Abstract
We report a catalog of the mouse embryonic pituitary gland transcriptome consisting of five cDNA libraries including wild type tissue from E12.5 and E14.5, Prop1df/df mutant at E14.5, and two cDNA subtractions: E14.5 WT-E14.5 Prop1df/df and E14.5 WT-E12.5 WT. DNA sequence information is assembled into a searchable database with gene ontology terms representing 12,009 expressed genes. We validated coverage of the libraries by detecting most known homeobox gene transcription factor cDNAs. A total of 45 homeobox genes were detected as part of the pituitary transcriptome, representing most expected ones, which validated library coverage, and many novel ones, underscoring the utility of this resource as a discovery tool. We took a similar approach for signaling-pathway members with novel pituitary expression and found 157 genes related to the BMP, FGF, WNT, SHH and NOTCH pathways. These genes are exciting candidates for regulators of pituitary development and function.
Keywords: Cap trapper, Homeobox gene, Prop1, Gene expression, Ames dwarf
Introduction
The pituitary gland is a pivotal component of the endocrine system due to its role in controlling a wide range of fundamental bodily activities including growth, pubertal development, thyroid gland function, and the capacity to cope with stress. These critical functions are regulated by six different hormones secreted by five specialized cell types within the anterior pituitary gland, in response to local stimulatory and inhibitory peptides released by the hypothalamus.
Pituitary development in the mouse begins at embryonic day (E) 9.5 and continues after birth. All of the hormone-secreting cells in the anterior lobe derive from a single primordial tissue, Rathke's pouch, which forms from the oral ectoderm adjacent to the base of the developing brain. Formation of Rathke's pouch occurs in contact with a specialized region of the neuroectoderm, the infundibulum, which will later form the posterior lobe of the pituitary gland.
The molecular mechanisms underlying pituitary organ growth and cell differentiation are complex. Signaling molecules including FGF, BMP, NOTCH, WNT and SHH family members have roles in establishing regional identity within Rathke's pouch and contribute to its growth [1; 2; 3; 4; 5] The overall effect of these signaling molecules is to induce the regional expression of numerous critical transcription factors, which control the differentiation of the hormone secreting cells between E13.5 and birth.
Failure or incomplete differentiation of anterior pituitary cells during embryonic development can result in the genetically heterogeneous disorder, Multiple Pituitary Hormone Deficiency (MPHD). Molecular genetic analysis of spontaneous and genetically engineered mouse mutants with pituitary defects has revealed a functional role for many homeobox transcription factors in the etiology of MPHD in mouse and man. These include Prop1, Pou1f1, Hesx1, Lhx3, Lhx4, Otx2, Pitx1, and Pitx2 [reviewed by[6; 7; 8]. Lesions in these genes only account for approximately half of the known genetic MPHD, suggesting that many of the key factors involved in the process of pituitary cell proliferation and differentiation remain unknown.
Mutations in PROP1 are the most common known genetic cause of MPHD in humans [9]. Patients with PROP1 mutations commonly exhibit deficiencies in Growth Hormone (GH), Prolactin (PRL), Thyroid Stimulating Hormone (TSH) and gonadotropins, with evolving adrenocorticotropin deficiency (ACTH). Similarly, the spontaneous Ames dwarf mouse model (Prop1df/df), that harbors a homeodomain mutation in Prop1 displays a phenotype that includes hypoplasia of the anterior pituitary accompanied by nearly complete loss of the hormone-secreting cell types that produce GH (somatotropes), PRL (lactotropes), and TSH (thyrotropes) [10; 11; 12; 13]. Interestingly, genetic background has a more significant impact on the viability of Prop1 deficient mice than any functional difference between the Prop1df and Prop1null alleles, suggesting that genetic modifiers may influence the clinical features of some MPHD patients [13].
Prop1 expression is first detectable in Rathke's pouch at E10 - 10.5. It reaches a peak at E12.5 after which time its expression declines rapidly [14]. A time of intense cell proliferation occurs in the developing pituitary from E12.5 - E14.5, which precedes the appearance of the differentiated pituitary cell types. The effects of mutations in Prop1 and other critical transcription factors including Pitx2, Lhx3 and Hesx1 are evident at this time [15; 16; 17]. This suggests that potential Prop1 targets and other important as yet unknown transcription factors may be expressed around E12.5 and/or E14.5.
At E12.5 the anterior pituitary is composed primarily of proliferating, undifferentiated cells. There is a very small group of non-proliferating cells that have begun to differentiate and express Foxl2 and the alpha subunit of the gonadotropins and thyrotropin, Cga [18]. At this developmental time Prop1df/df and WT pituitaries are indistinguishable in size and morphology [13; 19]. By E14.5 there are more cells in the anterior lobe of normal mice, and they appear loosely packed and glandular. In contrast, the pituitary glands of Prop1df/df mutants are highly dysmorphic at E14.5 because cells apparently fail to migrate away from the dorsal aspect of the gland and colonize the anterior lobe. They exit the cell cycle but fail to differentiate [13; 19; 20].
We exploited the Prop1df/df model to gain insights into the embryonic pituitary transcriptome. We used cDNA subtractive hybridization between Prop1df/df and wild type pituitary gland primordia at E14.5 to identify differentially expressed genes in mutant and normal embryonic pituitary [21]. We took a differential display approach to define an expression profile for the developing anterior pituitary at E12.5 and E14.5 by comparing Rathke's pouch tissue from wild type samples taken at these times [22]. Collectively, these studies represented the first expression profiles of the developing pituitary gland and also paved the way for demonstrating the role of the Wnt signaling pathway in regulating pituitary growth. More specifically, these studies identified Wnt5a, Aes and Tcf7l2 in the developing pituitary. We have now demonstrated functional roles for each of these genes in pituitary development through the analysis of corresponding mutant mouse models [2; 23; 24; 25].
Numerous gene expression profiling studies have been carried out using adult pituitary tissue and cell lines, but only a few studies have addressed developmental changes in pituitary gene expression aside from those with the Prop1df/df model [26; 27]. Alterations due to aging and caloric restriction have been investigated [28] and some studies with cell lines have implicated transcription factors as potential regulators of development and cell proliferation [29; 30]. The expression studies with adult pituitary tissue and cell lines identified collections of potentially pituitary-specific genes [31; 32], candidates for tumor biomarkers and mechanistic involvement in pituitary adenoma formation [reviewed in [33], and pathways induced by hormonal or growth factor treatment [34; 35; 36; 37].
Our studies with the Prop1df/df model have proven valuable for novel gene discovery and dissecting the genetic differences between the wild type and mutant pituitary; although, a major limitation for functional studies has been the lack of full-length cDNAs available. In addition, previous approaches have not addressed the depth and complexity of the developing pituitary transcriptome specifically. To overcome these limitations we generated a full-length cDNA encyclopedia of the mouse embryonic pituitary gland comprised of 5 transcriptome volumes including three samples and genotypes: E12.5 wild type pituitary (E12.5 WT), E14.5 wild type pituitary (E14.5 WT) and E14.5 Prop1df/df pituitary [38], and two subtracted libraries: E14.5 WT - E14.5 Prop1df/df, WT E14.5 – WT E12.5. We selected these times and genotypes because we expected E14.5 Prop1df/df mutants to continue to express genes that are normally expressed at E12.5 and wane in expression by E14.5 such as Hesx1 [12]. We also expected to find genes normally activated by Prop1, and expressed at E14.5 in WT mice, but absent in the E14.5 Prop1df/df mutant, such as Pou1f1 [10] and Notch2 [3]. Using the Cap-trapper method to enrich for full-length clones [39; 40; 41; 42; 43], followed by aggressive subtraction and normalization, we produced full-length cDNA libraries and generated DNA sequence from 71,649 cDNA clones. The comparison and global analysis of the transcripts contained within each library reveals expression of 12,009 unique genes that we have integrated into a comprehensive searchable database. We demonstrate the integrity, validity, utility and novelty of the database by identifying 45 homeobox genes expressed in the developing anterior pituitary gland and 157 genes related to the BMP, FGF, WNT, NOTCH or SHH signaling pathways, many of which were not previously known to be part of the developing pituitary transcriptome.
Results
The embryonic pituitary encyclopedia is a rich source for gene discovery
We sought to investigate the depth and complexity of the developing pituitary transcriptome, while maximizing novel gene discovery and developing full-length clones that could be exploited for functional studies of the developing pituitary gland. ‘Phase I’ involved end sequencing of clones from three embryonic pituitary libraries (E12.5 WT, E14.5 WT and E14.5 Prop1df/df) until less than 10% of the new sequences produced novel information [38]. Approximately 20% of the 32,476 sequences were obtained from the 5′ end and the remainders were sequenced from the 3′ end. Here, we present the identification and characterization of the unique cDNAs from these three libraries. We also present two additional libraries that were generated by a subtractive hybridization analysis between E14.5 WT and E14.5 Prop1df/df (Sub1) and between E14.5 WT and E12.5 WT (Sub2).
We conducted single pass end-sequencing of 56,716 unique cDNA clones from these five libraries. The sequences were each queried using BLAST analysis against the UniGene unique subset of the NCBI Mus musculus database (Build #159, www.ncbi.nlm.nih.gov/UniGene/). The same sequences were also subjected to BLAST analysis against the mouse-specific Reference Sequence (RefSeq) (Release #20, www.ncbi.nlm.nih.gov/RefSeq/). Clones were identified based on the first BLAST identity recorded with an expect value of 1.0-e5 or lower. This analysis identified 51,352 clones present in the encyclopedia. 5,364 clones were not identified by BLAST analysis. While some of these clones have poor sequence quality through the polyA stretch at the 3′ end that interferes with BLAST analysis, we anticipate that some of these clones will be identified when compared to more recent UniGene and Refseq builds. Further analysis to determine the number of unique genes represented revealed 12,009 unique cDNA clones distributed between the five libraries.
Table 1 lists the total number of cDNA clones sequenced from each library, the number of library-specific genes represented, and the distribution of cDNA clones relative to each of the five libraries in different pair-wise combinations. In terms of library-specific transcripts, 1246 genes in the encyclopedia were unique to E12.5 WT, 642 to E14.5 WT, 1256 to E14.5 Prop1df/df, 629 to the subtraction of E14.5 Prop1df/df from E14.5 WT, and 1264 from the subtraction of E12.5 WT from E14.5 WT. 3029 genes were expressed universally in all libraries.
Table 1. Number and distribution of cDNA clones in the embryonic pituitary encyclopedia.
| E12.5 WT | E14.5 WT | E14.5 Prop1df/df | Sub1 E14.5 WT-E14.5 Prop1df/df |
Sub2 E14.5 WT-E12.5 WT |
|
|---|---|---|---|---|---|
| Total UniGene entries/library | 4192 | 4339 | 4268 | 4710 | 5191 |
| E12.5 WTa | 1246b | 544 | 837 | 610 | 821 |
| E14.5 WT | 544 | 642 | 517 | 539 | 854 |
| E14.5 Prop1df/df | 837 | 517 | 1256 | 563 | 847 |
| Sub1 | 610 | 539 | 563 | 629 | 1160 |
| Sub2 | 821 | 854 | 847 | 1160 | 1264 |
| Genes represented in all five libraries | 3029 | ||||
Example interpretation: Clones sequenced from the E12.5 WT library matched 4192 different UniGene entries, 1246 were unique to this library, 3029 were found in all libraries.
Unique library-specific transcripts are on the diagonal, shaded in gray.
The majority of genes are represented by a small number of clones
The underlying principle of our approach was to develop a tool for novel gene discovery that addressed the depth and complexity of the embryonic pituitary transcriptome. Our sequencing effort was designed to maximize gene identification and enrich for those genes that were rare rather than abundant. The extremely high ranking of these libraries with regard to novelty in a pool consisting of 246 libraries generated from 71 different tissues is evidence of success [38]. The E14.5 Prop1df/df library ranked 1/246, where 24% of the unique clones sequenced in this library were found to be novel, and the libraries from E14.5 WT and E12.5 WT also ranked highly, 8/246 and 16/246, respectively. Another measure of success is to ascertain the distribution of clone representation per unique gene with the expectation that the majority will have few replicates. This trend was observed in all five libraries. Figure 1 shows the results for the E14.5 WT library following analysis. The same trend was observed in all of the other libraries (data not shown).
Figure 1. Distribution of clone representation per unique transcript for the E14.5 WT library.
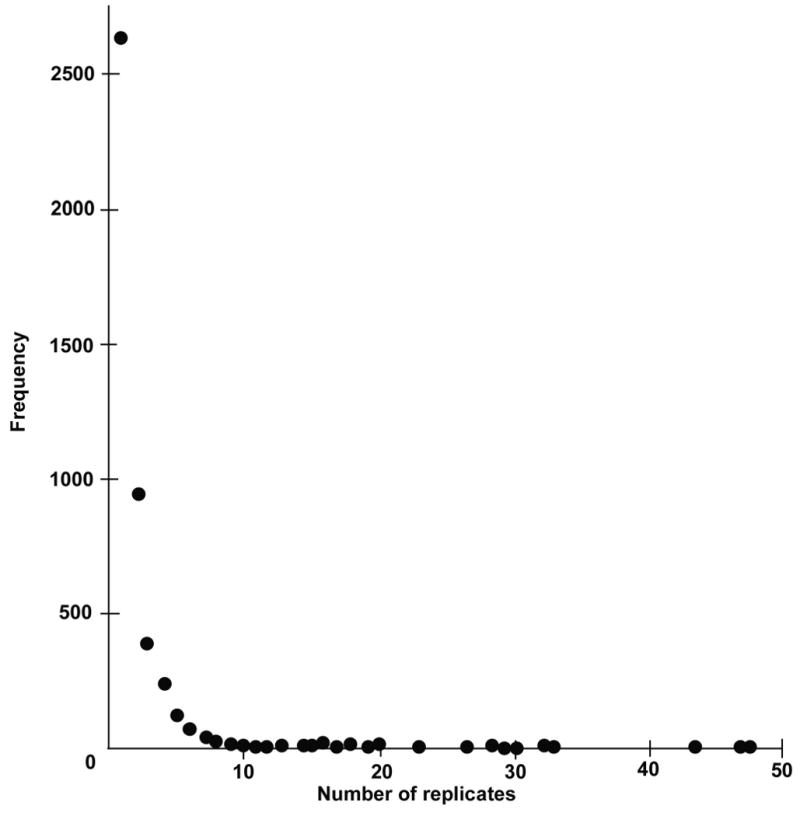
4339 unique UniGene entries are contained in the E14.5 WT library (Table 1). Over half of these (2644) are represented by a single cDNA clone (shown in the first column). A continually decreasing trend is observed, such that as the number of replicates or cDNA clones increases, the number of genes represented by these clones decreases. Number of Replicates (X axis) refers to the number of clones representing each unique UniGene entry. Frequency (Y axis) refers to the number of unique UniGene entries represented.
