Abstract
Adaptor proteins are likely to modulate spatially and temporally the trafficking of a number of membrane proteins, including neuronal nicotinic acetylcholine receptors (nAChRs). A yeast two-hybrid screen identified a novel UBX-containing protein, UBXD4, as one of the cytosolic proteins that interact directly with the α3 and α4 nAChR subunits. The function of UBX-containing proteins is largely unknown. Immunoprecipitation and confocal microscopy confirmed the interaction of UBXD4 with α3-containing nAChRs (α3* nAChRs) expressed in HEK293 cells, PC12 cells, and rat cortical neurons. Overexpression of UBXD4 in differentiated PC12 cells (dPC12) increased nAChR cell surface expression, especially that of the α3β2 subtype. These findings were corroborated by electrophysiology, immunofluorescent staining, and biotinylation of surface receptors. Silencing of UBXD4 led to a significant reduction of α3* nAChRs in rat cortical neurons and dPC12 cells. Biochemical and immunofluorescence studies of endogenous UBXD4 showed that the protein is located in both the ER and cis-Golgi compartments. Our investigations also showed that the α3 subunit is ubiquitinated and that UBXD4 can interfere with its ubiquitination and consequent degradation by the proteasome. Our data suggest that UBXD4 modulates the distribution of α3* nAChRs between specialized intracellular compartments and the plasma membrane. This effect is achieved by controlling the stability of the α3 subunit and, consequently, the number of receptors at the cell surface.
Introduction
nAChRs are pentameric ligand-gated ion channels composed of α and β or α-only subunits (Le Novère et al., 2002). The α3 subunit coassembles with the β2, β4, and/or α5 subunits to yield α3β2, α3α5β2, α3β4, or α3α5β4 nAChR subtypes (Jensen et al., 2005).
α3* nAChRs are highly expressed in the autonomic nervous system (ANS), where they mediate acetylcholine-induced fast excitatory ganglionic transmission (De Biasi, 2002). In addition to their wide expression in the ANS, α3* nAChRs are present in regions of the CNS including cortical deep layer IV, entorhinal cortex, medial habenula, interpeduncular nucleus, basolateral nucleus of the amygdala, locus ceruleus, and hypothalamus (Wada et al., 1989). The presence of α3* nAChRs in those CNS regions could influence the behavioral effects of nicotine and some manifestations of nicotine withdrawal (Salas et al., 2004a,b).
nAChR subunits comprise an extracellular N-terminal domain followed by three transmembrane domains (M1–M3), a large cytoplasmic domain, a fourth transmembrane domain (M4), and a short extracellular C terminus (Jensen et al., 2005). Several of those protein domains are involved in nAChR trafficking. For example, the M1 transmembrane domain contains an ER retention signal that is masked when the receptor is fully assembled, allowing the receptors to pass the ER compartment (Wang et al., 2002; Pons et al., 2004). nAChR levels are also influenced by a degradation signal contained in the large intracellular loop (Jeanclos et al., 2001; Lin et al., 2002; Conroy et al., 2003; Ficklin et al., 2005). The masking of degradation signals—possibly by an adaptor protein—seems a required step for nAChR escape from the checkpoint at the endoplasmic reticulum-associated degradation (ERAD) level (Keller et al., 2001; Ren et al., 2005). It is plausible that adaptor proteins act as chaperones to stabilize α3* receptors at the ER and cis-Golgi compartment levels where the ubiquitin–proteasome system (UPS) regulates protein trafficking (Braun et al., 2002; Haynes et al., 2002; Jentsch and Rumpf, 2007).
Several adaptor proteins have been shown previously to regulate nAChR trafficking. RIC-3 is necessary for proper folding/assembly of nAChRs (Lansdell et al., 2005), particularly those containing the α7 subunit (Williams et al., 2005). Rapsyn escorts muscle and neuronal AChRs between different cellular compartments (Kassner and Berg, 1997; Marchand et al., 2002), whereas 14-3-3η regulates the expression levels of α4β2 nAChRs (Jeanclos et al., 2001). The adaptor protein ubiquilin-1 (Plic-1), participates in the trafficking of α3* nAChRs by promoting their degradation by the proteasome (Ficklin et al., 2005).
To further identify proteins that associate with α3* nAChRs and might be involved in their trafficking, we used the large cytoplasmic domain of the α3 subunit as bait to screen a mouse brain cDNA library via yeast two-hybrid (Y2H) screen. Here we describe the isolation of a novel UBX-containing protein, UBXD4, and show that UBXD4 is a specific adaptor protein for α3* nAChRs.
Materials and Methods
Antibodies.
Polyclonal antibodies against α3 (rabbit clone #sc-5590 against amino acids 346–445, and goat clone #sc-1771, against amino acids 450–467, 1:200–500 dilution), β2 nAChR (clone H-92, 1:500 dilution), and monoclonal HA-antibody (1:500) were purchased from Santa Cruz Biotechnology. Rabbit polyclonal antibodies against the C terminus of α3 nAChR (against amino acids 496–503, 1:200 dilution) and the C terminus of β4 nAChR (1:500 dilution) were purchased from Biodesign International. Mouse monoclonal anti-TYG raised against the synthetic peptide EVHTNQDPLD (Brookman et al., 1995) was a gift from Dr. John Mayer (University of Nottingham, Nottingham, UK) (1:1000). Pacific Immunology generated a rabbit polyclonal antibody against a conserved 14 aa peptide, CIQRLQKTAEPFRKL, located at the C terminus of the mouse UBXD4 protein (residues 245–258). In immunoblots of total cell and tissue lysates, the rabbit anti-peptide antibody recognized a band of ∼28 kDa corresponding to the UBXD4 protein. To control for antibody specificity, we preabsorbed the UBXD4 antibody with the immunogenic peptide (100× excess compared with antibody molarity) followed by Western blot analysis. The peptide was able to eliminate binding of anti-UBXD4 to UBXD4. Monoclonal antibodies against GAL4 AD (1:1000) and GAL4 DNA-BD (1:1000) were purchased from Clontech. Anti-cadherin receptor [Sigma, 1:1000, plasma membrane (PM) marker], anti-Grp78 (anti-BiP, Stressgen, 1:1000, ER marker), anti-calnexin (Santa Cruz Biotechnology, 1:500, ER marker), anti-KDEL (Assay Designs, 1:400, ER/cis-Golgi marker), anti-GM130 (BD Biosciences, 1:250, Golgi marker), and anti-ERGIC-53 (Santa Cruz Biotechnology, 1:500, ERGIC marker) antibodies were used to locate different compartments. A goat anti-mouse IgG HRP (Santa Cruz Biotechnology, 1:2500), a mouse anti-rabbit IgG HRP (Santa Cruz Biotechnology, 1:2500), Alexa Fluor 488 donkey anti-rabbit IgG and Alexa Fluor 594-conjugated donkey anti-goat IgG (Invitrogen, 1:1000), and Cy3 552 donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, 1:1000) were used as secondary antisera.
Yeast two-hybrid screens.
A bait encoding the intracellular domain and transmembrane domains 3 (TM3) and 4 (TM4) of the mouse α3 nAChR subunit was amplified by PCR and cloned into the GAL4 DNA binding expression vector pGBKT7 DNA-BD (Clontech). The bait did not exhibit any intrinsic activation function in the AH109 strain. This bait was used to screen a mouse whole-brain cDNA library pooled from 9- to 12-week-old BALB/c mice and constructed in a pACT2 DNA-AD vector pretransformed into the yeast strain Y187 (Clontech). Approximately 1.8 × 107 transformants were screened and yielded 37 positive colonies. Positives clones were selected on triple dropout media (−Leu/−His/−Trp), and they were subsequently replated on quadruple dropout media (−Leu/−His/−Trp/−Ade) or on the triple dropout media with X-α-Gal for α-galactosidase activity. The Y187 and AH109 strains, pretransformed with plasmids encoding known interacting proteins (SV40 T-antigen and murine p53, respectively) were used as a positive two-hybrid control. The negative control was AH109 carrying the cloning vectors, pGBKT7 and pACT2.
Isolated plasmids were sequenced, and blast searches were performed against GenBank (NIH, genetic sequence database) to identify putative α3-interacting proteins. The sequencing of a positive isolated clone was identified as an 589 nt reading frame encoding the N terminus of UBXD4 (clone 42). To verify interactions, clone 42 as well as full-length UBXD4 (see plasmid constructs and Fig. 1a) cloned into the pACT2 DNA-AD vector were cotransformed with the empty pGBKT7 or the pGBKT7 DNA-BD vector containing the intracellular loop of the α3 subunit into yeast strain AH109. Baits encoding the large intracellular loop plus TM3 and TM4 of the nAChR subunits α4, α5, α7, β2, and β4 in a pGBKT7 DNA-BD vector were screened against clone 42 and full-length UBXD4 by one-on-one transformations. The fidelity of constructs was confirmed by automated sequencing. All template fusions were adequately expressed in yeast with expected sizes as determined by Western blot analysis of protein extracts using the TCA method described in the Yeast Protocols Handbook (Clontech).
Figure 1.
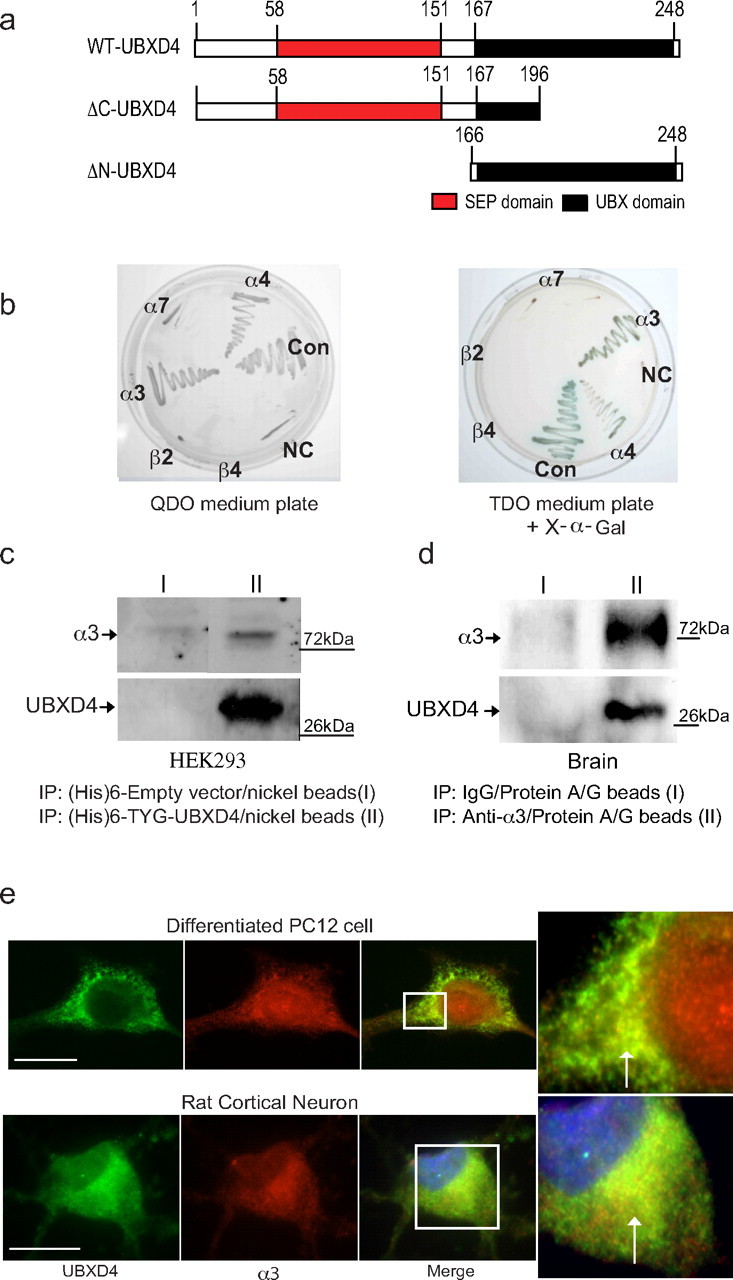
UBXD4 interacts with α3* and α4* nAChRs. A yeast two-hybrid screen showed that the large intracellular loop of the α3 nAChR subunit interacts with a novel UBX-containing protein named UBXD4. a, Schematic diagrams illustrating the structural location of the UBX and SEP domains in WT-UBXD4 (top panel). Middle and bottom panels represent the original clone 42 found in Y2H (ΔC-UBXD4, middle) and the generated ΔN-UBXD4 clone lacking the SEP domain (bottom). b, Interaction was verified with cotransformation of α3 and UBXD4 into the Y187 yeast strain. Further experiments in yeast showed that UBXD4 interacts with α4 but not with the α7, β2, and β4 nAChR subunits demonstrating that UBXD4 is a selective partner for the α3 and α4 subunits. Left panel, Quadruple dropout media (−Leu/−His/−Trp/−Ade). Right panel, Triple dropout media (−Leu/−His/−Trp) containing X-α-gal for α-galactosidase activity; CON, positive control; NC, negative control. c, In vitro experiments with charged Ni+ columns showed that the α3 subunit can bind to (His)6-TYG-tagged UBXD4 protein expressed in HEK293 cells. d, UBXD4 interacts with native α3 nAChR subunits from mouse brain. PFC lysates were incubated either with rabbit IgG (I, control) or the anti-α3 antibody (II) immobilized on protein A/G. Eluted proteins were subjected to SDS-PAGE and immunoblotted with the anti-α3 and anti-UBXD4 antibodies. e, Differentiated PC12 cells endogenously expressing α3-containing nAChRs (top) or rat cortical neurons (bottom) were cultured on coverslips, and processed for immunocytochemistry. Staining of the permeabilized cells for UBXD4 (green) and α3 (red) demonstrated partial overlapping of UBXD4 and α3 in the perinuclear region of the cells, as indicated by the white arrows in the magnified inset. Nonspecific binding or staining of the nucleoplasm in PC12 cells was observed by omitting the primary antibody during the incubation process (Fujiwara et al., 2006). Confocal scale bar, 10 μm.
