Abstract
We recently described ten peptides selected from a 16,384-member combinatorial library based on their ability to permeabilize synthetic lipid vesicles in vitro (Rathinakumar R and Wimley WC, J. Am. Chem. Soc. 2008, 130, 9849-9858). These peptides did not share a common sequence motif, length or net charge; nonetheless they shared a mechanism of action that is similar to the natural membrane permeabilizing antimicrobial peptides (AMP). To characterize the selected peptides and to compare the activity of AMPs in vivo and in vitro we report on the biological activity of the same selected peptides in bacteria, fungi, and mammalian cells. Each of the peptides has sterilizing activity against all classes of microbes tested, at 2-8 μM peptide, with only slight hemolytic or cytotoxicity against mammalian cells. Similar to many natural AMPs, bacteria are killed within a few minutes of peptide addition and the lethal step in vivo is membrane permeabilization. Single D-amino acid substitutions eliminated or diminished the secondary structure of the peptides and yet they retained activity against some microbes. Thus, secondary structure and biological activity are not coupled, consistent with the hypothesis that AMPs do not form pores of well defined structure in membranes, but rather destabilize membranes by partitioning into membrane interfaces and disturbing the organization of the lipids, a property that we have called “interfacial activity”. The observation that broad-spectrum activity, but not all antimicrobial activity, is lost by small changes to the peptides suggests that the in vitro screen is specifically selecting for the rare peptides that have broad-spectrum activity. We put forth the hypothesis that methods focusing on screening peptide libraries in vitro for members with the appropriate interfacial activity can enable the design, selection and discovery of novel, potent and broad-spectrum membrane-active antibiotics.
Introduction
Aggressively pathogenic microorganisms are a significant and growing medical problem worldwide1. Thus, antibiotic treatment of microbial infections is under scrutiny and discovery of new classes of antibiotics is warranted. One potentially useful, but underdeveloped, class of antibiotics is the antimicrobial peptides (AMP). AMPs have been isolated from the innate immune systems of many organisms, including humans, and understanding their activity will give insight into strategies for the design of new generations of antimicrobials. Natural AMPs typically have broad-spectrum activity against multiple classes of bacteria and fungi at micromolar concentrations including drug-resistant strains2, but are nontoxic to host cells3-6. There is considerable evidence that membrane permeabilization is usually the lethal activity. The probability of resistance is thus low because the fitness cost for a microbe to reconfigure its entire membrane is high. While partial resistance to AMPs has been described7, it is rare and not easily selected in the laboratory or in infected hosts. This provides an important advantage for AMPs over conventional antibiotics5, and therefore methods for identifying and designing new AMPs are desirable.
While AMPs permeabilize membranes in vivo and in vitro, the literature suggests that few AMPs function by a mechanism that is consistent with specific, stable, water-filled, transmembrane pores. We have proposed that AMP activity is dependent on interfacial activity: “the ability of a molecule to partition into in the membrane-water interface and to alter the packing and organization of lipids”8. For this reason, compelling structure-function relationships are not often observed for AMPs. Instead, their activity is best described by interfacial activity as exemplified by the ‘carpet model’ 9, ‘sinking raft model’10 and others11. The membrane permeabilizing activity of AMPs is recapitulated in synthetic bilayer vesicle systems12,13 which make experimentally convenient systems for structure-function studies. We show here that they also make useful systems for selecting potent AMPs from peptide libraries.
The lack of obvious structure-function relationships has hampered the bioengineering and rational design of novel AMPs. To select membrane-active peptides we recently designed a 16,384-member combinatorial library of short peptides in which the members' composition and amphipathicity was varied using a reduced “alphabet” of only 10 amino acids. This library was screened for membrane-permeabilization in vitro, with a liposome based, high throughput screen8 in which the lipid vesicles contained a mixture of zwitterionic and anionic lipids, similar to bacterial plasma membranes. We screened 20,000 peptides from the library and identified 10 water soluble, membrane permeabilizing peptides of 9-15 amino acids in length. The sequences are shown in Figure 1. In lipid vesicles, the mechanism of action of these peptides was consistent with the concept of interfacial activity8. In the work presented here, we show that all of the peptides selected in the vesicle-based high-throughput screen also have potent and broad-spectrum antimicrobial activity against bacteria and fungi, but have little toxic activity against mammalian cells. Furthermore we show that their biological activity is based on rapid, extensive permeabilization of the microbial membranes. Small changes to the peptide eliminated secondary structure without eliminating all antimicrobial activity, suggesting structure and activity are not strongly coupled. The observation that some antimicrobial activity is lost by small changes to the selected peptides suggests that the in vitro screen is specifically selecting for the rare peptides that have broad-spectrum activity. Based on these results we hypothesize that methods focusing on screening peptide libraries in vitro for members with the correct interfacial activity can enable the design, selection and discovery of novel, potent and broad-spectrum membrane-active antibiotics.
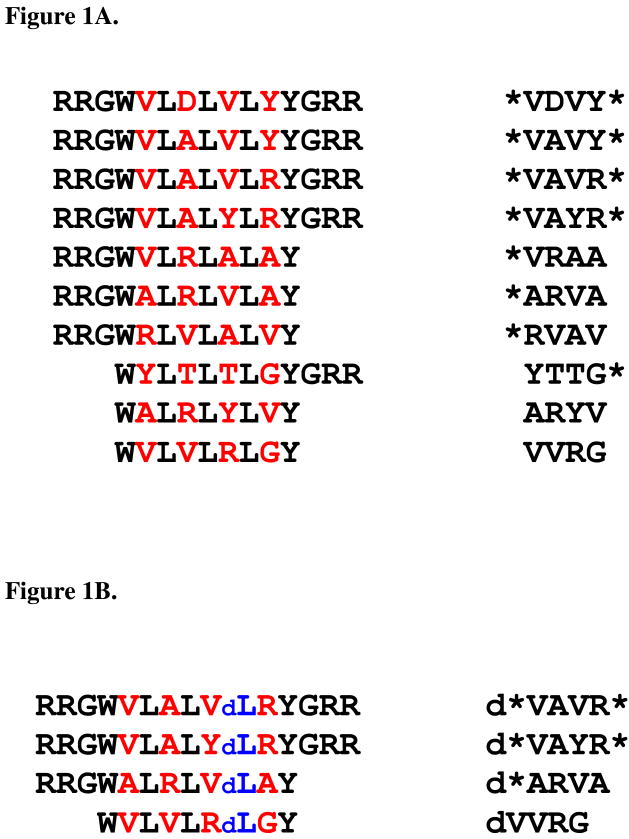
Figure 1.
Figure 1A. Membrane-active peptides from library
Figure 1B. D-amino acid substituted membrane-active peptides
A: The peptides used in this study. These peptides were selected from a 16,384 member combinatorial peptide library using an in vitro high-throughput screen for the ability to permeabilize synthetic lipid vesicles containing 90% phosphatidylcholine lipids and 10% phosphatidylglycerol lipids. Library design, peptide selection and characterization of the active peptides in vitro are described elsewhere8,14,15. The combinatorial sites in the peptides are highlighted in red. The RRG- and –GRR terminal cassettes were also varied combinatorially. On the right side, we give the four letter codes we use herein to represent the peptides. The code is based on the identity of the amino acids in the four combinatorial sites. The asterisk symbols (*) represents the terminal cassettes, when present. B: The D-amino acid substituted peptide homologs used in this study. Four of the peptides above were synthesized with a single D-leucine substitution near the C-terminus to study the coupling of structure to biological activity. The D-leucine is represented by ‘dL’ in blue.
