Abstract
Background: Because fragile X syndrome (FXS) is prevalent, it has become the subject of newborn and high-risk screening efforts. International screening, however, can be financially and logistically prohibitive, particularly in countries where resources may be scarce. Recently, we have developed a screening test on blood spot that can detect expanded alleles from the normal through the full mutation range in both males and females. It is accurate, rapid, inexpensive, and applicable on blood spots and therefore ideal for international screening. The use of this blood spot screening technique was piloted in “a high-risk screening” study of individuals in Guatemala. Methods: One hundred and five blood spots from subjects from Guatemala were screened for the Fragile X Mental Retardation 1 mutation. They were classified as “high-risk” through placement into one of the following five categories: (a) relatives of someone with a previous FXS diagnosis, (b) individuals with confirmed autism, (c) individuals with confirmed intellectual disability, (d) individuals with Parkinson's-like presentation, and (e) individuals with a family history of intellectual disability but no confirmed cases of FXS. Results: Fifteen of the individuals tested yielded an expanded allele, 10 premutations and 5 full mutations. All 15 expansions were found in individuals with a relative with a confirmed FXS diagnosis. No expansions were found in the other clinical groups. Conclusions: Blood spot polymerase chain reaction screening is an effective, cost-efficient method to conduct cascade testing in families with a known history of FXS, even in small screening cohorts.
Introduction
Fragile X Syndrome (FXS) is the leading inherited cause of intellectual disability (ID), and the leading known single-gene cause of autism (Hagerman et al., 2008). FXS is caused by a CGG repeat expansion of >200 repeats at the 5′ end of the Fragile X Mental Retardation 1 (FMR1) gene on the X chromosome which leads to methylation and silencing of the gene and a lack or deficiency of the FMR1 protein (FMRP). FMRP is an RNA-binding protein that regulates translation of many messenger RNAs. The absence of FMRP leads to upregulation of the translation of many proteins and suppression of others. The consequence of these changes leads to the fragile X phenotype characterized by cognitive, behavioral, and social deficits (Hagerman and Hagerman, 2002; Reiss and Dant, 2003; Loesch et al., 2004).
FXS has an estimated frequency of 1 in 4000 males and 1 in 2500 to 1 in 8000 females (Turner et al., 1996; De Vries et al., 1997; Morton et al., 1997; Pesso et al., 2000; Crawford et al., 2002). However, all screening studies were biased as they were conducted on populations with significant developmental problems. In a recent report (Hagerman, 2008), the estimated frequency of full mutation alleles in both males and females was determined to be 1/2355 individuals in the general population. The prevalence of the premutation allele (55–200 CGG repeats) is ∼1 in 130–250 females and 1 in 250–800 males (Toledano-Alhadef et al., 2001; Dombrowski et al., 2002; Hagerman, 2008; Fernandez-Carvajal et al., 2009). With such high rates of gene expansion (both the premutation and the full mutation), high-risk and newborn screening for FXS has moved into the forefront of current research efforts, both domestically and abroad (Bailey, 2007; Bailey et al., 2008). However, large-scale screening efforts can be logistically difficult because of the high cost and the technical difficulty of collecting, storing, and processing sufficient genetic sample to obtain accurate results. These obstacles are even more apparent and problematic in international screening efforts, particularly in poor countries where equipment, expertise, and financial support may be unavailable. Tassone et al. (2008) developed a blood spot DNA test that uses a chimeric CGG-targeted primer to quickly and reliably detect expanded alleles of the FMR1 gene from the normal to the full mutation range, in both males and females. This methodology, which has been recently applied to a pilot screening study on over 5000 anonymous newborn blood spots (no follow-up on family members) (Fernandez-Carvajal et al., 2009), is rapid and also inexpensive and allows for easier transportation and storage compared with whole blood samples. Therefore, the use of blood spots to identify expanded alleles is particularly suited for international screening programs. However, the blood spot screening will only identify expanded alleles without giving detailed information of the exact expansion size, particularly for the full mutation. This screening technique also does not provide information on the methylation status of the sample, which would have to be done with a follow-up test (i.e., Southern blot analysis). Thus, the CGG linker screening, by flagging those samples with an expanded allele, allows the screening of many samples rapidly and inexpensively (∼$5 supply cost for each sample), but follow-up of the positive samples is needed for confirmation by utilizing a Southern blot which is more expensive ($250–300 per sample).
