Summary
Autophagy is a cellular catabolic mechanism that plays an essential function in protecting multicellular eukaryotes from neurodegeneration, cancer and other diseases. However, we still know very little about mechanisms regulating autophagy under normal homeostatic conditions when nutrients are not limiting. In a genome-wide human siRNA screen, we demonstrate that under normal nutrient conditions up regulation of autophagy requires the type III PI3 kinase, but not inhibition of mTORC1, the essential negative regulator of starvation-induced autophagy. We show that a group of growth factors and cytokines inhibit the type III PI3 kinase through multiple pathways, including the MAPK-ERK1/2, Stat3, Akt/Foxo3 and CXCR4/GPCR, which are all known to positively regulate cell growth and proliferation. Our study suggests that the type III PI3 kinase integrates diverse signals to regulate cellular levels of autophagy, and that autophagy and cell proliferation may represent two alternative cell fates that are regulated in a mutually exclusive manner.
Introduction
Autophagy is an evolutionarily conserved catabolic process mediating turnover of intracellular constituents in a lysosome-dependent manner (Levine and Klionsky, 2004). In unicellular eukaryotes autophagy serves as a survival mechanism during periods of starvation by promoting intracellular recycling (Levine and Klionsky, 2004; Levine and Kroemer, 2008). In metazoa autophagy functions as an important intracellular catabolic mechanism involved in cellular homeostasis during development and adult life, by mediating the turnover of malfunctioning, aged or damaged proteins and organelles (Levine and Kroemer, 2008). Autophagy can also be activated in response to many forms of cellular stress beyond nutrient starvation, including DNA damage, ER stress, ROS and upon invasion by intracellular pathogens. On the organismal level, autophagy has been shown to participate in both innate and acquired immunity (Schmid and Munz, 2007), tumor suppression (Liang et al., 1999; Mathew et al., 2007a; Mathew et al., 2007b) and protection from neurodegeneration (Hara et al., 2006; Komatsu et al., 2006).
The signaling mechanisms leading to the activation of autophagy under nutrient starvation conditions have been extensively characterized. Inactivation of the mTORC1 kinase, a downstream effector of the type I PI3 kinase/Akt signaling, is critical for the activation of autophagy under these conditions (Levine and Klionsky, 2004; Levine and Kroemer, 2008). However, cells in complex multicellular eukaryotes such as mammals rarely experience nutrient deprivation under physiological conditions. Nevertheless, autophagy plays an essential role in maintenance of normal homeostasis at both cellular and organismal levels, as well as can be induced by a variety of cellular stresses under conditions when mTORC1 is known or expected to be active (Sakaki et al., 2008). Thus, there is an urgent need to understand the mechanisms that regulate autophagy under normal nutritional conditions.
In order to address this question and to understand the global regulation of mammalian autophagy, we performed a genome-wide image-based siRNA screen for genes involved in the regulation of autophagy under normal nutrient conditions. Additionally we developed and executed a series of high-throughput characterization assays and screens allowing us to characterize the hit genes and further the understanding of the global regulation of mammalian autophagy. Our data indicate that under normal nutrition autophagy is regulated by a wide array of extracellular factors, including growth factors, cytokines and chemokines. This response is mediated by a variety of cell surface receptor signalling pathways and, unlike during starvation, can be regulated in mTORC1 independent manner.
Results
Genome-wide siRNA screen for genes regulating autophagy
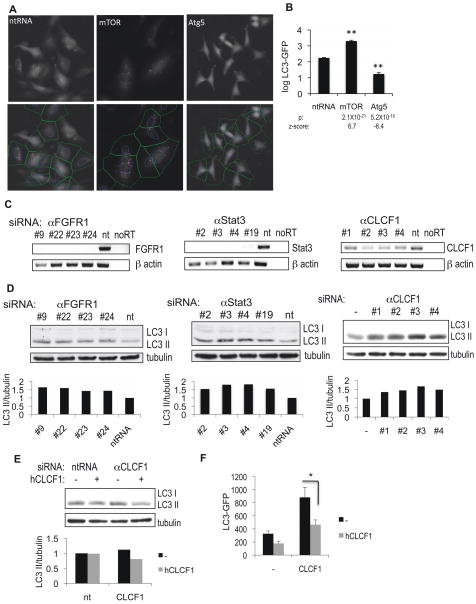
In order to identify new genes involved in the regulation of autophagy in mammals we screened a human genomic library containing siRNA pools targeting 21,121 genes, with 4 independent siRNA oligonucleotides for each gene. To quantify levels of autophagy, we used human neuroblastoma H4 cells stably expressing the LC3-GFP reporter (Shibata et al., 2006). In this system, transfection of siRNA targeting the essential autophagy mediator ATG5 led to significant down-regulation of autophagy, as assessed by a reduction in the number and intensity of LC3-GFP positive autophagosomes (Figure 1A–B) as well as a decrease in endogenous LC3II levels on a western blot (Supplemental Figure S1A) (Klionsky et al., 2008). Conversely, transfection of siRNA targeting subunits of mTORC1, mTOR or Raptor, led to increased levels of autophagy (Figure 1A–B, Figure S1B–C). The primary screen resulted in the identification of 574 genes (2.7%) which knock-down led to a significant (p<0.02) decrease or increase in LC3-GFP positive autophagosome formation. The candidate genes were then confirmed in a secondary screen in which the 4 pool siRNAs targeting different regions of the same gene, were evaluated separately. 236 of the candidate genes (41%) were confirmed with at least 2 independent siRNA oligonucleotides resulting in significant increase or decrease in the levels of autophagy as compared to non-targeting siRNA controls (Figure 2A, Table 1, Supplemental Table S1, p<0.01). The hit genes included 21 (9%) genes known or implicated in the regulation of autophagy, including ATG5, ATG7, IGF1, Rab7 and calpain-1 (Table S2). The ability to identify these known autophagy genes provides an important validation of our screening method.
Figure 1.