Experimental Procedures
Bacterial cultures, erythrocytes and mammalian cells
Strains of Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 25923), Pseudomonas aeruginosa (ATCC 27853) and Cryptococcus neoformans (ATCC 66031) were obtained from the American Type Culture Collection (Rockville, MD). The bacterial and yeast cultures were maintained in Trypticase soy broth (TSB) and YPD broth (Difco laboratories, Detroit, Mich.) respectively. Overnight cultures of bacteria were grown in 125 mL flasks shaken at 220 rpm at 37°C. Overnight cultures of yeast were grown at 30°C. Washed sheep erythrocytes and washed human erythrocytes (10%) were obtained from Lampire biological laboratories (Pipersville, PA). Erythrocytes were re-suspended in PBS-EDTA buffer pH 7.4. Human epithelial kidney cells (HEK293T) and mouse embryonic fibroblast NIH-3T3 cells were generously provided by Dr. Charles Hemenway. HEK293Tand NIH3T3 cells were grown with DMEM media.
Peptides
Details of peptide synthesis and purification are provided elsewhere.8,14,15 Stock solutions of 1-5 mM peptide were made in 0.025% acetic acid. Experimental solutions were made by diluting the appropriate amount of stock solution into buffer. As we showed previously8 these peptides in buffer contain some β-sheet secondary structure. Peptides remain soluble at least for a day and tryptophan fluorescence showed that the Trp residue is water-exposed. Thus we concluded that the peptides in buffer are small, weakly associated aggregates. Control experiments we have performed under a wide range of buffers, salt species, ionic strength and pH (not shown) have demonstrated little or no sensitivity of biological activity on buffer conditions.
Antimicrobial activity
E. coli, P. aeruginosa, S. aureus and C. neoformans were grown to mid-logorithmic phase and diluted to 103 colony forming units (CFU)/ml with minimal liquid test medium (LTM; 1% of growth broth in PBS). In sterile 96 well plates, 120 μl of cell suspension (1 × 103 cells/mL in minimal LTM) were added to 50 μl of 0.025% acetic acid containing peptide. Eight columns were prepared for each peptide in 2/3 fold serial dilution starting at 10 μM. Wells were incubated at 37°C for up to 3 hours and then 125 μl of 2× concentrated growth medium was added and cells were allowed to recover overnight. Wells were either opaque (OD600 > 0.5) indicating stationary phase growth or they were transparent (OD600 < 0.02) indicating no growth. Aliquots from wells with no apparent growth were spread on nutrient agar plates to verify sterility. In all cases there were few, if any, CFU in those wells compared to 108 CFU/mL in the opaque wells. For each experiment there were a number of sterile wells starting from the highest concentration. The lowest concentration of peptide that sterilized the cells is the minimum sterilizing concentration (MSC). All MSC measurements are the average of 3-5 independent experiments. Time course experiments were performed by reducing the pre-incubation time from the standard 3 hours to between 1 and 60 minutes before the addition of rich growth media. Little or no difference in MSC was observed with shortened pre-incubation except for the fungus Cryptococcus, as discussed below. Addition of rich growth medium to cells first followed by peptide gave similar MSC values. Also, the addition of 104 or 105 cell/ml had little effect on the measured MSC values.
Hemolysis
Red blood cells (RBC) from sheep and human were diluted to 7.7 × 106 cells/ml and 2 × 107 cells/ml respectively, and the assay was performed in 96 well plates in a final volume of 100 μl. Peptides at 0.5 μM, 5 μM and 15 μM were incubated with RBC at room temperature for 1hr. The plates were then centrifuged at 4,000 g for 2 minutes, and the release of hemoglobin was monitored by measuring the absorbance of the supernatant at 540 nm. Baseline was defined by RBC incubated with PBS-EDTA pH 7.4 only and complete lysis was normalized to measuring complete osmotic lysis in distilled water.
MTT cell viability assay
HEK293T and NIH3T3 cells were grown in DMEM media in Corning cellBIND® flasks. The confluent cells were then trypsonized and the cell counts were measured. Cell suspension in DMEM (5 × 105 cells/mL) was mixed with 0.5, 5 and 15 μM of peptide or an equivalent volume of buffer (positive control) in a 96-well cell culture plate. Following the addition of peptides, the cells were incubated for 72 hours at 37°C with 5% CO2 in a humidified incubator during which time the cells adhered to the plate. After the 72 hour incubation, 50 μg of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma Aldrich, St. Louis, MO) was added to each well and incubated for six hours. In living cells, mitochondrial reductases convert the MTT tetrazolium to formazan which precipitates. Formazan crystals are dissolved using acidified isopropanol (90% isopropanol : 10% Triton-X-100: 0.4% HCl) and the optical density at 550 nm was measured using a plate spectrophotometer. Melittin at 5 or 15 μM or 1% of the detergent TWEEN resulted in complete loss of cell viability.
SYTOX Green uptake
SYTOX Green is a cationic cyanine dye (∼ 900Da) that is not membrane permeable16. When a cell's plasma membrane integrity is compromised, influx of the dye and subsequent binding to DNA causes a large increase in fluorescence. For SYTOX Green assays, bacterial cells were grown to mid-logarithmic growth phase (OD600 ∼0.3), and then were centrifuged, washed and resuspended in PBS. Cell suspensions in PBS (2 × 107 cells/mL) were incubated with 1 μM SYTOX Green (Invitrogen, Carlsbad, CA) for 15 minutes in the dark prior to the influx assay. Similarly, mammalian cells (HEK293T and NIH3T3) were grown to confluent population in Corning cellBIND® flasks. The cells were trypsonized and resuspended in GIBCO DPBS buffer (5 × 105 cells/mL) followed by addition of 1 μM SYTOX Green. At 2-4 minutes after initiating data collection, 5 μM of peptides was added to the cell suspension, and the increase in SYTOX Green fluorescence was measured (Excitation wavelength at 485 nm and emission at 520 nm) for 40 minutes. Lysis of the cells with the detergent Triton X-100 or with the peptide melittin gives maximum fluorescence.
Membrane depolarization
Cytoplasmic membrane depolarization of E. coli was measured using a membrane potential-sensitive probe, 3,3′ - dipropylthiacarbocyanine [diSC3(5)] (Anaspec Inc, San Jose, CA). E. coli cells were grown to a mid-logarithmic phase (OD600=0.3), centrifuged, washed and re-suspended in a buffer containing 5 mM HEPES, 20 mM glucose and 100 mM KCl, pH 7.2. Measurements were made in a 1 mL total volume containing 2 × 107 cells/mL and 50 nM diSC3(5). The fluorescence of dye was monitored for 10 minutes at 25°C using an SLM spectrofluorometer at an excitation wavelength of 622 nm and an emission wavelength of 670 nm. Dye uptake, and resultant self-quenching, is modulated by the membrane potential. After reaching the maximum uptake of the dye by E. coli, which is indicated by a minimum in dye fluorescence, peptides were added to the cells and the decrease in potential was monitored by the increase in fluorescence. Complete collapse of the membrane potential was attained with 40 μM valinomycin, a potassium ionophore. Measurements were repeated at least twice under each condition.