Here, we pilot the use of this methodology in a high-risk population in Guatemala, through which we were able to demonstrate the feasibility of using it in international screening, particularly in third world countries where costs for supplies are limited.
Materials and Methods
Subjects
Blood spot samples were obtained from 105 Guatemalan individuals (63 males and 42 females). All individuals were identified through collaboration with La Asociacion Guatemalteca del Sindrome X Fragil (a parent support group based in the Guatemala City) and Integrame (a behavior therapy clinic located in the Guatemala City). Individuals were then classified as “high-risk” through placement into one of five different categories, described below:
Thirty-six relatives of someone with a preexisting diagnosis of FXS were screened (16 males and 20 females; mean age, 28.56 years). Included in this group were four individuals (three brothers and another unrelated boy) who received a clinical diagnosis of FXS from a pediatrician, but never received confirmation through genetic testing. All four of these individuals showed phenotypic features of fragile X, including facial dysmorphology (long face, prominent ears), and displayed several prototypical behavioral characteristics (gaze aversion, impulsivity, hand flapping). Additionally, 1 of the 36 people screened from this group had postural and intention tremor indicative of fragile X-associated tremor ataxia syndrome. These 36 individuals came from 8 separate families, 4 of which included multiple relatives outside of the proband's immediate family.
Thirty-four individuals with a previous clinical diagnosis of autism (29 males and 5 females; mean age, 7.62) were screened. These 34 individuals came from 30 separate families. Of these individuals, only 1 had dysmorphic and behavioral features typical of FXS.
Twenty-six individuals with confirmed developmental delay or ID (16 males and 10 females, mean age 9.89 years) were screened. These 26 individuals came from 26 separate families, 3 had dysmorphic features characteristic of FXS and 3 of them had Down syndrome, making FXS an unlikely codiagnosis.
Two individuals with a Parkinson's-like presentation including a resting tremor and difficulty walking (two individuals; one male and one female; mean age, 61.1 years) were screened.
Seven individuals with a family history of ID, but no confirmed instances of FXS (one male and six females; mean age, 32.40) were screened. Six of these individuals are related to individuals with autism who also underwent testing.
Following informed consent and in accordance with an institutional review board (IRB)-approved protocol, each subject gave a single blood spot on an FTA genetic sample card using a standard lancing device.
Molecular studies
About 2 × 1.2 mm bloodspot on FTA paper (Whatman) for each sample were punched and placed in a clean 0.5 μL polymerase chain reaction (PCR) tube and washed with 200 μL of Qiagen RBC Lysis Solution (Qiagen, Valencia, CA). The dry blood spots were used directly with the PCR mix containing primer c and f (Fu et al., 1991). The Roche FastStart PCR kit was used for the amplification, according to the manufacturer's instructions. The PCR conditions used were as follows: 10-min initial denaturation at 95°C; followed by 10 cycles of 95°C for 35 s, 64°C for 35 s, 68°C for 4 min; 25 cycles of 95°C for 35 s, 64°C for 35 s, 68°C for 4 min (with 20 s increase in each cycle); and a final extension of 10 min at 68°C. The PCR products were analyzed using the Qiaxcel Genetic Analyzer (Qiagen) as described by Fernandez-Carvajal et al. FMR1 alleles were classified as normal (6–44 CGG repeats), intermediate (45–54 CGG repeats), premutation (55–200 CGG repeats), and full mutation (>200 CGG repeats) (Maddalena et al., 2001). DNA samples from males that did not yield a band after the first-round PCR, or DNA samples from females that yielded only one normal band with primers c and f, were subjected to a secondary, CGG-primer–based PCR screening, as previously described (Tassone et al., 2008; Fernandez-Carvajal et al., 2009). The presence of a smear on a 2% agarose gel indicated the presence of an expanded allele, whereas the absence of a smear indicated the presence of a normal FMR1 allele. Individuals who were identified through the blood spot screening as having an FMR1 expanded allele were asked to send in a whole blood sample to confirm the blood spot results and obtain more precise genetic molecular information (CGG repeat size, methylation status).