All three non-subtracted cDNA libraries contribute unique genetic information
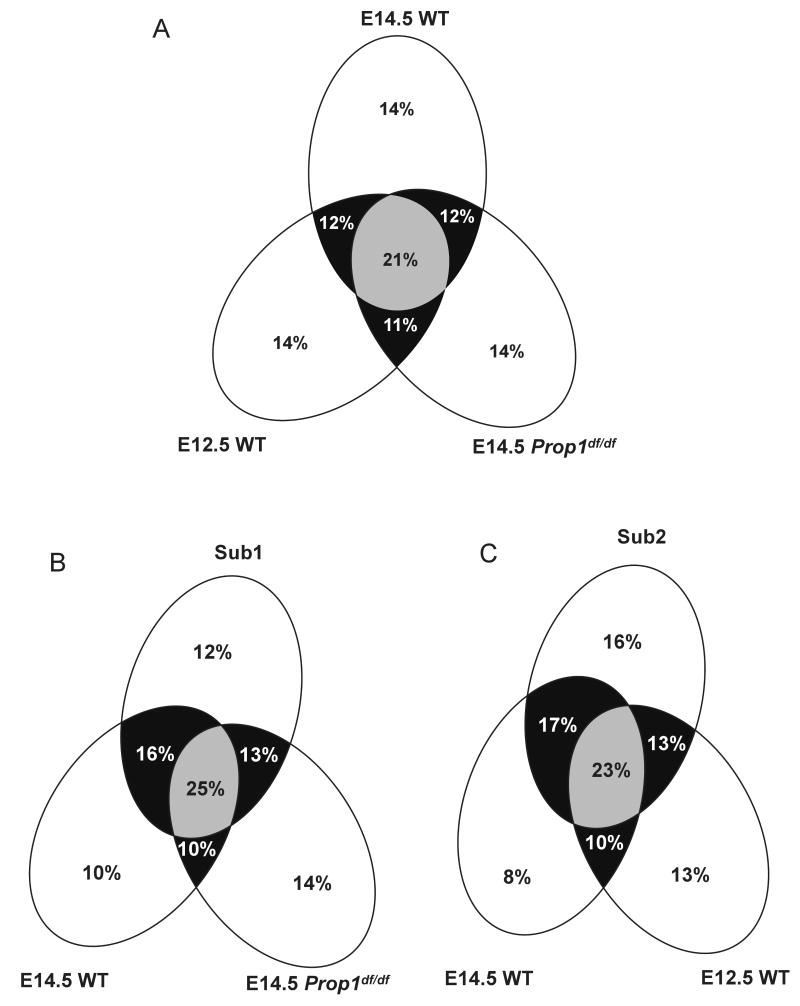
To gain greater insight into the unique cDNA distribution between the three non-subtracted libraries, we compared the relative distribution of 6,940 cDNA clones between the E12.5 WT, E14.5 WT and E14.5 Prop1df/df libraries (Figure 2a). Overall, there is similar overlap between transcripts detected in these three libraries, where any two of the three libraries considered share at least 11-12% clone representation. Each of the three libraries contains a significant proportion of apparently library-specific genes (14%), suggesting that each library contributes an important collection of unique genes to the encyclopedia that may be dependent on developmental stage and genotype.
Figure 2. Venn diagrams illustrate the relationships between the five libraries in the encyclopedia.
(A) Comparison of a total of 12,799 genes from the E12.5 WT, E14.5 WT and E14.5 Prop1df/df. (B) Comparison of a total of 5,765 genes from the E14.5 WT, E14.5 Prop1df/df and Sub1 libraries. (C) Comparison of a total of 7,590 genes from the E12.5 WT, E14.5 WT and Sub2 libraries.
The two subtracted libraries each add a dimension of genetic novelty to the encyclopedia
We compared the relative distribution of 5,765 cDNA clones between the E14.5 WT, E14.5 Prop1df/df and Sub1 libraries (Figure 2b). The Sub1 library (E14.5 WT minus E14.5 Prop1df/df) should theoretically represent those genes expressed at E14.5 in normal pituitaries but not in Prop1df/df mutants. This trend is observed, suggesting that the subtraction was effective. Gene representation in the E14.5 WT and Sub1 libraries overlaps by 16% compared to a 13% overlap between E14.5 Prop1df/df and Sub1. The percentage of genes expressed in both E14.5 WT and E14.5 Prop1df/df is also 10%. This implies that the Sub1 and E14.5 Prop1df/df libraries are as different as the E14.5 WT and E14.5 Prop1df/df libraries in terms of the expressed genes being detected or sampled. In addition to those genes that were detected in both E14.5 WT and Sub1, there are also genes uniquely detected in one library. Uniquely represented genes constitute 10-14% of the total expressed genes, which equates to 2527 genes in the case of the Sub1 library, demonstrating that it adds a dimension of genetic novelty to the encyclopedia.
We evaluated the E12.5 WT, E14.5 WT and Sub2 libraries, comparing 7,590 clones (Figure 2c). The Sub2 and E14.5 WT libraries should share expression of a larger subset of genes compared to E14.5 WT and E12.5 WT, and this trend was observed. E14.5 WT and Sub2 unique transcript expression overlaps by 17%, compared to an overlap of 10% between E14.5 WT and E12.5 WT. This overlap is equivalent to that observed between Sub2 and E12.5 WT (13%) suggesting that this subtraction was also effective. The Sub2 library adds new information to the encyclopedia as 1,264 (16%) unique cDNA clones present in the Sub2 library were not sampled in the E14.5 WT or E12.5 WT libraries.
Gene Ontology (GO) terminology reveals novel library-specific biological processes
We wanted to determine the unique biological processes represented in each library as a means for identifying the genes that contribute to the morphological and functional differences in pituitary development [13; 19]. Using Gene Ontology (GO) terms we probed the five sets of genes uniquely sampled from each library for enriched processes and also analyzed the set of genes that were detected universally. Twelve GO terms were significantly enriched (unadjusted p-value <0.01) in the universally expressed transcript set (Table 2). Enriched terms include ‘translation’, ‘RNA splicing’, ‘protein folding’ and ‘protein transport’, fundamental biological processes that are active in all tissues, irrespective of developmental stage or genotype.
Table 2. Enriched gene ontology (GO) terms.
| GO Terma | Description | P valueb |
|---|---|---|
| Universally expressed gene set | ||
| GO:0006412 | Translation | 0.0000012 |
| GO:0006457 | Protein folding | 0.0000027 |
| GO:0015671 | Oxygen transport | 0.000043 |
| GO:0008380 | RNA splicing | 0.00013 |
| GO:0006397 | mRNA processing | 0.00029 |
| GO:0009408 | Response to heat | 0.00143 |
| GO:0006986 | Response to unfolded protein | 0.00235 |
| GO:0015031 | Protein transport | 0.00242 |
| GO:0046907 | Intracellular transport | 0.00419 |
| GO:0006334 | Nucleosome assembly | 0.00629 |
| GO:0000398 | Nuclear mRNA splicing, via spliceosome | 0.00739 |
| GO:0030198 | Extracellular matrix organization and biogenesis | 0.00907 |
| E12.5 WT | ||
| GO:0019068 | Virus assembly | 0.0007 |
| GO:0050850 | Positive regulation of calcium-mediated signaling | 0.0077 |
| E14.5 WT | ||
| GO:0008216 | Spermidine metabolic process | 0.002 |
| GO:0042744 | Hydrogen peroxide catabolic process | 0.002 |
| GO:0042742 | Defense response to bacterium | 0.005 |
| E14.5 Prop1df/df | ||
| GO:0042742 | Defense response to bacterium | 0.0003 |
| GO:0006952 | Defense response | 0.0012 |
| GO:0048015 | Phosphoinositide-mediated signaling | 0.0062 |
| GO:0051046 | Regulation of secretion | 0.0062 |
| GO:0000305 | Response to oxygen radical | 0.0083 |
| GO:0006111 | Regulation of gluconeogenesis | 0.0083 |
| Sub1 (E14.5 WT - E14.5 Prop1df/df) | ||
| GO:0030174 | Regulation of DNA replication initiation | 0.002 |
| GO:0007264 | Small GTPase mediated signal transduction | 0.005 |
| Sub2 (E14.5 WT - E12.5 WT) | ||
| GO:0009268 | Response to pH | 0.002 |
| GO:0006887 | Exocytosis | 0.004 |
GO terms with an unadjusted p-value < 0.01 are listed
The E14.5 WT library contains three significantly over-represented GO terms; ‘spermidine metabolic process’, ‘hydrogen peroxide catabolic process’, and ‘defense response to bacterium’ (Table 2). While these were unexpected, it is not difficult to envision how genes in these categories could be engaged in other known processes that occur predominantly at this time. For example, spermidine markedly increases DNA binding activity of certain transcription factors in the pituitary gland [44]. It may, therefore, play an important role in the changing transcription characteristic of the developing pituitary at this time. The ‘defense response to bacterium’ GO term is also significantly enriched in the Prop1df/df library (Table 2) and is represented by more genes, compared to the E14.5 library (data not shown).
The E14.5 Prop1df/df library sampling was enriched for six GO terms (Table 2), five of which are not over-represented in any of the other library samples, providing clues regarding the types of processes occurring in the Prop1df/df pituitary that could differ from the normal state. For example, ‘Response to oxygen radical’ was enriched, a process that is involved in apoptosis, which is elevated in Prop1df/df [19; 45]. Two significantly over-represented GO terms emerged from the E12.5 WT library sample (Table 2). ‘Positive regulation of calcium signaling’ precedes the catabolism of spermidine in the pituitary, which was represented at E14.5, and the role for calcium signaling is well documented [46; 47]. The Sub1 and Sub2 library samples collectively contain four over-represented GO terms, which do not appear in any of the other samples (Table 2). ‘Exocytosis’ and ‘response to pH’ may provide a platform on which to launch future functional studies for some of the genes represented.
Validation of the embryonic pituitary encyclopedia
To validate the five libraries that comprise the embryonic pituitary encyclopedia we queried the database for the presence of several ‘knowns’, consisting of some homeobox genes with well-characterized roles in the developing pituitary gland and related gene family members. Searches for gene name, UniGene ID, and DNA or peptide sequences identified nine homeobox genes in the database (Table 3). Pou1f1, a target of Prop1 [14] was detected in the E14.5 WT library, and not in the E14.5 Prop1df/df library, as expected. Lhx3, Pitx1 and Pitx2, and two members of the Six gene family, were identified in several libraries [6; 48; 49; 50; 51]. Other expected genes detected included Otx2, Prop1, and Pax6 [6; 7; 52]. Three of the expected homeobox genes, Lhx4, Hesx1, and Pou3f4, were not detected in the database following in silico searching. Absence from the database can be an artifact of the sampling strategy for sequencing. For example, Hesx1 was detected by PCR at the expected developmental stages, suggesting that deeper sequencing would have revealed the presence of these genes (data not shown).
Table 3. Libraries contain homeobox genes implicated in pituitary development.
| Gene Symbol | Gene Name | Unigene IDa | NUMBER OF CLONES IDENTIFIEDb Pituitary cDNA Source |
||||
|---|---|---|---|---|---|---|---|
| E12.5 WT | E14.5 WT | E14.5 Prop1df/df | Sub1c | Sub2d | |||
| Lhx3 | LIM homeobox protein 3 | Mm.386765 | 5 | 3 | 2 | 5 | |
| Otx2 | Orthodenticle homolog 2 | Mm.134516 | 1 | ||||
| Pax6 | Paired box gene 6 | Mm.3608 | 1 | ||||
| Pitx1 | Paired-like homeodomain transcription factor 1 | Mm.135195 | 1 | 1 | 2 | 3 | |
| Pitx2 | Paired-like homeodomain transcription factor 2 | Mm.246804 | 2 | 1 | 2 | 5 | |
| Pou1f1 | POU domain, class 1, transcription factor 1 | Mm.457971 | 3 | ||||
| Prop1 | Paired like homeodomain factor 1 | Mm.1386 | 2 | 2 | 4 | ||
| Six3 | Sine oculis-related homeobox 3 homolog | Mm.370208 | 1 | 1 | 1 | 3 | |
| Six6 | Sine oculis-related homeobox 6 homolog | Mm.57138 | 4 | 8 | 5 | 1 | 2 |
NCBI UniGene Accession Number
number of cDNA clones identified in each library corresponding to indicated gene. Blank indicates none.
E14.5 WT - E14.5 Prop1df/df
E14.5 WT - E12.5 WT
To confirm the identity and integrity of the clones identified computationally from end sequence, at least one clone representing each homeobox gene identified in Table 3 was sequenced from both ends and examined to determine whether the libraries were enriched for full-length clones. In most cases the clones were full-length compared to the NCBI Mus musculus UniGene database (Build #159) validating the Cap-trapper technique for producing full-length clones. This validation strategy revealed that the high throughput first pass sequencing correctly identifies greater than 95% of the clones accurately. The misidentified clones tended to be ones with poor expect values from the initial BLAST analysis.
Identification of additional homeobox genes in the embryonic pituitary encyclopedia
Given the critical roles of numerous homeobox genes in the developing pituitary gland, we used the database as a tool for discovering additional homeobox genes expressed in the pituitary gland. The database is searchable using a combination of text searching with the terms ‘homeo’, ‘homeobox’ and ‘homeodomain’ or querying the database using the 60 amino acid homeodomain consensus sequence [53]. The most comprehensive method was to search using gene names that are included in a list of homeodomain containing proteins at Prosite (http://ca.expasy.org/prosite/). We recovered numerous additional homeobox genes expressed in the encyclopedia (Table 4).