Results
Antimicrobial activity of the selected peptides
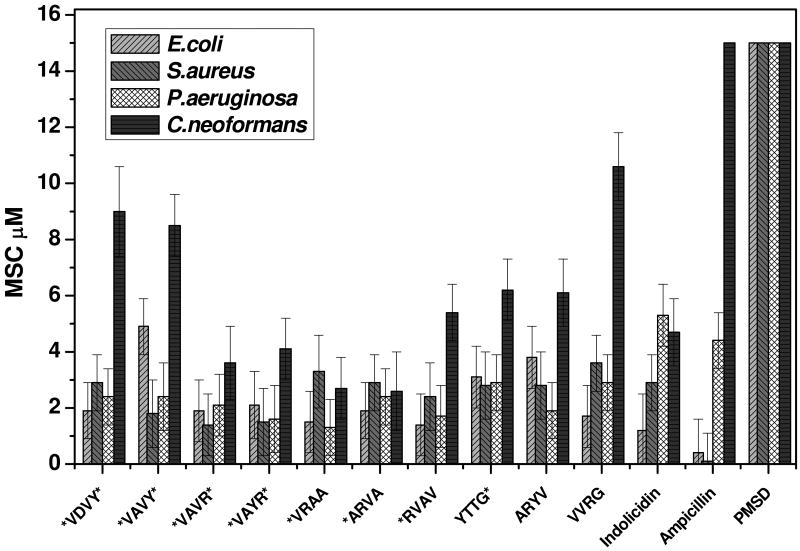
The antimicrobial activity of the selected peptides was assessed by their ability to sterilize a culture of bacteria or fungi in rich growth media such that no growth occurred in a 24 hour incubation. We used Gram positive bacteria (Staphylococcus aureus), Gram negative bacteria (Escherichia coli and Pseudomonas aeruginosa) and a fungus (Cryptococcus neoformans). The chemical antibiotic ampicillin and the antimicrobial peptide indolicidin from bovine neutrophils17 served as positive controls for the microbes and gave the expected minimal inhibitory concentrations (MICs) (Figure 2). The perfringolysin O membrane spanning domain (PMSD) which is an anionic membrane spanning beta-hairpin peptide in the context of a large protein toxin18,19 but does not cause leakage as a free peptide (unpublished observation) served as a negative control. The data shown in figure 2 indicates that all of the selected peptides have potent antimicrobial activity against all of the organisms. We refer to the minimum effective concentration as a minimum sterilizing concentration (MSC) rather than a minimum inhibitory concentration (MIC) because wells with no growth after 24 hours showed few, if any, colony forming units (CFU) when spread on nutrient agar plates compared to 108 CFU/ml in peptide free wells. In fact, sterilized wells remained sterile indefinitely on a bench top at room temperature, thus the microbes are being killed rather than just inhibited. We recently tested these peptides against other microorganisms including the Gram positive bacterium Bacillus anthracis and the fungus Candida albicans and we find the same low micromolar sterilizing activity. The low micromolar, sterilizing antimicrobial activity of these peptides is comparable to the natural antimicrobial peptide indolicidin (Fig. 2) and to other natural antimicrobial peptides20,21. Therefore, the peptides that we selected from the simple combinatorial library using an in vitro liposome leakage assay are potent, broad-spectrum antimicrobial agents.
Figure 2. Antimicrobial activity.
The sterilizing antimicrobial activity of the ten peptides against Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Cryptococcus neoformans. Each row in a 96-well plate contained one peptide in serial dilution from 10 μM to 0.1 μM. Cells at 103 cells/ml suspended in minimal media were added and incubated for 3 hrs, and then 2× growth media is added to the cells which were allowed to recover overnight at 37 °C. Wells containing less than a threshold peptide concentration were opaque, indicating stationary phase growth, while wells containing above the threshold peptide concentration were transparent, indicating no growth at all. The lowest concentration of peptide that prevented growth is the minimal sterilizing concentration (MSC). MSC values in the figure are the average of 3-5 independent measurements with standard errors.
Time-course of antimicrobial activity
To help elucidate the killing mechanism, we varied the pre-incubation time and order of addition of peptide, microbes and growth media. The results (not shown) reveal only small changes, if any, in bacterial MSC values for pre-incubation times ranging from the standard three hours to just a few seconds. In fact, even reverse addition (growth media added to cells first, followed immediately by peptide) did not change MSC values substantially. We conclude from these experiments that the lethal step for bacteria occurs immediately and renders the cells incapable of recovering after only a few moments of contact with peptide. This result is consistent with the membrane permeabilization experiments described below which show immediate effects of these peptides on the integrity on microbial membranes.
The fungus Cryptococcus required more than one hour of pre-incubation for complete sterilization, suggesting a different mode of action. Addition of rich growth medium to fungi that had been preincubated with peptides for intermediate times resulted in a reduction in growth of C. neoformans after overnight incubation, but not sterilization. Consistent with these observations, we show below that these peptides permeabilize fungal membranes much more slowly than bacterial membranes.
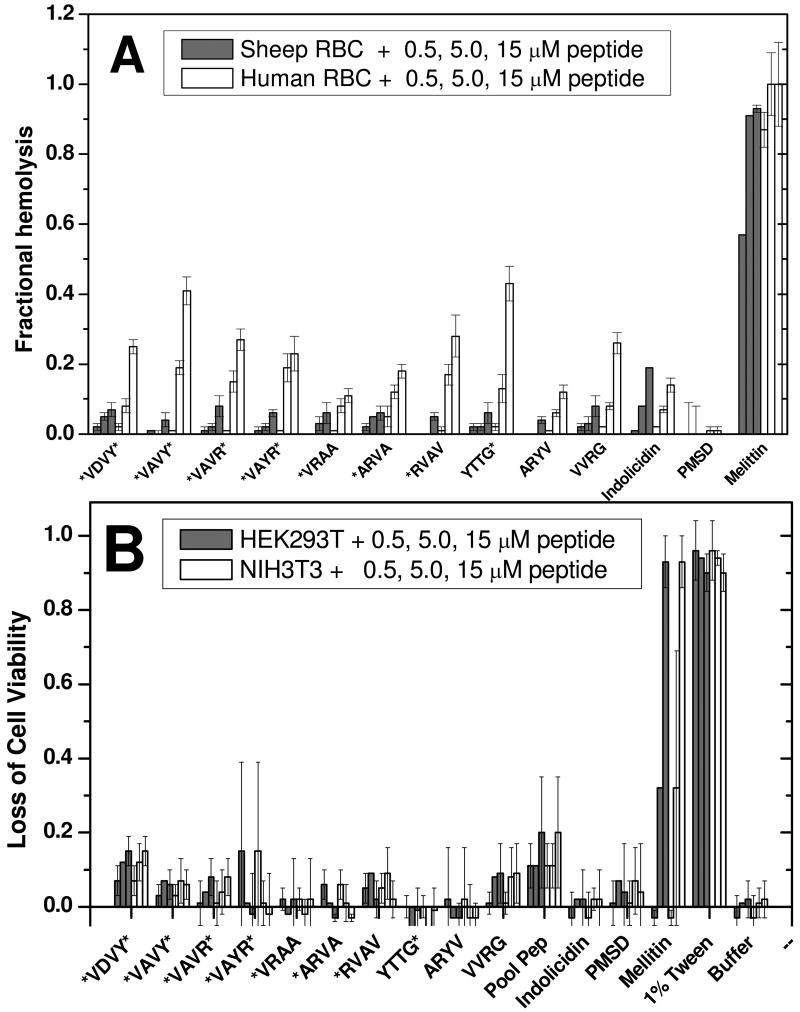
Selective biological activity
To distinguish selective antimicrobial activity from non-selective lytic activity, we measured the toxicity and lytic activity of the selected peptides using erythrocytes and mammalian cultured cells. In Figure 3A we show the hemolytic activity of the selected peptides against sheep and human erythrocytes. Buffer served as a negative control and 100% hemolysis of cells was achieved by suspending the cells in distilled water for 1 hr. The non-specific lytic peptide melittin gave 100% hemolysis at 5 μM or above. The negative control peptide, PMSD, gave <5% hemolysis at 15 μM. The selected peptides have little hemolytic activity against the more robust sheep erythrocytes. Some have activity against human erythrocytes, but only at the highest concentration. The hemolytic activity is comparable to that caused by the natural antimicrobial peptide indolicidin. Similarly, the membrane active peptides were tested for cytotoxicity against living mammalian cells lines. Cell viability was measured using the MTT assay, which measures the activity of mitochondrial reductases (Figure 3B). The selected peptides have little toxic effect on the mammalian HEK293T and NIH3T3 cells even at 15 μM, which is well above the MSC levels. The non-selective lytic peptide melittin is always 100% lethal at 5 or 15 μM. Among the selected peptides, the rank order of the slight MTT cytotoxicity is similar to the rank order of slight hemolytic activity.