Results
Of the 105 samples collected, 104 were successfully processed. One sample did not contain sufficient genetic material to confidently ascertain results. Of the 104 samples processed, an expanded allele was observed in 15 samples (6 males and 9 females). Of the 15 positive results, 5 were fully expanded (4 males and 1 female). The other 10 samples yielded an expansion within the premutation range (2 males and 8 females); 3 of these being in the high-end range with greater than 100 CGG repeats (1 of 2 males and 2 of 8 females). However, the PCR technique used to process the blood spots can neither distinguish between a true premutation carrier and an individual with prefull mosaicism, nor distinguish between a high-end premutation and full mutation alleles, which is why each positive blood spot has to have followed-up DNA testing (Southern blot) on whole blood.
All 15 expansions were found in the group of 36 individuals with a fragile X relative. Additionally, of the eight families that constitute this group, only four came back with expansions, leaving four families in which all tested individuals were negative (Table 1). There were no cases of FXS or premutation in any of the other clinical screening groups. Peripheral blood leukocytes were obtained from the subjects who showed an expanded allele during the blood spot screening.
Table 1.
Results from Blood Spot Screening in Individuals with Known Family History of Fragile X Syndrome
| Family | n screened (M, F) | n premutation (M, F) | n expanded (M, F) | n normal (M, F) |
|---|---|---|---|---|
| A | 8 (4, 4) | 6 (2, 4) | 0 | 2 (2, 0) |
| B | 7 (4, 3) | 1 (0, 1) | 3 (3, 0) | 3 (1, 2) |
| C | 8 (2, 6) | 3 (0, 3) | 0 | 5 (2, 3) |
| D | 2 (1, 1) | 0 | 2 (1, 1) | 0 |
| E | 3 (3, 0) | 0 | 0 | 3 (3, 0) |
| F | 2 (1, 1) | 0 | 0 | 2 (1, 1) |
| G | 1 (1, 0) | 0 | 0 | 1 (1, 0) |
| H | 5 (0, 5) | 0 | 0 | 5 (0, 5) |
| Total | 36 (16, 20) | 10 (2, 8) | 5 (4, 1) | 21(10, 11) |
Family A
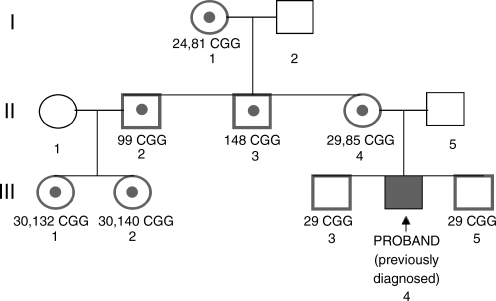
Six individuals with an expanded FMR1 allele were identified (see pedigree in Fig. 1). The mother of the proband was already aware of her status as an obligate carrier (which was confirmed through the screening; 31 and 87 repeats), but the other five individuals had no signs of neurological involvement and therefore no clinical indication of the premutation. The mother and proband were found to be carriers of an expanded allele, whereas the two proband's brothers were found to have a normal allele, using the CGG-primer–based PCR screening as shown in Figure 2. The proband's cousins and maternal uncles were found to be carriers of an expanded allele (132, 140, 99, and 148 CGG repeats, respectively) in addition to the grandmother of the proband (24 and 81 repeats). Southern blot analysis confirmed the results obtained using the blood spot screening (Fig. 3).
FIG. 1.
Pedigree analysis of family A.