Table 4. Identification of additional homeobox genes in the embryonic pituitary encyclopedia.
| Gene Symbol | Gene Name | NUMBER OF CLONES IDENTIFIEDa Pituitary cDNA Source |
Homeobox Classb | |||||
|---|---|---|---|---|---|---|---|---|
| E12.5 WT | E14.5 WT | E14.5 Prop1df/df | Sub1 | Sub2 | ||||
| Adnp | Activity-dependent neuroprotective protein | 1 | 3 | 3 | POU-like | |||
| Dlx1 | Distal-less homeobox 1 | 1 | 3 | Dll | ||||
| Dlx3 | Distal-less homeobox 3 | 1 | Dll | |||||
| Emx2 | Empty spiracles homolog 2 | 3 | 1 | Ems | ||||
| Hlx1 | H2.0-like homeo box | 1 | Hlx | |||||
| Homez | Homeodomain leucine zipper-encoding | 2 | 1 | ZF | ||||
| Isl1 | Islet 1 Tanscription factor | 2 | 1 | LIM | ||||
| Lass2 | LAG1 homolog, ceramide synthase 2 | 3 | 2 | |||||
| Lass3 | LAG1 homolog, ceramide synthase 3 | 1 | ||||||
| Lass4 | LAG1 homolog, ceramide synthase 4 | 3 | 3 | |||||
| Lass5 | LAG1 homolog, ceramide synthase 5 | 1 | 1 | 2 | ||||
| Lass6 | LAG1 homolog, ceramide synthase 6 | 3 | 2 | |||||
| Lhx2 | LIM homeobox protein 2 [69] | 1 | LIM | |||||
| Meis1 | Myeloid ecotropic viral integration site 1 | 1 | TALE | |||||
| Meox2 | Mesenchyme homeobox 2 | 1 | TALE | |||||
| Mrg1 | Myeloid ecotropic viral integration site-related gene 1 [66; 71] | 2 | 1 | TALE | ||||
| Msx1 | Homeo box, msh-like 1 [67; 86] | 1 | 1 | 1 | MSH | |||
| Nkx2-4 | NK2 transcription factor related, locus 4 | 2 | NK | |||||
| Nkx3-1 | NK-3 transcription factor, locus 1 | 3 | NK | |||||
| Obox3 | Oocyte specific homeobox 3 | 2 | OBOX | |||||
| Pax1 | Paired box gene 1 | 2 | 2 | Prd | ||||
| Pbx2 | Pre B-cell leukemia transcription factor 2 | 1 | 3 | TALE | ||||
| Pknox1 | Pbx/knotted 1 homeobox 1 [64] | 1 | TALE | |||||
| Pknox2 | Pbx/knotted 1 homeobox 2 | 1 | TALE | |||||
| Pou4f1 | POU domain class 4, transcription factor 1 [37] | 1 | 1 | POU | ||||
| Pou6f1 | POU domain class 4 transcription factor 1 [70] | 1 | POU | |||||
| Prrx2 | Paired related homeobox 2 | 1 | Prd | |||||
| Prrxl1 | Paired related homeobox protein-like 1 | 2 | Prd | |||||
| Rax | Retina and anterior neural fold homeobox [63] | 2 | 2 | 1 | Prd | |||
| Satb1 | Special AT-rich sequence binding protein 1 | 2 | 1 | 3 | Cut | |||
| Six1 | Sine oculis-related homeobox 1 homolog | 5 | 2 | 4 | 10 | 6 | Six | |
| Six4 | Sine oculis-related homeobox 4 homolog | 1 | 3 | 2 | Six | |||
| Tgif1 | TGFB-induced factor homeobox 1 | 1 | 1 | TALE | ||||
| Tgif2 | TGFB-induced factor homeobox 2 | 2 | TALE | |||||
| Zeb2 | Zinc finger E-box binding homeobox 2 [60] | 1 | ZF | |||||
| Zfhx3 | Zinc finger homeobox 3 | 1 | 2 | ZF | ||||
| Zhx1 | Zinc fingers and homeoboxes protein 1 | 2 | ZF | |||||
Number of cDNA clones identified in each library corresponding to indicated gene.
Homeobox class is indicated where known. None known for Lass3.
Differential expression analysis confirms transcription of novel homeobox genes
We analyzed 23 of these genes with respect to expression in the developing pituitary gland at E12.5, E14.5, and E18.5 using a combination of in situ hybridization analysis, RT-PCR, and quantitative PCR. Figure 3 shows a subset of the in situ hybridization results. Dlx1 expression was confirmed at E12.5 (Figure 3a) and Dlx3, Rax1, and Zhx1 at E14.5 (Figure 3b-d). For PCR analyses cDNA was generated from E12.5, E14.5, and E18.5 WT, and E14.5 Prop1df/df pituitary primordia. The quality of these cDNAs was confirmed using a series of primers specific for Prop1, Pou1f1, Hesx1, and Hprt (Figure 4, data not shown). Using standard RT-PCR expression of 12 genes was confirmed during pituitary gland development. Meis1, Adnp, Prrx2, Lhx2, Atbf1, Pknox2, Pbx2, and Mrg1 were expressed at similar levels at all time points analyzed in wild type and in Prop1df/dfRathke's pouch cDNA. Meox2, Emx2, Otx2, and Zeb2 appeared to exhibit developmentally regulated expression, with the peak expression of Meox2 at E12.5, Emx2 at E14.5, and Zeb2 at E18.5 (Figure 4a). In addition, expression of Meox2 and Otx2 appeared elevated in Prop1df/df mutant Rathke's pouch compared to wild type controls. Quantitative Real Time PCR was used to confirm the results suggested by standard RT-PCR (Figure 4b). cDNA was generated from additional E12.5, E14.5, E18.5 WT, and E14.5 Prop1df/df pituitary primordia and the quality confirmed using primer sets for Prop1 and Pou1f1. All samples were done in triplicate and normalized to Hprt. The fold activation was determined by comparison to E14.5 WT, which was set to 1. Both Meox2 and Otx2 peak in expression at E12.5 and are elevated 7.9 and 6 fold, respectively, relative to E14.5 WT. The peak expression of Emx2 is between E12.5 and E14.5, while Zeb2 peaks later in development at E18.5. Meox2 and Otx2 are slightly elevated in the E14.5 Prop1df/dfmutants relative to E14.5 WT by 1.59 fold and 2.06 fold, respectively. Zeb2, Adnp, and Meis1 are decreased in the Prop1df/dfmutants relative to E14.5 WT by 2.09, 2.15, and 2.39 fold, respectively. Prrx2 does not exhibit developmental regulation or differences between wild type and Prop1df/df. Taken together these results suggest that Meox2, Otx2, Zeb2, Adnp, and Meis1 could merit further evaluation as downstream targets of Prop1.
Figure 3. In situ hybridization of four homeobox genes with novel expression in the developing pituitary gland.
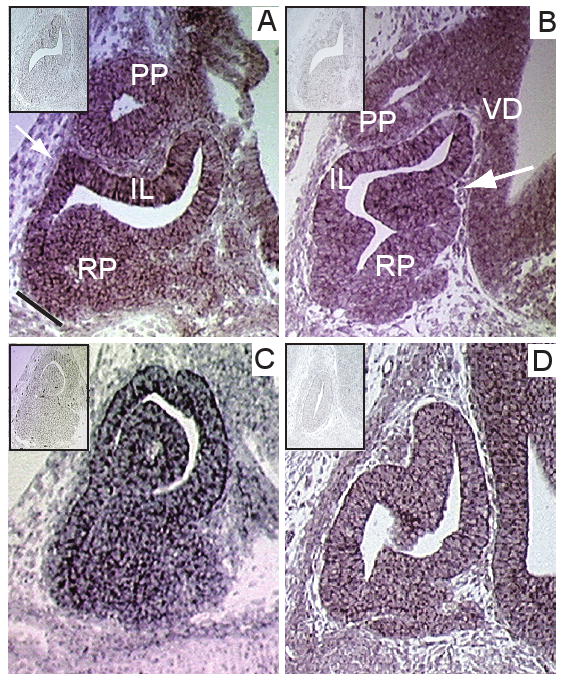
In situ hybridization analysis of Dlx3 at E12.5 (A) and Dlx1 (B), Rax (C) and Zhx1 (D) at E14.5 in the developing pituitary indicates robust expression. (A) At E12.5 Dlx3 is expressed strongly in the intermediate lobe (IL), shown by the white arrow, as well as in the posterior pituitary (PP) and throughout the expanding Rathke's pouch (RP), though predominantly in the dorsal aspect. The dorsal aspect is indicated by the black diagonal line. (B) At E14.5 Dlx1 is expressed throughout Rathke's pouch and the intermediate lobe. Expression appears concentrated in the ventral aspect of the pouch, as shown by the white arrow. Dlx1 expression is also present in the ventral diencephalon (VD). Rax (C) and Zhx1 (D) are expressed throughout the developing Rathke's pouch (RP). Sense controls corresponding to each gene are shown in the top left hand corner of each panel.
Figure 4. Expression of homeobox genes in the developing wild type and Prop1df/df pituitary.
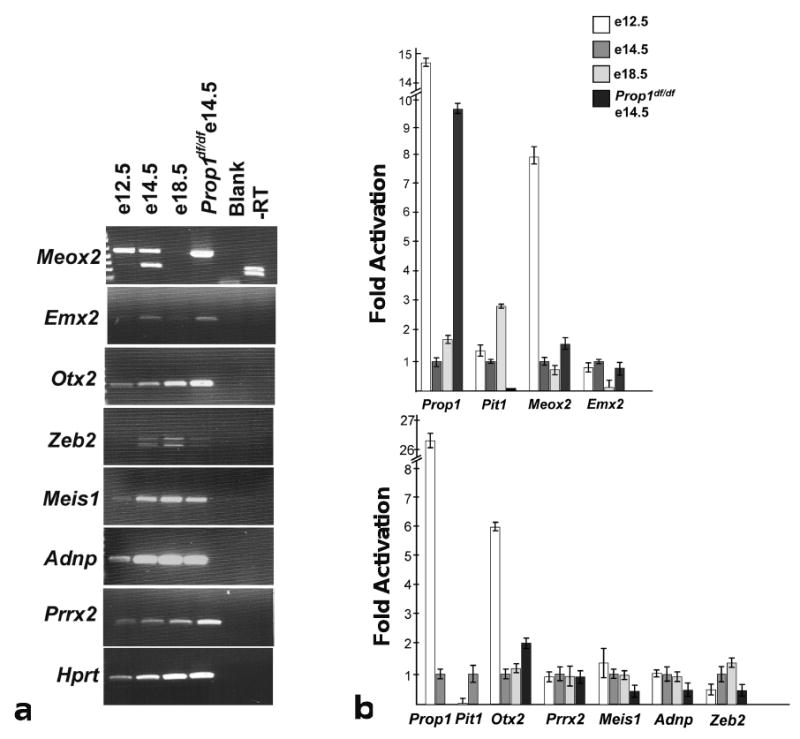
(A) Pituitary expression of a set of homeobox genes identified in the embryonic pituitary encyclopedia was validated by RT-PCR using cDNA generated from pituitary primordia at E12.5, E14.5, E18.5, and Prop1df/df E14.5. The homeobox genes Meox2, Emx2, Otx2, and Zeb2 show differential expression between E12.5-E18.5 and in the Prop1df/df E14.5 pituitary, while Meis1, Adnp, and Prrx2 showed similar expression in all samples. Hprt is a control for RNA and cDNA quality. Differential expression was quantitated using Real Time PCR (B). The quality of the cDNA samples was confirmed by demonstrating the expected pattern of Prop1 and Pit1 expression. Meox2, Emx2, Otx2, Prrx2, Meis1 and Adnp expression levels were validated using pituitary cDNA from E12.5 WT (white bars), E14.5 WT (dark gray bars), E18.5 WT (light gray bars). Different E12.5 cDNA preparations were used in the top and bottom panels of B. All samples were done in triplicate and standardized to Hprt. Fold activation and standard deviations were calculated relative to E14.5 WT based on the ΔCt values of each sample compared to the ΔCt of Hprt.
The identification of individual homeobox genes in the specific libraries does not necessarily correlate with the actual expression profile of that gene. For instance, Meis1 was only identified in the E12.5 library; however, our RT-PCR data show that it is expressed at E12.5, E14.5, and E14.5 Prop1df/df embryos. Therefore, it is misleading to assume that the identification of a gene in a specific library or even a subtracted library provides accurate differential expression information.
Database analysis implicates additional signaling molecules in pituitary development
Fundamental to the current understanding of the genetic mechanisms driving pituitary development has been the identification of numerous signaling pathways that exert their effects on the pituitary primordia. Previous studies have demonstrated the importance of individual members of the BMP, FGF, NOTCH, SHH and WNT signaling pathways in directing normal pituitary development [1; 2; 3; 5; 23; 24; 25; 54; 55]. A continuing challenge in the field is to identify additional members of these pathways and to determine how these various signaling pathways interact with each other during pituitary organogenesis to promote development of the organ.
We used Genomatix software (www.genomatix.de/) to build functional genetic networks for signaling pathways known to affect pituitary development and queried the database for the expression of these genes. For example, in the case of the BMP pathway, a group of 78 Bmp-related genes emerged, and we identified 20 of them in the encyclopedia. This analysis was conducted for additional pathways, revealing a total of 61 genes expressed in the developing pituitary gland related to the BMP (20), FGF (13), NOTCH (2), SHH (1) or WNT (25) signaling pathways. A subset corresponding to the FGF pathway is shown in Table 5, and the complete list is available in Supplementary Table 1. 17 additional genes were identified that are involved in functional crosstalk between two or more signaling pathways (Table 5 and Supplementary Table 1).
Table 5. Identification of signaling-related genes in the embryonic pituitary encyclopediaa.
| Gene Symbol | Gene Name |
|---|---|
| Fgf-relatedb | |
| Fgf10 | Fibroblast growth factor 10 |
| Fgf13 | Fibroblast growth factor 13 |
| Fgf14 | Fibroblast growth factor 14 |
| Fgf17 | Fibroblast growth factor 17 |
| Fgfr1 | Fibroblast growth factor receptor 1 |
| Fgfr2 | Fibroblast growth factor receptor 2 |
| Fibp | Fibroblast growth factor intracellular binding protein |
| Frs3 | Fibroblast growth factor receptor substrate 3 |
| Spry1 | Sprouty homolog 1 (Drosophila) |
| Spry2 | Sprouty homolog 2 (Drosophila) |
| Spred2 | Sprouty-related, EVH1 domain containing 2 |
| Sp3 | Trans-acting transcription factor 3 |
| Tbx3 | T-box 3 |
| BMP-relatedc | |
| Id2 | Inhibitor of DNA binding 2 |
| Id3 | Inhibitor of DNA binding 3 |
| Tgfβi | Transforming growth factor, beta induced |
| Functional Cross Talkd | |
| Ascl1* | Achaete-scute complex homolog-like 1 (Drosophila) |
| Id1* | Inhibitor of DNA binding 1 |
| Jun* | Jun oncogene |
| Neurod1* | Neurogenic differentiation 1 |
A subset of the 63 genes identified is shown. The complete list is available in Supplementary Figure 1.