High-throughput image-based screen for genes regulating autophagy. A, H4 cells stably expressing LC3-GFP were transfected with non-targeting siRNA (ntRNA) or siRNA against mTOR or Atg5 for 72h, fixed, counterstained with Hoechst and imaged on a high-throughput fluorescent microscope (10×). The bottom panels demonstrate results of image segmentation used for quantification of the ratio of soluble (diffuse cytosolic) versus autophagosome-associated (punctate) LC3-GFP: blue – nuclei, green – cell segmentation, pink – autophagosomal LC3-GFP. B, Quantification of data from (A). C, Confirmation of knock-down of selected screen hits by RT-PCR. H4 cells were transfected with indicated siRNAs for 72h. D, Confirmation of changes in levels of autophagy following knock-down of FGFR1, Stat3 or CLCF1 by western blot with antibodies against LC3. 10 μg/mL E64d was added to the media for 8 hours before cells were harvested. Quantification of LC3 II/tubulin ratio is shown. E–F, Exogenous CLCF1 can suppress increase in autophagy induced by knockdown of CLCF1. H4 (E) or H4 LC3-GFP (F) cells were transfected with non-targeting siRNA (nt) or siRNA against CLCF1 and grown in the absence or presence of 100 ng/mL human CLCF1. Levels of autophagy were assessed by western blot (E) or by quantification of autophagosomal LC3-GFP (F). All error bars are s.e.m. * p=0.05 based on two-tailed student t-test. n≥10
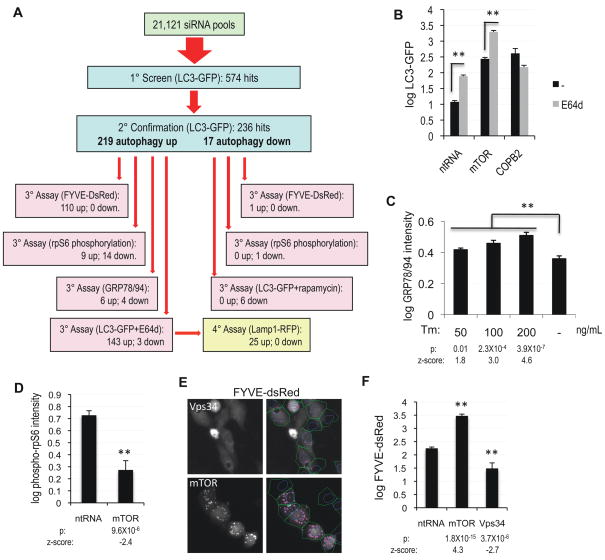
Figure 2.
High-throughput characterization of the autophagy screen hits. A, Summary of all screen and characterization assays and their results. B, Quantification of levels of autophagy in H3 LC3-GFP cells transfected with non-targeting siRNA or siRNA against mTOR or COPB2. 10 μg/mL lysosomal protease inhibitor E64d was added for 8h before fixation. p(mTOR)=6.5×10−17 C, Quantification of ER stress in H4 cells treated with indicated levels of tumicamycin (Tm) for 24h by in-cell-western assay with antibody against GRP78 and GRP94. D, Quantification of mTORC1 activity by in-cell-western assay with antibody against phospho-rpS6 (Ser235/236) in H4 cells transfected with non-targeting siRNA or siRNA against mTOR for 72h. E, To assess the type III PI3 kinase activity, H4 cells stably expressing the PtdIns3P reporter FYVE-dsRed were transfected with siRNA against mTOR or Vps34. Right-hand panels represent the results of image segmentation used for quantification of the ratio of soluble (diffuse cytosolic) versus vesicle-associated (punctate) FYVE-dsRed: blue – nuclei, green – cell segmentation, pink – vesicular FYVE-dsRed. F, Quantification of data from panel (E). All error bars are s.e.m. n≥6
Table 1.
Genes regulating and mediating autophagy identified in the screen.
| Genes which knock-down increases autophagy flux | |||||||
|---|---|---|---|---|---|---|---|
| Gene symbol | Gene ID | Gene symbol | Gene ID | Gene symbol | Gene ID | Gene symbol | Gene ID |
| ADMR | 11318 | FANCC | 2176 | MMP17 | 4326 | SCOTIN | 51246 |
| ADRA1A | 148 | FBXL20 | 84961 | MUC3A | 4584 | SDHB | 6390 |
| AGER | 177 | FGFR1 | 2260 | MYL3 | 4634 | SEMA4B | 10509 |
| ARSE | 415 | FRAG1 | 27315 | NAGK | 55577 | SETDB1 | 9869 |
| ATG16L2 | 89849 | GABBR2 | 9568 | NCR3 | 259197 | SF3A2 | 8175 |
| AURKA | 6790 | GHSR | 2693 | NFIL3 | 4783 | SIDT1 | 54847 |
| BAI3 | 577 | GJA4 | 2701 | NNMT | 4837 | SIX2 | 10736 |
| BAIAP2 | 10458 | GNAI1 | 2770 | NPTX1 | 4884 | SLC25A19 | 60386 |
| C18orf8 | 29919 | GNG11 | 2791 | NUDT1 | 4521 | SMARCD1 | 6602 |
| CAMKV | 79012 | GNG5 | 2787 | OGDH | 4967 | SOD1 | 6647 |
| CAPN1 | 823 | GNRH2 | 2797 | P2RX1 | 5023 | SORCS2 | 57537 |
| CARKL | 23729 | GPX2 | 2877 | PA2G4 | 5036 | SP140 | 11262 |
| CASP1 | 834 | GRK6 | 2870 | PAK6 | 56924 | STAT3 | 6774 |
| CDCA8 | 55143 | GTF2IRD2 | 84163 | PAQR6 | 79957 | TACR2 | 6865 |
| CDK5RAP3 | 80279 | GTPBP4 | 23560 | PCGF1 | 84759 | TGFBI | 7045 |
| CDKN2D | 1032 | HIST2H3C | 126961 | PDCD5 | 9141 | THBS2 | 7058 |