Figure 3. Hemolytic and cytotoxic activity.
Effect of selected peptides on mammalian cell membranes. A: Hemolytic activity of the selected peptides against sheep and human erythrocytes. Sheep erythrocytes (7.7 × 106 cells/ml) and human erythrocytes (2 × 107 cells/ml) in phosphate-sodium buffer solution (PBS) pH 7.4, were incubated with peptides at 0.5, 5 and 15 μM for 1 hour. For each peptide six values are shown, 0.5, 5 and 15 μM peptide for sheep and human erythrocytes, left to right. Cells were then centrifuged at 4000g for 2 minutes. The optical absorbance of hemoglobin at 540nm is used to measure hemolysis. Activity is expressed as percent hemolysis where zero is a buffer only control and 100% hemolysis is the value for osmotic lysis with distilled water. B: The cytotoxicity activity of the selected peptides against mammalian HEK293T and NIH3T3 cells. The cells at 5 × 105 cells/mL were treated with peptide at 0.5, 5 and 15 μM and incubated for 72 hours. Cell viability was measured by using the MTT assay in which mitochondrial reductases in living cells produce formazan crystals after the addition of MTT. The crystals were dissolved in isopropanol and the optical density measured at 550 nm. Activity is expressed as percent cytotoxicity where zero is a buffer-only control and 100% is the value for treatment with 1% of the detergent TWEEN.
In some of these experiments, we also used a random pooled peptide stock from the original library to compare selected peptides to the average library peptide. We pooled about 200 randomly chosen peptides from the library and used this peptide pool to evaluate the potential of the library overall prior to selection, on permeabilization in living microorganisms and mammalian cells. In figure 3B we show that the random pooled peptides cause slight toxicity which is at the same level as the selected antimicrobial peptides.
Biological membrane permeabilization
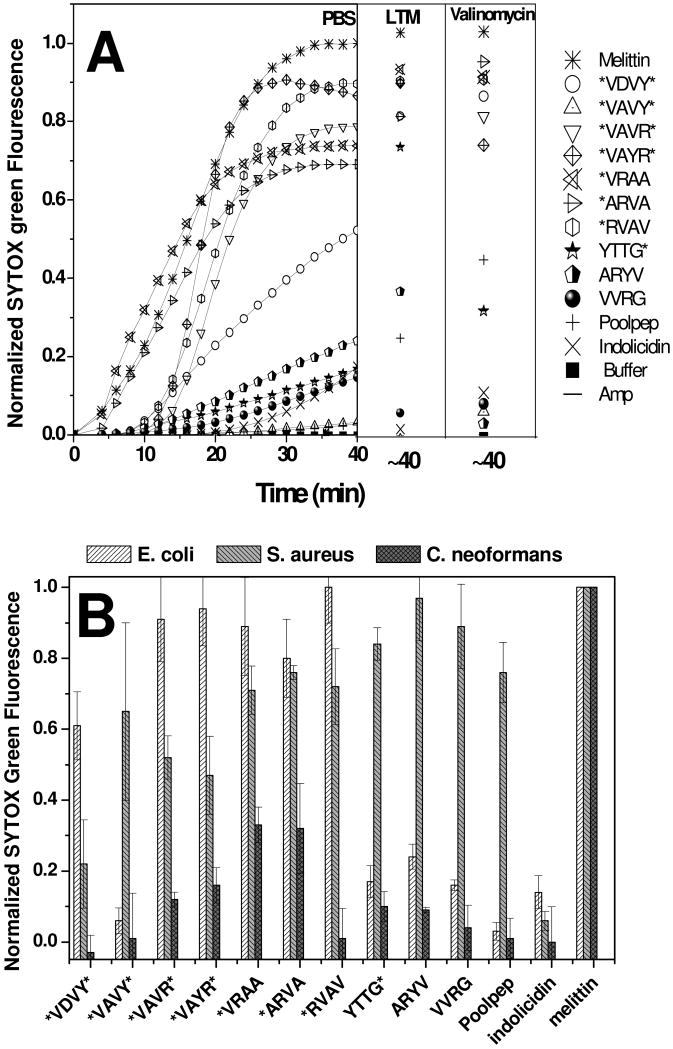
In order to correlate the selective killing of microbes by these peptides to their selection using an in vitro liposome leakage assay, we tested whether killing of bacterial cells is due to membrane permeabilization. To study the effect of these peptides on the membranes of living microbial cells, we used the fluorescence of SYTOX Green, a membrane impermeant, DNA binding dye. Membrane permeabilization allows entry of the dye, as indicated by an increase in fluorescence. In Figure 4A SYTOX Green fluorescence is monitored after addition of 5 μM peptide to E. coli cells suspended in PBS buffer. Treatment with melittin shows an accelerating activity in which 50% of the maximum SYTOX Green uptake occurred by about 15 minutes and complete SYTOX Green leakage occurred by 25 min. The selected peptides all showed significant SYTOX Green influx. The control AMP indolicidin causes only a very small amount of dye influx, suggesting a different mechanism of antimicrobial activity. The antibiotic ampicillin has no effect in the influx of SYTOX Green into the cells. The selected peptides have a direct effect on the living bacterial inner cell membrane.
Figure 4. Mitochondrial membrane permeabilization.
Microbial membrane permeabilization caused by the selected peptides. A: Cytoplasmic membrane permeabilization of E. coli measured by entry of the DNA binding dye SYTOX Green. Dye (1 μM) was added to 2 × 107 cells/ml suspended in PBS and the time course of fluorescence was monitored after addition of 5 μM peptide. Fluorescence, monitored with excitation at 485nm and emission at 520 nm, increases only when the dye can cross the membrane and bind DNA. The pore-forming peptide melittin served as a positive control for membrane permeabilization. The dye uptake is measured at three different assay conditions of E. coli. PBS: metabolically dormant cells in PBS buffer alone; LTM: metabolically active cells in “liquid test medium”, containing 1% growth medium in PBS; and valinomycin: cells treated with 40 μM valinomycin to dissipate the membrane potential. SYTOX Green fluorescence is shown relative to that caused by complete lysis with melittin at 40 minutes. B: Membrane permeabilization of different microorganisms caused by the selected peptides. Membrane permeabilization was measured using the SYTOX Green DNA binding dye as described above. Cells in PBS (2 × 107 cells/mL) were incubated with 1 μM SYTOX Green and 5 μM peptide. After 40 minutes, the net uptake of the dye through the plasma membrane was monitored by fluorescence and normalized to the fluorescence in the presence of the same amount of melittin.
To help elucidate the mechanism of action of these peptides in cells and to establish their versatility, we repeated the SYTOX Green experiments using cell suspensions under three different conditions. Cells re-suspended in PBS (above) will be metabolically dormant due to the lack of nutrients, but they may still have a residual transmembrane electrochemical potential and ATP store. Cells supplemented with growth media (LTM) will have metabolic activity, including a transmembrane potential, and cells treated with 40 μM valinomycin will have fully dissipated transmembrane potential and no ATP stores. We observed little difference in the SYTOX Green influx under these different conditions, as shown in Figure 4A, suggesting that the peptide-membrane interactions and membrane disruption are independent of the metabolic state of the cells. The negative-inside electrochemical potential of bacterial plasma membranes has been suggested to be an important factor in AMP activity and selectivity for microbes22,23. The data presented here, however, shows that these peptides cause the rapid and extensive permeabilization of microbial membranes independent of the membrane potential.