Of the Fgf genes detected, only Fgf10 was previously known to be expressed in the pituitary gland.
The expression patterns of these were studied by in situ hybridization.
The asterisk
indicates genes in the basic helix-loop-helix family.
As an alternative strategy we searched the library for GO terms related to signaling and for specific gene names for genes known to be involved in the BMP, FGF, NOTCH, SHH, and WNT pathways. This search identified an additional 72 genes (Supplement Table 1). This list has not been verified by end sequencing yet.
Spatial expression of three Bmp-related genes in the developing pituitary
We searched the set of 61 signaling genes described above for those with previously demonstrated roles in the developing pituitary. Twelve “known” genes are in the set including Aes and Tle4 [23], Bmpr1a and Bmpr1b [56], Chrd [55], Fgf10 [1] Titf1 [4], Hey1 and Notch2 [3], Wnt5a [2; 25; 55], Axin2 [2; 57] and Fzd5 [21] (Supplementary Table 1). Previously we presented the expression pattern for the BMP signaling related genes Nbl1 and Fstl1, found in our developmental library, at E12.5 and E14.5, and for Id3 at E10.5 [54]. We expanded the analysis of Id3 and examined the expression of Id2 and Tgfbi (Figure 5). Id2 expression was detected in the dorsal aspect of Rathke's pouch at E12.5 (Figure 5a). Expression persists at E14.5 in the expanding anterior pituitary (Figure 5b) but appears restricted to those cells closest to the lumen of the pouch. Id2 does not appear to be expressed in the proliferating cells of the expanding Rathke's pouch. By E16.5 Id2 expression is barely detectable in the pituitary (data not shown). Id3 expression was detected in Rathke's pouch at E12.5, enriched in the ventral part of the pouch and in the intermediate lobe (Figure 5c). Similar to Id2, Id3 expression does not appear in the proliferating cells of the expanding Rathke's pouch. Expression of Id3 continues at E14.5 (Figure 5d) in similar pattern, restricted to the dorsal pouch and intermediate lobe and is barely detectable by E16.5 (data not shown). Tgfbi showed a striking expression pattern. While there are no transcripts in Rathke's pouch itself, the gene is expressed in the mesenchymal tissue immediately surrounding the pouch at E12.5 (Figure 5e). At E14.5, this expression continues more strongly and is also detectable surrounding the posterior pituitary (Figure 5f). Like Id2 and Id3, Tgfbi expression wanes by E16.5. These studies confirm expression of these genes in the pituitary and support the encyclopedia as reflection of pituitary gene expression.
Figure 5. Developmental expression of 3 novel Bmp-related genes in the embryonic pituitary gland.
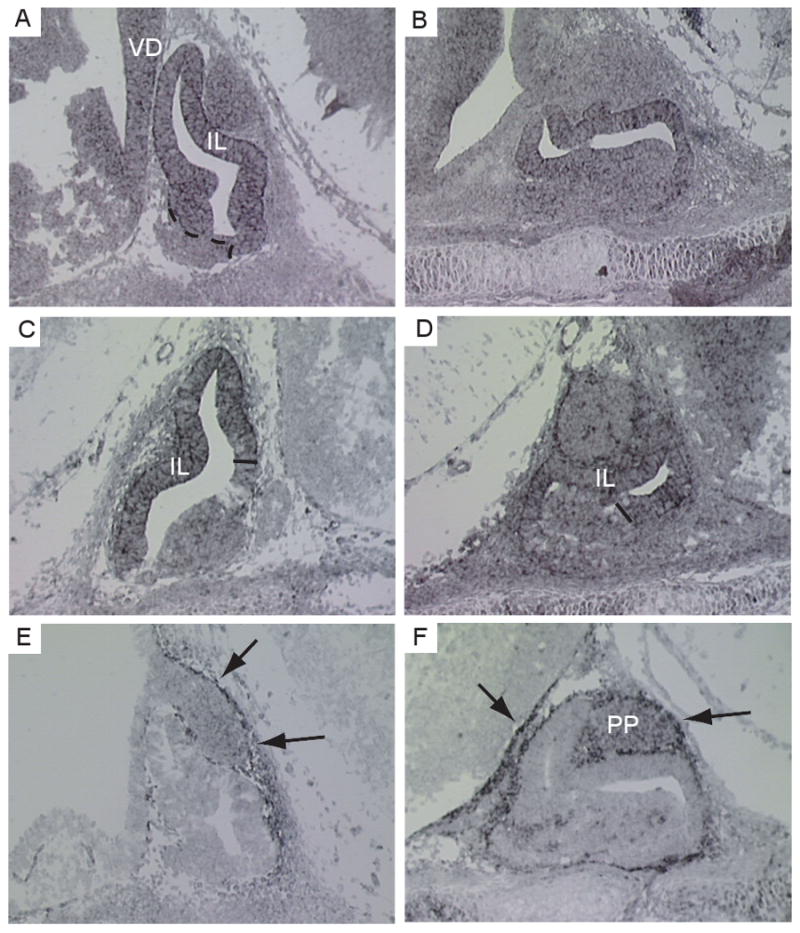
(A) Expression of Id2 is predominantly in the intermediate lobe (IL) and around the lumen of the pituitary. Expression appears restricted to the cells that are not in the expanding proliferating Rathke's pouch (delineated by the black dashed line). Expression is also detected in the ventral diencephalon (VD). This expression pattern persists at E14.5 (B). (C) At E12.5 Id3 is expressed in the IL and dorsal part of the cells surrounding the pituitary lumen (indicated by the horizontal black line) and this expression pattern continues at E14.5 (D). (E) At E12.5 Tgfbi was detected in the mesenchymal tissue immediately surrounding the developing pituitary (shown by the black arrows). By E14.5 (F), Tgfbi expression becomes stronger in the mesenchyme and is also detected surrounding the posterior pituitary (PP) (shown by the black arrows).
Interplay of signaling pathways in the developing pituitary
We surveyed the signaling genes represented in the encyclopedia using Genomatix software to identify those genes that displayed a functional overlap between two or more of the five pathways defined in Supplementary Table 1. This analysis revealed the presence of an additional 16 genes in our encyclopedia that are involved in functional cross talk during organ development, including pituitary (Supplementary Table 1). A subset of these (4/16) is shown in Table 5.
4/16 genes of the pathway interaction genes identified belong to the helix-loop-helix transcription factor family (Table 5). These include Ascl1 and Id1. The zebrafish Ascl1 orthologue (Ascl1a) is a critical gene for pituitary cell differentiation and cell survival [58]. Zebrafish Fgf3 mutants fail to express Ascl1a, suggesting that Ascl1a acts downstream of Fgf. Zebrafish Fgf3 is orthologous to mammalian Fgf10, known for its function in mammalian pituitary development, implying a conserved mechanism for FGF signaling and Ascl1 expression [1]. Additionally, Id1 mediated expression of Bmp2 leads to reduced Ascl1 expression [59]. Ascl1, Id1, Id2, Id3, and Id4 are all implicated as differentially expressed in pituitary tumors [28; 60; 61; 62]. Together these studies suggest a functional link between FGF and BMP signaling and identify the Id gene family and Ascl1 as interesting candidates in mediating cell differentiation in the developing pituitary. This example demonstrates the utility of the embryonic pituitary encyclopedia in providing a foundation for furthering our understanding of signaling in the developing pituitary.
Discussion
In previous studies we described a partial expression profile of the developing pituitary gland and paved the way for understanding the role of WNT signaling in pituitary development using a combination of differential display PCR [22] and subtractive hybridization [21]. The datasets generated in these studies were fairly small (<400 genes in each), but the gene discovery was significant, providing proof of principle that generation of a more comprehensive pituitary transcriptome profile would likely enable significant gene discovery and provide insight into the transcription factors and signaling molecules expressed in the developing pituitary.
In this study we generated a full-length cDNA encyclopedia of the developing pituitary at wild type E12.5 and E14.5 and in the Prop1df/df pituitary at E14.5 using an approach designed to enrich for full-length clones, maximize gene discovery, and permit high throughput gene recovery [38; 40; 41; 42; 43]. Using sequence data from these libraries we built a comprehensive database through which to access transcript information. It is searchable by Gene Ontology (GO) terminology; direct sequence query, including nucleic and amino acid sequence; gene name or text-string options; motifs for transcription factor binding; and UniGene ID and database-specific clone number. This encyclopedia contains 12,009 unique transcripts, represented by 56,716 cDNA clones, and accounts for over half the known genes in the mouse genome (22,723) (www.ensembl.org/Mus_musculus/index.html). Thus, the developing pituitary transcriptome has depth and complexity, and our study provides clones ideal for use in functional studies such as cell culture transfection and the generation of transgenic mice.
Each of the libraries contained within the encyclopedia includes a set of library-specific cDNA clones representing between 630 (Sub1) and 1328 (Sub2) unique transcripts. Exclusivity in any one library is not a true readout of the pituitary transcriptome, but rather, a reflection of the sequencing approach taken. Novelty and enrichment for rare transcripts were favored in place of the complete sequencing of any one library. Despite this limitation, the GO terminology analysis suggests that certain biological processes are significantly enriched in a library-specific manner. The genes represented by these processes could provide the basis for future novel mechanistic studies of embryonic pituitary development.
We identified a total of 45 homeobox genes that are expressed in the developing pituitary gland, expanding significantly on the subset of homeobox genes with well-characterized roles. Transcripts for seven of these newer genes have been detected in pituitary cell lines (Lhx2, Mrg1, Pknox1, Pou6f1), pituitary adenomas (Pou4f1, Zeb2), or other developing vertebrates (Rax) [37; 60; 63; 64; 65; 66; 67; 68; 69; 70; 71]. These genes correspond to over fifteen different homeodomain classes including the LIM, Paired (Prd), Paired-like (Prd-like) and POU homeobox families [reviewed by [72]. Given the importance of known genes from these gene families in pituitary organogenesis such as Prop1, Pou1f1, Pitx1, and Pitx2, the identification of Adnp (POU-like), Prrx2 (Prd), and Prrxl1 (Prd), suggests that these genes may also play important roles in driving cell proliferation and/or differentiation during the formation of the pituitary gland. Mutations in some of these genes may underlie some cases of MPHD, where the current genetic cause is unknown.
Interestingly, Meox2, Otx2, Emx2, and Zeb2 are differentially expressed in the developing Rathke's pouch between E12.5 and E18.5. No obvious differences in pituitary cell specification are noted in Emx2 mutant mice (Potok, unpublished observation). Meox2 and Otx2 are elevated in the Prop1df/df E14.5 pituitary relative to E14.5 wild type, and Prop1df/df newborns have ectopic, elevated expression of Otx2 compared to wild type littermates (Mortensen, unpublished observation). Meis1 and Adnp are reduced in the Prop1df/df E14.5 pituitary relative to E14.5 WT. The dynamic expression patterns of these homeobox genes in the developing pituitary raise the question of whether they are regulated by Prop1 and whether they have functional roles in pituitary development.
Some of the remaining homeobox genes identified represent novel classes of homeobox genes, not previously found in the developing pituitary. For example, three zinc finger (ZF) homeobox genes were identified in the encyclopedia (Table 4). Zeb2 expression was detected at low levels at E12.5, expression levels increased at both E14.5 and E18.5 in normal mice and expression was reduced in the Prop1df/df E14.5 pituitary (Figure 4), and Zhx1 has been detected at e12.5 by in situ hybridization (data not shown). Mutations in one of these genes, ZEB2 or ZFHX1B, causes Mowat Wilson Syndrome, a human disorder characterized by a distinctive facial phenotype in association with mental retardation, microcephaly, and short stature [73]. Because this gene is expressed in human pituitary, it is intriguing to hypothesize that affected individuals with this syndrome have a pituitary defect [60].
Several genes belonging to the TALE family of homeobox genes were identified in the embryonic pituitary (Pbx2, Meox2, Meis1, Mrg1, Pknox2, Tgif1, Tgif2). Genes from this family are required to specify cell fate in numerous organs including the spleen, and they may play a similar role in the developing pituitary [74]. The TALE gene Pbx1 regulates expression of the Fshb gene in pituitary gonadotrope-like cells and Gnrh in the hypothalamus [64; 75]. MRG1 interacts with LHX2 to regulate Cga expression in pituitary gonadotrope-like cells [66]. Our expression analysis of Meox2 suggests that Prop1 may repress its expression during pituitary development.
One of the current challenges in the field is to determine how the various signaling pathways interact with each other during pituitary organogenesis. BMP and FGF signals influence anterior pituitary cell differentiation [1; 55], and SHH signaling may participate in boundary formation with FGF and BMP signaling in the ventral diencephalon [54; 55]. Cross talk between the WNT, BMP, FGF and SHH pathways is evident in Tcf7l2, Wnt5a, and noggin mutants, which exhibit pituitary developmental abnormalities [2; 24; 54]. Using the embryonic pituitary encyclopedia we identified 79 additional BMP, FGF, NOTCH, SHH or WNT pathway members with novel expression in the developing pituitary. One such example is Fgf13, which is critical for neural differentiation in Xenopus [76]. Additionally, three Sprouty genes were identified in the encyclopedia (Spry1, Spry2 and Spred2). Spry1, Spry2, and Spry4 expression was previously reported in E14.5 Rathke's pouch [77]. Sprouty genes have highly conserved roles as antagonists of FGF signaling, which can affect differentiation [78; 79]. Thus, several members of the sprouty family may regulate FGF signaling during pituitary organogenesis.
An important application of our studies lies in the ability to utilize the full-length cDNA clones. Some examples are the full-length cDNA clones for the Fstl1, Id3, and Six6 genes that were used for expression analysis by in situ hybridization in pituitary development in demonstrating the important role of noggin [54]. Full-length cDNA clones for the forkhead transcription factor Foxl2 and numerous members of both the Wnt and Frizzled gene families, (Fzd6, Axin2 and Dvl2, see Supplementary Table 1) have been invaluable [2; 18]
In conclusion, the embryonic pituitary encyclopedia represents a valuable gene discovery tool that can be used to identify novel genes and pathways in the developing wild type and mutant pituitary (Prop1df/df) and to initiate functional studies. We have just scratched the surface and anticipate that this full-length cDNA encyclopedia will act as an ongoing resource for studying pituitary development and disease.