| CEND1 | 51286 | HIVEP2 | 3097 | PFDN2 | 5202 | TINP1 | 10412 |
| CENPJ | 55835 | HLA-DRB1 | 3123 | PFKL | 5211 | TLR3 | 7098 |
| CHAF1B | 8208 | HMGCL | 3155 | PHB2 | 11331 | TMPRSS5 | 80975 |
| CHID1 | 66005 | HRC | 3270 | PIGY | 84992 | TNF | 7124 |
| CHRND | 1144 | HSFY2 | 159119 | PLDN | 26258 | TNFRSF17 | 608 |
| CLCF1 | 23529 | ICT1 | 3396 | PLXNA2 | 5362 | TRIM69 | 140691 |
| CNKSR2 | 22866 | IGF1 | 3479 | POLR3G | 10622 | TRNT1 | 51095 |
| COL14A1 | 7373 | IRAK3 | 11213 | PPFIA4 | 8497 | TRPA1 | 8989 |
| COPE | 11316 | ITGAV | 3685 | PRAF2 | 11230 | TSPAN4 | 7106 |
| COX5A | 9377 | KIAA0133 | 9816 | PRKAG3 | 53632 | TUBGCP6 | 85378 |
| CPNE6 | 9362 | KIAA0196 | 9897 | PRKCZ | 5590 | UBE1L2 | 55236 |
| CXCL12 | 6387 | KRT18 | 3875 | PROSC | 11212 | UBE2D1 | 7321 |
| CYP27A1 | 1593 | LIF | 3976 | PSD | 5662 | UNC13B | 10497 |
| DBX1 | 120237 | LOC285643 | 285643 | PTGER2 | 5732 | USP19 | 10869 |
| DDX24 | 57062 | LOC388959 | 388959 | PTMA | 5757 | USP24 | 23358 |
| EP300 | 2033 | MAP3K7IP1 | 10454 | PTPRH | 5794 | WASF1 | 8936 |
| EPHA6 | 203806 | MATN3 | 4148 | PTPRU | 10076 | WFDC2 | 10406 |
| EVL | 51466 | MCCC1 | 56922 | RBBP8 | 5932 | XPO1 | 7514 |
| F12 | 2161 | MMACHC | 25974 | RFX1 | 5989 | ZFY | 7544 |
| FABP1 | 2168 | MMP10 | 4319 | SCAMP4 | 113178 | ||
| Genes which knock-down increases autophagy due to decreased lysosomal processing | |||||||
| ADM | 133 | DOCK8 | 81704 | NTN2L | 4917 | SORBS3 | 10174 |
| AMH | 268 | EGLN2 | 112398 | NUPR1 | 26471 | SSPN | 8082 |
| ARCN1 | 372 | ERH | 2079 | NUTF2 | 10204 | SUV39H1 | 6839 |
| ASMT | 438 | FCER1A | 2205 | PLAGL2 | 5326 | TAF2 | 6873 |
| BGN | 633 | FXYD2 | 486 | PLCH2 | 9651 | TH | 7054 |
| C11orf68 | 83638 | GJA3 | 2700 | PNKD | 25953 | TNFRSF19L | 84957 |
| C2orf13 | 200558 | HMBS | 3145 | PPARD | 5467 | TOM1 | 10043 |
| C5AR1 | 728 | HNRPK | 3190 | PRKAA2 | 5563 | TRAF1 | 7185 |
| CCT4 | 10575 | HNRPM | 4670 | PRKCA | 5578 | TRHR | 7201 |
| CD300C | 10871 | HNRPU | 3192 | PTCRA | 171558 | TRIM8 | 81603 |
| CD79A | 973 | HOXD11 | 3237 | QSCN6 | 5768 | TRIP6 | 7205 |
| CENPE | 1062 | IHPK3 | 117283 | RAB7A | 7879 | TUBB2A | 7280 |
| CHKA | 1119 | INTS4 | 92105 | RAGE | 5891 | USP54 | 159195 |
| CKAP5 | 9793 | MKLN1 | 4289 | RANGAP1 | 5905 | UTS2R | 2837 |
| CLCN1 | 1180 | NAT9 | 26151 | RELN | 5649 | ZBTB16 | 7704 |
| COPB2 | 9276 | NLK | 51701 | RORC | 6097 | ||
| Genes which knock-down increases autophagy (flux not determined) | |||||||
| BPGM | 669 | KIF11 | 3832 | NPTN | 27020 | TRPM3 | 80036 |
| FGF2 | 2247 | LOR | 4014 | RIPK1 | 8737 | ||
| GCM2 | 9247 | OA48-18 | 10414 | RNPEPL1 | 57140 | ||
| GFER | 2671 | NOXO1 | 124056 | TACC2 | 10579 | ||
| Genes which knock-down decreases autophagy | |||||||
| ATG5 | 9474 | KIF5C | 3800 | PDCL | 5082 | TCEB3 | 6924 |
| ATG7 | 10533 | LOC285647 | 285647 | PGGT1B | 5229 | TPR | 7175 |
| CATSPER4 | 378807 | MBP | 4155 | RELA | 5970 | ||
| GAB1 | 2549 | MEGF10 | 84466 | SMYD3 | 64754 | ||
| GPR18 | 2841 | NFKB1 | 4790 | STIM1 | 6786 | ||
As further validation of our hits, we confirmed knock-down of a selected group by RT-PCR and verified their ability to induce autophagy by western blot (Figure 1C–D). Importantly, stimulation of autophagy by knock-down of the cardiotrophin-like cytokine factor 1 (CLCF1), one of the cytokines identified in the screen, was suppressed in the presence of exogenous human CLCF1, demonstrating specificity of the observed phenotype (Figure 1E–F).
Although our screen was conducted under normal nutritional conditions when levels of autophagy are low, we were able to uncover a small set of hits (17. 7% of all confirmed genes) that resulted in the suppression of autophagy. Consistent with the function of these genes in mediation of autophagy, knock-down of a large fraction (35%) of these genes was able to down-regulate autophagy in the presence of rapamycin, a potent mTORC1 inhibitor and well-known inducer of autophagy (Figure 2A, Table S3) (Klionsky et al., 2008; Levine and Kroemer, 2008).