In Figure 4B we compare membrane permeabilization of other classes of microbes. Consistent with their broad-spectrum activity, the selected peptides permeabilize the membranes of the bacterium S. aureus. Against the fungus C. neoformans the peptides behave somewhat differently. Although all of the selected peptides are potently fungicidal (Fig. 2), some show little leakage of SYTOX Green into the fungal cells compared to the lytic peptide melittin which completely permeabilizes the fungal cells in just a few minutes. This is probably related to the observation, above, that sterilization of C. neoformans requires more than one hour, compared to only seconds for the bacteria. Similar sterilizing antifungal activity and slow activity is observed for these peptides against other fungal species (RR and WCW unpublished results). This observation suggests that the mode of antifungal activity is different from the mechanism of action in bacteria. The mechanism of antifungal activity is under further study.
We also measured the permeabilizing effect of these peptides on living mammalian cells, HEK293T and NIH293T (not shown). Treatment of these cells with 5 μM peptide as in Figure 4 caused no detectable influx of SYTOX Green into cells in 40 minutes. Mean SYTOX Green fluorescence after 40 minutes was -6 ± 8% (SD) of the buffer and melittin controls. Melittin at 5 μM concentration was used as a positive control and it caused maximum dye uptake by the cells in less than 10 minutes. The observation that the selected peptides have little or no permeabilizing effect on mammalian cells is consistent with their lack of hemolytic or cytotoxic activity (Fig. 3).
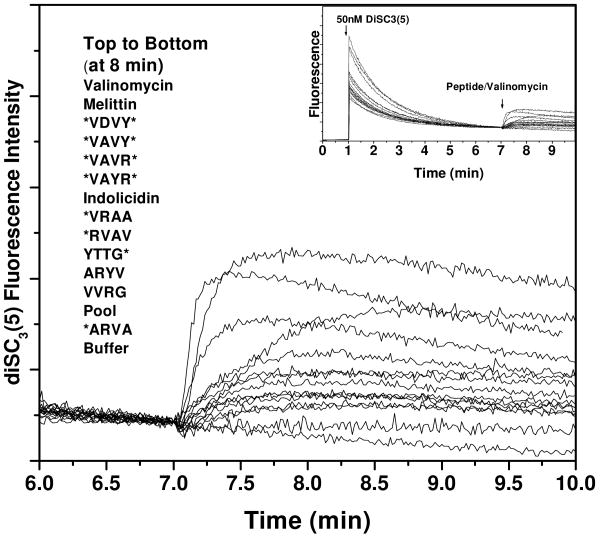
Peptide effect on membrane potential
The membrane potential sensitive dye diSC3(5) was used to monitor the cytoplasmic membrane depolarization of E. coli cells in the presence of peptides. In Figure 5 we show a typical set of experiments. In each experiment the dye is added to E. coli cells and the fluorescence decreases as the dye accumulates in the membranes and becomes self-quenched. Dissipation of the potential causes an increase in fluorescence due to dye de-quenching. Complete dissipation is given by the valinomycin curve. The potential dissipation caused by the lytic peptide melittin is comparable to that caused by valinomycin, and occurs in less than 30 seconds. Most of the selected peptides affect the membrane potential of cells in less than a minute, although not all to the same extent as melittin and valinomycin. Importantly, this experiment shows that peptide-induced dissipation of the membrane potential can occur in less than one minute, while the SYTOX Green influx requires 10-40 minutes, and often does not even begin for several minutes. This result suggests that a sequence of steps is occurring at the membrane, beginning with depolarization (small ion permeability) and followed by more significant membrane disruption leading to SYTOX Green influx.
Figure 5. Microbial membrane depolarization.
Cytoplasmic membrane depolarization of E. coli. A membrane potential-sensitive dye, 3,3′- dipropylthiacarbocyanine [diSC3(5)] was added to E. coli cells (2 × 107 cells/mL) at 50 nM and the change in dye fluorescence (excitation at 622 nm, emission at 670 nm) due to membrane binding and self quenching was monitored until it was stable (inset). Peptides (5 μM) were added at 7 min and the loss of membrane potential was measured. The fluorescence increase intensity for complete collapse of cytoplasm membrane potential is given by the valinomycin curve, which is closely matched by the positive control peptide melittin.
The importance of structure to interfacial activity
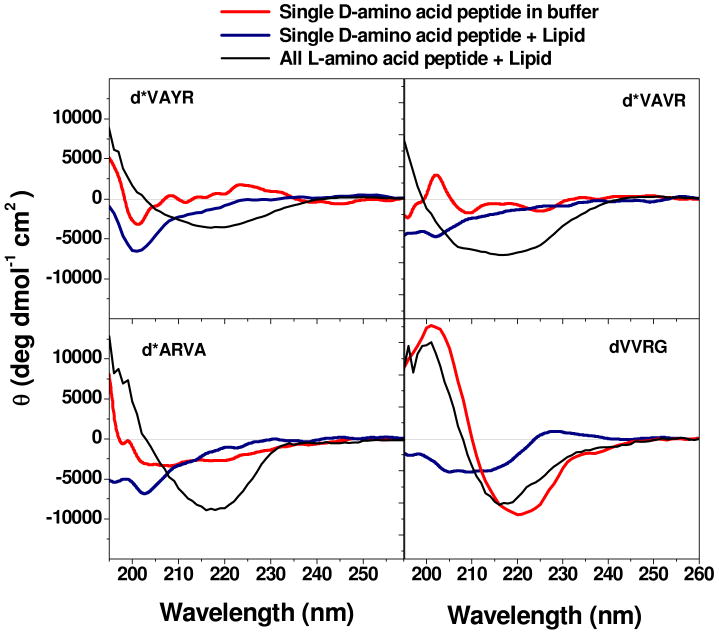
We have proposed that the rare active peptides selected from the library have the correct balance of solubility, hydrophobicity and amphipathicity (i.e. interfacial activity) such that they can partition into in the membrane-water interface and alter the packing and organization of lipids disruption to cause leakage of contents entrapped in vesicles8,2,15. In this work, we show that the same peptides also have potent, broad-spectrum antimicrobial activity. However the importance of peptide structure to interfacial activity remains poorly defined for antimicrobial peptides. To examine the importance of secondary structure in membrane disruption, we synthesized analogs of four of the active peptides in which a single D-amino acid was substituted at a position near the C-terminus (Fig. 1). This small change was expected to decrease the peptide secondary structure24. We compared the structure and activity of these peptides in living cell membranes as well as in liposomes (Table 1). In Figure 6 we show the circular dichroism (CD) spectra of the all L peptides and their single D-amino acid variants The all L peptides have β-sheet secondary structure in buffer and in bilayers8. The D-amino substituted peptides have reduced structure propensity. All four are mostly random coil in buffer. The longer peptides d*VAVR*, d*VAYR* and d*ARVA showed no increase in structure content when 2.5 mM lipid was added. The shorter peptide dVVRG gained significant β-sheet content when bound to membranes.
Table 1. Properties of D-substituted antimicrobial peptides.