Materials and Methods
Mice
Prop1df/+ mouse stocks were obtained from Dr. A. Bartke (Southern Illinois University, Carbondale, Ill.) in 1988 and maintained at the University of Michigan according to NIH guidelines. Heterozygote carriers were intercrossed to generate timed pregnancies and the morning after mating was designated as E0.5. Developing pituitary glands were dissected from E14.5 embryos with the aid of a dissecting microscope and were individually stored in RNA Later (Ambion, Austin, TX) at -20°C. Genomic DNA was extracted from the yolk sac, genotyping was performed as described [21], and pituitaries were pooled for RNA extraction and cDNA generation following genotyping. CD1 mice were purchased from Charles River Laboratories (Wilmington, MA) as timed pregnancies and pituitary glands were dissected from E12.5, E14.5, and E18.5 embryos. Pituitaries were pooled for RNA extraction and cDNA generation. The University of Michigan Committee on Use and Care of Animals approved all experiments.
Library generation
Pituitary tissue for library generation was prepared as described [38]. Briefly, starting from 50 micrograms of total RNA, first strand cDNA was prepared with an anchored oligo-dT primer adapter [80] and the cDNA was cap-selected [81] and subsequently cloned in a Lambda FLC I vector [41]. Libraries were excised into plasmids and sequenced. E14.5 WT was given the library ID K7, E14.5 Prop1df/df was K8, and E12.5 WT was K9 [38]. Additionally, subtracted libraries were prepared from E14.5 WT minus E14.5 Prop1df/df and E14.5 WT minus E12.5 WT, using established procedures [82].
Database generation
Mouse UniGene Build #159 (Mm.seq and Mm.seq.uniq) and Mouse RefSeq Release 20 were obtained from NCBI FTP servers, along with the ‘blastall’ program used to perform the BLAST identifications. Each cDNA sequence was first BLASTed against the Mm.seq.uniq dataset, and then against the full mouse UniGene set (Mm.seq). All cDNA sequences were also BLASTed against RefSeq to provide an alternative identification.
A database was constructed to contain the sequences themselves, along with Riken identification and a ‘library’ identification, indicative of the tissue source along with the UniGene and RefSeq identifications, where obtainable. Web-based search functions were developed to allow exploration of the contents of the various libraries by sequence, gene name, etc. From the GO Consortium (www.geneontology.org) we downloaded a database of GO identifiers in hierarchical format (10/31/06 release). This data, along with files from the NCBI ‘Gene’ database (‘gene2go’, ‘gene2unigene’) allowed us to query our UniGene identifications using GO terminology.
Gene Ontology (GO) terminology analysis
Over-represented Gene Ontology (GO) terms in the set of unique sequences from each library as well as the intersecting sequences for all libraries were detected using a modification of the standard Fisher's exact test [83]. Briefly, we selected those sequences that were unique to each library based on the UniGene ID and mapped these IDs to Entrez Gene IDs using the mouseLLMappings package of Bioconductor (http://www.bioconductor.org). We mapped the union of all sequences in each library to Entrez Gene to use as the ‘universe’ from which the unique sequences were selected. In other words, the usual heuristic explanation of a Fisher's exact test is the idea of selecting colored balls from an urn. In our case, the unique sequences from each library are the balls that were selected from the urn, and the union of all sequences in each library is the ‘universe’ of balls in the urn. We computed over-representation of GO terms using the topGO package of Bioconductor, which uses the directed acyclic graph (DAG) structure of the GO ontologies to decorrelate significant subordinate terms from consideration when estimating the significance of higher-order terms.
In situ hybridization
RNA in situ hybridization was performed as described previously [84]. Dlx1, Dlx3, Rax, Zhx1, Id2, Id3 and Tgfbi cDNAs were isolated from the embryonic pituitary encyclopedia [38]. Id2, Id3 and Tgfbi were linearized with NotI, and Dlx1, Dlx3, Rax and Zhx1 were linearized with SacII. T3 polymerase was used in all cases to generate antisense probes.
RT-PCR and PCR
RNA was extracted from pituitary tissue dissected from CD1 mouse embryos at E12.5, E14.5, and E18.5 and stored in RNAlater (Ambion, Foster City, CA) following the recommended commercial protocol. RNA was further purified via RNAqueous 4PCR (Ambion, Foster City, CA). cDNA was prepared from total RNA using the Superscript First Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). Briefly, 5μg total RNA was incubated at 42°C for 50 minutes in a 10μl reaction containing 2μl cDNA buffer, 4μl MgCl2, 2μl 0.1μl DTT, and 1μl of Superscript II RT enzyme. Standard PCR reaction components at final concentrations were 1 X PCR Reaction buffer (Roche, Indianapolis, IN), 1.5mM MgCl2 (Perkin Elmer, Waltham, MA), 200mM dNTP mix (Roche), and 0.4U GoTaq polymerase (Perkin Elmer) per 25μl reaction. Standard reactions contained 25-50ng template cDNA.
All PCR reactions were performed using the following conditions: 94°C; 4 minutes, followed by 30 cycles of 94°C; 30 seconds, PCR-specific annealing temperature; 30 seconds and 72°C; 30 seconds, followed by 1 cycle of 72°C; 10 minutes. PCR-specific annealing temperatures and primers are listed in Supplementary Table 2.
Quantitative Real Time PCR was performed using TaqMan Gene Expression Assays On Demand (Applied Biosystems, Foster City, CA). Briefly 25-50ng of template cDNA was added to 10μl Taqman Universal PCR Master Mix (Applied Biosystems), 8 μl double distilled H2O, and 1 μl TaqMan gene specific primer set. All samples were done in triplicate and standardized to Hprt. Real Time PCR was performed using the Prism 7000 Sequence Detection System (Applied Biosystems). Fold activation and standard deviations were determined as previously described [85].
Supplementary Material
Acknowledgments
This work was supported by the following grants: National Institutes of Health R37HD30428 and HD34283 (SAC), NIH Grant R01GM72007 (JM, DG), University of Michigan Center for Computational Medicine and Biology (SAC, RHL, DG), University of Michigan DNA Sequencing Core Facility (RHL) and Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (YH) and a grant for the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology, Japan (YH). We thank A.H. Mortensen, M.A. Potok, and N.M. Solomon for contributions to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ericson J, Norlin S, Jessell TM, Edlund T. Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development. 1998;125:1005–15. doi: 10.1242/dev.125.6.1005. [DOI] [PubMed] [Google Scholar]
- 2.Potok MA, Cha KB, Hunt A, Brinkmeier ML, Leitges M, Kispert A, Camper SA. WNT signaling affects gene expression in the ventral diencephalon and pituitary gland growth. Dev Dyn. 2008;237:1006–20. doi: 10.1002/dvdy.21511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raetzman LT, Ross SA, Cook S, Dunwoodie SL, Camper SA, Thomas PQ. Developmental regulation of Notch signaling genes in the embryonic pituitary: Prop1 deficiency affects Notch2 expression. Dev Biol. 2004;265:329–40. doi: 10.1016/j.ydbio.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 4.Takuma N, Sheng HZ, Furuta Y, Ward JM, Sharma K, Hogan BL, Pfaff SL, Westphal H, Kimura S, Mahon KA. Formation of Rathke's pouch requires dual induction from the diencephalon. Development. 1998;125:4835–40. doi: 10.1242/dev.125.23.4835. [DOI] [PubMed] [Google Scholar]
- 5.Treier M, O'Connell S, Gleiberman A, Price J, Szeto DP, Burgess R, Chuang PT, McMahon AP, Rosenfeld MG. Hedgehog signaling is required for pituitary gland development. Development. 2001;128:377–86. doi: 10.1242/dev.128.3.377. [DOI] [PubMed] [Google Scholar]
- 6.Kelberman D, Dattani MT. The role of transcription factors implicated in anterior pituitary development in the aetiology of congenital hypopituitarism. Ann Med. 2006;38:560–77. doi: 10.1080/07853890600994963. [DOI] [PubMed] [Google Scholar]
- 7.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–58. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 8.Szeto DP, Rodriguez-Esteban C, Ryan AK, O'Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisua-Belmonte JC, Rosenfeld MG. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–94. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mody S, Brown MR, Parks JS. The spectrum of hypopituitarism caused by PROP1 mutations. Best Pract Res Clin Endocrinol Metab. 2002;16:421–31. doi: 10.1053/beem.2002.0218. [DOI] [PubMed] [Google Scholar]
- 10.Andersen B, Pearse RV, 2nd, Jenne K, Sornson M, Lin SC, Bartke A, Rosenfeld MG. The Ames dwarf gene is required for Pit-1 gene activation. Dev Biol. 1995;172:495–503. doi: 10.1006/dbio.1995.8040. [DOI] [PubMed] [Google Scholar]
- 11.Gage PJ, Brinkmeier ML, Scarlett LM, Knapp LT, Camper SA, Mahon KA. The Ames dwarf gene, df, is required early in pituitary ontogeny for the extinction of Rpx transcription and initiation of lineage-specific cell proliferation. Mol Endocrinol. 1996;10:1570–81. doi: 10.1210/mend.10.12.8961267. [DOI] [PubMed] [Google Scholar]
- 12.Gage PJ, Roller ML, Saunders TL, Scarlett LM, Camper SA. Anterior pituitary cells defective in the cell-autonomous factor, df, undergo cell lineage specification but not expansion. Development. 1996;122:151–60. doi: 10.1242/dev.122.1.151. [DOI] [PubMed] [Google Scholar]
- 13.Nasonkin IO, Ward RD, Raetzman LT, Seasholtz AF, Saunders TL, Gillespie PJ, Camper SA. Pituitary hypoplasia and respiratory distress syndrome in Prop1 knockout mice. Hum Mol Genet. 2004;13:2727–35. doi: 10.1093/hmg/ddh311. [DOI] [PubMed] [Google Scholar]
- 14.Sornson MW, Wu W, Dasen JS, Flynn SE, Norman DJ, O'Connell SM, Gukovsky I, Carriere C, Ryan AK, Miller AP, Zuo L, Gleiberman AS, Andersen B, Beamer WG, Rosenfeld MG. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–33. doi: 10.1038/384327a0. [DOI] [PubMed] [Google Scholar]
- 15.Charles MA, Suh H, Hjalt TA, Drouin J, Camper SA, Gage PJ. PITX genes are required for cell survival and Lhx3 activation. Mol Endocrinol. 2005;19:1893–903. doi: 10.1210/me.2005-0052. [DOI] [PubMed] [Google Scholar]
- 16.Ellsworth BS, Butts DL, Camper SA. Mechanisms underlying pituitary hypoplasia and failed cell specification in Lhx3-deficient mice. Dev Biol. 2008;313:118–29. doi: 10.1016/j.ydbio.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suh H, Gage PJ, Drouin J, Camper SA. Pitx2 is required at multiple stages of pituitary organogenesis: pituitary primordium formation and cell specification. Development. 2002;129:329–37. doi: 10.1242/dev.129.2.329. [DOI] [PubMed] [Google Scholar]
- 18.Ellsworth BS, Egashira N, Haller JL, Butts DL, Cocquet J, Clay CM, Osamura RY, Camper SA. FOXL2 in the pituitary: molecular, genetic, and developmental analysis. Mol Endocrinol. 2006;20:2796–805. doi: 10.1210/me.2005-0303. [DOI] [PubMed] [Google Scholar]
- 19.Ward RD, Raetzman LT, Suh H, Stone BM, Nasonkin IO, Camper SA. Role of PROP1 in pituitary gland growth. Mol Endocrinol. 2005;19:698–710. doi: 10.1210/me.2004-0341. [DOI] [PubMed] [Google Scholar]
- 20.Ward RD, Stone BM, Raetzman LT, Camper SA. Cell Proliferation and Vascularization in Mouse Models of Pituitary Hormone Deficiency. Mol Endocrinol. 2006 doi: 10.1210/me.2005-0409. [DOI] [PubMed] [Google Scholar]
- 21.Douglas KR, Brinkmeier ML, Kennell JA, Eswara P, Harrison TA, Patrianakos AI, Sprecher BS, Potok MA, Lyons RH, Jr, MacDougald OA, Camper SA. Identification of members of the Wnt signaling pathway in the embryonic pituitary gland. Mamm Genome. 2001;12:843–51. doi: 10.1007/s00335-001-2076-0. [DOI] [PubMed] [Google Scholar]
- 22.Douglas KR, Camper SA. Partial transcriptome of the developing pituitary gland. Genomics. 2000;70:335–46. doi: 10.1006/geno.2000.6400. [DOI] [PubMed] [Google Scholar]
- 23.Brinkmeier ML, Potok MA, Cha KB, Gridley T, Stifani S, Meeldijk J, Clevers H, Camper SA. TCF and Groucho-related genes influence pituitary growth and development. Mol Endocrinol. 2003;17:2152–61. doi: 10.1210/me.2003-0225. [DOI] [PubMed] [Google Scholar]
- 24.Brinkmeier ML, Potok MA, Davis SW, Camper SA. TCF4 deficiency expands ventral diencephalon signaling and increases induction of pituitary progenitors. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cha KB, Douglas KR, Potok MA, Liang H, Jones SN, Camper SA. WNT5A signaling affects pituitary gland shape. Mech Dev. 2004;121:183–94. doi: 10.1016/j.mod.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Ellestad LE, Carre W, Muchow M, Jenkins SA, Wang X, Cogburn LA, Porter TE. Gene expression profiling during cellular differentiation in the embryonic pituitary gland using cDNA microarrays. Physiol Genomics. 2006;25:414–25. doi: 10.1152/physiolgenomics.00248.2005. [DOI] [PubMed] [Google Scholar]