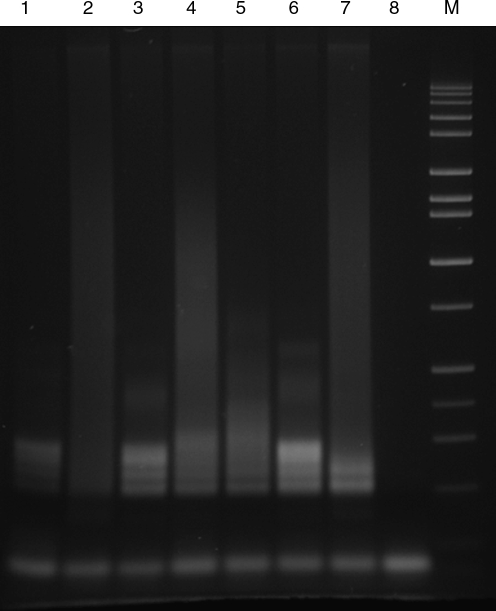
FIG. 2.
Polymerase chain reaction analysis on agarose gel showing expanded alleles obtained by the CGG linker screening of blood spots. Samples were loaded from lanes 1 to 7 as follows: III-3, III-4, III-5, II-4, C1 (gray zone control 30, 54 CGG), C2 (normal control), C3 (full mutation control). Lane 8 was a negative control (no DNA). A DNA size marker is shown in lane 9.
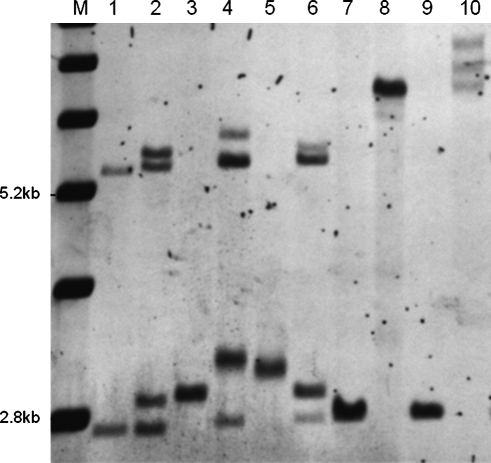
FIG. 3.
Southern blot analysis of genomic DNA isolated from members of family A (in Fig. 1). A DNA 1-kb ladder is shown in lane 1. The normal unmethylated band (2.8 kb) and the normal methylated band (5.2 kb) are indicated. A normal female control is shown in lane 2. Full mutation control is shown in lane 10. The samples were loaded from lanes 2 to 9 as follows: I-1, II-2, III-2, II-3, II-4, III-3, III-4, III-5. DNA sample from III-1 was not available for Southern blot analysis.
Family B
Full mutation status was confirmed in three individuals who had previously received a clinical diagnosis of FXS from their pediatrician. Further, the blood spot screening confirmed the premutation status in their mother and ruled out carrier status in their maternal aunt and two cousins. Although the three boys had previously received a diagnosis of FXS from their pediatrician, they lacked financial resources to obtain genetic testing to confirm this clinical assessment, or to carry out cascade testing of the extended family. Southern blot analysis of samples from the members of this family confirmed the FMR1 mutation status determined by the blood spot screening.
Family C
Three individuals were identified as having the premutation, including the proband's mother. The remaining five people had normal alleles. In the case of this family, the PCR screening of blood spots was able to rule out fragile X, thereby alleviating the family's concern for future generations. These results were confirmed by Southern blot analysis.
Family D
Only two members of this family were tested, a mother and a son. Both individuals had expanded alleles upon analysis of the blood spot sample, including the boy who had previously received a clinical diagnosis of FXS from his pediatrician, and the results were confirmed by Southern blot analysis.
Blood spot PCR screening from six individuals (families E, F, and G in Table 1) did not yield any expanded alleles and therefore were not investigated any further.
