Abstract
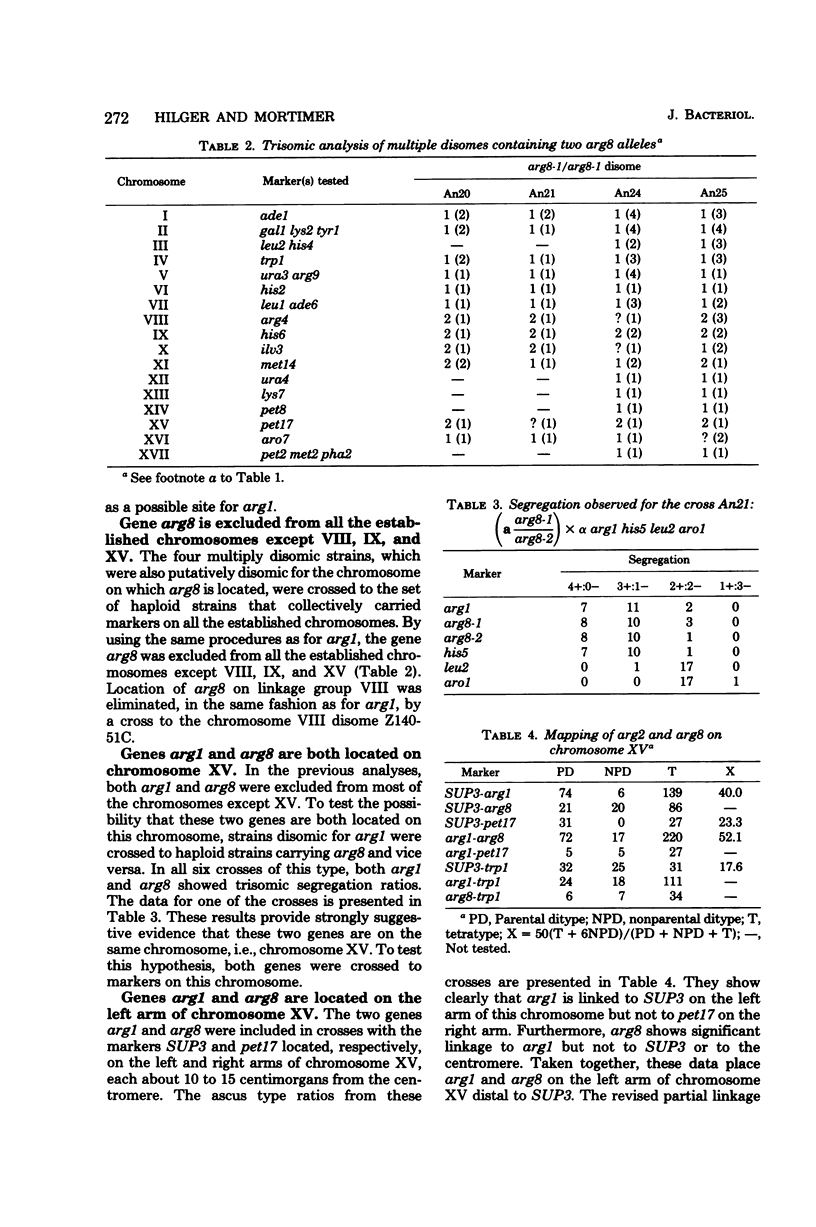
Through use of multiply disomic strains, the genes arg1 and arg8 were excluded from all of chromosomes I to XVII except (i) XV and (ii) IX and XV, respectively. Further aneuploid analyses showed that these two genes were on the same chromosome. By tetrad analysis, arg1 was shown to be linked to SUP3 on the left arm of chromosome XV (parental ditype:nonparental ditype:tetratype = 74; 6:139) and arg8 was shown to be loosely linked to arg1 (parental ditype:nonparental ditype:tetratype 72:17:220) on the same arm. The sequence of the genes on this chromosome arm is centromere-SUP3-arg8. Because arg1 had previously been used to define an 18th chromosome, these results reestablished the minimum chromosome number in Saccharomyces cerevisiae as 17.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cohn M. S., Tabor C. W., Tabor H. Isolation and characterization of Saccharomyces cerevisiae mutants deficient in S-adenosylmethionine decarboxylase, spermidine, and spermine. J Bacteriol. 1978 Apr;134(1):208–213. doi: 10.1128/jb.134.1.208-213.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D C, Mortimer R K. Chromosome Mapping in Saccharomyces: Centromere-Linked Genes. Genetics. 1960 Aug;45(8):1085–1110. doi: 10.1093/genetics/45.8.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D. C., Mortimer R. K. Genetic mapping of nonsense suppressors in yeast. Genetics. 1968 Dec;60(4):735–742. doi: 10.1093/genetics/60.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger F., Simon J. P., Stalon V. Yeast argininosuccinate synthetase. Purification; structural and kinetic properties. Eur J Biochem. 1979 Feb 15;94(1):153–163. doi: 10.1111/j.1432-1033.1979.tb12882.x. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Hawthorne D. C. Genetic Mapping in Saccharomyces IV. Mapping of Temperature-Sensitive Genes and Use of Disomic Strains in Localizing Genes. Genetics. 1973 May;74(1):33–54. doi: 10.1093/genetics/74.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Hawthorne D. C. Genetic mapping in Saccharomyces. Genetics. 1966 Jan;53(1):165–173. doi: 10.1093/genetics/53.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Hawthorne D. C. Genetic mapping in yeast. Methods Cell Biol. 1975;11:221–233. doi: 10.1016/s0091-679x(08)60325-8. [DOI] [PubMed] [Google Scholar]
- Parry E. M., Cox B. S. The tolerance of aneuploidy in yeast. Genet Res. 1970 Dec;16(3):333–340. doi: 10.1017/s0016672300002597. [DOI] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]