High-throughput characterization of the hit genes
Since knock-down of the vast majority of hits (219, 93% of all confirmed) led to the induction of autophagy, our subsequent analyses concentrated on this group of genes. To elucidate the molecular pathways involved in regulation of autophagy by the candidate genes, we developed and performed a series of additional high-throughput assays and screens to characterize the hits (Figure 2A). Accumulation of autophagosomal LC3-GFP can be due to either increased initiation of autophagy or to a block in degradation of autophagosomes. In order to distinguish between these possibilities, we evaluated induction of autophagy in the presence of the inhibitor of lysosomal proteases E64d. As expected, treatment with E64d led to a significant increase in levels of autophagy following knock-down of mTOR, but did not affect levels of autophagy induced by knock-down of the coatamer subunit COPB2, which is predicted to inhibit general vesicular trafficking (Figure 2B). Knock-down of 143 genes (69% of 206 tested) led to significantly higher induction of autophagy in the presence than in the absence of E64d (Table 1, Table S1), suggesting that similarly to starvation or inhibition of mTORC1, loss of these genes results in enhanced initiation of autophagy. On the other hand, the 63 (31%) genes which knockdown was unable to induce autophagy in the presence of E64d, likely cause the accumulation of autophagosomal LC3-GFP by blocking maturation and/or degradation of autophagosomes. For 24 of those genes this may be due to a general defect in lysosomal proteolysis as induction of autophagy upon loss of these genes was accompanied by significant expansion of the lysosomal compartment, as assessed by the accumulation of lysosomal reporter protein Lamp1-RFP (Figure S2A–B, Table S4).
Autophagy is often induced in response to various forms of cellular stress, including ER stress (Ogata et al., 2006; Yorimitsu et al., 2006). In our screen we observed that the accumulation of LC3-GFP was sometimes associated with decreased cell viability (Table S1), raising the question as to whether these cases may reflect a general response to cellular stress rather than a specific function in the regulation of autophagy. To assess that, we performed an in-cell-western assay to measure the expression levels of GRP78 and GRP94, specific markers of ER stress. As expected, treatment with the inducer of ER stress, tunicamycin, led to a dose-dependent up-regulation of GRP78 and GRP94 (Figure 2C) as well as an increase in autophagy (data not shown). However, with just a few exceptions (6 out of 188 genes tested, 3%, Table S5), we were unable to detect significant up-regulation of ER stress following knockdown of the hit genes. Therefore, ER stress does not appear to be a major contributor to the induction of autophagy observed in our screen.
We evaluated the contribution of mTORC1, an essential suppressor of starvation-induced autophagy (Levine and Klionsky, 2004), by an in-cell-western assay of the phosphorylation status of a downstream target of mTORC1 signaling, the ribosomal protein S6 (rpS6) (Hoffman et al., 2010). As expected, transfection of siRNA against mTOR led to a significant decrease in the levels of rpS6 phosphorylation as compared to non-targeting siRNA (Figure 2D). Surprisingly, for only 14 (6%) out of the 219 confirmed genes induction of autophagy was strongly correlated with down-regulation of mTORC1 activity (Figure 2A, Table S1). Importantly, out of the 143 genes, which similarly to starvation were able to enhance initiation of autophagy based on the E64d data, only 7 (5%) led to the suppression of mTORC1 activity, suggesting that only this small subset may be involved in the regulation of mTORC1. Conversely, knock down of 4 out of these genes (3%) led to both up-regulation of autophagy and of mTORC1 activity (Table S1). Therefore, high mTORC1 activity may not be always incompatible with the induction of autophagy. The fact that knock down of the vast majority of the screen hits did not affect mTORC1 activity suggests that these genes act either downstream or independently of this kinase. Since our hits include a large group of known upstream regulators of cellular signaling such as cytokines, growth factors and their receptors (see bioinformatics analysis below), our data implicate the existence of mTORC1 independent pathways regulating initiation of autophagy under normal nutritional conditions.
To characterize the contribution of the type III PI3 kinase, an important mediator of autophagy which regulation remains poorly understood (Levine and Klionsky, 2004), we used H4 cells stably expressing FYVE-dsRed reporter, which specifically binds to its product, PtdIns3P. Confirming the major role of the type III PI3 kinase in regulation of PtdIns3P levels in our system, transfection of siRNA against the catalytic component of this kinase, Vps34, led to a significant decline in FYVE-dsRed vesicle recruitment (Figure 2E–F). The relationship between mTORC1 and the type III PI3 kinase remains poorly understood. Although in some contexts Vps34 can play a positive role in the mediation of mTORC1 activation in response to amino acid stimulation (Byfield et al., 2005; Nobukuni et al., 2005), we did not observe significant down-regulation of mTORC1 activity following knock-down of Vps34 (Figure S2C–D). On the other hand, Vps34 is known to function downstream of TORC1 during induction of autophagy in Drosophila (Juhasz et al., 2008), and inhibition of mTORC1 due to either starvation or rapamycin treatment of mammalian cells can lead to accumulation of vesicle associated PtdIns3P (Takahashi et al., 2007; Tassa et al., 2003; Zhang et al., 2007). Consistent with the data placing Vps34 downstream of mTORC1 in the regulation of autophagy, in our system knock-down of either mTOR or Raptor increased FYVE-dsRed vesicle recruitment (Figure 2E–F, Figure S2E). Using this system we determined that knockdown of 110 (47%) out of the 236 confirmed genes led to a significant (p<0.05) alteration in PtdIns3P levels, which positively correlated with the change in autophagosome formation (Table S1), suggesting that these genes act upstream of the type III PI3 kinase in the regulation of autophagy. These data confirm that the type III PI3 kinase plays an essential function in the regulation of autophagy under normal nutritional conditions and provide new insights into the mechanisms that regulate cellular levels of PtdIns3P.
