0)] was added to each well. After adding 3 mol/l H2SO4, the absorbance was read at 490 nm.
Western blot analysis
293T cells were transiently transfected with plasmids using Lipofectamine 2000 (Invitrogen). The lysate of transfectants and SK-MEL-23 was prepared with RIPA Lysis Buffer (Santa Cruz Biotechnology, CA, USA). The cell lysate was separated by electrophoresis under reducing conditions and transferred to a polyvinylidene difluoride (PVDF) membrane (Hybond-P; Amersham Biosciences). The membrane was incubated with TI-35 mAb (1 µg/ml) or anti-FLAG M2 mAb (Sigma-Aldrich) (1 µg/ml) for 1 h at room temperature. Bound antibody was detected with alkaline phosphatase-conjugated second antibody (1:1000; Pierce, IL, USA) using an AP Conjugate Substrate kit (Bio-Rad Laboratories, Hercules, CA, USA).
Immunofluorescence staining
Transfected 293T or COS7 cells growing on glass slides were fixed with 4% paraformaldehyde. For intracellular staining, cells were permeabilized by treatment with 0.5% Triton-X. Staining was performed using TI-35 mAb (1 µg/ml) or anti-FLAG M2 mAb (1 µg/ml). Bound antibody was detected with FITC-labeled anti-mouse IgG F(ab')2 (Sigma-Aldrich) at a concentration of 20 µg/ml. Rhodamine-conjugated wheat germ agglutinin (WGA) (Vector Laboratories, Burlingame, CA, USA) was used for staining of the cell membrane. Nuclei were stained with DAPI (VECTASHIELD, Vector Laboratories). Slides were visualized using a fluorescence microscope (Biozero BZ-8000, Keyence, Osaka, Japan).
Flow cytometry analysis
The FACScan (Becton Dickinson, Mountain View, CA, USA) was used according to the manufacturer's instructions.
Immunohistochemistry
Tumor specimens were fixed with buffered formalin and embedded in paraffin. Five-micrometer sections were placed on glass slides, heated at 60˚C overnight, and deparaffinized with xylene and ethanol. Glass slides were microwave-heated in antigen retrieval buffer (10 mmol/l citrate buffer, pH 6.0) using a pressure cooker for 20 min. After the inactivation of endogenous peroxidase with 0.3% H2O2 for 5 min, specimens were preincubated with serum-free blocking solution (DakoCytomation, Kyoto, Japan). TI-35 mAb was added at a concentration of 5 µg/ml and incubated for 2 h at room temperature. After washing, DAKO EnVision+ horseradish peroxidase-conjugated goat anti-mouse IgG (Dako Cytomation) was applied. After incubation for 30 min at room temperature, the specimens were visualized with 3,3'-diaminobenzidine in H2O2 and counterstained with hematoxylin solution.