Plasmid constructs and RT-PCR.
PCR was used to generate desired constructs by using two synthetic oligonucleotide primers (Integrated DNA Technologies) and Pfu Turbo polymerase (Stratagene). The cDNA sequence corresponding to the large cytoplasmic domain (amino acids 305–490) of the mouse α3 subunit was amplified using the forward primer 5′-GGA ATT CCA TAT GCT CCT CTT CAC TAT GAT TTT TGT CAC-3′ and the reverse primer 5′-ACG CGT CGA CCA GAA ATA ATC CTG CAG TTC CTA AAA TG-3′ by PCR and subcloned into the NdeI-SalI sites of the pGBKT7 DNA-BD vector (Clontech) to form the α3 bait. Restriction enzyme sites are underlined for all primers.
To generate a complementary DNA (cDNA) copy of UBXD4 RNA, we performed RT-PCR using mRNA isolated from a habenula of C57BL/6J mouse males using the RNAgents total RNA isolation kit (Promega). After first-strand cDNA synthesis, the templates were amplified using one set of primers flanking the N and C termini of UBXD4 cDNA with a NotI restriction site at both sides: forward primer 5′-GCG GCC GCC ATG AAA GAA GTA GAT AAT CTT GAC AG-3′; reverse primer 5′-GCG GCC GCT CAA AGT TTT CTA AAA GGC TCG GCA G-3′. The PCR product was ligated into the pGEM-T vector (Promega). Following digestion with NotI, the PCR product was ligated in-frame into the NotI site of pcDNA3.1 Zeo+ vector (Invitrogen) downstream of the (His)6-TYG sequence using T4 DNA ligase (New England Biolabs). Primers yielded a PCR product with the predicted sizes corresponding to the mouse UBXD4 sequence (777 bp). Another full-length UBXD4 PCR product with SfiI and EcoRI restriction sites was cloned into the pACT2 DNA-AD (for expression in yeast cells) and the pCMV-HA vectors (for expression in mammalian cells; Clontech) using the following primers: forward primer 5′-GGC CAT GGA GGC CAT GAA AGA AGT AGA TAA TCT TGA CAG-3′; reverse primer 5′-GGA ATT CGA ATG AAA GAA GTA GAT AAT CTT GAC AG-3′. Truncated forms of UBXD4 were generated by PCR: for ΔC-UBXD4 (amino acids 1–196), we used the forward primer for full-length UBXD4 and reverse primer 5′-GCG GCC GCT CAC ACC CTA TGA GAA ACA TTA AAT C-3′. For ΔN-UBXD4 (amino acids 166–258), we used the reverse primer for full-length UBXD4 and the forward primer 5′-GCG GCC GCC GTT TCA CTG AAC AAC TTG GAG CCC A-3′. The PCR products of ΔC-UBXD4 and ΔN-UBXD4 were subsequently subcloned into the NotI site of the pcDNA 3.1 (Z+). These constructs were used to transfect HEK293 and PC12 cells. The identity of PCR products was additionally confirmed by DNA sequencing.
Cell culture.
Human embryonic kidney HEK293 cells stably expressing α3β2 or α3β4 were originally generated in Dr. J. Lindstrom's laboratory. Cells were maintained in DMEM as described previously (Wang et al., 1998). Cytotoxic selection antibiotics were added to the media to ensure integrity of AChR subunit expression. Zeocin (0.5 mg/ml; Invitrogen) was used for α3 subunit selection and 0.6 mg/ml G-418 (Invitrogen) was used for β2 or β4 subunit selections. PC12, a rat pheochromocytoma cell line (American Tissue Culture Collection, lot number 060603), was cultured in F12 K media containing 5% fetal bovine serum and 10% horse serum in a humidified 95% air–5% CO2 incubator. Depending on the experiment, the cells were used either in the undifferentiated or the differentiated state.
HEK cells or PC12 cells were harvested in buffer A (in mm: Na2HPO4·NaH2PO4, 50; pH 7.5; NaCl, 50; EDTA, 5; EGTA, 5; benzamidine, 5; iodoacetamide, 15; phenylmethylsulfonyl fluoride, 2) and pelleted by centrifugation. The pellets were rinsed 3 times with buffer A before adding buffer B (buffer A + 1% Triton X-100). Samples were incubated at 4°C for 1 h followed by centrifugation at 13,000 RPM for 10 min in a refrigerated bench-top centrifuge. All cell extracts were subjected to Western blot analysis after protein concentrations were determined with a BCA Protein Assay Kit (Pierce Biotechnology). To prepare cell lysates for the p62 pull down and the iodixanol gradient fractionation, the pellets were rinsed 3 times with phosphate buffer serum (PBS, pH = 7.4) before addition of lysis buffer (20 mm Tris-HCl, pH 7.2; 1 ml/100 mm plate) containing EDTA (1 mm), NaN3 (1 mm), β-mercaptoethanol (1 mm), NP40 (0.1% v/v), glycerol (10% v/v), and a tissue extracts protease inhibitor cocktail (Sigma). Cell extracts were prepared by repeated freeze/thaw in liquid nitrogen or by sonicating on ice. Extracts were centrifuged for 10 min at 13,000 RPM.
Primary cultures of cortical neurons.
Primary rat neuronal cultures were derived from the cortices of E18 Long–Evans rats. Multiple cortices were dissected in Hank's saline and dissociated by trypsinization followed by mechanical dissociation. Cell suspension were plated in equal amounts in serum-free Neurobasal media with B27 supplement (Invitrogen), penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mm glutamine on laminin- and poly-d-lysine-treated coverslips. More than 95% of cells in the preparations were neurons.
Transfection and drug treatments.
HEK293 cells (80–90% confluent) were transiently transfected with expression vectors by calcium phosphate precipitation. Two days after transfection, cells were harvested and cell lysates were used for desired assays. Because the transfection efficiency was ∼40% in PC12 cells, as others have reported (Silverman et al., 2004), stably transfected PC12 cells were generated. cDNA containing the UBXD4, truncated forms of UBXD4, or the empty vector were transfected into PC12 cells by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. Forty-eight hours after transfection, the cells were treated with Zeocin at a concentration of 500 μg/ml. Cells were kept on Zeocin for 1 month to ensure selection of a stable cell line (Fukuda et al., 2002). To obtain differentiated PC12 cells (dPC12 cells), stably transfected PC12 cells were treated for 2–7 d with 50 ng/ml nerve growth factor (NGF, Sigma). The differentiated cells were either harvested for protein studies or used for patch-clamp experiments. For the confocal experiments in Figure 4a, PC12 cells transfected transiently with a pCMV-HA vector containing UBXD4. For degradation assays the proteasomal inhibitors MG132 (10 μm; Biomol) and PS-341 (0.5 μm; a gift from Dr John F. de Groot, MD Anderson Cancer Center, Houston, TX) or the lysosomal inhibitor E64 (10 μm; Research Products International) were used in the presence of the protein synthesis blocker emetine (150 μm, EMD Biosciences).
Figure 4.
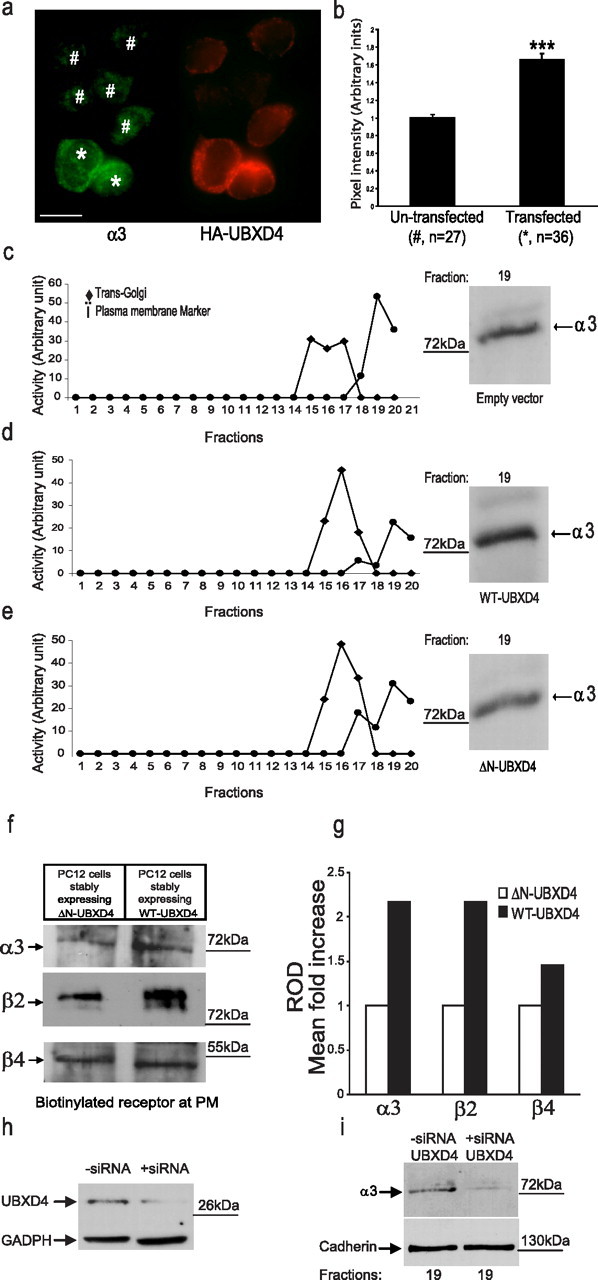
UBXD4 upregulates plasma membrane levels of α3* nAChRs. a, The fluorescent signal for α3 (green) increased when differentiated PC12 cells were transiently transfected with HA-tagged UBXD4 (red). The left panel shows the levels of α3 signal at the PM of UBXD4-overexpressing cells (*) compared with those of cells expressing endogenous levels of UBXD4 (#). Expression levels of UBXD4 where detected in the same cells with an anti-HA antibody after permeabilization (right panel). Confocal scale bar, 10 μm. b, Densitometric analysis of green pixel density in 63 cells (27 untransfected and 36 transfected) showed a significant increase in the total levels of α3 (p < 0.001). c–e, Differentiated PC12 cells stably expressing the corres-ponding empty vector (c), WT-UBXD4 (d), or ΔN-UBXD4 (e) were subjected to 8–34% linear iodixanol gradient fractionation. Enzymatic activity assays following iodixanol gradient fractionation were used to identify the fractions containing the Golgi and PM compartments (left panels). The fraction containing the highest levels of PM (fraction 19) showed increased levels of α3 upon overexpression of UBXD4 (right panels). ΔN-UBXD4 overexpression had no effect on α3 PM levels. f, Biotinylation experiments confirmed that expression of WT-UBXD4 but not ΔN-UBXD4 (control) greatly increases the surface levels of the α3 and β2 nAChR subunits. g, Quantitation of the data shown in f indicated that α3 and β2 levels increased 2.2-fold while β4 levels increased 1.4-fold compared with control. h, Stable expression of UBXD4 siRNA in PC12 cells decreased UBXD4 expression. i, The reduction in UBXD4 levels led to a significant decrease in α3 levels in PM fractions. The above experiments were conducted twice with similar results each time.
In vitro protein interaction (nickel pull down). For pull-down experiments, HEK293 cells stably expressing nAChRs were transfected with (His)6-TYG-tagged UBXD4. Cells lysates were diluted with 1 ml of wash buffer (0.5 m NaCl, 50 mm imidazole, 20 mm Tris, pH 7.0) and loaded onto Ni-NTA agarose (Qiagen) at 4°C, and then incubated overnight. Proteins were eluted with elution buffer (0.5 m NaCl, 0.2 m imidazole, 20 mm Tris, pH 7.0). Each eluted sample was concentrated with a microsep centrifugal concentrator (Pall Life Sciences) to give final volumes of 20 μl. Eluted samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Bound proteins were detected by Western blotting using Supersignal West Femto (Pierce Biotechnology).
p62 pull-down assays and immunoprecipitation.