Southern blot analysis did not confirm the results obtained with the blood spot screening in the sister of a known male proband (who was not screened). Although the presence of a long light smear was observed on the agarose gel, it was not as dark as the usual positives and so we requested the DNA studies for follow-up. Surprisingly, the proband's mother (who was assumed to be the obligate carrier), grandmother, and maternal aunt tested negative for the blood spot screening. Southern blot analysis was then run on samples from all members of the families, including the male proband who was thought to have FXS. The results showed that none of the members of this family was a carrier of FMR1 mutation as we obtained a normal pattern for everyone including the male proband (family H in Table 1). Thus, the sister who showed a hint of a smear on the original blood spot was not confirmed to have an expanded allele. This points out the need to always carryout a Southern blot/PCR DNA test as a follow-up of this blood spot screening test, especially if there is any doubt about a positive result.
Discussion
This is the first report of international screening and cascade testing utilizing the new fragile X PCR-based screening of blood spots, a technique developed by Tassone et al. (2008). Of the 104 samples successfully processed, 15 were positive and confirmed by Southern blot 14.42%. All of the confirmed positives were members of families with a known proband, including the three brothers with ID who had never been tested for fragile X DNA, although their pediatrician thought that they clinically had FXS. They were detected with our screening and confirmed by Southern blot analysis.
Although no cases of FXS were found in any of the nonfragile X-related screening groups, these findings are within the range of expected results, given the relatively small sample size in each group. Based on the literature, it is expected that roughly 2–6% of individuals with autism will have FXS as the genetic etiology. Given a sample size of 34 children, it is expected that 0–2 individuals should test positive for fragile X. Similarly, it is expected that between 1% and 3% of individuals with developmental delay or intellectual deficiency has FXS, which, given a sample size of 24 people, is likely to yield no positive results. For the other two groups (those with Parkinson's-like presentation and those with a family history of ID) the expected percentages are much smaller, as was the tested sample size, so again a lack of any positive results is within the expected range. Therefore, though high-risk blood spot screening would be beneficial to these nonfragile X-related clinical groups, its utility would be better demonstrated with larger sample sizes involving hundreds of individuals.
For the group of individuals with a known family history of FXS, blood spot screening effectively identified fragile X full and premutation alleles. In doing so, the screening provided these individuals with a presumptive diagnosis that was made definitive with Southern blot/PCR follow-up testing. No DNA testing for the fragile X gene is currently available in Guatemala. Follow-up efforts have provided these families with genetic counseling via phone from the M.I.N.D. Institute and have placed them in contact with local fragile X parent support groups.
High-risk blood spot screening in Guatemala yielded success on two fronts. On the one hand, 36 individuals with a known family history of FXS (and thus a known risk of inheriting an expanded allele) were given a diagnosis. Fifteen of these individuals were correctly diagnosed with an expanded allele; a diagnosis they would likely not have received if they had to rely on more expensive, logistically difficult testing. Therefore, blood spot screening is an effective tool to use for cascade screening, particularly in populations where the screening cohort may be small and genetic testing resources may be scarce.
Further, although the screening cohorts of the other clinical groups (autism, DD/ID, Parkinson's-like, family history of ID with unconfirmed etiology) were small in number and did not result in any expanded alleles, being screened allowed each of these individuals to rule out FXS as the cause of their disabilities. None of these individuals would have been screened had it not been for the low cost and feasibility of the blood spot PCR screening technique. Therefore, although larger screening cohorts are necessary to identify cases of FXS in these groups, high-risk blood spot screening is a cost-efficient and an easy way to rule out a FXS mutation.
Acknowledgments
The authors are grateful to the research participants and their families; Louise Gane for her genetic counseling and assistance in contacting the families; Selina and Jose Molina and Integrame for the use of their clinics to carry out the screening; and the National Fragile X Foundation for the provision of informational documents and for organizing the contact with the families. This study was supported by the National Institutes of Child Health and Development Grant R01 HD02274 (to F.T. and R.H.).
Disclosure Statement
No competing financial interests exist.