Comparison of the biological and biophysical activity of D-amino acid substituted peptides with the normal peptides. Minimum sterilizing concentrations (MSC), % hemolysis, and % Sytox Green leakage were measured as described in the text. Leakage of terbium from lipid vesicles was done as described elsewhere44. MSC values in bold signify peptides that retain good antimicrobial activity MSC < 7 μM.
| Peptide | Antimicrobial activity MSC (μM) | % of Hemolysis at 15 μM peptide | % of Sytox Green leakage | % of Tb leakage | ||||
|---|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | P. Aeuriginosa | C. Neoformans | Human RBC | sheep RBC | E. coli | P:L 1:100 | |
| *VAVR* | 2.2 | 1.9 | 1.6 | 3.9 | 24 ± 1 | 15 ± 16 | 71 ± 23 | 14 |
| d*VAVR* | 4.0 | 1.5 | 9.7 | 5.7 | 5 ± 6 | 0.0 | 30 ± 23 | 5 |
| *VAYR* | 2.2 | 1.4 | 1.7 | 4.1 | 23 ± 10 | 0 | 77 ± 19 | 53 |
| d*VAYR* | 8.5 | 2.1 | >10 | 9.7 | 5 ± 3 | 15 ± 20 | 12 ± 7 | 7 |
| *ARVA | 2.0 | 1.6 | 1.8 | 3.0 | 20 ± 6 | 3 ± 2 | 117 ± 26 | 13 |
| d*ARVA | 9.7 | 8.8 | >10 | 6.5 | 4.0 | 10 ± 12 | 10 ± 5 | 7 |
| VVRG | 1.8 | 3.4 | 3.9 | >10 | 21 ± 5 | 16 ± 17 | 38 ± 14 | 39 |
| dVVRG | >10 | >10 | >10 | >10 | 12 ± 13 | 11 ± 9 | 2 ± 1 | 6 |
| Indolicidin | 1.8 | 3.6 | 2.4 | 3.6 | 23 ± 6 | 4 ± 10 | 14 ± 14 | 8 |
| Melittin | 1.5 | 1.3 | 0.9 | 1.1 | 109 ± 6 | 86 ± 17 | 100.0 | 100 |
Figure 6. Circular dichroism spectroscopy of D-amino acid substituted peptides.
Change in secondary structure content of D-leucine substituted peptides. Circular dichroism spectra of the single D-amino acid substituted peptides at 50 μM concentration in phosphate buffer, pH 7.0. Spectra were taken before and after the addition of 2.5 mM phospholipid vesicles composed of 90% POPC and 10% POPG. The CD spectra of the same all L-peptide in phosphate buffer with 2.5 mM phospholipid vesicles are also shown for comparison. The ellipticity values are given as molar, or mean residue, ellipticities.
Generally, D-substituted peptides had at least slightly lower antimicrobial activity, lower hemolysis and lower cell permeabilizing and vesicle permeabilizing activity as shown in Table 1. However the quantitative effect of D-amino acid substitutions on biological activity is mixed, and does not correlate completely with loss or retention of secondary structure. For example, d*VAVR*, although completely unstructured in buffer and in membranes compared to all L*VAVR*, which has significant β-sheet content, retained good antimicrobial activity against E. coli, S. aureus and C. neoformans, while losing most activity against P. aeruginosa. The peptide d*VAYR* retained its activity against S. aureus while losing activity against the other three microorganisms and d*ARVA* lost activity against the three bacteria, but retained activity against the fungus. The one peptide that showed “normal” β-sheet secondary structure in membranes, dVVRG, lost all activity against all four microbes. In all cases, the broad-spectrum antimicrobial activity that is the hallmark of these selected peptides is lost upon a single L to D amino acid substitution; however the pattern of species-specific activity is unpredictable and some potent antimicrobial activity is retained. These results suggest that the coupling between peptide secondary structure and biological or interfacial activity is incomplete. Importantly, by showing that broad-spectrum activity is always lost, even when some species activity is retained, this result supports our hypothesis that the universal broad-spectrum antimicrobial activity of selected peptides is a specific property that is selected for in the liposome-based high throughput screen.
Discussion
The lack of compelling structure-function-activity relationships has made it difficult to rationally design novel peptide antibiotics. While systematic engineering of AMPs has given active analogs that are equal to or better than the parent peptides25-28, random sequence scrambling26 and other drastic changes29,30 have had the same result. We and others8,11,14,15,31,32 have hypothesized that structure-activity relationships in antimicrobial peptides are not obvious and more importantly for bioengineering are not easily predictable because the biological activity of antimicrobial peptides is derived more from “interfacial activity” than from the formation of specific structures in membranes. Membrane permeabilization is dependent on a peptide having the correct balance of hydrophobicity, solubility, amphipathicity and perhaps propensity to self assemble on membranes into peptide-rich domains. A peptide with the appropriate interfacial activity can bind to membranes, perturb the packing and organization of the lipids in a membrane to cause permeation. This function is not necessarily dependent on a peptide's ability to adopt a particular three-dimensional structure such as a barrel-stave or toroidal pore.
The concept of interfacial activity is based on physical chemical principles. The minimum elements are membrane binding, penetration into the interfacial region of the bilayer33, and disruption of the lipid packing to break down the permeability barrier. Binding of peptides to membranes can be driven by electrostatic or hydrophobic interactions34 or both, but only hydrophobic interactions drive penetration into the hydrophobic core of the bilayer33. Binding and penetration into the bilayer are necessary, but not sufficient to cause permeability changes. For example, we have described small hydrophobic peptides that bind well and partition deeply into bilayers and self-assemble into oligimeric β-sheets, but do not cause leakage even at very high peptide concentrations35. The final element of interfacial activity required for bilayer permeabilization is the disruption of the lipid packing. We hypothesize that membrane disruption is specifically driven by AMPs because most of them are amphipathic, but imperfectly so. AMPs have hydrophobic surfaces/sequences, but the hydrophobic surfaces that penetrate into the hydrophobic portion of the bilayer either contain or are closely bounded by polar or charged moieties. This is one of the only conserved features of the peptides selected from our libraries (Figure 1); most have an imperfectly amphipathic nature, containing either one charged residue or two polar residues within the otherwise hydrophobic core of 9 amino acids. The work presented here supports the hypothesis that interfacial activity also accounts for biological activity. First, the selected peptides, which function by interfacial activity in synthetic membranes8,14,15, all have antimicrobial activity as well. Second, disruption of secondary structure did not eliminate all biological activity of the peptides studies here, suggesting that peptide composition is as important to activity as secondary or tertiary structure. Numerous studies exist in the literature that cite a similar lack of clear structure-function correlation for AMPs (e.g. 23,26,36,37).
Here we found that the rare membrane-active peptides selected from our library also have low micromolar broad-spectrum, sterilizing antimicrobial activity with little lytic activity against mammalian cells. These results match our observations for a completely different 27 amino acid library screened using the same high throughput method14, thus we hypothesize that the power of this method may be generalizable, at least for β-sheet peptides. The potency and breadth of activity of these in vitro selected peptides is equal to the biological activity of the best natural antimicrobial peptides38,39. This property, coupled with the compositional simplicity of the selected peptides, and reduced amino acid alphabet, makes them potentially useful starting points for bioengineering of new antibiotics. More importantly these results demonstrate the utility of our composition-based libraries and vesicle-based high-throughput screening for selection and design of antimicrobial peptides.
Mechanism of Biological Activity
Most antimicrobial peptides, given sufficient time and concentration, will physically destroy bacterial membranes, as shown by leakage of membrane-impermeant compounds, membrane blebbing, fragmentation and, ultimately, gross disruption or total destruction of cell morphology40. But whether these latter observables are causes or effects of cell death is sometimes difficult to establish, especially for observations are made hours after peptide addition when killing requires only a few minutes or less. To determine if membrane permeabilization is directly responsible for the microbicidal activity of these peptides, we characterized the real-time physical disruption of bacterial membranes using two methods; by monitoring the fluorescence of a membrane-bound, potential-sensitive dye and by monitoring the entry of a larger membrane-impermeant DNA binding dye into cells. Most of the selected peptides begin dissipating the membrane potential of the bacterial inner membranes within seconds of addition, presumably due to proton and small ion (e.g. Na+, K+) leakage. Larger-scale membrane disruption, evidenced by entry of the larger SYTOX Green dye into cells, takes longer beginning within 2-10 minutes of peptide addition, and reaching completion by 30-40 minutes. These results support a model of biological activity in which membrane integrity is successively compromised to different degrees as peptides bind to and accumulate on the microbial plasma membrane.