Pull-down assays were conducted with the agarose-immobilized p62-derived UBA domain (Biomol) as previously described (Rezvani et al., 2007). Immunoprecipitations were conducted as described previously (Wiser and Schweiger, 1986; Karan et al., 2005). For the IP experiment shown in Figures 1d and 3c, proteins were yielded from beads with sample buffer without β-mercaptoethanol to eliminate the presence of heavy and light chains.
Figure 3.
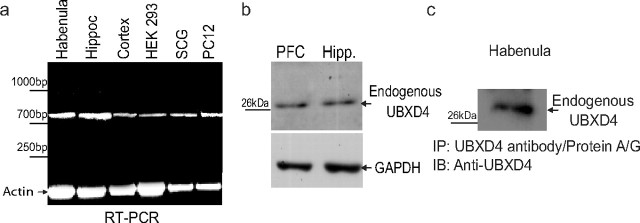
UBXD4 is expressed in neuronal and non-neuronal tissues. a, RT-PCR experiments showed that UBXD4 mRNA is expressed in all the examined tissues including cortex, hippocampus, habenula, peripheral ganglia, and two different cell lines (HEK293 and PC12 cells). b, c, Analysis of UBXD4 at the protein level with anti-UBXD4 antibodies demonstrated that the prefrontal cortex, hippocampus, and habenula express UBXD4 endogenously. Due to the small size of the habenula, we used IP techniques to enrich for UBXD4 by pulling down UBXD4 with anti-UBXD4 antibodies immobilized on protein A/G beads followed by probing with anti-UBXD4 (see Materials and Methods for details).
Confocal microscopy.
HEK293 and dPC12 cells, plated on coverslips, were washed and fixed with 2% paraformaldehyde in PBS for 10–20 min (Wang et al., 1998) and either permeabilized before antibodies incubation or stained first with the anti-α3 (1:200) antibody and then permeabilized for staining with anti-HA (1:400) or anti-UBXD4 (1:50) antibodies. Polyclonal anti-α3, monoclonal anti-HA, monoclonal anti-KDEL (1:400), and polyclonal anti-UBXD4 antibodies were used in conjunction with Alexa488-conjugated donkey anti-rabbit IgG, Alexa Fluor 594-conjugated donkey anti-goat IgG, Alexa Fluor 350-conjugated donkey anti-rabbit IgG (Invitrogen), and Cys543-conjugated donkey anti-mouse (Jackson ImmunoResearch Laboratories), respectively. Cells were imaged using a Zeiss LSM 510 laser scanning confocal microscope (Carl Zeiss MicroImaging) at 63× and 100×.
Biotinylation and isolation of surface nAChR subunits.
Surface expression of the α3, β2, and β4 nAChR subunits in PC12 cells was assayed with a Cell Surface Protein Biotinylation and Purification Kit (Pierce Biotechnology) according to manufacturer's instruction. Biotinylated plasma membrane proteins were then subjected to Western blot analysis.
Iodixanol gradient analysis.
We used iodixanol for gradient fractionation because it maintains relatively constant osmolality and viscosity despite changes in the density of the gradient. Because of the mild iso-osmotic conditions, all organelles and endosomes can be fractionated intact, without loss of water, as the density of the gradient increases. One to 2 ml of the cell lysates were layered onto preformed, precooled discontinuous 8, 16, 28, and 38% iodixanol Optiprep gradients (Greiner Bio-One) layered stepwise in centrifuge tubes using a gradient maker. Tubes were centrifuged for 18 h at 28,500 rpm (∼100,000 g) using a Beckman SW41i rotor at 4°C (Koulen et al., 2002). Sequential 20–21 equal-volume fractions (∼500 μl) were collected from the bottom of the tube with a Labconco Auto Densi-flow gradient collector (Labconco Corporation) and analyzed by Western blotting.
Leucine aminopeptidase activity.
Leucine aminopeptidase activity was measured as a trans-Golgi marker (Chi and Lodish, 2000) with the fluorogenic peptide substrate l-leucine-4-methyl-7-coumarinylamide hydrochloride (Sigma). Iodixanol gradient fractions (20 μl) were added to 3 ml of 50 mm Tris-HCl buffer, pH 7.5 containing 0.5 mm substrate in dimethyl sulfoxide (0.5% v/v). Samples were incubated at 25°C for 30 min. Fluorescence was measured at an excitation wavelength of 380 nm and an emission wavelength of 440 nm.
Alkaline phosphodiesterase activity.
Alkaline phosphodiesterase activity was measured as a PM marker (Touster et al., 1970; Wibo et al., 1981) in iodixanol gradient fractions (20 μl) with 500 μl of p-nitrophenyl 5′-thymidylate (1 mm) in 0.02 m Tris (hydroxymethyl) amino methane-HCl buffer, pH 9.0, supplied as ready to use reagent (pNPP, Sigma). Samples were incubated at 37°C for 15 min and the reaction terminated by the addition of 3N NaOH (1.5 ml). Optical densities were measured in a DU 640B Beckman Coulter spectrophotometer at a wavelength of 405 nm.
Whole-cell patch clamp recording and drug application.
dPC12 cells were continuously superfused with an external solution of the following composition (in mm): 150 NaCl, 5 KCl, 1.1 MgCl2, 2 CaCl2, 10 glucose, and 10 HEPES, pH 7.4. All the recordings were obtained at room temperature (22–25°C). The recording pipettes were filled with the following solution (in mm): 140 CsCl, 10 HEPES, 2 ATP (sodium salt), and 10 EGTA, adjusted to pH 7.3 with CsOH. The recording pipette had a resistance of 3–5 MΩ. Data were acquired with an Axopatch-1B amplifier at 5 kHz, and the membrane potential was held at −70 mV. The series resistance was usually in a range of 5–15 MΩ and was uncompensated. Data were accepted if series resistance did not change by >30% during the experiment.
Solutions were gravity fed into a linear array of glass barrels (380 μm ID, 500 μm OD, Garner glass) that were positioned ∼100 μm from the cell. The barrels were mounted on a 3-D manipulator that could be repositioned by a motorized driver (Newport Corporation). Cells were exposed for 2 s to the agonist, acetylcholine (ACh, 300 μm), which was controlled by a computer-operated magnetic valve. ACh was prepared every day. To isolate α3* nAChR currents, α-Bungarotoxin (25 nm) was present in the external solution. α-Conotoxin MII (α-CtxMII) was a generous gift from Dr. J. M. McIntosh. The nAChR antagonist, methyllycaconitine (MLA, 5 nm), or α-Ctx MII (100 nm) were also applied via rapid perfusion. PClampfit 8.0 software was used to measure peak and steady-state current amplitudes, and to fit the desensitization kinetics of α3-containing currents with a single exponential function.
siRNA design.
Three siRNA duplexes (clones A71, A295, and A689) were designed using online siRNA software provided at http://sfold.wadsworth.org (Ding and Lawrence, 2003; Ding et al., 2005). All siRNA sequences were subjected to BLAST to exclude sequence similarity to other genes. Oligonucleotides were synthesized by Integrated DNA Technologies followed by cloning into a siRNA expression vector, pSilencer 2.1-U6 neo vector (Ambion). Lipofectamine 2000 (Invitrogen) was used to transfect PC12 cells with pSilencer 2.1-U6 neo when they were 50% confluent. To create a population of cells stably expressing the siRNA, cells were incubated with 0.6 mg/ml G-418 for 2 weeks to eliminate cells that were not transfected. After 2 weeks the selection was complete and the cells were grown without antibiotics. The surviving cell population was maintained and assessed for reduction of endogenous UBXD4 gene expression. Stably transfected PC12 cells with clone A689 showed >90% silencing of the protein levels of endogenous UBXD4. For immunofluorescence experiment in supplemental Figure 5 (available at www.jneurosci.org as supplemental material), pSilencer 2.1-U6 neo vectors containing UBXD4 siRNA were transiently transfected into dPC12 cells followed by staining for α3* nAChRs at the PM and endogenous UBXD4. A set of 4 pLKO.1 lentiviral shRNA expression vector containing shRNA constructs against human and mouse UBXD4 proteins was purchased from Open Biosystems. Silencing efficiency was characterized in PC12 cells and the effective clone was used in rat cortical neurons to silence UBXD4.
Animals and brain slices.
Male C57BL/6J mice (250–350 g) were decapitated under anesthesia with a mixture of xylazine, acepromazine, and ketamine, and their brains were removed immediately for sectioning. The prefrontal cortex, hippocampus, and habenula were separated in ice-cold PBS. Tissue homogenization was performed in 500 μl of ice-cold homogenization buffer (0.32 m sucrose, 10 mm HEPES buffer, and 2 mm EDTA, pH 7.4) containing a protease inhibitor cocktail (Sigma) or a RNAgents total RNA isolation kit (Promega), as described before. Homogenized tissues were solubilized by repeated freeze/thaw in liquid nitrogen for protein assay. Supernatants were analyzed by Western blotting. All procedures were approved by the Institutional Animal Care and Use Committee in accordance with federal guidelines.
Densitometric quantification of Western blot bands.
Quantification of the intensity of Western blot bands was carried by using an automated digitizing system (UN-Scan-it gel, version 6.1). The UN-Scan-it software enabled us to quantify our Western blot data with two different methods. The segmental analysis method provided pixel summation and band analysis for individual segments (data presented in Fig. 4g), while lane analysis provided density profile extraction and band analysis for entire lanes (data presented in Fig. 7e). Developed films were scanned as JPEG in 8 bit grayscale format at 600 dpi, and the pixel intensities within a band were measured. Pixel intensities were corrected for background. After exporting into Excel, the digitized gel data (pixel total) were used to calculate the relative optical densities of each gel band for further analysis.
Figure 7.
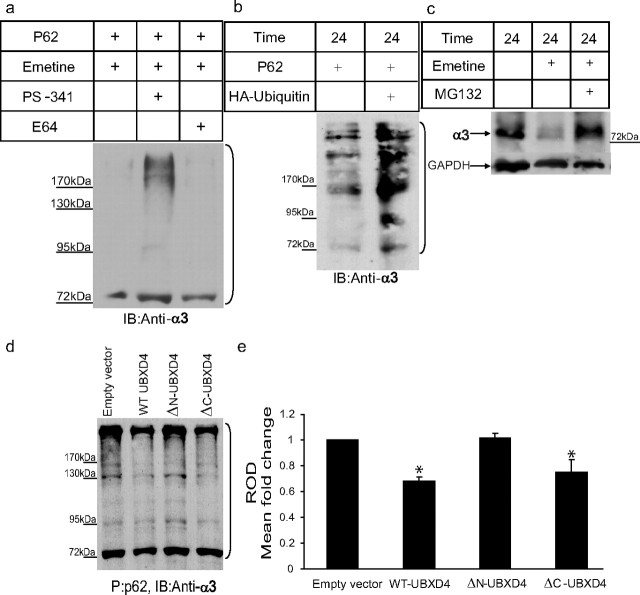
UBXD4 decreases the levels of ubiquitinated α3. Differentiated PC12 cells were treated for 24 h with PS-341 (a selective proteasome inhibitor) or E64 (an inhibitor of the lysosome) in the presence of emetine (a protein synthesis blocker). Cell lysates were probed for α3. a, A ladder of various polyubiquitinated forms of α3 (bracket) could be pulled down by the p62-derived UBA domain, suggesting that α3 is ubiquitinated and can be degraded by the proteasome. In untreated cells (left lane) and cells treated with E64 (right lane), the majority of α3 receptors was degraded by the proteasome after ubiquitination. b, In another set of experiments, the level of ubiquitination of α3 in the presence of overexpressed HA-ubiquitin was determined. Cell lysates of PC12 cells stably expressing empty vector or HA-ubiquitin were subjected to p62 pull down followed by immunoblotting with anti-α3 antibodies. Overexpression of HA-ubiquitin led to an increase in the levels of ubiquitinated α3. c, The degradation of the α3 nAChR subunit was decreased dramatically in the presence of the proteasome inhibitor MG132 in PC12 cells. d, P62 pull down was performed on cell lysates from dPC12 cells stably transfected with the corresponding empty vector, WT-UBXD4, ΔN-UBXD4, or ΔC-UBXD4 followed by Western blot analysis with anti-α3 antibodies. A ladder of ubiquitinated α3 could be detected in each sample. Quantification of the α3 signal showed that WT-UBXD4 and ΔC-UBXD4, but not ΔN-UBXD4, significantly decrease the ubiquitinated levels of α3 (e; p < 0.05). The brackets on the right (a, b, d) mark a ladder of bands corresponding to polyubiquitinated α3.
Statistics.
Statistical analysis was conducted using either the Student's t test for normally distributed variables to conclude the significant differences between control and subjects or by one-way ANOVA and Tukey multiple comparison post tests when appropriate. A p value ≤0.05 was considered statistically significant. All data are reported as mean ± SEM.
Results
A UBX-containing protein binds to α3* nAChRs
To identify proteins involved in the trafficking of α3* nAChRs to the PM, we performed Y2H screen against a mouse brain cDNA library. cDNA encoding the large intracellular domain of the α3 nAChR subunit and the flanking transmembrane (TM) regions 3 and 4 (amino acids 305–490) was used as bait. The TM regions were included in the bait because intracellular domains can adopt a more precise conformation when expressed with TM regions on either end (Bedford et al., 2001).