References
- Bailey DB., Jr Family adaptation to intellectual and developmental disabilities. Ment Retard Dev Disabil Res Rev. 2007;13:291–292. doi: 10.1002/mrdd.20168. [DOI] [PubMed] [Google Scholar]
- Bailey DB., Jr Skinner D. Davis AM, et al. Ethical and social concerns about expanded newborn screening: fragile X syndrome as a prototype for emerging issues. Ment Retard Dev Disabil Res Rev. 2008;121:e693–e704. doi: 10.1542/peds.2007-0820. [DOI] [PubMed] [Google Scholar]
- Crawford DC. Meadows KL. Newman JL, et al. Prevalence of the fragile X syndrome in African Americans. Am J Med Genet. 2002;110:226–233. doi: 10.1002/ajmg.10427. [DOI] [PubMed] [Google Scholar]
- De Vries B. van den Ouweland A. Mohkamsking S, et al. Screening and diagnosis for the fragile X syndrome among the mentally retarded: an Epidemiological and Psychological Survey. Am Med Genet. 1997;3(61):660–667. doi: 10.1086/515496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski C. Levesque S. Morel ML, et al. Premutation and intermediate-size FMR1 alleles in 10 572 males from the general population: loss of an AGG interruption is a late event in the generation of fragile X syndrome alleles. Hum Mol Genet. 2002;61:660–667. doi: 10.1093/hmg/11.4.371. [DOI] [PubMed] [Google Scholar]
- Fernandez-Carvajal I. Walichiewicz P. Xiaosen X, et al. Screening for expanded alleles for the FMR1 gene in blood spots from newborn males in a Spanish population. J Mol Diagn. 2009;11:324–329. doi: 10.2353/jmoldx.2009.080173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH. Kuhl DP. Pizzuti A, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Hagerman PJ. The fragile X prevalence paradox. J Med Genet. 2008;45:498–499. doi: 10.1136/jmg.2008.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ. Hagerman PJ. Fragile X Syndrome: Diagnosis, Treatment, and Research, third edition. The Johns Hopkins University Press; Baltimore: 2002. [Google Scholar]
- Hagerman RJ. Rivera SM. Hagerman PJ. The fragile X family of disorders: a model for autism and targeted treatments. Curr Pediatr Rev. 2008;1(4):40–52. [Google Scholar]
- Loesch DZ. Huggins RM. Hagerman RJ. Phenotypic variation and FMRP levels in fragile X. Ment Retard Dev Disabil Res Rev. 2004;10:31–41. doi: 10.1002/mrdd.20006. [DOI] [PubMed] [Google Scholar]
- Maddalena A. Richards CS. McGinniss MJ, et al. Technical standards and guidelines for fragile X: the first of a series of diease-specific supplements to the standards and guidelines for Clinical Genetics Laboratories of the American College of Medical Genetics. Quality Assuarance Subcommittee of the Laboratory Practice Committee. Genet Med. 2001;3:200–205. doi: 10.1097/00125817-200105000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JE. Bundey S. Webb TP, et al. Fragile X syndrome is less common than previously estimated. J Med Genet. 1997;34:1–5. doi: 10.1136/jmg.34.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesso R. Berkenstadt M. Cuckle H, et al. Screening for fragile X syndrome in women of reproductive age. Prenat Diagn. 2000;20:611–614. doi: 10.1002/1097-0223(200008)20:8<611::aid-pd881>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Reiss AL. Dant CC. The behavioral neurogenetics of fragile X syndrome: analyzing gene–brain–behavior relationships in child developmental psychopathologies. Dev Psychopathol. 2003;15:927–968. doi: 10.1017/s0954579403000464. [DOI] [PubMed] [Google Scholar]
- Tassone F. Pan R. Amiri K, et al. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 2008;10:43–49. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano-Alhadef H. Basel-Vanagaite L. Magal N, et al. Fragile-X carrier screening and the prevalence of premutation and full-mutation carriers in Israel. Am J Hum Genet. 2001;69:351–360. doi: 10.1086/321974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G. Webb T. Wake S. Robinson H. Prevalence of fragile X syndrome. Am J Med Genet. 1996;64:196–197. doi: 10.1002/(SICI)1096-8628(19960712)64:1<196::AID-AJMG35>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]