Extracellular factors regulate autophagy under normal nutrition
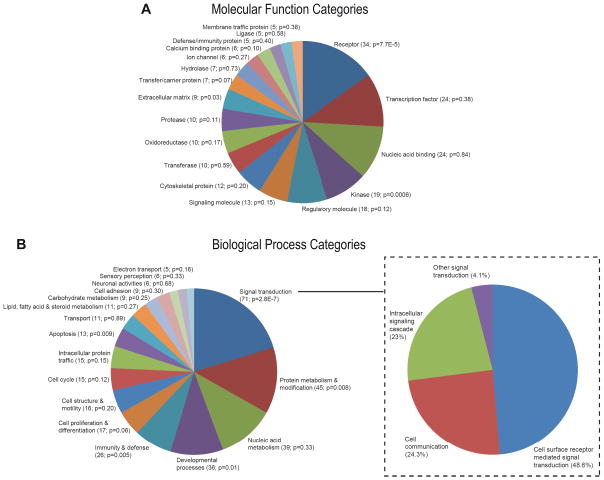
To gain further insight into the mechanisms by which the identified genes regulate cellular levels of autophagy, we used bioinformatics analysis. Molecular function analysis of the 236 confirmed hits using Gene Ontology (GO) revealed a highly significant enrichment in kinases (p=0.0006), proteins with receptor activity (p=7.7×10−5) and extracellular matrix proteins (p=0.03) (Figure 3A, Table S6). The latter categories suggest that extracellular environment including the presence of growth factors, hormones and cytokines, as well as ECM and cell-cell contacts, may play especially important function in the regulation of autophagy under normal nutritional conditions. The results of GO biological process analysis also demonstrated significant enrichment in signaling molecules (p=2.8×10−7) (Figure 3B, Table S7). In agreement with the function of extracellular clues in regulation of autophagy, further subdivision of these molecules revealed that the largest sub-group (49%) is involved in cell surface receptor signal transduction.
Figure 3.
Autophagy is regulated in response to extracellular stimuli through multiple receptor mediated signaling pathways. A–B, Classification of the autophagy screen hits into molecular function (A) and biological process (B) categories according to the PANTHER system. Panel B includes sub-divisions of the biological process ‘Signal transduction’ category. Categories with at least 5 genes are displayed. Categories with p<0.05 (hyper geometric distribution) are considered enriched.
Cytokines regulate autophagy independently of mTORC1
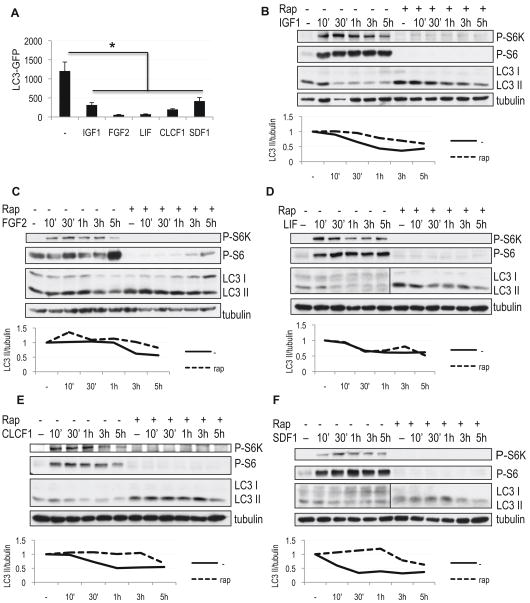
As an siRNA independent confirmation of the function of extracellular molecules in the regulation of autophagy, we treated cells with several of the cytokines and growth factors identified as hits in our screen. Based on the results of the characterization assays, knock-down of IGF1, FGF2, LIF, CLCF1 and the chemokine SDF1 (CXCL12) resulted in mTORC1 independent increase in initiation of autophagy (Table S1). In agreement, treatment of H4 LC3-GFP cells grown in a serum-free medium with any of these cytokines led to a significant down-regulation of autophagy as measured by LC3-GFP translocation (Figure 4A, Figure S3A). This data was furthermore confirmed in multiple cell lines (H4, HEK293, HeLa and MCF7) by western blot (Figure 4B–F, Figure S3B–L and data not shown). In agreement with the proposed function of cytokines in the regulation of autophagy, cells cultured in their absence displayed high basal levels of autophagy as assessed by accumulation of LC3II, which was partially suppressed by the addition of even single cytokines identified in the screen. Thus, we have identified a group of cytokines and growth factors that are both necessary and sufficient for the regulation of autophagy.
Figure 4.
Cytokines identified in the screen suppress autophagy independently of mTORC1 activity. A, Quantification of autophagy in H4 LC3-GFP cells grown in serum-free medium supplemented with indicated cytokines for 24h. All error bars are s.e.m. * p<0.05 n≥8 B–F, Cytokines are able to suppress autophagy in the absence and presence of rapamycin. H4 cells were grown in serum-free medium, followed by addition of 100 ng/mL IGF1 (B), 50 ng/mL FGF2 (C), 50 ng/mL LIF (D) or 50 ng/mL CLCF1 (F) and 10 μg/mL E64d. Where indicated, cells were pre-treated with 50 nM rapamycin 1 hour prior to the addition of cytokines. Levels of autophagy were assessed by western blot using antibody against LC3; mTORC1 activity was evaluated with antibodies against phospho-S6 (Ser235/236, P-S6) and phospho-S6 kinase (Thr389, P-S6K). Quantification of LC3 II/tubulin ratio is shown.
To determine whether down-regulation of autophagy by these cytokines is due to re-activation of mTORC1 signaling, we inhibited mTORC1 by addition of rapamycin. As expected, treatment with rapamycin led to increases in the overall levels of autophagy (Figure 4). Despite efficient inhibition of mTORC1 as evidenced by dephosphorylation of both rpS6 and the S6 kinase, treatment with IGF1, FGF2, LIF, CLCF1 or SDF1 was able to reduce autophagy induced by rapamycin (Figure 4B–F and Figure S4A). Similar results were obtained using an alternative mTORC1 inhibitor, Torin 1 (Figure S4B) (Thoreen et al., 2009). Additionally, siRNA mediated knock-down of mTOR also failed to prevent down-regulation of autophagy by any of the cytokines (Figure S4C–H). Therefore, cytokines and growth factors identified in the screen are able to regulate autophagy via mTORC1 dependent as well as mTORC1 independent pathways. Additionally, only partial suppression of autophagy in the presence of rapamycin suggests that the effects of the Akt/mTORC1 and the cytokine-mediated mTORC1 independent pathways on the levels of autophagy may be additive, rather than mutually exclusive.