The Y2H screen yielded an interaction between the large intracellular loop of the α3 nAChR subunit and a novel protein. Sequencing deciphered a 589 nt reading frame corresponding to the truncated N terminus of the UBXD4 protein (Fig. 1a, middle panel, clone 42; accession number: NM_145441). The SMART database (Schultz et al., 1998; Letunic et al., 2006) identified UBXD4 as the corresponding 258 aa protein (Fig. 1a, top) and pinpointed two domains of interest contained therein: SEP and UBX. SEP (Saccharomyces cerevisiae, Drosophila melanogaster eyes closed gene and vertebrate p47) is a reversible competitive inhibitor of cathepsin L (Chu et al., 1995; Soukenik et al., 2004); UBX (ubiquitin regulatory X), consisting of 80 amino acid residues, can typically be found at the C terminus of a variety of eukaryotic proteins of differing functions (Buchberger et al., 2001; Buchberger, 2002; Bogan et al., 2003; Schuberth and Buchberger, 2008). A search in GenBank revealed sequences of the human (accession number: NM_181713) and rat homologs (accession number: XR_007472) to UBXD4. Comparison of all three sequences indicated that UBXD4 is highly conserved between the three species (85.7%, 95.5%, and 86.0% identical between mouse and human, mouse and rat, and rat and human, respectively), suggesting that it may possess an important function in mammals.
To examine the binding specificity of UBXD4 for other nAChR subunits, the intracellular loop plus the respective TM flanking regions of α4, α7, β2, and β4 were used as probes in additional Y2H screens against the original UBXD4 clone (clone 42). Clone 42 bound to the intracellular loop of both the α3 and α4 nAChR subunits, but not to the corresponding loops of α7, β2, and β4 (Fig. 1b). Due to the toxicity of the α5 subunit to yeast, we were not able to test its interaction with UBXD4. These results suggest that UBXD4 might be an exclusive non-nicotinic partner for the α3 and α4 nAChR subunits.
Because clone 42 for which we found the initial interaction with α3 was a ΔC-UBXD4 version of the full-length wild-type (WT) UBXD4, we also tested and confirmed the interaction of WT-UBXD4 with the intracellular loops of α3 and α4 by Y2H. Because clone 42 does not contain a complete UBX domain (Fig. 1a, middle panel), the SEP domain, and not the UBX domain is probably necessary for the interaction with the two α nAChR subunits.
UBXD4 interacts with intracellular α3* nAChRs
To confirm our Y2H observations, we further tested the interaction of UBXD4 with the α3 nAChR subunit using an affinity purification assay (nickel pull down) and Western blotting. (His)6-TYG-tagged UBXD4, but not (His)6-TYG alone, was able to pull down the α3 nAChR subunit from cell extracts of HEK293 cells stably expressing α3β2 nAChRs (Wang et al., 1998) (Fig. 1c). The same result was obtained in HEK293 cells stably expressing α3β4 nAChRs (data not shown).
To investigate the physiological relevance of the α3/UBXD4 interaction found in vitro, we determined whether UBXD4 associates with native α3* nAChRs immunoprecipitated from the mouse brain. The prefrontal cortex (PFC) of C57BL/6J male mice was dissected and homogenized, followed by immunoprecipitation of α3* nAChRs with an anti-α3 antibody. As a control, homogenized PFCs were also incubated with beads coupled to rabbit IgG. The interaction of UBXD4 with the α3 nAChR subunit was then detected by immunoblotting with an anti-UBXD4 rabbit polyclonal antibody. As already shown in cultured cells, UBXD4 was found to be associated with native α3* nAChRs (Fig. 1d), suggesting a physiological interaction between UBXD4 and α3* nAChRs in vivo.
The localization of UBXD4 with respect to α3* nAChRs was also examined by confocal microscopy. dPC12 cells endogenously expressing α3* nAChRs (Rogers et al., 1992) and cortical rat neurons were cultured on coverslips and processed for immunocytochemistry. Before staining with the corresponding fluorescent secondary antibodies, permeabilized cells were incubated with a rabbit polyclonal anti-UBXD4 antibody and a goat polyclonal antibody against a peptide mapping near the C terminus of α3. Antibody specificity was determined by blocking with the identical peptide sequence used to generate the polyclonal antibodies for UBXD4 and α3 [supplemental Fig. 1a,b (available at www.jneurosci.org as supplemental material), respectively]. The signal for UBXD4 (green) was visible throughout the cytoplasm in patchy structures (Fig. 1e), possibly reflecting the presence of UBXD4 in complexes with p97 (Alexandru et al., 2008). The signal for α3 (red) was mostly seen in perinuclear structures and adjacent compartments, possibly reflecting the large pools of α3 localized either in the ER (Ren et al., 2005) or the trans-Golgi compartments (Jacob et al., 1986). Overlapping of UBXD4 with the α3 nAChR subunit occurred mostly in puncta around the nucleus in both PC12 cells and rat cortical neurons (Fig. 1e, magnified inset). Similar results were achieved when PC12 cells were transiently transfected with HA-tagged UBXD4 (data not shown). The immunocytochemistry data, together with the Y2H, and immunoprecipitation data provide evidence for the direct interaction between UBXD4 and the α3 nAChR subunit.
Characterization of UBXD4
We next examined the subcellular localization of UBXD4 using iodixanol gradient fractionation in undifferentiated PC12 cells. In particular, we used a high-resolution 8–34% linear iodixanol gradient optimized for endoplasmic reticulum (ER), Golgi, and PM separation (Koulen et al., 2002). The collected iodixanol gradient fractions were tested with markers indicative of the presence of the ER, Golgi Intermediate Compartment (ERGIC), cis-, middle-, and trans-Golgi, or PM compartments. Enzymatic activity of markers for the trans-Golgi (leucine aminopeptidase activity) and PM (alkaline phosphodiesterase activity) (Fig. 2a) and Western blotting of collected fractions (Fig. 2b) revealed that UBXD4 particularly overlaps with fractions enriched in the cis- and middle-Golgi subcompartments (GM130 (Tompkins et al., 2003), the ERGIC compartment (ERGIC-53), and the ER compartment (BiP and calnexin). Together, these results indicate that compartmentalization of UBXD4 may be selective to certain substructures in the ER and Golgi compartment. Confocal microscopy in dPC12 cells also showed that UBXD4 (green) can colocalize with KDEL receptors (red), which are known to be enriched in the ER and cis-Golgi compartment (Fig. 2c) (Hayes et al., 2003; Hamada et al., 2004; Le-Niculescu et al., 2005).
Figure 2.
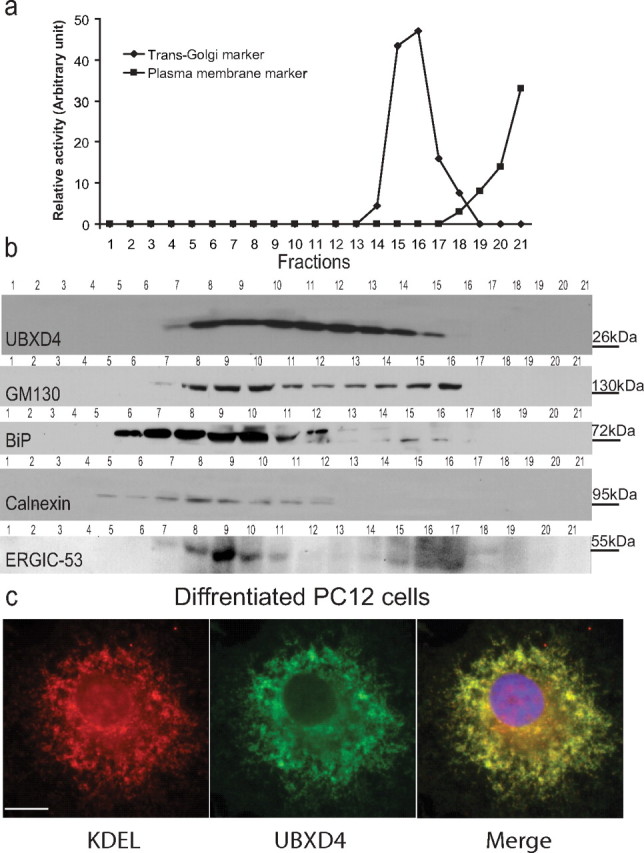
UBXD4 is located in the ER/Golgi compartments. a, b, Naive PC12 cells were subjected to 8–34% linear iodixanol gradient analysis. According to the leucine aminopeptidase (trans-Golgi marker) and alkaline phosphodiesterase (PM marker) activities and staining with anti-BiP and anti-calnexin (ER markers), anti-GM130 (cis- and middle Golgi marker), and ERGIC-53 (ERGIC marker) antibodies, UBXD4 is preferentially located in the ER/Golgi compartments (fractions 7–15). c, The predominant colocalization of UBXD4 with KDEL protein (an ER and cis-Golgi marker) in differentiated PC12 cells confirms that UBXD4 is an ER/Golgi protein. Confocal scale bar is 10 μm.
RT-PCR revealed that UBXD4 mRNA has a broad pattern of tissue expression in the CNS, including prominent expression in habenula, hippocampus, and cortex (Fig. 3a). UBXD4 mRNA is also present in peripheral ganglia (Fig. 3a) as well as non-neural tissues such as the lungs, liver, heart, kidney, ileum, pancreas, and spleen (data not shown). In addition, UBXD4 is also expressed in HEK293 and PC12 cells (Fig. 3a). A similar gene expression profile for human UBXD4 mRNA has been reported by OriGene Technologies (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). In addition to detecting UBXD4 mRNA, we also studied the expression of UBXD4 protein with anti-UBXD4 antibodies. For example, Figure 3, b and c, shows that an ∼28 kDa band, corresponding to WT-UBXD4, could be detected in the PFC, hippocampus, and habenula of male C57BL/6J mice.
Figure 3c describes experiments performed in the habenula, which expresses high levels of α3. Due to the small size of the habenula, we used immunoprecipitation techniques (Wiser and Schweiger, 1986; Ficklin et al., 2005) to enrich our preparations for UBXD4. We first used an anti-UBXD4 antibody immobilized on protein A/G beads to pull down UBXD4 from 3 habenula tissue punches. The immunoprecipitates were then probed with the anti-UBXD4 antibody to reveal the presence of UBXD4-bound UBXD4.
UBXD4 modulates α3* nAChR cell surface expression
To investigate the functional consequences of the interaction between UBXD4 and α3* nAChRs, we conducted a confocal microscopy experiment in dPC12 cells transiently transfected with HA-tagged UBXD4. Nonpermeabilized cells were first fixed briefly with paraformaldehyde and stained for the α3 subunit. Subsequently, cells were permeabilized and stained for HA-tagged UBXD4. These experiments showed that WT-UBXD4 facilitates the trafficking of α3* nAChRs to the PM (Fig. 4a). Densitometric analysis was performed using ImageJ software (NIH, Bethesda, MD, USA) to quantitate pixel intensity from fluorescence images like those shown in Figure 4a. The analysis of green pixel density of 63 dPC12 cells (27 untransfected and 36 transfected) showed a significant increase in the total levels of α3 (untransfected vs transfected: p < 0.001). Similar results were obtained in HEK293 cells stably expressing α3β2 nAChRs that were transiently transfected with UBXD4 (supplemental Fig. 3a, available at www.jneurosci.org as supplemental material).
To further investigate the mechanisms underlying the ability of UBXD4 to increase the cell surface expression of α3* nAChRs, we generated three PC12 cell lines stably expressing WT-UBXD4, the corresponding empty vector, or a truncated N terminus UBXD4 (ΔN-UBXD4, amino acids 166–258) (Fig. 1a, bottom). The ΔN-UBXD4 construct was used as an additional control besides the empty vector. Due to the absence of the SEP domain, we anticipated that the ΔN-UBXD4 construct may not interact with the α3 subunit. After differentiation with NGF, cell lysates of dPC12 cells stably expressing WT-UBXD4, the empty vector, or ΔN-UBXD4 were subjected to iodixanol gradient fractionation. A PM marker indicated that fraction 19 contained the highest purified PM compartment (Fig. 4c–e, left panels). WT-UBXD4 increased the level of α3* nAChRs in fraction 19 compared with the empty vector and ΔN-UBXD4, confirming that UBXD4 promotes the translocation of α3* nAChRs to the PM compartment (Fig. 4c–e, right panels). Quantification of the corresponding bands showed an ∼40% increase in surface α3* nAChRs in cells expressing WT-UBXD4 compared with control cells (Fig. 4, compare c, d), while cells expressing the ΔN-UBXD4 did not show significant increase in α3 levels (Fig. 4, compare c, e). The increase in PM α3 in the presence of overexpressed UBXD4 was paralleled by an increase in the total levels of α3 and β2 in dPC12 cells (data not shown). To confirm the observation made in PC12 cells, HEK293 cells stably expressing α3β2 nAChRs were transiently transfected with (His)6-TYG-tagged UBXD4 or (His)6-TYG-tagged empty vector. Cell lysates were subjected to glycerol gradient fractionation followed by Western blot analysis. According to the localization of Golgi and PM markers, UBXD4 increased both total levels and PM levels of α3* nAChRs (supplemental Fig. 3b, available at www.jneurosci.org as supplemental material).