- 27.Mortensen AH, MacDonald JW, Ward RD, Camper SA. Endo 2007. Toronto, ON: 2007. Comparative Gene Expression PROP1 and PIT1 Newborn Pituitaries. [Google Scholar]
- 28.Chen H. Gene expression by the anterior pituitary gland: effects of age and caloric restriction. Mol Cell Endocrinol. 2004;222:21–31. doi: 10.1016/j.mce.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Mohammad HP, Seachrist DD, Quirk CC, Nilson JH. Reexpression of p8 contributes to tumorigenic properties of pituitary cells and appears in a subset of prolactinomas in transgenic mice that hypersecrete luteinizing hormone. Mol Endocrinol. 2004;18:2583–93. doi: 10.1210/me.2004-0163. [DOI] [PubMed] [Google Scholar]
- 30.Quirk CC, Seachrist DD, Nilson JH. Embryonic expression of the luteinizing hormone beta gene appears to be coupled to the transient appearance of p8, a high mobility group-related transcription factor. J Biol Chem. 2003;278:1680–5. doi: 10.1074/jbc.M209906200. [DOI] [PubMed] [Google Scholar]
- 31.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka S, Tatsumi K, Okubo K, Itoh K, Kawamoto S, Matsubara K, Amino N. Expression profile of active genes in the human pituitary gland. J Mol Endocrinol. 2002;28:33–44. doi: 10.1677/jme.0.0280033. [DOI] [PubMed] [Google Scholar]
- 33.Qian X, Scheithauer BW, Kovacs K, Lloyd RV. DNA microarrays: recent developments and applications to the study of pituitary tissues. Endocrine. 2005;28:49–56. doi: 10.1385/ENDO:28:1:049. [DOI] [PubMed] [Google Scholar]
- 34.Abbud RA, Kelleher R, Melmed S. Cell-specific pituitary gene expression profiles after treatment with leukemia inhibitory factor reveal novel modulators for proopiomelanocortin expression. Endocrinology. 2004;145:867–80. doi: 10.1210/en.2003-0897. [DOI] [PubMed] [Google Scholar]
- 35.Kerr JM, Gordon DF, Woodmansee WW, Sarapura VD, Ridgway EC, Wood WM. Growth arrest of thyrotropic tumors by thyroid hormone is correlated with novel changes in Wnt-10A. Mol Cell Endocrinol. 2005;238:57–67. doi: 10.1016/j.mce.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network organization. J Biol Chem. 2001;276:47195–201. doi: 10.1074/jbc.M108716200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Bailey JS, Coss D, Lin B, Tsutsumi R, Lawson MA, Mellon PL, Webster NJ. Activin modulates the transcriptional response of LbetaT2 cells to gonadotropin-releasing hormone and alters cellular proliferation. Mol Endocrinol. 2006;20:2909–30. doi: 10.1210/me.2006-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carninci P, Waki K, Shiraki T, Konno H, Shibata K, Itoh M, Aizawa K, Arakawa T, Ishii Y, Sasaki D, Bono H, Kondo S, Sugahara Y, Saito R, Osato N, Fukuda S, Sato K, Watahiki A, Hirozane-Kishikawa T, Nakamura M, Shibata Y, Yasunishi A, Kikuchi N, Yoshiki A, Kusakabe M, Gustincich S, Beisel K, Pavan W, Aidinis V, Nakagawara A, Held WA, Iwata H, Kono T, Nakauchi H, Lyons P, Wells C, Hume DA, Fagiolini M, Hensch TK, Brinkmeier M, Camper S, Hirota J, Mombaerts P, Muramatsu M, Okazaki Y, Kawai J, Hayashizaki Y. Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res. 2003;13:1273–89. doi: 10.1101/gr.1119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, Muramatsu M, Hayashizaki Y, Schneider C. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–36. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- 40.Carninci P, Nishiyama Y, Westover A, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Hayashizaki Y. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc Natl Acad Sci U S A. 1998;95:520–4. doi: 10.1073/pnas.95.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carninci P, Shibata Y, Hayatsu N, Itoh M, Shiraki T, Hirozane T, Watahiki A, Shibata K, Konno H, Muramatsu M, Hayashizaki Y. Balanced-size and long-size cloning of full-length, cap-trapped cDNAs into vectors of the novel lambda-FLC family allows enhanced gene discovery rate and functional analysis. Genomics. 2001;77:79–90. doi: 10.1006/geno.2001.6601. [DOI] [PubMed] [Google Scholar]
- 42.Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000;10:1617–30. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibata Y, Carninci P, Watahiki A, Shiraki T, Konno H, Muramatsu M, Hayashizaki Y. Cloning full-length, cap-trapper-selected cDNAs by using the single-strand linker ligation method. Biotechniques. 2001;30:1250–4. doi: 10.2144/01306st01. [DOI] [PubMed] [Google Scholar]
- 44.Kuramoto N, Inoue K, Gion K, Takano K, Sakata K, Ogita K, Yoneda Y. Modulation of DNA binding of nuclear transcription factors with leucine-zipper motifs by particular endogenous polyamines in murine central and peripheral excitable tissues. Brain Res. 2003;967:170–80. doi: 10.1016/s0006-8993(02)04268-3. [DOI] [PubMed] [Google Scholar]
- 45.Machiavelli LI, Poliandri AH, Quinteros FA, Cabilla JP, Duvilanski BH. Reactive oxygen species are key mediators of the nitric oxide apoptotic pathway in anterior pituitary cells. Nitric Oxide. 2007;16:237–46. doi: 10.1016/j.niox.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Stojilkovic SS. Pituitary cell type-specific electrical activity, calcium signaling and secretion. Biol Res. 2006;39:403–23. doi: 10.4067/s0716-97602006000300004. [DOI] [PubMed] [Google Scholar]
- 47.van den Hurk MJ, Ouwens DT, Scheenen WJ, Limburg V, Gellekink H, Bai M, Roubos EW, Jenks BG. Expression and characterization of the extracellular Ca(2+)-sensing receptor in melanotrope cells of Xenopus laevis. Endocrinology. 2003;144:2524–33. doi: 10.1210/en.2003-0014. [DOI] [PubMed] [Google Scholar]
- 48.Drouin J, Lamolet B, Lamonerie T, Lanctot C, Tremblay JJ. The PTX family of homeodomain transcription factors during pituitary developments. Mol Cell Endocrinol. 1998;140:31–6. doi: 10.1016/s0303-7207(98)00026-4. [DOI] [PubMed] [Google Scholar]
- 49.Gaston-Massuet C, Andoniadou CL, Signore M, Sajedi E, Bird S, Turner JM, Martinez-Barbera JP. Genetic interaction between the homeobox transcription factors HESX1 and SIX3 is required for normal pituitary development. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–54. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297:1180–3. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- 52.Bentley CA, Zidehsarai MP, Grindley JC, Parlow AF, Barth-Hall S, Roberts VJ. Pax6 is implicated in murine pituitary endocrine function. Endocrine. 1999;10:171–7. doi: 10.1385/ENDO:10:2:171. [DOI] [PubMed] [Google Scholar]
- 53.Burglin TR. Guidebook to the Homeobox Genes. Oxford University Press, Inc.: New York; 1994. [Google Scholar]
- 54.Davis SW, Camper SA. Noggin regulates Bmp4 activity during pituitary induction. Dev Biol. 2007;305:145–60. doi: 10.1016/j.ydbio.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Treier M, Gleiberman AS, O'Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 1998;12:1691–704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McNatty KP, Juengel JL, Wilson T, Galloway SM, Davis GH, Hudson NL, Moeller CL, Cranfield M, Reader KL, Laitinen MP, Groome NP, Sawyer HR, Ritvos O. Oocyte-derived growth factors and ovulation rate in sheep. Reprod Suppl. 2003;61:339–51. [PubMed] [Google Scholar]
- 57.Olson LE, Tollkuhn J, Scafoglio C, Krones A, Zhang J, Ohgi KA, Wu W, Taketo MM, Kemler R, Grosschedl R, Rose D, Li X, Rosenfeld MG. Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125:593–605. doi: 10.1016/j.cell.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 58.Pogoda HM, von der Hardt S, Herzog W, Kramer C, Schwarz H, Hammerschmidt M. The proneural gene ascl1a is required for endocrine differentiation and cell survival in the zebrafish adenohypophysis. Development. 2006;133:1079–89. doi: 10.1242/dev.02296. [DOI] [PubMed] [Google Scholar]
- 59.Vinals F, Reiriz J, Ambrosio S, Bartrons R, Rosa JL, Ventura F. BMP-2 decreases Mash1 stability by increasing Id1 expression. Embo J. 2004;23:3527–37. doi: 10.1038/sj.emboj.7600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moreno CS, Evans CO, Zhan X, Okor M, Desiderio DM, Oyesiku NM. Novel molecular signaling and classification of human clinically nonfunctional pituitary adenomas identified by gene expression profiling and proteomic analyses. Cancer Res. 2005;65:10214–22. doi: 10.1158/0008-5472.CAN-05-0884. [DOI] [PubMed] [Google Scholar]
- 61.Mould AW, Duncan R, Serewko-Auret M, Loffler KA, Biondi C, Gartside M, Kay GF, Hayward NK. Global expression profiling of murine MEN1-associated tumors reveals a regulatory role for menin in transcription, cell cycle and chromatin remodelling. Int J Cancer. 2007;121:776–83. doi: 10.1002/ijc.22734. [DOI] [PubMed] [Google Scholar]
- 62.Ruebel KH, Leontovich AA, Jin L, Stilling GA, Zhang H, Qian X, Nakamura N, Scheithauer BW, Kovacs K, Lloyd RV. Patterns of gene expression in pituitary carcinomas and adenomas analyzed by high-density oligonucleotide arrays, reverse transcriptase-quantitative PCR, and protein expression. Endocrine. 2006;29:435–44. doi: 10.1385/ENDO:29:3:435. [DOI] [PubMed] [Google Scholar]
- 63.Asbreuk CH, van Schaick HS, Cox JJ, Smidt MP, Burbach JP. Survey for paired-like homeodomain gene expression in the hypothalamus: restricted expression patterns of Rx, Alx4 and goosecoid. Neuroscience. 2002;114:883–9. doi: 10.1016/s0306-4522(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 64.Bailey JS, Rave-Harel N, McGillivray SM, Coss D, Mellon PL. Activin regulation of the follicle-stimulating hormone beta-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1. Mol Endocrinol. 2004;18:1158–70. doi: 10.1210/me.2003-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ermakova GV, Solovieva EA, Martynova NY, Zaraisky AG. The homeodomain factor Xanf represses expression of genes in the presumptive rostral forebrain that specify more caudal brain regions. Dev Biol. 2007;307:483–97. doi: 10.1016/j.ydbio.2007.03.524. [DOI] [PubMed] [Google Scholar]
- 66.Glenn DJ, Maurer RA. MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha-subunit gene expression. J Biol Chem. 1999;274:36159–67. doi: 10.1074/jbc.274.51.36159. [DOI] [PubMed] [Google Scholar]
- 67.Mizokami Y, Egashira N, Takekoshi S, Itoh J, Itoh Y, Osamura RY, Matsumae M. Expression of MSX1 in Human Normal Pituitaries and Pituitary Adenomas. Endocr Pathol. 2008 doi: 10.1007/s12022-008-9021-7. [DOI] [PubMed] [Google Scholar]
- 68.Nolen LD, Amor D, Haywood A, St Heaps L, Willcock C, Mihelec M, Tam P, Billson F, Grigg J, Peters G, Jamieson RV. Deletion at 14q22-23 indicates a contiguous gene syndrome comprising anophthalmia, pituitary hypoplasia, and ear anomalies. Am J Med Genet A. 2006;140:1711–8. doi: 10.1002/ajmg.a.31335. [DOI] [PubMed] [Google Scholar]
- 69.Roberson MS, Schoderbek WE, Tremml G, Maurer RA. Activation of the glycoprotein hormone alpha-subunit promoter by a LIM-homeodomain transcription factor. Mol Cell Biol. 1994;14:2985–93. doi: 10.1128/mcb.14.5.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toda K, Yamamoto D, Fumoto M, Ikeshita N, Herningtyas EH, Iida K, Takahashi Y, Kaji H, Chihara K, Okimura Y. Involvement of mPOU (Brn-5), a class VI POU protein, in the gene expression of Pit-1 as well as PRL. Mol Cell Endocrinol. 2008;280:20–9. doi: 10.1016/j.mce.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–6. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watkins-Chow DE, Camper SA. How many homeobox genes does it take to make a pituitary gland? Trends Genet. 1998;14:284–90. doi: 10.1016/s0168-9525(98)01476-0. [DOI] [PubMed] [Google Scholar]
- 73.Mowat DR, Croaker GD, Cass DT, Kerr BA, Chaitow J, Ades LC, Chia NL, Wilson MJ. Hirschsprung disease, microcephaly, mental retardation, and characteristic facial features: delineation of a new syndrome and identification of a locus at chromosome 2q22-q23. J Med Genet. 1998;35:617–23. doi: 10.1136/jmg.35.8.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brendolan A, Ferretti E, Salsi V, Moses K, Quaggin S, Blasi F, Cleary ML, Selleri L. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development. 2005;132:3113–26. doi: 10.1242/dev.01884. [DOI] [PubMed] [Google Scholar]