Growth-promoting pathways negatively regulate autophagy
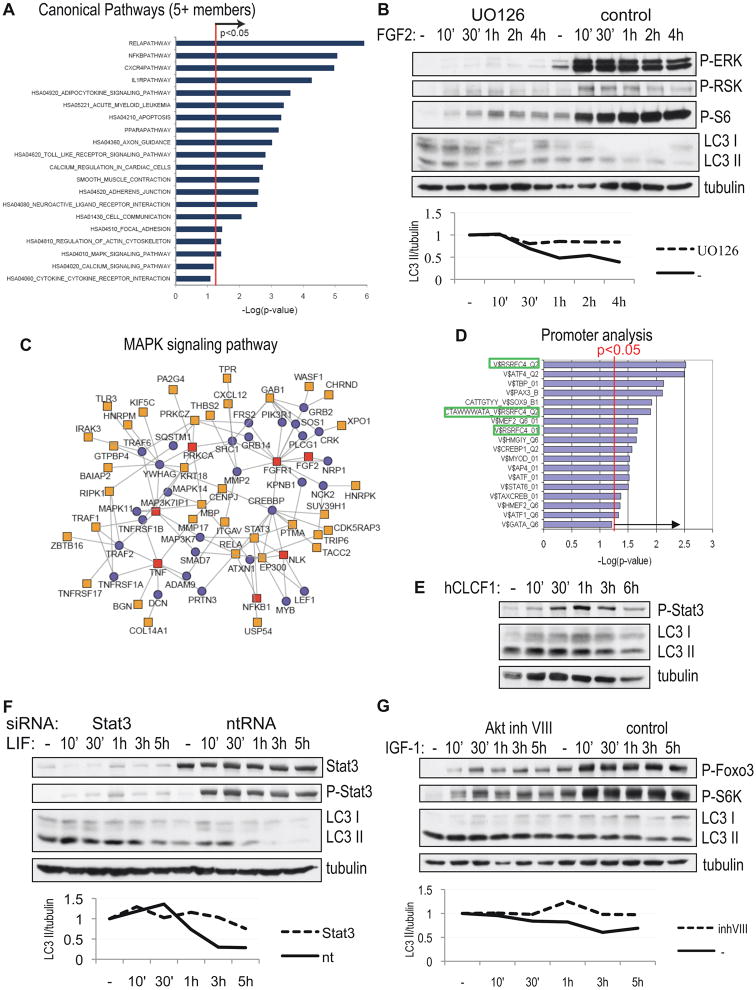
Bioinformatics analysis of the autophagy screen hits indicated significant enrichment for several canonical pathways known to mediate signaling from cell surface receptors (Figure 5A, Table S8). These pathways included the MAPK (p=0.039), Stat3 (p=0.008) and CXCR4 (p=1.1×10−5) pathways regulated by the cytokines identified in the screen. FGF2 is known to activate the MAPK pathway. Indeed, we observed an increased level of phospho-ERK1/2 and phospho-RSK following treatment with FGF2 (Figure 5B). Confirming the essential function of the MAPK pathway, pre-treatment with UO126, an inhibitor of MEK, attenuated inhibition of autophagy following addition of FGF2 (Figure 5B). Interestingly, two of the genes identified in the screen, vinnexin (SORBS3) and tyrosine hydrolase (TH) (Table S1) are known ERK1/2 phosphorylation targets, suggesting that they may play a function in regulation of autophagy downstream of the MAPK pathway. Further confirming the important function of this pathway in the regulation of autophagy, we were able to construct a protein-protein interaction networks anchored by the hit genes belonging to the MAPK pathway (Figure 5C), which encompasses 42 (18%) of all the hit genes and directly connects to the known autophagy machinery through the interaction of the RIP kinase 1 (RIPK1) and PKCζ (PRKCZ) with p62/sequestrosome (SQSTM1). Additionally, analysis of the promoter regions of all the hit genes revealed significant enrichment in consensus sites for several transcription factors (Figure 5D), including 3 enriched sites for RSRFC4, a member of the serum response factor (SRF) family and a known downstream target of MAPK signaling (Pollock and Treisman, 1991), suggesting additional involvement of transcriptional regulation by the MAPK pathway in control of autophagy under normal growth conditions.
Figure 5.
Growth signaling pathways negatively regulate autophagy in response to cytokines. A, Enrichment analysis of canonical pathways (MSigDB) among the hit genes relative to all genes examined in the screen. A p-value<0.05 (hyper geometric distribution) is considered significant. Only categories with at least five genes are displayed. B, Down-regulation of autophagy by 50 ng/mL FGF2 is prevented by addition of MEK inhibitor UO126. H4 cells were grown in serum-free media, levels of autophagy were assessed in the presence of 10 μg/mL E64d, with antibodies against LC3, inhibition MEK with phospho-ERK 1/2, phospho-RSK and phospho-S6 (Ser235/236). Quantification of LC3 II/tubulin ratio is shown. C, Network extensions of the canonical MAPK pathway. Using human interactome data, this pathway-centric network was constructed by anchoring on canonical pathway components and extended by establishing connections with other hit genes, including at most one intervening component. Red squares - screen hits that are part of the MAPK pathway, yellow squares – other screen hits, blue circles – intervening proteins. D, Enrichment analysis of cis-regulatory elements/transcription factor (TF)-binding sites in the promoters of the hit genes, using motif-based gene sets from MSigDB and TF-binding sites defined in the TRANSFAC database. SRF sites are highlighted. E, Phosphorylation of Stat3 following treatment with 50 ng/mL CLCF1. F, Down-regulation of autophagy by 50 ng/mL LIF is prevented by siRNA mediated knock-down of Stat3. H4 cells were transfected with indicated siRNAs for 72h, than cells were treated as in (B). Protein levels and phosphorylation of Stat3 are shown. G, Suppression of autophagy by 100 ng/mL IGF1 is prevented by Akt inhibitor VIII. Cells were treated as in (B); Akt activity was assessed with antibodies against phospho-Foxo3a and phospho-rpS6.