To verify that heterologous protein expression in PC12 cells allows to detect changes in nAChR trafficking and compartmentalization, dPC12 cells were stably transfected with Plic-1 (ubiquilin-1), an adaptor protein involved in GABAA receptor trafficking and nAChRs degradation (Bedford et al., 2001; Ficklin et al., 2005). After iodixanol gradient fractionation, an anti-β2 nAChR subunit antibody was used to detect the location of α3β2* nAChRs in ER/Golgi and PM fractions. As already shown (Ficklin et al., 2005), Plic-1 overexpression decreased α3* nAChRs levels in ER but not Golgi fractions (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). A similar compartment-specific effect has been described for the effects of Plic-1 with respect to GABAA receptors (Bedford et al., 2001).
Finally, to better estimate the percentage of upregulation for cell surface α3β2 and α3β4 nAChRs, we performed surface biotinylation assays using a membrane impermeable biotin. The experiment was conducted in dPC12 cells stably expressing WT-UBXD4 or ΔN-UBXD4. Western blotting followed by quantitative analysis of the data indicated that WT-UBXD4 significantly augments the levels of α3* nAChRs found at the PM (Fig. 4f). In particular, α3β2* nAChRs were markedly upregulated (∼2.2-fold), while those of the α3β4 nAChRs had only a 1.4-fold increase (Fig. 4g).
In another set of experiments we generated a stable PC12 cell line expressing UBXD4-siRNA to knockdown endogenous UBXD4. Transfection with UBXD4-siRNA resulted in a >80% reduction in endogenous UBXD4 protein levels (Fig. 4h). After differentiation with NGF, cell lysates of UBXD4-silenced cells and controls were subjected to iodixanol gradient fractionation. Immunoblotting of PM-enriched fractions showed that the decrease in UBXD4 levels coincided with a significant (≥90%) reduction in the levels of α3* nAChRs at the PM (Fig. 4i). Anti-cadherin receptor antibodies were used as loading control for the PM compartment. This result further confirmed our hypothesis that UBXD4 facilitates the trafficking of α3* nAChRs to the PM. The expression of α3 in dPC12 cells transiently transfected with UBXD4 siRNA was also determined by confocal microscopy (supplemental Fig. 5a,b, available at www.jneurosci.org as supplemental material). In these experimental conditions UBXD4 siRNA treatment reduced pixel density of α3* receptors at the PM compared with untransfected cells by 40.85% ±2.15 (p < 0.005, n = 28).
Because rat cortical neurons express α3-containing nAChRs (Yeh et al., 2001), we studied the effect of UBXD4 silencing in those neurons with confocal microscopy techniques. To functionally silence UBXD4, we obtained from Open Biosystems four pLKO.1 lentiviral shRNA expression vectors containing shRNA constructs targeting human and mouse UBXD4. To check the efficiency of silencing, the four vectors were transiently transfected into rat PC12 cells. Clone 99 lentiviral-based vector showed the maximum silencing effect (Fig. 5a). Embryonic day 18 rat cortical neurons were transfected with clone 99 shRNA along with a GFP-empty vector using the calcium phosphate method (Arikkath et al., 2008). GFP allows easy identification of the transfected neurons by fluorescence microscopy. Cells were fixed and processed for immunofluorescence with the appropriate primary antibodies and fluorescent secondary antibodies. The percentage of cortical neurons expressing GFP 72 h after lentiviral transfection was 1–3%. Figure 5b shows that, in GFP-positive cells, UBXD4 shRNA decreased the levels of UBXD4 protein, which in turn resulted in a reduction in total levels of α3. The nAChR subunit was decreased in the soma, predominantly in the perinuclear compartment. Images quantified with ImageJ software revealed a significant, 30% reduction of α3 signals in GFP-positive cells (n = 6) compared with untransfected cells (GFP negative, n = 11) (supplemental Fig. 5c, available at www.jneurosci.org as supplemental material).
Figure 5.
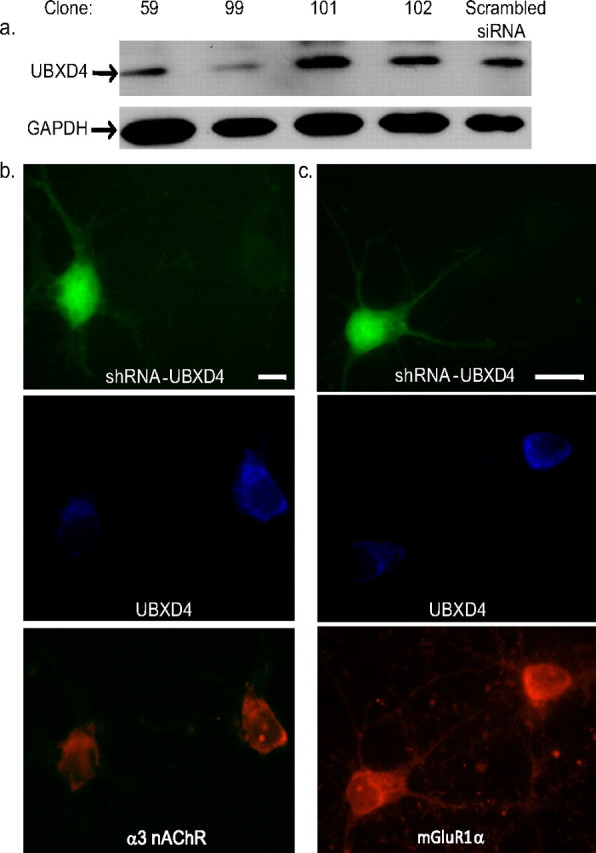
UBXD4 shRNA selectively decreases α3 nAChRs signal in rat cortical neurons. a, Western blot analyses of PC12 cells transfected with four lentiviral-based shRNA showed that clone 99 can efficiently silence endogenous UBXD4. b, Confocal microscopy of rat cortical neurons transfected with clone 99 against UBXD4 demonstrated a significant reduction of α3 signal in permeabilized, shRNA-transfected cells. The reduction of signal for α3-containing nAChRs by UBXD4 siRNA suggests that UBXD4 regulates trafficking of α3. c, shRNA against UBXD4 had no significant effect on mGluR1α receptor levels detected in rat cortical neuron. See supplemental Figure 5, c and d (available at www.jneurosci.org as supplemental material), for the quantification of the confocal data.
To determine whether UBXD4 can regulate other transmembrane receptors besides nAChRs, we studied the effect of UBXD4 on metabotropic glutamate receptors 1α (mGluR1α). As mGluRs are expressed in the prefrontal cortex (Gupta et al., 2005), we examined the effects of clone 99 lentiviral-based UBXD4 shRNA vector on mGluR1α levels (Fig. 5c). Analyses of the immunostaining experiments conducted with anti-mGluR1α antibodies showed lack of a significant effect of UBXD4 shRNA on total mGluR1α levels (supplemental Fig. 5d, available at www.jneurosci.org as supplemental material).
Interaction of UBXD4 with the ubiquitin–proteasome system
It has been suggested that UBX domains may associate with ubiquitin (Hofmann and Bucher, 1996; Buchberger, 2002). To determine whether the UBX domain of UBXD4 can indeed interact with ubiquitin, we conducted Y2H screening of a mouse brain cDNA library with UBXD4 as bait. The Y2H experiment yielded 94 positive clones. Subsequent DNA sequencing of the positive clones showed that one clone encoded Mus musculus ubiquitin C (accession number: BC025894) containing 2.5 tandem ubiquitins. Ubiquitin C is a ubiquitin precursor composed of four tandem repeats of the ubiquitin unit (Kim et al., 1998). Ubiquitin monomer in many species can be generated by processing of a polyubiquitin precursor protein in which several exact repeats of the ubiquitin amino acid sequence are joined in head to tail repeats (Ozkaynak et al., 1984; Shiba et al., 2004; Tanaka et al., 2004).
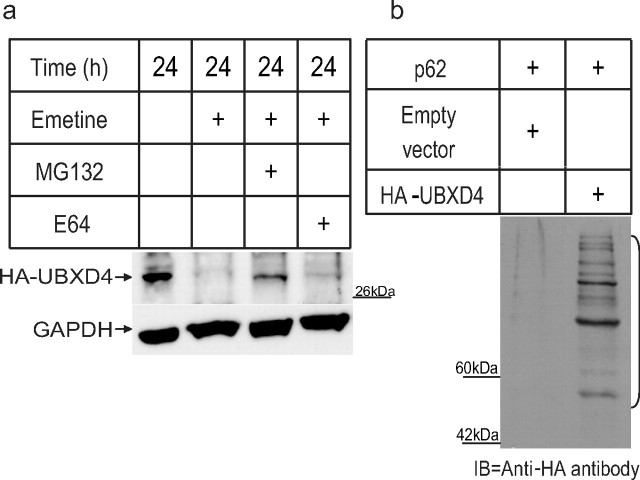
To examine whether UBXD4 is a target of the ubiquitin–proteasome pathway, we measured the half-life of UBXD4 in HEK293 cells in the presence of MG132, a known proteasome inhibitor (Crawford et al., 2006). A degradation assay showed that exposure to MG132 in the absence of protein synthesis increased the half-life of UBXD4 (Fig. 6a), while the lysosome inhibitor E64 did not, suggesting that UBXD4 is degraded by the proteasome. To determine the relative half-life of UBXD4, time course experiments were performed in HEK293 cells transiently transfected with HA-tagged UBXD4. A 12 h time course experiment revealed that the half-life of UBXD4 is ∼6 h (data not shown).
Figure 6.
UBXD4 is ubiquitinated and degraded by the 26S proteasome. a, A 24 h exposure to the proteasome blocker MG132 in the presence of the protein synthesis blocker, emetine, decreased the rate of degradation of UBXD4 in HEK293 cells transiently transfected with HA-tagged UBXD4. Exposure to the lysosome inhibitor E64 did not alter the degradation rate of UBXD4. b, A ladder of ubiquitinated, HA-tagged UBXD4 could be pulled down with p62 from α3β2 expressing HEK293 cells transiently transfected with HA-tagged UBXD4, but not with the HA-empty vector. The bracket on the right marks a ladder of bands corresponding to polyubiquitinated UBXD4. The above experiments were conducted twice with similar results each time.
To confirm the fact that UBXD4 turnover is regulated by the UPS, we used the p62-derived UBA domain to pull down multiubiquitinated UBXD4 from HEK293 cells transiently transfected with either HA-tagged UBXD4 or the corresponding empty vector. P62 is a cytoplasmic protein that specifically interacts noncovalently with ubiquitin (Vadlamudi et al., 1996). Its sequence contains a motif—the ubiquitin associated (UBA) domain—that can, by itself, bind to ubiquitinated proteins (Raasi et al., 2005; Rezvani et al., 2007). The p62 pull down assay showed that UBXD4 is ubiquitinated (Fig. 6b) as revealed by the ladder of various forms of ubiquitinated UBXD4 detected by an anti-HA antibody against HA-tagged UBXD4.
UBXD4 enhances the stability of the α3 nAChR subunit
Ubiquitin-related proteins such as UBXD4 have been suggested to have a number of functions. Such functions include linking binding partners to scaffolding/cytoskeletal proteins, modulation of the degradation of the proteins they interact with, and regulation of the proteasome (Buchberger, 2002; Welchman et al., 2005; Hurley et al., 2006; Schuberth and Buchberger, 2008). To investigate the possible involvement of UBXD4 in UPS-related mechanisms, we first investigated whether the UPS interacts with α3. To this end, we demonstrated in dPC12 cells that the α3 subunit is ubiquitinated (Fig. 7a,b) and degraded by the 26S proteasome complex (Fig. 7c). Second, to test the role of UBXD4 in controlling α3* nAChRs stability, we examined the ubiquitination state of α3* nAChRs in dPC12 cells stably expressing either WT-UBXD4, ΔN-UBXD4, ΔC-UBXD4 (the original clone 42, amino acids 1–196) (Fig. 1a), or the empty vector. After p62 pull down, ubiquitinated proteins were detected with anti-α3 antibodies (Fig. 7d). Quantitative analysis of the ladders of ubiquitinated α3 [for details, see Materials and Methods and Rezvani et al. (2007)] revealed that WT-UBXD4 and ΔC-UBXD4 significantly decreased the ubiquitinated levels of α3 compared with ΔN-UBXD4 and empty vector [empty vector vs WT-UBXD4: p < 0.05, empty vector vs ΔN-UBXD4: p > 0.05 (nonsignificant), empty vector vs ΔC-UBXD4: p ≤ 0.05, WT-UBXD4 vs ΔN-UBXD4: p < 0.05, WT-UBXD4 vs ΔC-UBXD4: p > 0.05 (nonsignificant), ΔN-UBXD4 vs ΔC-UBXD4: p < 0.05; n = 3 for each group] (Fig. 7e). The fact that UBXD4 decreases the amount of ubiquitinated α3 provides a mechanism for the upregulation of α3* nAChRs levels in dPC12 cells.