- 75.Rave-Harel N, Givens ML, Nelson SB, Duong HA, Coss D, Clark ME, Hall SB, Kamps MP, Mellon PL. TALE homeodomain proteins regulate gonadotropin-releasing hormone gene expression independently and via interactions with Oct-1. J Biol Chem. 2004;279:30287–97. doi: 10.1074/jbc.M402960200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nishimoto S, Nishida E. Fibroblast growth factor 13 is essential for neural differentiation in Xenopus early embryonic development. J Biol Chem. 2007;282:24255–61. doi: 10.1074/jbc.M704277200. [DOI] [PubMed] [Google Scholar]
- 77.Zhang S, Lin Y, Itaranta P, Yagi A, Vainio S. Expression of Sprouty genes 1, 2 and 4 during mouse organogenesis. Mech Dev. 2001;109:367–70. doi: 10.1016/s0925-4773(01)00526-3. [DOI] [PubMed] [Google Scholar]
- 78.Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–63. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 79.Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–75. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- 80.Carninci P, Shiraki T, Mizuno Y, Muramatsu M, Hayashizaki Y. Extra-long first-strand cDNA synthesis. Biotechniques. 2002;32:984–5. doi: 10.2144/02325bm01. [DOI] [PubMed] [Google Scholar]
- 81.Carninci P, Westover A, Nishiyama Y, Ohsumi T, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Schneider C, Hayashizaki Y. High efficiency selection of full-length cDNA by improved biotinylated cap trapper. DNA Res. 1997;4:61–6. doi: 10.1093/dnares/4.1.61. [DOI] [PubMed] [Google Scholar]
- 82.Hirozane-Kishikawa T, Shiraki T, Waki K, Nakamura M, Arakawa T, Kawai J, Fagiolini M, Hensch TK, Hayashizaki Y, Carninci P. Subtraction of cap-trapped full-length cDNA libraries to select rare transcripts. Biotechniques. 2003;35:510–6. 518. doi: 10.2144/03353st04. [DOI] [PubMed] [Google Scholar]
- 83.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–7. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 84.Cushman LJ, Watkins-Chow DE, Brinkmeier ML, Raetzman LT, Radak AL, Lloyd RV, Camper SA. Persistent Prop1 expression delays gonadotrope differentiation and enhances pituitary tumor susceptibility. Hum Mol Genet. 2001;10:1141–53. doi: 10.1093/hmg/10.11.1141. [DOI] [PubMed] [Google Scholar]
- 85.Vesper AH, Raetzman LT, Camper SA. Role of prophet of Pit1 (PROP1) in gonadotrope differentiation and puberty. Endocrinology. 2006;147:1654–63. doi: 10.1210/en.2005-1080. [DOI] [PubMed] [Google Scholar]
- 86.Sarapura VD, Wood WM, Woodmansee WW, Haakinson DJ, Dowding JM, Gordon DF, Ridgway EC. Pituitary tumors arising from glycoprotein hormone alpha-subunit-deficient mice contain transcription factors and receptors present in thyrotropes. Pituitary. 2006;9:11–8. doi: 10.1007/s11102-006-7865-8. [DOI] [PubMed] [Google Scholar]
- 87.Ge G, Fernandez CA, Moses MA, Greenspan DS. Bone morphogenetic protein 1 processes prolactin to a 17-kDa antiangiogenic factor. Proc Natl Acad Sci U S A. 2007;104:10010–5. doi: 10.1073/pnas.0704179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoshikawa SI, Aota S, Shirayoshi Y, Okazaki K. The ActR-I activin receptor protein is expressed in notochord, lens placode and pituitary primordium cells in the mouse embryo. Mech Dev. 2000;91:439–44. doi: 10.1016/s0925-4773(99)00320-2. [DOI] [PubMed] [Google Scholar]
- 89.Pizarro CB, Oliveira MC, Pereira-Lima JF, Leaes CG, Kramer CK, Schuch T, Barbosa-Coutinho LM, Ferreira NP. Evaluation of angiogenesis in 77 pituitary adenomas using endoglin as a marker. Neuropathology. 2008 doi: 10.1111/j.1440-1789.2008.00937.x. [DOI] [PubMed] [Google Scholar]
- 90.Canibano C, Rodriguez NL, Saez C, Tovar S, Garcia-Lavandeira M, Borrello MG, Vidal A, Costantini F, Japon M, Dieguez C, Alvarez CV. The dependence receptor Ret induces apoptosis in somatotrophs through a Pit-1/p53 pathway, preventing tumor growth. EMBO J. 2007;26:2015–28. doi: 10.1038/sj.emboj.7601636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Japon MA, Urbano AG, Saez C, Segura DI, Cerro AL, Dieguez C, Alvarez CV. Glial-derived neurotropic factor and RET gene expression in normal human anterior pituitary cell types and in pituitary tumors. J Clin Endocrinol Metab. 2002;87:1879–84. doi: 10.1210/jcem.87.4.8383. [DOI] [PubMed] [Google Scholar]
- 92.Yoshimoto K, Tanaka C, Moritani M, Shimizu E, Yamaoka T, Yamada S, Sano T, Itakura M. Infrequent detectable somatic mutations of the RET and glial cell line-derived neurotrophic factor (GDNF) genes in human pituitary adenomas. Endocr J. 1999;46:199–207. doi: 10.1507/endocrj.46.199. [DOI] [PubMed] [Google Scholar]
- 93.Fedele M, Battista S, Kenyon L, Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R, Pierantoni GM, Outwater E, Santoro M, Croce CM, Fusco A. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene. 2002;21:3190–8. doi: 10.1038/sj.onc.1205428. [DOI] [PubMed] [Google Scholar]
- 94.Fedele M, Pierantoni GM, Visone R, Fusco A. E2F1 activation is responsible for pituitary adenomas induced by HMGA2 gene overexpression. Cell Div. 2006;1:17. doi: 10.1186/1747-1028-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fedele M, Pierantoni GM, Visone R, Fusco A. Critical role of the HMGA2 gene in pituitary adenomas. Cell Cycle. 2006;5:2045–8. doi: 10.4161/cc.5.18.3211. [DOI] [PubMed] [Google Scholar]
- 96.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, Helin K, Croce CM, Fusco A. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–71. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 97.Finelli P, Pierantoni GM, Giardino D, Losa M, Rodeschini O, Fedele M, Valtorta E, Mortini P, Croce CM, Larizza L, Fusco A. The High Mobility Group A2 gene is amplified and overexpressed in human prolactinomas. Cancer Res. 2002;62:2398–405. [PubMed] [Google Scholar]
- 98.Pierantoni GM, Finelli P, Valtorta E, Giardino D, Rodeschini O, Esposito F, Losa M, Fusco A, Larizza L. High-mobility group A2 gene expression is frequently induced in non-functioning pituitary adenomas (NFPAs), even in the absence of chromosome 12 polysomy. Endocr Relat Cancer. 2005;12:867–74. doi: 10.1677/erc.1.01049. [DOI] [PubMed] [Google Scholar]
- 99.Lasorella A, Rothschild G, Yokota Y, Russell RG, Iavarone A. Id2 mediates tumor initiation, proliferation, and angiogenesis in Rb mutant mice. Mol Cell Biol. 2005;25:3563–74. doi: 10.1128/MCB.25.9.3563-3574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hendy GN, Kaji H, Sowa H, Lebrun JJ, Canaff L. Menin and TGF-beta superfamily member signaling via the Smad pathway in pituitary, parathyroid and osteoblast. Horm Metab Res. 2005;37:375–9. doi: 10.1055/s-2005-870152. [DOI] [PubMed] [Google Scholar]
- 101.Nudi M, Ouimette JF, Drouin J. Bone morphogenic protein (Smad)-mediated repression of proopiomelanocortin transcription by interference with Pitx/Tpit activity. Mol Endocrinol. 2005;19:1329–42. doi: 10.1210/me.2004-0425. [DOI] [PubMed] [Google Scholar]
- 102.Bernard DJ. Both SMAD2 and SMAD3 mediate activin-stimulated expression of the follicle-stimulating hormone beta subunit in mouse gonadotrope cells. Mol Endocrinol. 2004;18:606–23. doi: 10.1210/me.2003-0264. [DOI] [PubMed] [Google Scholar]
- 103.Lau MT, Ge W. Cloning of Smad2, Smad3, Smad4, and Smad7 from the goldfish pituitary and evidence for their involvement in activin regulation of goldfish FSHbeta promoter activity. Gen Comp Endocrinol. 2005;141:22–38. doi: 10.1016/j.ygcen.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 104.Ruebel KH, Leontovich AA, Tanizaki Y, Jin L, Stilling GA, Zhang S, Coonse K, Scheithauer BW, Lombardero M, Kovacs K, Lloyd RV. Effects of TGFbeta1 on gene expression in the HP75 human pituitary tumor cell line identified by gene expression profiling. Endocrine. 2008;33:62–76. doi: 10.1007/s12020-008-9060-3. [DOI] [PubMed] [Google Scholar]
- 105.Bilezikjian LM, Corrigan AZ, Blount AL, Chen Y, Vale WW. Regulation and actions of Smad7 in the modulation of activin, inhibin, and transforming growth factor-beta signaling in anterior pituitary cells. Endocrinology. 2001;142:1065–72. doi: 10.1210/endo.142.3.8028. [DOI] [PubMed] [Google Scholar]
- 106.Gore AJ, Philips DP, Miller WL, Bernard DJ. Differential regulation of follicle stimulating hormone by activin A and TGFB1 in murine gonadotropes. Reprod Biol Endocrinol. 2005;3:73. doi: 10.1186/1477-7827-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ikeda H, Yoshimoto T, Shida N, Miyoshi I, Nakayama K, Oshima M, Taketo MM. Morphologic and molecular analysis of estrogen-induced pituitary tumorigenesis in targeted disruption of transforming growth factor-beta receptor type II and/or p27 mice. Endocrine. 2001;16:55–65. doi: 10.1385/ENDO:16:1:55. [DOI] [PubMed] [Google Scholar]
- 108.Shida N, Ikeda H, Yoshimoto T, Oshima M, Taketo MM, Miyoshi I. Estrogen-induced tumorigenesis in the pituitary gland of TGF-beta(+/-) knockout mice. Biochim Biophys Acta. 1998;1407:79–83. doi: 10.1016/s0925-4439(98)00024-6. [DOI] [PubMed] [Google Scholar]
- 109.Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–9. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 110.Bhagavath B, Layman LC. The genetics of hypogonadotropic hypogonadism. Semin Reprod Med. 2007;25:272–86. doi: 10.1055/s-2007-980221. [DOI] [PubMed] [Google Scholar]
- 111.Cristina C, Diaz-Torga G, Gongora A, Guida MC, Perez-Millan MI, Baldi A, Becu-Villalobos D. Fibroblast growth factor-2 in hyperplastic pituitaries of D2R knockout female mice. Am J Physiol Endocrinol Metab. 2007;293:E1341–51. doi: 10.1152/ajpendo.00260.2007. [DOI] [PubMed] [Google Scholar]
- 112.Fukui S, Otani N, Nawashiro H, Yano A, Nomura N, Miyazawa T, Ohnuki A, Tsuzuki N, Katoh H, Ishihara S, Shima K. Subcellular localization of basic fibroblast growth factor and fibroblast growth factor receptor 1 in pituitary adenomas. Brain Tumor Pathol. 2002;19:23–9. doi: 10.1007/BF02482452. [DOI] [PubMed] [Google Scholar]
- 113.Gonzalez AM, Logan A, Ying W, Lappi DA, Berry M, Baird A. Fibroblast growth factor in the hypothalamic-pituitary axis: differential expression of fibroblast growth factor-2 and a high affinity receptor. Endocrinology. 1994;134:2289–97. doi: 10.1210/endo.134.5.8156932. [DOI] [PubMed] [Google Scholar]
- 114.Komisarczuk AZ, Topp S, Stigloher C, Kapsimali M, Bally-Cuif L, Becker TS. Enhancer detection and developmental expression of zebrafish sprouty1, a member of the fgf8 synexpression group. Dev Dyn. 2008;237:2594–603. doi: 10.1002/dvdy.21689. [DOI] [PubMed] [Google Scholar]
- 115.Xu N, Qin Y, Reindollar RH, Tho SP, McDonough PG, Layman LC. A mutation in the fibroblast growth factor receptor 1 gene causes fully penetrant normosmic isolated hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2007;92:1155–8. doi: 10.1210/jc.2006-1183. [DOI] [PubMed] [Google Scholar]
- 116.De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–92. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 117.Zhu X, Lee K, Asa SL, Ezzat S. Epigenetic silencing through DNA and histone methylation of fibroblast growth factor receptor 2 in neoplastic pituitary cells. Am J Pathol. 2007;170:1618–28. doi: 10.2353/ajpath.2007.061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lombardi MS, Kavelaars A, Cobelens PM, Schmidt RE, Schedlowski M, Heijnen CJ. Adjuvant arthritis induces down-regulation of G protein-coupled receptor kinases in the immune system. J Immunol. 2001;166:1635–40. doi: 10.4049/jimmunol.166.3.1635. [DOI] [PubMed] [Google Scholar]
- 119.Vines CR, Weigent DA. Identification of SP3 as a negative regulatory transcription factor in the monocyte expression of growth hormone. Endocrinology. 2000;141:938–46. doi: 10.1210/endo.141.3.7381. [DOI] [PubMed] [Google Scholar]
- 120.Wang Z, Melmed S. Characterization of the murine pituitary tumor transforming gene (PTTG) and its promoter. Endocrinology. 2000;141:763–71. doi: 10.1210/endo.141.2.7294. [DOI] [PubMed] [Google Scholar]
- 121.Pontecorvi M, Goding CR, Richardson WD, Kessaris N. Expression of Tbx2 and Tbx3 in the developing hypothalamic-pituitary axis. Gene Expr Patterns. 2008;8:411–7. doi: 10.1016/j.gep.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 122.Sbrogna JL, Barresi MJ, Karlstrom RO. Multiple roles for Hedgehog signaling in zebrafish pituitary development. Dev Biol. 2003;254:19–35. doi: 10.1016/s0012-1606(02)00027-1. [DOI] [PubMed] [Google Scholar]
- 123.Vila G, Papazoglou M, Stalla J, Theodoropoulou M, Stalla GK, Holsboer F, Paez-Pereda M. Sonic hedgehog regulates CRH signal transduction in the adult pituitary. FASEB J. 2005;19:281–3. doi: 10.1096/fj.04-2138fje. [DOI] [PubMed] [Google Scholar]
- 124.Altenberger T, Bilban M, Auer M, Knosp E, Wolfsberger S, Gartner W, Mineva I, Zielinski C, Wagner L, Luger A. Identification of DLK1 variants in pituitary- and neuroendocrine tumors. Biochem Biophys Res Commun. 2006;340:995–1005. doi: 10.1016/j.bbrc.2005.12.094. [DOI] [PubMed] [Google Scholar]