Another hit gene pulled out of the screen as a negative regulator of autophagy was the transcription factor Stat3 (Table S1), a known mediator of LIF and CLCF1 signaling. Indeed, treatment with either LIF or CLCF1 increased activating phosphorylation of Stat3 (Figure 5E–F). Consistent with the essential function of Stat3, its siRNA mediated knock-down attenuated down-regulation of autophagy in response to LIF (Figure 5F). Therefore, LIF and CLCF1 regulate autophagy through the Stat3 pathway. Analysis of the promoters of the core autophagy genes uncovered the presence of a consensus Stat3 site in the promoter of human Atg3 gene (V$STAT3_02, p=0.041), raising the possibility that Stat3 may also contribute to the transcriptional regulation of the core autophagy machinery.
In addition to activating mTORC1, Akt is known to directly phosphorylate and inhibit Foxo3a, a transcription factor previously shown to positively regulate autophagy during muscle degeneration (Zhao et al., 2007). Indeed, phosphorylation of both Akt and Foxo3a was increased following IGF-1 treatment in both the absence and presence of rapamycin (Figure 5G, Figure S3B and data not shown). Inhibition of Akt by treatment with Akt inhibitor VIII attenuated phosphorylation of both Foxo3a and the mTORC1 target S6 kinase, as well as prevented inhibition of autophagy by IGF1 (Figure 5G). Therefore, under normal nutrient conditions IGF-1 regulates autophagy in a type I PI3 kinase/Akt dependent manner, likely through both the mTORC1 and Foxo3a pathways.
Cytokine signaling converges on the type III PI3 kinase
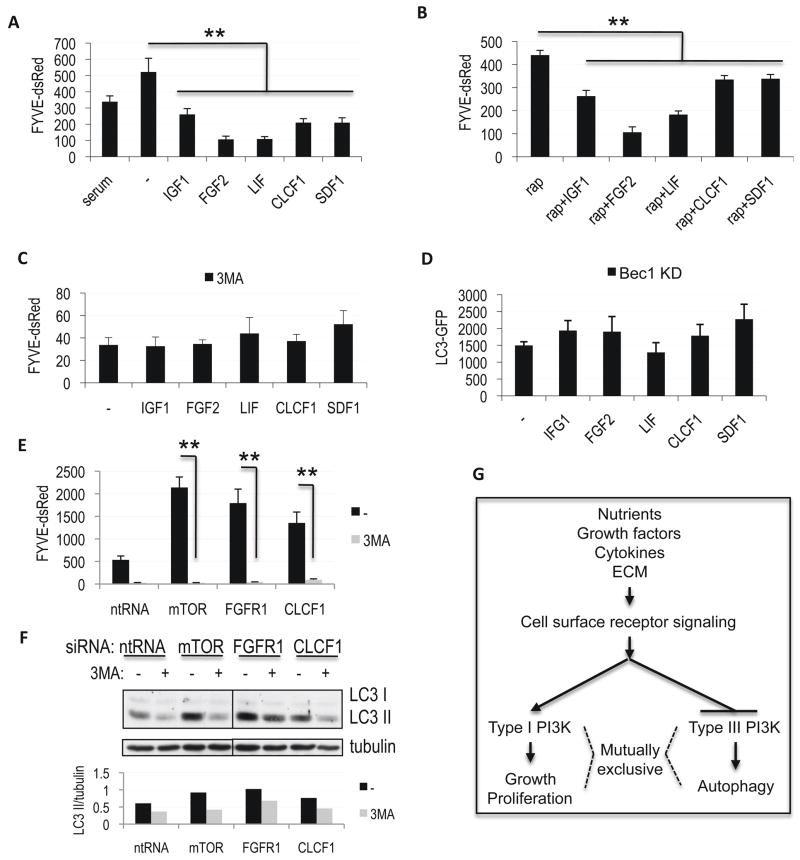
To further characterize the mechanism by which the cytokine dependent signaling pathways regulate autophagy, we treated FYVE-dsRed expressing H4 cells grown in serum-free media with IGF1, FGF2, LIF, CLCF1 or SDF1. All of the cytokines inhibited vesicular recruitment of FYVE-dsRed (Figure 6A, Figure S5A). Consistent with their ability to suppress autophagy in an mTORC1 independent manner, treatment with cytokines was able to suppress FYVE-dsRed in the presence of rapamycin (Figure 6B). This suggests that irrespective of the upstream signaling pathway, cytokines identified in the screen may regulate autophagy by modulating the levels of PtdIns3P. As treatment with 3MA, an inhibitor of the type III PI3 kinase, prevented further down-regulation of PtdIns3P following treatment with cytokines (Figure 6C), this is dependent on the function of the type 3 PI3 kinase rather than other cellular sources of PtdIns3P. Confirming a crucial role of the type III PI3 kinase in regulation of autophagy downstream of cytokine signaling, treatment with any of the five cytokines was unable to further decrease levels of autophagy in H4 LC3-GFP cells deficient for Beclin 1 (Figure 6D, Figure S5B), the regulatory subunit of this kinase (Shibata et al., 2006).
Figure 6.
Regulation of autophagy by cytokines is dependent on the type III PI3 kinase. A–B, Quantification of PtdIns3P levels in H4 FYVE-dsRed cells grown in serum-free medium supplemented for 24h with 100 ng/mL IGF1, 50 ng/mL FGF2, 50 ng/mL LIF, 50 ng/mL CLCF1 or 50 ng/mL SDF1, in the absence (A) or presence (B) of 50 nM rapamycin. C, Quantification of PtdIns3P levels in H4 FYVE-dsRed cells treated with indicated cytokines in the presence of the type III PI3 kinase inhibitor 3MA (10 mM). D, Quantification of levels of autophagy following cytokine treatment of Beclin 1 knock-down H4 LC3-GFP cells. E–F, Induction of PtdIns3P levels in H4 FYVE-dsRed cells (E) and up-regulation of autophagy in H4 cells (F) following siRNA mediated knock-down of indicated genes is attenuated in the presence of 3MA. Cells were transfected with indicated siRNAs for 72h, 10 mM 3MA was added for 8h before cells were processed for analysis. For western blots 10 μg/mL E64d was added and quantification of LC3 II/tubulin ratio is shown. **p<0.01, n≥6 All error bars are s.e.m. G, Model for opposing regulation of cell growth and autophagy by the type I and type III PI3 kinases in response to changes in extracellular environment.