Because it has been reported that overexpression of nonproteasomal multiubiquitin binding proteins can inhibit proteasomal activity (Raasi and Pickart, 2003), we conducted experiments in which proteasomal chymotrypsin-like activity was measured in dPC12 cells overexpressing UBXD4. In those experiments, UBXD4 overexpression did not interfere with proteasomal activity (supplemental Fig. 3c,d, available at www.jneurosci.org as supplemental material), a result that suggests that the effect of UBXD4 on α3 nAChRs stability is probably due to a direct interaction of the two proteins rather than the inhibition of the proteasome complex.
UBXD4 regulates the size of α3β2 nAChR currents
The stably transfected dPC12 cells used to study the ubiquitination state of the α3 subunit in the presence of UBXD4 and its truncated forms were also used in electrophysiology experiments. Transfection with WT- and ΔC-UBXD4, but not with ΔN-UBXD4, significantly increased the peak amplitude of ACh-gated nAChR currents compared with the empty vector (Fig. 8a,b, top left panel). Peak current densities were also higher in WT- and ΔC-UBXD4 cells versus the empty vector, indicating that the changes in measured currents were not due to transfection-induced changes in cell size and morphology (Fig. 8b, bottom left panel). While WT- and ΔC-UBXD4 increased peak currents, they had no significant effect on the levels of steady-state currents measured in the same cells (Fig. 8b, top and bottom right panels). Furthermore, transfection with WT- and ΔC-UBXD4 yielded nAChR currents with faster inactivation kinetics compared with the empty vector- or ΔN-UBXD4-transfected cells.
Figure 8.
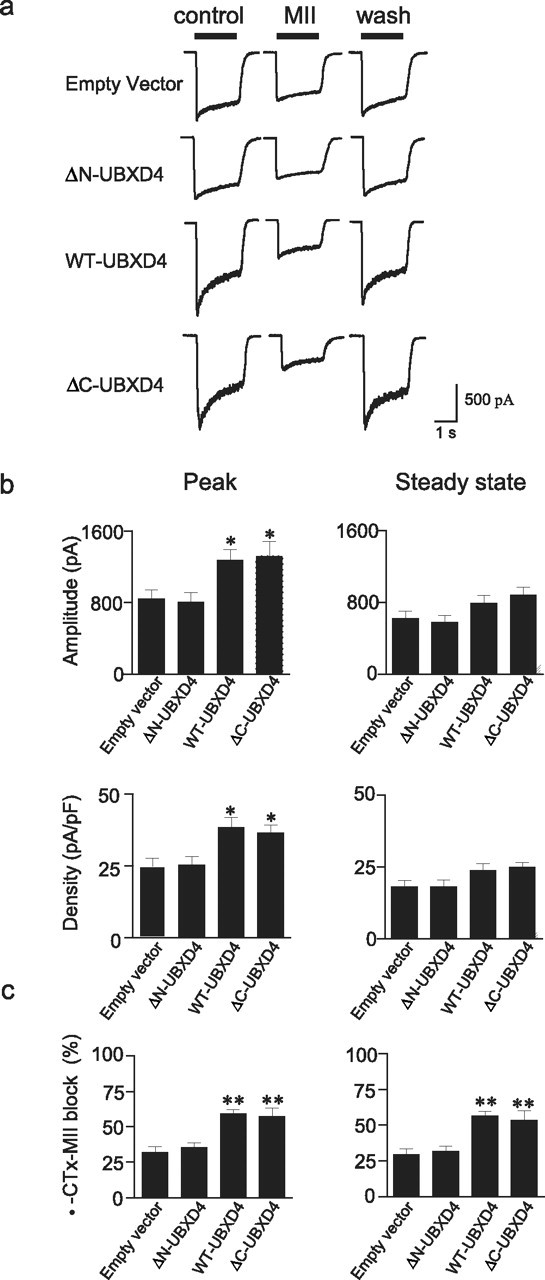
Functional regulation of α3* nAChRs by UBXD4. a, Currents elicited by 300 μm ACh were measured before, during, and after a 5 min exposure to 100 nm α-CTx-MII in dPC12 cells stably transfected with empty vector, ΔN-UBXD4, WT-UBXD4, or ΔC-UBXD4. b shows peak (left panels) and steady-state (right panels) current amplitude and density in the various cell lines described in a. Overexpression of WT-UBXD4 and ΔC-UBXD4 produced a significant increase (p < 0.05) in peak amplitude and current density compared with empty vector or ΔN-UBXD4. c, Block by α-CTx-MII of peak (left panel) and steady-state (right panel) α3* nAChR currents. The α-CTx-MII-sensitive component was greater in cells overexpressing WT-UBXD4 and ΔC-UBXD4 than in cells expressing the corresponding empty vector or ΔN-UBXD4 (p < 0.01). Values represent mean ± SEM values from cells expressing empty vector (n = 16), ΔN-UBXD4 (n = 12), WT-UBXD4 (n = 34), and/or ΔC-UBXD4 (n = 8), respectively.
Changes in current kinetics and the fact that our biotinylation data suggested a preferential effect of UBXD4 on α3β2* nAChRs encouraged us to test the effects of the α-conotoxin MII (MII) on the currents. MII can block α3β2* (Cartier et al., 1996), β3* (Cui et al., 2003), and α6* (Vailati et al., 1999; Champtiaux et al., 2002) nAChRs. As shown in Figure 8, MII was able to block a larger proportion of ACh-gated currents in the dPC12 cells overexpressing WT-UBXD4 or ΔC-UBXD4 (Fig. 8a,c). The effects of the toxin on ΔN-UBXD4 were comparable to those observed in empty vector-transfected cells suggesting that the UBX domain is not necessary for the effects of UBXD4 on α3* nAChR levels. Interestingly, we found that steady-state nAChR currents were also more sensitive to MII in WT- and ΔC-UBXD4-transfected cells, suggesting that UBXD4 might promote changes in the overall nAChR subunit composition.
Finally, to further confirm that UBXD4 selectively influences α3* nAChRs, we also measured α7* nAChRs currents, which were identified by their fast kinetics and their sensitivity to methyllycaconitine (MLA) (supplemental Fig. 6a, available at www.jneurosci.org as supplemental material). As indicated in supplemental Figure 6, b and c (available at www.jneurosci.org as supplemental material), transfection with WT-UBXD4 or the truncated forms of UBXD4 did not alter the size or the kinetics of α7 currents.
Discussion
To address the mechanisms underlying α3* nAChR trafficking, we searched for unidentified protein partners for the α3 subunit. Through a Y2H screen we identified a UBX-containing protein, UBXD4, that interacts with the large cytoplasmic loop of α3 and α4. This work focused on the interaction between UBXD4 and α3* nAChRs. Our data indicate that UBXD4 participates in the trafficking of α3* nAChRs through the ER and Golgi compartments, possibly protecting α3* nAChRs from proteasomal degradation (Fig. 9). UBXD4 overexpression led to a greater insertion of α3* nAChRs in the PM, while silencing of UBXD4 reduced it. Biotinylation experiments revealed that UBXD4 might preferentially facilitate the insertion of α3β2 nAChRs in the PM. Indeed the α-conotoxin MII, a selective antagonist of α3β2 nAChRs (Cartier et al., 1996; Vailati et al., 1999; Champtiaux et al., 2002; Cui et al., 2003) blocked a greater percentage of ACh-gated currents in dPC12 cells overexpressing UBXD4 than in control cells. The preferential “upregulation” of α3β2 receptors might be explained by conformational differences between α3β2 and α3β4 nAChRs or by the fact that dPC12 cells already contain a large intracellular pool of α3β4 receptors (Gerzanich et al., 1995; Wang et al., 1996, 1998; Arias and Bhumireddy, 2005; Srivatsan et al., 2006). The selective nature of UBXD4's influence on the PM insertion of α3* nAChRs was further confirmed by the fact that UBXD4 overexpression had no effect on the amplitude or density of α7 nAChRs currents or mGluR1α receptor levels.
Figure 9.
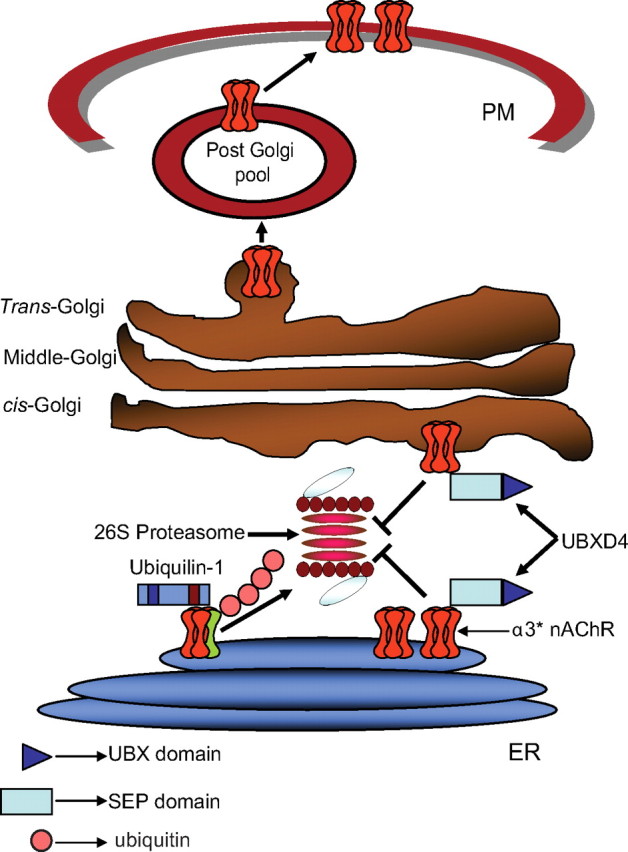
UBXD4 facilitates the trafficking of α3* nAChRs. Model showing the putative role of UBXD4 in the trafficking of α3* nAChRs after assembly in the endoplasmic reticulum (ER). Unfolded subunits and unmasked receptors are recognized by the ER-associated degradation system (ERAD) with the aid of chaperons like Plic-1. Association of other adaptor proteins such as UBXD4 can mask the degradation signal allowing nAChRs to bypass the checkpoints at the ER and cis-Golgi level before insertion in the PM.
UBXD4 comprises two domains of interest: UBX, at the C terminus, and SEP, at the N terminus. The original partial UBXD4 clone isolated in the Y2H experiments only contained the SEP domain suggesting that the binding site for α3 is located at the N terminus of UBXD4. This hypothesis was confirmed by experiments conducted with various truncated forms of UBXD4 indicating that UBXD4 needs the N terminus, but not the C terminus, to exert its effects on the levels of α3* nAChRs. Another protein in the UBXD family, TUG (tether, containing a UBX domain, for GLUT4), which is involved in the trafficking of the glucose transporter GLUT4 to the PM in response to insulin (Bogan et al., 2003), behaves similarly to UBXD4. The N terminus and a binding site located in the center of TUG, but not the UBX domain, bind to the large intracellular loop of GLUT4 to increase its PM levels (Tettamanzi et al., 2006; Yu et al., 2007).
The UBX domain is found in a variety of proteins with differing functions and seems to play a role in modulating the UPS. Examples of these proteins are p47 (Kondo et al., 1997), TUG (Bogan et al., 2003), FAF1 (Chu et al., 1995), and UBXD2 (Liang et al., 2006; Wilson et al., 2006). Despite differences in amino-acid sequence, the UBX domain is structurally very similar to ubiquitin, displaying characteristic ubiquitin-like folds (Buchberger et al., 2001; Yuan et al., 2001).
Our results revealed for the first time that the α3 nAChR subunit is ubiquitinated and degraded by the UPS. We also showed that UBXD4 can slow down the degradation of α3. In fact, interaction of α3 with either WT or ΔC-UBXD4 decreased the ubiquitination levels of the nAChR subunit in dPC12 cells while increasing its half-life. The ability of UBXD4 to protect α3 from degradation resembles that of other adaptor proteins. Neurofilament-light protein interferes with the ubiquitination and proteolysis of the NR1 subunit of NMDA receptors (Bedford et al., 2001; Ratnam and Teichberg, 2005; Saliba et al., 2008), while TUG blocks the proteolysis of the GLUT4 transporter (Yu et al., 2007).