Whether or not the cells were metabolically active did not make a large difference in membrane permeabilizing activity. Even dissipation of the transmembrane potential with valinomycin did not alter the membrane permeabilization to SYTOX Green, disproving, at least for these peptides, the long-standing idea that the bacterial transmembrane potential is an important factor in the activity and selectivity of cationic antimicrobial peptides, such as these. While some antimicrobial peptides may have other mechanisms of actionfoornote 1 our results suggest strongly that this family of peptides acts by passively binding to bacterial cytoplamsic membranes and successively disrupting the membrane to allow the leakage of small ions followed by larger solutes. These observations are consistent with the hypothesis that interfacial activity is responsible for the biological activity of these peptides.
Structure-Function Relationships and the Interfacial Activity Model
Based on the hypothesis that peptide composition is more important than structure for the biological activity of antimicrobial peptides, we synthesized and characterized a set of peptide homologs in which one L-leucine residue near the C-terminus was replaced by a D-leucine. This small change decreased or eliminated structure propensity without altering hydrophobicity, charge or peptide length. Blazyk and colleagues reported that single D-amino acid replacements caused modulation of the activity of linear amphipathic β-sheet peptides through structural perturbation36 without eliminating antimicrobial activity. Others have shown that antimicrobial activity can even be retained even in scrambled peptides26. The observations in Table 1 support the importance of interfacial activity by showing that peptide secondary structure and biological activity are not strictly coupled. These results highlight the fact that detailed relationship between the physical chemistry of binding and membrane perturbation and the biological activity of AMPs are only poorly describable for interfacially active AMPs. For example, we cannot yet explain why the sequences in Figure 1 were highly active while closely related sequences in the library were not as active. To discuss AMP activity, Bechinger11 recently described a molecular shape-based concept for antimicrobial/interfacial activity, and suggested that the physical chemistry of AMPs in membranes would best be described using phase diagrams, as one might use to describe mixtures of lipids and detergents. Almeida and colleagues have also described AMP activity in terms of binding, insertion and perturbation10,41-43 using kinetic models. Like the concept of interfacial activity, such physical-chemical descriptions of AMP activity could perhaps allow for predictions of activity to be made from sequence or composition, if they could be parametrized, although this is currently not feasible. In contrast, the frequently cited phenomenological models such the “carpet model” or “detergent model” do not offer physical-chemical explanations or prediction of the important parameters but rather serve to classify the observed mechanism of AMP action. We suggest here that physical chemical concepts such as interfacial activity can be used to explain the “carpet model” or detergent-like activity of AMPs.
What we have done in this work is to embrace the idea of interfacial activity and use it to design compositionally-varied peptide libraries that were screened for the most potent membrane permeabilizing peptides without regard to structure or mechanism. Using vesicle-based high-throughput screens of two very different peptide libraries8,14,15 we have now observed that 100% of about 20 peptides selected for membrane-permeabilizing activity in our vesicle-based system also have broad-spectrum antimicrobial activity against Gram-positive and Gram-negative bacteria and against fungi8,14,15. The fact that the four peptides with single C-terminal D-amino acid substitutions no longer have broad-spectrum activity but retain some species-specific activity, suggests that broad-spectrum activity results from a precise balance of factors that we are selecting for in the vesicle based screen which is lost upon D-amino acid substitution. Taken together, these results show that biological activity is not directly coupled to secondary or tertiary structure in these peptides and that the activities against the different microbes are not necessarily coupled to each other. Thus the selection of broad-spectrum antimicrobial activity in the vesicle screens is, thus, not an inevitable result of interfacial activity, but instead is a unique and powerful aspect of our high throughput screen.
Conclusion
In summary, these findings demonstrate the power of designing simplified peptide libraries based on the idea that interfacial activity is an important factor in biological activity of antimicrobial peptides. This work also supports the hypothesis that our methods for screening such libraries in vitro selects for rare members that have potent, broad-spectrum antimicrobial activity with little hemolytic or cytotoxic activity. We hypothesize here that methods focusing on screening for the proper balance of physical chemical interactions between peptides membranes and could be powerful ways to identify many new compounds, including non-peptide compounds that are active against membranes in vitro and in vivo. The flexibility and power of this approach will enable better design and section of novel membrane active antibiotics.
Acknowledgments
We appreciate the technical assistance of Christopher M. Bishop for peptide synthesis and purification, Drs. Sidhartha Hazari and Cecily Bennett for mammalian cell culture techniques, and Phuong Vo and Rachel Hahn for the antimicrobial time course experiments. Mammalian cell lines were a gift from Dr. Charles Hemenway. We thank Aram Krauson for the TOC image. This work was funded by the National Institutes of Health NIGMS-060000 (to WCW) and also by the Louisiana Board of Regents RC/EEP program (WCW and WFW) and the Louisiana Board of Regents ENH-PKSFI program to Dr. Frank Jordan, Loyola University.
The abbreviations used are
- AMP
antimicrobial peptide
- PBS
phosphate buffered saline
- CFU
colony forming units
- LTM
liquid test medium for antimicrobial assays (1% medium in PBS)
- MSC
minimum sterilizing concentration
- RBC
red blood cell (erythrocyte)
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- CD
circular dichroism spectroscopy
- PMSD
prefringolysin-O membrane spanning domain peptide (negative control for permeabilization)
Footnotes
Reference List
- 1.D'Agata EM. Rapidly rising prevalence of nosocomial multidrag-resistant, Gram-negative bacilli: a 9-year surveillance study. Infect Control Hosp Epidemiol. 2004;25(10):842–846. doi: 10.1086/502306. [DOI] [PubMed] [Google Scholar]
- 2.Hancock RE. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;1(3):156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 3.Avrahami D, Shai Y. Conjugation of a magainin analogue with lipophilic acids controls hydrophobicity, solution assembly, and cell selectivity. Biochemistry. 2002;41(7):2254–2263. doi: 10.1021/bi011549t. [DOI] [PubMed] [Google Scholar]
- 4.Makovitzki A, Shai Y. pH-dependent antifungal lipopeptides and their plausible mode of action. Biochemistry. 2005;44(28):9775–9784. doi: 10.1021/bi0502386. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature (London) 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Debono M, Gordee RS. Antibiotics that inhibit fungal cell wall development. Annu Rev Microbiol. 1994;48:471–497. doi: 10.1146/annurev.mi.48.100194.002351. [DOI] [PubMed] [Google Scholar]