- 125.Ansell PJ, Zhou Y, Schjeide BM, Kerner A, Zhao J, Zhang X, Klibanski A. Regulation of growth hormone expression by Delta-like protein 1 (Dlk1) Mol Cell Endocrinol. 2007;271:55–63. doi: 10.1016/j.mce.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nakakura T, Sato M, Suzuki M, Hatano O, Takemori H, Taniguchi Y, Minoshima Y, Tanaka S. The spatial and temporal expression of delta-like protein 1 in the rat pituitary gland during development. Histochem Cell Biol. 2008 doi: 10.1007/s00418-008-0494-8. [DOI] [PubMed] [Google Scholar]
- 127.Chen J, Crabbe A, Van Duppen V, Vankelecom H. The notch signaling system is present in the postnatal pituitary: marked expression and regulatory activity in the newly discovered side population. Mol Endocrinol. 2006;20:3293–307. doi: 10.1210/me.2006-0293. [DOI] [PubMed] [Google Scholar]
- 128.Raetzman LT, Wheeler BS, Ross SA, Thomas PQ, Camper SA. Persistent expression of Notch2 delays gonadotrope differentiation. Mol Endocrinol. 2006;20:2898–908. doi: 10.1210/me.2005-0394. [DOI] [PubMed] [Google Scholar]
- 129.Bottner M, Christoffel J, Jarry H, Wuttke W. Effects of long-term treatment with resveratrol and subcutaneous and oral estradiol administration on pituitary function in rats. J Endocrinol. 2006;189:77–88. doi: 10.1677/joe.1.06535. [DOI] [PubMed] [Google Scholar]
- 130.Rasier G, Parent AS, Gerard A, Lebrethon MC, Bourguignon JP. Early maturation of gonadotropin-releasing hormone secretion and sexual precocity after exposure of infant female rats to estradiol or dichlorodiphenyltrichloroethane. Biol Reprod. 2007;77:734–42. doi: 10.1095/biolreprod.106.059303. [DOI] [PubMed] [Google Scholar]
- 131.Heckert LL, Schultz K, Nilson JH. The cAMP response elements of the alpha subunit gene bind similar proteins in trophoblasts and gonadotropes but have distinct functional sequence requirements. J Biol Chem. 1996;271:31650–6. doi: 10.1074/jbc.271.49.31650. [DOI] [PubMed] [Google Scholar]
- 132.Mayer SI, Dexheimer V, Nishida E, Kitajima S, Thiel G. Expression of the transcriptional repressor ATF3 in gonadotrophs is regulated by Egr-1, CREB and ATF2 following GnRH receptor stimulation. Endocrinology. 2008 doi: 10.1210/en.2008-0251. [DOI] [PubMed] [Google Scholar]
- 133.Lee EY, Cam H, Ziebold U, Rayman JB, Lees JA, Dynlacht BD. E2F4 loss suppresses tumorigenesis in Rb mutant mice. Cancer Cell. 2002;2:463–72. doi: 10.1016/s1535-6108(02)00207-6. [DOI] [PubMed] [Google Scholar]
- 134.Mazhawidza W, Winters SJ, Kaiser UB, Kakar SS. Identification of gene networks modulated by activin in LbetaT2 cells using DNA microarray analysis. Histol Histopathol. 2006;21:167–78. doi: 10.14670/HH-21.167. [DOI] [PubMed] [Google Scholar]
- 135.Asa SL, Bamberger AM, Cao B, Wong M, Parker KL, Ezzat S. The transcription activator steroidogenic factor-1 is preferentially expressed in the human pituitary gonadotroph. J Clin Endocrinol Metab. 1996;81:2165–70. doi: 10.1210/jcem.81.6.8964846. [DOI] [PubMed] [Google Scholar]
- 136.Barnhart KM, Mellon PL. The orphan nuclear receptor, steroidogenic factor-1, regulates the glycoprotein hormone alpha-subunit gene in pituitary gonadotropes. Mol Endocrinol. 1994;8:878–85. doi: 10.1210/mend.8.7.7527122. [DOI] [PubMed] [Google Scholar]
- 137.Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development. 2001;128:147–54. doi: 10.1242/dev.128.2.147. [DOI] [PubMed] [Google Scholar]
- 138.Dilley WG, Kalyanaraman S, Verma S, Cobb JP, Laramie JM, Lairmore TC. Global gene expression in neuroendocrine tumors from patients with the MEN1 syndrome. Mol Cancer. 2005;4:9. doi: 10.1186/1476-4598-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Elston MS, Gill AJ, Conaglen JV, Clarkson A, Shaw JM, Law AJ, Cook RJ, Little NS, Clifton-Bligh RJ, Robinson BG, McDonald KL. Wnt pathway inhibitors are strongly down-regulated in pituitary tumors. Endocrinology. 2008;149:1235–42. doi: 10.1210/en.2007-0542. [DOI] [PubMed] [Google Scholar]
- 140.Castillo AI, Jimenez-Lara AM, Tolon RM, Aranda A. Synergistic activation of the prolactin promoter by vitamin D receptor and GHF-1: role of the coactivators, CREB-binding protein and steroid hormone receptor coactivator-1 (SRC-1) Mol Endocrinol. 1999;13:1141–54. doi: 10.1210/mend.13.7.0320. [DOI] [PubMed] [Google Scholar]
- 141.Cohen LE, Hashimoto Y, Zanger K, Wondisford F, Radovick S. CREB-independent regulation by CBP is a novel mechanism of human growth hormone gene expression. J Clin Invest. 1999;104:1123–30. doi: 10.1172/JCI7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ferry AL, Locasto DM, Meszaros LB, Bailey JC, Jonsen MD, Brodsky K, Hoon CJ, Gutierrez-Hartmann A, Diamond SE. Pit-1beta reduces transcription and CREB-binding protein recruitment in a DNA context-dependent manner. J Endocrinol. 2005;185:173–85. doi: 10.1677/joe.1.05738. [DOI] [PubMed] [Google Scholar]
- 143.Hashimoto K, Yamada M, Monden T, Satoh T, Wondisford FE, Mori M. Thyrotropin-releasing hormone (TRH) specific interaction between amino terminus of P-Lim and CREB binding protein (CBP) Mol Cell Endocrinol. 2005;229:11–20. doi: 10.1016/j.mce.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 144.Hashimoto K, Zanger K, Hollenberg AN, Cohen LE, Radovick S, Wondisford FE. cAMP response element-binding protein-binding protein mediates thyrotropin-releasing hormone signaling on thyrotropin subunit genes. J Biol Chem. 2000;275:33365–72. doi: 10.1074/jbc.M006819200. [DOI] [PubMed] [Google Scholar]
- 145.Kishimoto M, Okimura Y, Yagita K, Iguchi G, Fumoto M, Iida K, Kaji H, Okamura H, Chihara K. Novel function of the transactivation domain of a pituitary-specific transcription factor, Pit-1. J Biol Chem. 2002;277:45141–8. doi: 10.1074/jbc.M202991200. [DOI] [PubMed] [Google Scholar]
- 146.Xu L, Lavinsky RM, Dasen JS, Flynn SE, McInerney EM, Mullen TM, Heinzel T, Szeto D, Korzus E, Kurokawa R, Aggarwal AK, Rose DW, Glass CK, Rosenfeld MG. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–6. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 147.Zanger K, Cohen LE, Hashimoto K, Radovick S, Wondisford FE. A novel mechanism for cyclic adenosine 3′,5′-monophosphate regulation of gene expression by CREB-binding protein. Mol Endocrinol. 1999;13:268–75. doi: 10.1210/mend.13.2.0245. [DOI] [PubMed] [Google Scholar]
- 148.Gardner S, Maudsley S, Millar RP, Pawson AJ. Nuclear stabilization of beta-catenin and inactivation of glycogen synthase kinase-3beta by gonadotropin-releasing hormone: targeting Wnt signaling in the pituitary gonadotrope. Mol Endocrinol. 2007;21:3028–38. doi: 10.1210/me.2007-0268. [DOI] [PubMed] [Google Scholar]
- 149.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–85. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 150.Salisbury TB, Binder AK, Grammer JC, Nilson JH. Maximal activity of the luteinizing hormone beta-subunit gene requires beta-catenin. Mol Endocrinol. 2007;21:963–71. doi: 10.1210/me.2006-0383. [DOI] [PubMed] [Google Scholar]
- 151.Semba S, Han SY, Ikeda H, Horii A. Frequent nuclear accumulation of beta-catenin in pituitary adenoma. Cancer. 2001;91:42–8. doi: 10.1002/1097-0142(20010101)91:1<42::aid-cncr6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 152.Sun C, Yamato T, Kondo E, Furukawa T, Ikeda H, Horii A. Infrequent mutation of APC, AXIN1, and GSK3B in human pituitary adenomas with abnormal accumulation of CTNNB1. J Neurooncol. 2005;73:131–4. doi: 10.1007/s11060-004-4597-3. [DOI] [PubMed] [Google Scholar]
- 153.Kakar SS, Roy D. Curcumin inhibits TPA induced expression of c-fos, c-jun and c-myc proto-oncogenes messenger RNAs in mouse skin. Cancer Lett. 1994;87:85–9. doi: 10.1016/0304-3835(94)90413-8. [DOI] [PubMed] [Google Scholar]
- 154.Ferretti E, Di Stefano D, Zazzeroni F, Gallo R, Fratticci A, Carfagnini R, Angiulli S, Santoro A, Minniti G, Tamburrano G, Alesse E, Cantore G, Gulino A, Jaffrain-Rea ML. Human pituitary tumours express the bHLH transcription factors NeuroD1 and ASH1. J Endocrinol Invest. 2003;26:957–65. doi: 10.1007/BF03348192. [DOI] [PubMed] [Google Scholar]
- 155.Fratticci A, Grieco FA, Spilioti C, Giangaspero F, Ventura L, Esposito V, Piccirilli M, Santoro A, Gulino A, Cantore G, Alesse E, Jaffrain-Rea ML. Differential expression of neurogenins and NeuroD1 in human pituitary tumours. J Endocrinol. 2007;194:475–84. doi: 10.1677/JOE-07-0020. [DOI] [PubMed] [Google Scholar]
- 156.Lamolet B, Poulin G, Chu K, Guillemot F, Tsai MJ, Drouin J. Tpit-independent function of NeuroD1(BETA2) in pituitary corticotroph differentiation. Mol Endocrinol. 2004;18:995–1003. doi: 10.1210/me.2003-0127. [DOI] [PubMed] [Google Scholar]
- 157.Poulin G, Turgeon B, Drouin J. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol Cell Biol. 1997;17:6673–82. doi: 10.1128/mcb.17.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pulichino AM, Lamolet B, Vallette-Kasic S, Poulin G, Chu K, Gillemot F, Tsai MJ, Drouin J. Tpit-/-NeuroD1-/- mice reveal novel aspects of corticotroph development. Endocr Res. 2004;30:551–2. doi: 10.1081/erc-200043625. [DOI] [PubMed] [Google Scholar]
- 159.Tahara S, Kurotani R, Ishii Y, Sanno N, Teramoto A, Osamura RY. A case of Cushing's disease caused by pituitary adenoma producing adrenocorticotropic hormone and growth hormone concomitantly: aberrant expression of transcription factors NeuroD1 and Pit-1 as a proposed mechanism. Mod Pathol. 2002;15:1102–5. doi: 10.1097/01.MP.0000030451.28828.00. [DOI] [PubMed] [Google Scholar]
- 160.Donangelo I, Marcos HP, Araujo PB, Marcondes J, Filho PN, Gadelha M, Chimelli L. Expression of retinoblastoma protein in human growth hormone-secreting pituitary adenomas. Endocr Pathol. 2005;16:53–62. doi: 10.1385/ep:16:1:053. [DOI] [PubMed] [Google Scholar]
- 161.Honda S, Tanaka-Kosugi C, Yamada S, Sano T, Matsumoto T, Itakura M, Yoshimoto K. Human pituitary adenomas infrequently contain inactivation of retinoblastoma 1 gene and activation of cyclin dependent kinase 4 gene. Endocr J. 2003;50:309–18. doi: 10.1507/endocrj.50.309. [DOI] [PubMed] [Google Scholar]
- 162.Loffler KA, Biondi CA, Gartside MG, Serewko-Auret MM, Duncan R, Tonks ID, Mould AW, Waring P, Muller HK, Kay GF, Hayward NK. Lack of augmentation of tumor spectrum or severity in dual heterozygous Men1 and Rb1 knockout mice. Oncogene. 2007;26:4009–17. doi: 10.1038/sj.onc.1210163. [DOI] [PubMed] [Google Scholar]
- 163.Pearce SH, Trump D, Wooding C, Sheppard MN, Clayton RN, Thakker RV. Loss of heterozygosity studies at the retinoblastoma and breast cancer susceptibility (BRCA2) loci in pituitary, parathyroid, pancreatic and carcinoid tumours. Clin Endocrinol (Oxf) 1996;45:195–200. doi: 10.1046/j.1365-2265.1996.d01-1561.x. [DOI] [PubMed] [Google Scholar]
- 164.Simpson DJ, Hibberts NA, McNicol AM, Clayton RN, Farrell WE. Loss of pRb expression in pituitary adenomas is associated with methylation of the RB1 CpG island. Cancer Res. 2000;60:1211–6. [PubMed] [Google Scholar]
- 165.Smith AP, Henze M, Osborn KG, Lee JA, Bortner DM, Rosenberg MP, Reed SI. Switching of melanocyte pigmentation associated with pituitary pars intermedia tumors in Rb+/- and p27-/- female mice with yellow pelage. Comp Med. 2003;53:75–80. [PubMed] [Google Scholar]
- 166.Kelberman D, de Castro SC, Huang S, Crolla JA, Palmer R, Gregory JW, Taylor D, Cavallo L, Faienza MF, Fischetto R, Achermann JC, Martinez-Barbera JP, Rizzoti K, Lovell-Badge R, Robinson IC, Gerrelli D, Dattani MT. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. J Clin Endocrinol Metab. 2008;93:1865–73. doi: 10.1210/jc.2007-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kelberman D, Rizzoti K, Avilion A, Bitner-Glindzicz M, Cianfarani S, Collins J, Chong WK, Kirk JM, Achermann JC, Ross R, Carmignac D, Lovell-Badge R, Robinson IC, Dattani MT. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest. 2006;116:2442–55. doi: 10.1172/JCI28658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sato N, Kamachi Y, Kondoh H, Shima Y, Morohashi K, Horikawa R, Ogata T. Hypogonadotropic hypogonadism in an adult female with a heterozygous hypomorphic mutation of SOX2. Eur J Endocrinol. 2007;156:167–71. doi: 10.1530/EJE-06-0606. [DOI] [PubMed] [Google Scholar]
- 169.Bousquet C, Melmed S. Critical role for STAT3 in murine pituitary adrenocorticotropin hormone leukemia inhibitory factor signaling. J Biol Chem. 1999;274:10723–30. doi: 10.1074/jbc.274.16.10723. [DOI] [PubMed] [Google Scholar]
- 170.Bousquet C, Zatelli MC, Melmed S. Direct regulation of pituitary proopiomelanocortin by STAT3 provides a novel mechanism for immuno-neuroendocrine interfacing. J Clin Invest. 2000;106:1417–25. doi: 10.1172/JCI11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.