How UBXD4 stabilizes α3* nAChRs remains to be established. UBXD4 might mask a putative degradation signal needed for the targeting of α3 to the proteasome. Degradation of proteins is normally mediated by selective regions within protein known as “degron” (Varshavsky, 1992). A degron structure can be a linear sequence (Gilon et al., 2000), a PEST domain (Belizario et al., 2008), or distributed motifs (Gardner and Hampton, 1999). PEST domains are rich in proline (P), glutamic acid (E), serine (S), and threonine (T) (Rogers et al., 1986). For example, masking by Homer-1A of the PEST domain located at the C terminus of the mGluR1α receptor (1121–1156 aa) can prevent mGluR1α internalization and proteolysis (Soloviev, 2000). The large intracellular loop of α3 also contains two weak PEST regions (304–327 and 391–404 aa) which might represent the degradation signal that is masked by UBXD4 (Gomes and Barnes, 1997). It is also possible that UBXD4 interferes with more upstream or downstream processes by modulating E3 ligase activity or by interfering with the binding of ubiquitinated α3 to other proteins within the degradation pathway. A third hypothesis could be that UBXD4 works as a scaffold to bring together a deubiquitinating enzyme and α3, which would consequently help α3 to escape proteasomal degradation (Wang et al., 2004; Millard and Wood, 2006). Further experiments will be necessary to determine how UBXD4 interferes with α3 ubiquitination and degradation.
In addition to requiring ER chaperone proteins for proper assembly/maturation, nAChRs seem to also need adaptor proteins for an efficient delivery from the ER to the PM (Keller and Taylor, 1999; Millar, 2003; Wanamaker et al., 2003; Wanamaker and Green, 2005, 2007). Because iodixanol gradient fractionation showed that UBXD4 cofractionates with the ER as well as the Golgi compartment, UBXD4 might also escort nAChRs through the Golgi apparatus in addition to protecting them from ERAD-associated degradation. Interestingly, an ERAD-like system has been shown to exist next to the cis-Golgi compartment (Haynes et al., 2002).
The fact that UBXD4 is itself regulated by the UPS adds an additional level of complexity to the regulation of α3* nAChR trafficking. Because UBXD4 is degraded by the proteasome, any intervention capable of changing proteasomal activity might impinge on α3* nAChR levels directly, by controlling nAChR degradation and indirectly, by affecting the amount of UBXD4 in the cytoplasm. This observation suggests the presence of an autoregulatory feedback loop between the UPS, UBXD4, and nAChRs that controls the receptors cellular abundance. Interestingly, we recently showed that nicotine acts as a partial proteasomal inhibitor that can prevent proteins from being degraded by the UPS (Rezvani et al., 2007). Nicotine-induced upregulation could increase the levels of α3* (and possibly α4*) nAChRs by both preventing nAChR degradation and increasing the cellular levels of UBXD4. Finally, the discovery of UBXD4 might be relevant to the pathophysiology of a human disorder, the megacystis-microcolon-intestinal hypoperistalsis (MMIH) syndrome. The clinical manifestations of the MMIH syndrome are similar to the phenotypes we described for mice null for the α3 nAChR subunit (Berdon et al., 1976; Xu et al., 1999). Because no loss-of-function mutations in the α3 gene could be found in the MMIH patients examined (Lev-Lehman et al., 2001), other protein deficits could underlie the syndrome. UBXD4, with its ability to regulate the levels of α3* nAChRs at the PM is a potential candidate.
In summary, we identified a protein, UBXD4, that selectively regulates the levels of α3* nAChRs at the PM by facilitating their maturation and/or regulating their degradation. UBXD4 might influence the expression of α3* nAChRs at the synapse in normal conditions as well as during smoking.
Footnotes
This work was supported by grants from the National Institute on Drug Abuse (DA017173) and Philip Morris USA and Philip Morris International to M.D.B. and National Institute of Mental Health Grant MH 53631 to J.M.M. We thank Sanam Sane and Miguel Alejandro Marin for excellent technical support, Dr. Jerry A. Stitzel for providing the mouse α3 and β4 cDNAs, and Dr. Eric J. Brown for providing the Plic-1 cDNA. We also thank Dr. Francois Clemente and Dr. Yen-Ping Kuo for providing the mouse α5 cDNA. We thank the Mental Retardation and Developmental Disabilities Research Center core at Baylor College of Medicine for their support and Dr. Kimberley R. Tolias and Feng Liu for providing the rat culture cells.
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias HR, Bhumireddy P. Anesthetics as chemical tools to study the structure and function of nicotinic acetylcholine receptors. Curr Protein Pept Sci. 2005;6:451–472. doi: 10.2174/138920305774329331. [DOI] [PubMed] [Google Scholar]
- Arikkath J, Israely I, Tao Y, Mei L, Liu X, Reichardt LF. Erbin controls dendritic morphogenesis by regulating localization of δ-catenin. J Neurosci. 2008;28:7047–7056. doi: 10.1523/JNEUROSCI.0451-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford FK, Kittler JT, Muller E, Thomas P, Uren JM, Merlo D, Wisden W, Triller A, Smart TG, Moss SJ. GABA(A) receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat Neurosci. 2001;4:908–916. doi: 10.1038/nn0901-908. [DOI] [PubMed] [Google Scholar]
- Belizario JE, Alves J, Garay-Malpartida M, Occhiucci JM. Coupling caspase cleavage and proteasomal degradation of proteins carrying PEST motif. Curr Protein Pept Sci. 2008;9:210–220. doi: 10.2174/138920308784534023. [DOI] [PubMed] [Google Scholar]
- Berdon WE, Baker DH, Blanc WA, Gay B, Santulli TV, Donovan C. Megacystis-microcolon-intestinal hypoperistalsis syndrome: a new cause of intestinal obstruction in the newborn. Report of radiologic findings in five newborn girls. AJR Am J Roentgenol. 1976;126:957–964. doi: 10.2214/ajr.126.5.957. [DOI] [PubMed] [Google Scholar]
- Bogan JS, Hendon N, McKee AE, Tsao TS, Lodish HF. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature. 2003;425:727–733. doi: 10.1038/nature01989. [DOI] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookman JL, Stott AJ, Cheeseman PJ, Adamson CS, Holmes D, Cole J, Burns NR. Analysis of TYA protein regions necessary for formation of the Ty1 virus-like particle structure. Virology. 1995;212:69–76. doi: 10.1006/viro.1995.1454. [DOI] [PubMed] [Google Scholar]
- Buchberger A. From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol. 2002;12:216–221. doi: 10.1016/s0962-8924(02)02269-9. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Howard MJ, Proctor M, Bycroft M. The UBX domain: a widespread ubiquitin-like module. J Mol Biol. 2001;307:17–24. doi: 10.1006/jmbi.2000.4462. [DOI] [PubMed] [Google Scholar]
- Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM. A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors. J Biol Chem. 1996;271:7522–7528. doi: 10.1074/jbc.271.13.7522. [DOI] [PubMed] [Google Scholar]
- Champtiaux N, Han ZY, Bessis A, Rossi FM, Zoli M, Marubio L, McIntosh JM, Changeux JP. Distribution and pharmacology of α6-containing nicotinic acetylcholine receptors analyzed with mutant mice. J Neurosci. 2002;22:1208–1217. doi: 10.1523/JNEUROSCI.22-04-01208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- Chu K, Niu X, Williams LT. A Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc Natl Acad Sci U S A. 1995;92:11894–11898. doi: 10.1073/pnas.92.25.11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy WG, Liu Z, Nai Q, Coggan JS, Berg DK. PDZ-containing proteins provide a functional postsynaptic scaffold for nicotinic receptors in neurons. Neuron. 2003;38:759–771. doi: 10.1016/s0896-6273(03)00324-6. [DOI] [PubMed] [Google Scholar]
- Crawford LJ, Walker B, Ovaa H, Chauhan D, Anderson KC, Morris TC, Irvine AE. Comparative selectivity and specificity of the proteasome inhibitors BzLLLCOCHO, PS-341, and MG-132. Cancer Res. 2006;66:6379–6386. doi: 10.1158/0008-5472.CAN-06-0605. [DOI] [PubMed] [Google Scholar]
- Cui C, Booker TK, Allen RS, Grady SR, Whiteaker P, Marks MJ, Salminen O, Tritto T, Butt CM, Allen WR, Stitzel JA, McIntosh JM, Boulter J, Collins AC, Heinemann SF. The β3 nicotinic receptor subunit: a component of α-conotoxin MII-binding nicotinic acetylcholine receptors that modulate dopamine release and related behaviors. J Neurosci. 2003;23:11045–11053. doi: 10.1523/JNEUROSCI.23-35-11045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi M. Nicotinic mechanisms in the autonomic control of organ systems. J Neurobiol. 2002;53:568–579. doi: 10.1002/neu.10145. [DOI] [PubMed] [Google Scholar]
- Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficklin MB, Zhao S, Feng G. Ubiquilin-1 regulates nicotine-induced up-regulation of neuronal nicotinic acetylcholine receptors. J Biol Chem. 2005;280:34088–34095. doi: 10.1074/jbc.M506781200. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Mori Y, Chu DL, Koyama Y, Miyata S, Tanaka H, Yachi K, Kubo T, Yoshikawa H, Tohyama M. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol Cell Biol. 2006;26:2273–2285. doi: 10.1128/MCB.26.6.2273-2285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Ogata Y, Saegusa C, Kanno E, Mikoshiba K. Alternative splicing isoforms of synaptotagmin VII in the mouse, rat and human. Biochem J. 2002;365:173–180. doi: 10.1042/BJ20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Hampton RY. A ‘distributed degron’ allows regulated entry into the ER degradation pathway. EMBO J. 1999;18:5994–6004. doi: 10.1093/emboj/18.21.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerzanich V, Peng X, Wang F, Wells G, Anand R, Fletcher S, Lindstrom J. Comparative pharmacology of epibatidine: a potent agonist for neuronal nicotinic acetylcholine receptors. Mol Pharmacol. 1995;48:774–782. [PubMed] [Google Scholar]
- Gilon T, Chomsky O, Kulka RG. Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol. 2000;20:7214–7219. doi: 10.1128/mcb.20.19.7214-7219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AV, Barnes JA. Protein phosphatases are pest containing proteins. Biochem Mol Biol Int. 1997;41:65–73. doi: 10.1080/15216549700201071. [DOI] [PubMed] [Google Scholar]
- Gupta DS, McCullumsmith RE, Beneyto M, Haroutunian V, Davis KL, Meador-Woodruff JH. Metabotropic glutamate receptor protein expression in the prefrontal cortex and striatum in schizophrenia. Synapse. 2005;57:123–131. doi: 10.1002/syn.20164. [DOI] [PubMed] [Google Scholar]
- Hamada H, Suzuki M, Yuasa S, Mimura N, Shinozuka N, Takada Y, Suzuki M, Nishino T, Nakaya H, Koseki H, Aoe T. Dilated cardiomyopathy caused by aberrant endoplasmic reticulum quality control in mutant KDEL receptor transgenic mice. Mol Cell Biol. 2004;24:8007–8017. doi: 10.1128/MCB.24.18.8007-8017.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes DM, Braud S, Hurtado DE, McCallum J, Standley S, Isaac JT, Roche KW. Trafficking and surface expression of the glutamate receptor subunit, KA2. Biochem Biophys Res Commun. 2003;310:8–13. doi: 10.1016/j.bbrc.2003.08.115. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Caldwell S, Cooper AA. An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J Cell Biol. 2002;158:91–101. doi: 10.1083/jcb.200201053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob MH, Lindstrom JM, Berg DK. Surface and intracellular distribution of a putative neuronal nicotinic acetylcholine receptor. J Cell Biol. 1986;103:205–214. doi: 10.1083/jcb.103.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanclos EM, Lin L, Treuil MW, Rao J, DeCoster MA, Anand R. The chaperone protein 14–3-3eta interacts with the nicotinic acetylcholine receptor alpha 4 subunit. Evidence for a dynamic role in subunit stabilization. J Biol Chem. 2001;276:28281–28290. doi: 10.1074/jbc.M011549200. [DOI] [PubMed] [Google Scholar]
- Jensen AA, Frølund B, Liljefors T, Krogsgaard-Larsen P. Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. J Med Chem. 2005;48:4705–4745. doi: 10.1021/jm040219e. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Karan G, Yang Z, Howes K, Zhao Y, Chen Y, Cameron DJ, Lin Y, Pearson E, Zhang K. Loss of ER retention and sequestration of the wild-type ELOVL4 by Stargardt disease dominant negative mutants. Mol Vis. 2005;11:657–664. [PubMed] [Google Scholar]