- 7.Scott MG, Yan H, Hancock RE. Biological properties of structurally related alpha-helical cationic antimicrobial peptides. Infect Immun. 1999;67(4):2005–2009. doi: 10.1128/iai.67.4.2005-2009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rathinakumar R, Wimley WC. Biomolecular engineering by combinatorial design and high-throughput screening: small, soluble peptides that permeabilize membranes. J Am Chem Soc. 2008;130(30):9849–9858. doi: 10.1021/ja8017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shai Y, Oren Z. From “carpet” mechanism to de-novo designed diastereomeric cell-selective antimicrobial peptides. Peptides. 2001;22(10):1629–1641. doi: 10.1016/s0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- 10.Pokorny A, Almeida PF. Kinetics of dye efflux and lipid flip-flop induced by delta-lysin in phosphatidylcholine vesicles and the mechanism of graded release by amphipathic, alpha-helical peptides. Biochemistry. 2004;43(27):8846–8857. doi: 10.1021/bi0497087. [DOI] [PubMed] [Google Scholar]
- 11.Bechinger B. Rationalizing the membrane interactions of cationic amphipathic antimicrobial peptides by their molecular shape. Current Opinionin Colloid & Interface Science. 2009 in press. [Google Scholar]
- 12.White SH, Wimley WC, Selsted ME. Structure, function, and membrane integration of defensins. Cur Opinion Struc Biol. 1995;5(4):521–527. doi: 10.1016/0959-440x(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 13.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: Evidence for the formation of multimeric pores. Protein Sci. 1994;3(9):1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rausch JM, Marks JR, Rathinakumar R, Wimley WC. Beta-sheet pore-forming peptides selected from a rational combinatorial library: mechanism of pore formation in lipid vesicles and activity in biological membranes. Biochemistry. 2007;46(43):12124–12139. doi: 10.1021/bi700978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rausch JM, Marks JR, Wimley WC. Rational combinatorial design of pore-forming beta-sheet peptides. Proc Natl Acad Sci U S A. 2005;102(30):10511–10515. doi: 10.1073/pnas.0502013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth BL, Poot M, Yue ST, Millard PJ. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl Environ Microbiol. 1997;63(6):2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selsted ME, Novotny MJ, Morris WL, Tang YQ, Smith W, Cullor JS. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J Biol Chem. 1992;267:4292–4295. [PubMed] [Google Scholar]
- 18.Ramachandran R, Heuck AP, Tweten RK, Johnson AE. Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nat Struct Biol. 2002;9(11):823–827. doi: 10.1038/nsb855. [DOI] [PubMed] [Google Scholar]
- 19.Shepard LA, Heuck AP, Hamman BD, Rossjohn J, Parker MW, Ryan KR, Johnson AE, Tweten RK. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an alpha-helical to beta-sheet transition identified by fluorescence spectroscopy. Biochemistry. 1998;37(41):14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- 20.Van Abel RJ, Tang YQ, Rao VSV, Dobbs CH, Tran D, Barany G, Selsted ME. Synthesis and characterization of indolicidin, a tryptophan-rich antimicrobial peptide from bovine neutrophils. Int J Pept Protein Res. 1995;45(5):401–409. doi: 10.1111/j.1399-3011.1995.tb01055.x. [DOI] [PubMed] [Google Scholar]
- 21.Hancock RE, Brown KL, Mookherjee N. Host defence peptides from invertebrates--emerging antimicrobial strategies. Immunobiology. 2006;211(4):315–322. doi: 10.1016/j.imbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 22.Yount NY, Bayer AS, Xiong YQ, Yeaman MR. Advances in antimicrobial peptide immunobiology. Biopolymers. 2006;84(5):435–458. doi: 10.1002/bip.20543. [DOI] [PubMed] [Google Scholar]
- 23.Matsuzaki K, Yoneyama S, Fujii N, Miyajima K, Yamada KI, Kirino Y, Anzai K. Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry. 1997;36(32):9799–9806. doi: 10.1021/bi970588v. [DOI] [PubMed] [Google Scholar]
- 24.Lee DL, Powers JP, Pflegerl K, Vasil ML, Hancock RE, Hodges RS. Effects of single D-amino acid substitutions on disruption of beta-sheet structure and hydrophobicity in cyclic 14-residue antimicrobial peptide analogs related to gramicidin S. J Pept Res. 2004;63(2):69–84. doi: 10.1046/j.1399-3011.2003.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Won HS, Jung SJ, Kim HE, Seo MD, Lee BJ. Systematic peptide engineering and structural characterization to search for the shortest antimicrobial peptide analogue of gaegurin 5. J Biol Chem. 2004;279(15):14784–14791. doi: 10.1074/jbc.M309822200. [DOI] [PubMed] [Google Scholar]
- 26.Hilpert K, Volkmer-Engert R, Walter T, Hancock RE. High-throughput generation of small antibacterial peptides with improved activity. Nat Biotechnol. 2005;23(8):1008–1012. doi: 10.1038/nbt1113. [DOI] [PubMed] [Google Scholar]
- 27.Makovitzki A, Avrahami D, Shai Y. Ultrashort antibacterial and antifungal lipopeptides. Proc Natl Acad Sci U S A. 2006;103(43):15997–16002. doi: 10.1073/pnas.0606129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blazyk J, Wiegand R, Klein J, Hammer J, Epand RM, Epand RF, Maloy WL, Kari UP. A novel linear amphipathic beta-sheet cationic antimicrobial peptide with enhanced selectivity for bacterial lipids. J Biol Chem. 2001;276(30):27899–27906. doi: 10.1074/jbc.M102865200. [DOI] [PubMed] [Google Scholar]
- 29.Lee DL, Powers JP, Pflegerl K, Vasil ML, Hancock RE, Hodges RS. Effects of single D-amino acid substitutions on disruption of beta-sheet structure and hydrophobicity in cyclic 14-residue antimicrobial peptide analogs related to gramicidin S. J Pept Res. 2004;63(2):69–84. doi: 10.1046/j.1399-3011.2003.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuzaki K, Nakayama M, Fukui M, Otaka A, Funakoshi S, Fujii N, Bessho K, Miyajima K. Role of disulfide linkages in tachyplesin-lipid interactions. Biochemistry. 1993;32(43):11703–11710. doi: 10.1021/bi00094a029. [DOI] [PubMed] [Google Scholar]
- 31.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66(4):236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 32.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 33.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 34.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nature Struct Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 35.Wimley WC, Hristova K, Ladokhin AS, Silvestro L, Axelsen PH, White SH. Folding of β-sheet membrane proteins: A hydrophobic hexapeptide model. J Mol Biol. 1998;277:1091–1110. doi: 10.1006/jmbi.1998.1640. [DOI] [PubMed] [Google Scholar]
- 36.Pate M, Blazyk J. Methods for assessing the structure and function of cationic antimicrobial peptides. Methods Mol Med. 2008;142:155–173. doi: 10.1007/978-1-59745-246-5_13. [DOI] [PubMed] [Google Scholar]
- 37.Papo N, Shai Y. Effect of drastic sequence alteration and D-amino acid incorporation on the membrane binding behavior of lytic peptides. Biochemistry. 2004;43(21):6393–6403. doi: 10.1021/bi049944h. [DOI] [PubMed] [Google Scholar]
- 38.Yount NY, Yeaman MR. Multidimensional signatures in antimicrobial peptides. Proc Natl Acad Sci. 2004;101(19):7363–7368. doi: 10.1073/pnas.0401567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55(1):27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 40.Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J. 1999;341(Pt 3):501–513. [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory SM, Pokorny A, Almeida PF. Magainin 2 Revisited: A Test of the Quantitative Model for the All-or-None Permeabilization of Phospholipid Vesicles. Biophys J. 2009;96(1):116–131. doi: 10.1016/j.bpj.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pokorny A, Kilelee EM, Wu D, Almeida PF. The activity of the amphipathic peptide delta-lysin correlates with phospholipid acyl chain structure and bilayer elastic properties. Biophys J. 2008;95(10):4748–4755. doi: 10.1529/biophysj.108.138701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory SM, Cavenaugh A, Journigan V, Pokorny A, Almeida PF. A quantitative model for the all-or-none permeabilization of phospholipid vesicles by the antimicrobial peptide cecropin A. Biophys J. 2008;94(5):1667–1680. doi: 10.1529/biophysj.107.118760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rausch JM, Wimley WC. A high-throughput screen for identifying transmembrane pore-forming peptides. Anal Biochem. 2001;293(2):258–263. doi: 10.1006/abio.2001.5137. [DOI] [PubMed] [Google Scholar]