- Kassner PD, Berg DK. Differences in the fate of neuronal acetylcholine receptor protein expressed in neurons and stably transfected cells. J Neurobiol. 1997;33:968–982. doi: 10.1002/(sici)1097-4695(199712)33:7<968::aid-neu8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Keller SH, Taylor P. Determinants responsible for assembly of the nicotinic acetylcholine receptor. J Gen Physiol. 1999;113:171–176. doi: 10.1085/jgp.113.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SH, Lindstrom J, Ellisman M, Taylor P. Adjacent basic amino acid residues recognized by the COP I complex and ubiquitination govern endoplasmic reticulum to cell surface trafficking of the nicotinic acetylcholine receptor alpha-subunit. J Biol Chem. 2001;276:18384–18391. doi: 10.1074/jbc.M100691200. [DOI] [PubMed] [Google Scholar]
- Kim NS, Yamaguchi T, Sekine S, Saeki M, Iwamuro S, Kato S. Cloning of human polyubiquitin cDNAs and a ubiquitin-binding assay involving its in vitro translation product. J Biochem (Tokyo) 1998;124:35–39. doi: 10.1093/oxfordjournals.jbchem.a022093. [DOI] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- Koulen P, Cai Y, Geng L, Maeda Y, Nishimura S, Witzgall R, Ehrlich BE, Somlo S. Polycystin-2 is an intracellular calcium release channel. Nat Cell Biol. 2002;4:191–197. doi: 10.1038/ncb754. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Gee VJ, Harkness PC, Doward AI, Baker ER, Gibb AJ, Millar NS. RIC-3 enhances functional expression of multiple nicotinic acetylcholine receptor subtypes in mammalian cells. Mol Pharmacol. 2005;68:1431–1438. doi: 10.1124/mol.105.017459. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Niesman I, Fischer T, DeVries L, Farquhar MG. Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J Biol Chem. 2005;280:22012–22020. doi: 10.1074/jbc.M501833200. [DOI] [PubMed] [Google Scholar]
- Le Novère N, Corringer PJ, Changeux JP. The diversity of subunit composition in nAChRs: evolutionary origins, physiologic and pharmacologic consequences. J Neurobiol. 2002;53:447–456. doi: 10.1002/neu.10153. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Lehman E, Bercovich D, Xu W, Stockton DW, Beaudet AL. Characterization of the human beta4 nAChR gene and polymorphisms in CHRNA3 and CHRNB4. J Hum Genet. 2001;46:362–366. doi: 10.1007/PL00010921. [DOI] [PubMed] [Google Scholar]
- Liang J, Yin C, Doong H, Fang S, Peterhoff C, Nixon RA, Monteiro MJ. Characterization of erasin (UBXD2): a new ER protein that promotes ER-associated protein degradation. J Cell Sci. 2006;119:4011–4024. doi: 10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- Lin L, Jeanclos EM, Treuil M, Braunewell KH, Gundelfinger ED, Anand R. The calcium sensor protein visinin-like protein-1 modulates the surface expression and agonist sensitivity of the alpha 4beta 2 nicotinic acetylcholine receptor. J Biol Chem. 2002;277:41872–41878. doi: 10.1074/jbc.M206857200. [DOI] [PubMed] [Google Scholar]
- Marchand S, Devillers-Thiéry A, Pons S, Changeux JP, Cartaud J. Rapsyn escorts the nicotinic acetylcholine receptor along the exocytic pathway via association with lipid rafts. J Neurosci. 2002;22:8891–8901. doi: 10.1523/JNEUROSCI.22-20-08891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar NS. Assembly and subunit diversity of nicotinic acetylcholine receptors. Biochem Soc Trans. 2003;31:869–874. doi: 10.1042/bst0310869. [DOI] [PubMed] [Google Scholar]
- Millard SM, Wood SA. Riding the DUBway: regulation of protein trafficking by deubiquitylating enzymes. J Cell Biol. 2006;173:463–468. doi: 10.1083/jcb.200602082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkaynak E, Finley D, Varshavsky A. The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein. Nature. 1984;312:663–666. doi: 10.1038/312663a0. [DOI] [PubMed] [Google Scholar]
- Pons S, Sallette J, Bourgeois JP, Taly A, Changeux JP, Devillers-Thiéry A. Critical role of the C-terminal segment in the maturation and export to the cell surface of the homopentameric alpha 7–5HT3A receptor. Eur J Neurosci. 2004;20:2022–2030. doi: 10.1111/j.1460-9568.2004.03673.x. [DOI] [PubMed] [Google Scholar]
- Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Ratnam J, Teichberg VI. Neurofilament-light increases the cell surface expression of the N-methyl-D-aspartate receptor and prevents its ubiquitination. J Neurochem. 2005;92:878–885. doi: 10.1111/j.1471-4159.2004.02936.x. [DOI] [PubMed] [Google Scholar]
- Ren XQ, Cheng SB, Treuil MW, Mukherjee J, Rao J, Braunewell KH, Lindstrom JM, Anand R. Structural determinants of α4β2 nicotinic acetylcholine receptor trafficking. J Neurosci. 2005;25:6676–6686. doi: 10.1523/JNEUROSCI.1079-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani K, Teng Y, Shim D, De Biasi M. Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J Neurosci. 2007;27:10508–10519. doi: 10.1523/JNEUROSCI.3353-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rogers SW, Mandelzys A, Deneris ES, Cooper E, Heinemann S. The expression of nicotinic acetylcholine receptors by PC12 cells treated with NGF. J Neurosci. 1992;12:4611–4623. doi: 10.1523/JNEUROSCI.12-12-04611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas R, Pieri F, De Biasi M. Decreased signs of nicotine withdrawal in mice null for the β4 nicotinic acetylcholine receptor subunit. J Neurosci. 2004a;24:10035–10039. doi: 10.1523/JNEUROSCI.1939-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas R, Cook KD, Bassetto L, De Biasi M. The alpha3 and beta4 nicotinic acetylcholine receptor subunits are necessary for nicotine-induced seizures and hypolocomotion in mice. Neuropharmacology. 2004b;47:401–407. doi: 10.1016/j.neuropharm.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Saliba RS, Pangalos M, Moss SJ. The ubiquitin-like protein Plic-1 enhances the membrane insertion of GABAA receptors by increasing their stability within the endoplasmic reticulum. J Biol Chem. 2008;283:18538–18544. doi: 10.1074/jbc.M802077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y, Katoh Y, Shiba T, Yoshino K, Takatsu H, Kobayashi H, Shin HW, Wakatsuki S, Nakayama K. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J Biol Chem. 2004;279:7105–7111. doi: 10.1074/jbc.M311702200. [DOI] [PubMed] [Google Scholar]
- Silverman E, Frödin M, Gammeltoft S, Maller JL. Activation of p90 Rsk1 is sufficient for differentiation of PC12 cells. Mol Cell Biol. 2004;24:10573–10583. doi: 10.1128/MCB.24.24.10573-10583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloviev MM. Detection of a surface-exposed PEST like sequence in the metabotropic glutamate receptor mGluR1 alpha. Bioinformatics. 2000;16:837–838. doi: 10.1093/bioinformatics/16.9.837. [DOI] [PubMed] [Google Scholar]
- Soukenik M, Diehl A, Leidert M, Sievert V, Büssow K, Leitner D, Labudde D, Ball LJ, Lechner A, Nägler DK, Oschkinat H. The SEP domain of p47 acts as a reversible competitive inhibitor of cathepsin L. FEBS Lett. 2004;576:358–362. doi: 10.1016/j.febslet.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Srivatsan M, Treece J, Shotts EE. Nicotine alters nicotinic receptor subunit levels differently in developing mammalian sympathetic neurons. Ann N Y Acad Sci. 2006;1074:505–513. doi: 10.1196/annals.1369.051. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yeh ET, Kamitani T. NUB1-mediated targeting of the ubiquitin precursor UbC1 for its C-terminal hydrolysis. Eur J Biochem. 2004;271:972–982. doi: 10.1111/j.1432-1033.2004.03999.x. [DOI] [PubMed] [Google Scholar]
- Tettamanzi MC, Yu C, Bogan JS, Hodsdon ME. Solution structure and backbone dynamics of an N-terminal ubiquitin-like domain in the GLUT4-regulating protein, TUG. Protein Sci. 2006;15:498–508. doi: 10.1110/ps.051901806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins MM, Gai WP, Douglas S, Bunn SJ. Alpha-synuclein expression localizes to the Golgi apparatus in bovine adrenal medullary chromaffin cells. Brain Res. 2003;984:233–236. doi: 10.1016/s0006-8993(03)03040-3. [DOI] [PubMed] [Google Scholar]
- Touster O, Aronson NN, Jr, Dulaney JT, Hendrickson H. Isolation of rat liver plasma membranes. Use of nucleotide pyrophosphatase and phosphodiesterase I as marker enzymes. J Cell Biol. 1970;47:604–618. doi: 10.1083/jcb.47.3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- Vailati S, Hanke W, Bejan A, Barabino B, Longhi R, Balestra B, Moretti M, Clementi F, Gotti C. Functional alpha6-containing nicotinic receptors are present in chick retina. Mol Pharmacol. 1999;56:11–19. doi: 10.1124/mol.56.1.11. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- Wada E, Wada K, Boulter J, Deneris E, Heinemann S, Patrick J, Swanson LW. Distribution of alpha 2, alpha 3, alpha 4, and beta 2 neuronal nicotinic receptor subunit mRNAs in the central nervous system: a hybridization histochemical study in the rat. J Comp Neurol. 1989;284:314–335. doi: 10.1002/cne.902840212. [DOI] [PubMed] [Google Scholar]
- Wanamaker CP, Green WN. N-linked glycosylation is required for nicotinic receptor assembly but not for subunit associations with calnexin. J Biol Chem. 2005;280:33800–33810. doi: 10.1074/jbc.M501813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanamaker CP, Green WN. ER chaperones stabilize nicotinic receptor subunits and regulate receptor assembly. J Biol Chem. 2007;282:31113–31123. doi: 10.1074/jbc.M705369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanamaker CP, Christianson JC, Green WN. Regulation of nicotinic acetylcholine receptor assembly. Ann N Y Acad Sci. 2003;998:66–80. doi: 10.1196/annals.1254.009. [DOI] [PubMed] [Google Scholar]
- Wang F, Gerzanich V, Wells GB, Anand R, Peng X, Keyser K, Lindstrom J. Assembly of human neuronal nicotinic receptor alpha5 subunits with alpha3, beta2, and beta4 subunits. J Biol Chem. 1996;271:17656–17665. doi: 10.1074/jbc.271.30.17656. [DOI] [PubMed] [Google Scholar]
- Wang F, Nelson ME, Kuryatov A, Olale F, Cooper J, Keyser K, Lindstrom J. Chronic nicotine treatment up-regulates human alpha3 beta2 but not alpha3 beta4 acetylcholine receptors stably transfected in human embryonic kidney cells. J Biol Chem. 1998;273:28721–28732. doi: 10.1074/jbc.273.44.28721. [DOI] [PubMed] [Google Scholar]
- Wang JM, Zhang L, Yao Y, Viroonchatapan N, Rothe E, Wang ZZ. A transmembrane motif governs the surface trafficking of nicotinic acetylcholine receptors. Nat Neurosci. 2002;5:963–970. doi: 10.1038/nn918. [DOI] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97–p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Wibo M, Thinès-Sempoux D, Amar-Costesec A, Beaufay H, Godelaine D. Analytical study of microsomes and isolated subcellular membranes from rat liver VIII. Subfractionation of preparations enriched with plasma membranes, outer mitochondrial membranes, or Golgi complex membranes. J Cell Biol. 1981;89:456–474. doi: 10.1083/jcb.89.3.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Burton B, Urrutia A, Shcherbatko A, Chavez-Noriega LE, Cohen CJ, Aiyar J. Ric-3 promotes functional expression of the nicotinic acetylcholine receptor alpha7 subunit in mammalian cells. J Biol Chem. 2005;280:1257–1263. doi: 10.1074/jbc.M410039200. [DOI] [PubMed] [Google Scholar]
- Wilson JD, Liu Y, Bentivoglio CM, Barlowe C. Sel1p/Ubx2p participates in a distinct Cdc48p-dependent endoplasmic reticulum-associated degradation pathway. Traffic. 2006;7:1213–1223. doi: 10.1111/j.1600-0854.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- Wiser MF, Schweiger HG. Increased sensitivity in antigen detection during immunoblot analysis resulting from antigen enrichment via immunoprecipitation. Anal Biochem. 1986;155:71–77. doi: 10.1016/0003-2697(86)90226-5. [DOI] [PubMed] [Google Scholar]
- Xu W, Gelber S, Orr-Urtreger A, Armstrong D, Lewis RA, Ou CN, Patrick J, Role L, De Biasi M, Beaudet AL. Megacystis, mydriasis, and ion channel defect in mice lacking the alpha3 neuronal nicotinic acetylcholine receptor. Proc Natl Acad Sci U S A. 1999;96:5746–5751. doi: 10.1073/pnas.96.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JJ, Yasuda RP, Dávila-García MI, Xiao Y, Ebert S, Gupta T, Kellar KJ, Wolfe BB. Neuronal nicotinic acetylcholine receptor alpha3 subunit protein in rat brain and sympathetic ganglion measured using a subunit-specific antibody: regional and ontogenic expression. J Neurochem. 2001;77:336–346. doi: 10.1046/j.1471-4159.2001.t01-1-00259.x. [DOI] [PubMed] [Google Scholar]
- Yu C, Cresswell J, Löffler MG, Bogan JS. The glucose transporter 4-regulating protein TUG is essential for highly insulin-responsive glucose uptake in 3T3–L1 adipocytes. J Biol Chem. 2007;282:7710–7722. doi: 10.1074/jbc.M610824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Shaw A, Zhang X, Kondo H, Lally J, Freemont PS, Matthews S. Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly. J Mol Biol. 2001;311:255–263. doi: 10.1006/jmbi.2001.4864. [DOI] [PubMed] [Google Scholar]