In agreement with the cytokine data, knock-down of IGF1 or CLCF1 increased vesicular accumulation of FYVE-dsRed (Table S1). Additionally, we confirmed that inhibition of the Akt/TORC1, MAPK and Stat3 pathways by knock down of their components (mTOR, FGFR1 and Stat3/CLCF1, respectively) induced accumulation of PtdIns3P as assessed by increased levels of vesicular FYVE-dsRed (Figure 6E). Both, the accumulation of FYVE-dsRed and induction of autophagy by knock-down of mTOR, FGFR1 or CLCF1 was significantly attenuated following treatment with 3MA (Figure 6E–F) and in Beclin 1 and/or Vps34 knock-down cells (Figure S5C–E), confirming the critical involvement of the type III PI3 kinase in the regulation of autophagy downstream of the Akt/mTOR, MAPK and Stat3 pathways.
Discussion
We developed and executed an innovative high-throughput image-based screening and analysis approach, which allowed us to identify and characterize a group of 236 genes involved in the regulation of basal levels of autophagy under normal nutritional conditions. Further analysis revealed that the screen hits are highly enriched for genes involved in receiving and mediating extracellular signalling. Our data demonstrate that in higher eukaryotes autophagy is regulated in a more versatile manner consistent with its expanded function in the maintenance of tissue and cell homeostasis and protection of organisms from cancer, neurodegeneration and other diseases. The nutritional cues important for regulating autophagy in unicellular eukaryotes have been expanded to include an extensive array of environmental factors acting via multiple receptor-mediated signaling pathways, acting in an additive manner. Unlike starvation, the majority of these pathways are able to regulate autophagy in mTORC1 independent manner, instead converging on the type III PI3 kinase (see Graphical Abstract). Therefore, in addition to playing a necessary function during execution of autophagy, the type III PI3 kinase serves to integrate signals from many pathways to allow flexible adjustment of cellular levels of autophagy in response to diverse extracellular clues.
Interestingly, all of these pathways, including Akt/mTORC1, Akt/Foxo3, MAPK, Stat3 and CXCR4, in addition to negatively regulating autophagy as we show here, are well known to positively regulate cell growth and proliferation. Therefore, while autophagy is operating constantly at low levels under physiological conditions, induction of autophagy and cell growth may represent two alternative cell fates, which are regulated in a mutually exclusive manner (Figure 6G). This suggests that reducing levels of autophagy under normal nutritional condition may be advantageous in actively proliferating cells, including tumors. Conversely, high levels of autophagy may be incompatible with cell growth and proliferation, and their reduction could stimulate cell proliferation. Consistent with this hypothesis, mutations in at least 6 of the autophagy genes identified in the screen are causally implicated in human cancers and cancer syndromes (Table S9) (Futreal et al., 2004). We suggest that the mutually exclusive regulation of cell growth and autophagy is at least in part achieved through the differential activation of the type I PI3 kinase, which is known to suppress autophagy (Levine and Kroemer, 2008), and the type III PI3 kinase, in response to growth factors, cytokines and other environmental cues. Thus, our study implicates the type III PI3 kinase as a tumor suppressor important for the negative control of cell growth and proliferation, providing a potential mechanism for increased rates of cancer incidence as a result of Beclin1 deficiency (Liang et al., 1999).
Taken together our data provide important insights into the global regulation of autophagy as well as provide a rich resource and point out novel avenues for further study of this essential cellular catabolic pathway. Lastly, our study highlights the utility of combining multi-level screening, characterization and bioinformatics analysis to elucidate global regulation of a complex biological process.
Methods
Cell lines and culture conditions
H4 human neuroblastoma cells (Krex et al., 2001) were cultured under standard TC conditions in DMEM media supplemented with 10% normal calf serum, 1X penicillin/streptomycin and 1X Na pyruvate (Invitrogen). LC3-GFP, FYVE-dsRed and LC3-GFP pSRP-Beclin1 knock-down H4 cells have been previously described (Shibata et al., 2006; Zhang et al., 2007). To create a stable line expressing Lamp1, H4 cells were transfected with Lamp1-RFP plasmid, followed by selection with 0.4 mg/mL G418.
For the cytokine assays cells were seeded at .5×105 in full medium. After 24h cells were transferred to serum-free medium (DMEM or OptiMEM, Invitrogen); growth factors were added for 6–24h: human LIF (GenScript Corporation, 50 ng/mL), human FGF2 (ProSpec, 50 ng/mL), human IGF1 (ProSpec, 100–200 ng/mL), human CLCF1 (R&D Systems, 50 ng/mL) or human SDF1 (ProSpec, 50–100 ng/mL). Where indicated, rapamycin or Torin 1 (a gift from Dr. Nathaniel Gray, Harvard Medical School, Boston, MA) was added at the same time at 50 nM. For western assays, cells were cultured over night in serum-free media, followed by addition of indicated cytokines and 10 μg/mL lysosomal protease inhibitor E64d (Sigma). For signaling pathway analysis cells were pre-treated with indicated inhibitors for 1 hour before addition of cytokines and E64d, or transfected with indicated siRNAs at the time of initial plating. For CLCF1 complementation cells were transfected with indicated siRNAs and plated in full media supplemented with 100 ng/mL human CLCF1 and incubated for 72 hours. E64d was added for the last 6–8 hours before cell lysis.
siRNA transfection
For the primary screen we used an arrayed library of 21,121 siRNA pools covering the vast majority of the human genome (Dharmacon siARRAY siRNA library (Human Genome, G-005000-05), Thermo Fisher Scientific, Lafayette, CO). Each pool consisted of 4 oligonucleotides targeting a different region of the same gene. Each assay plate included the following controls: non-targeting siRNA, mTOR, ATG5 and PLK1 (transfection efficiency control) siRNA. siRNAs were transiently transfected in triplicate into H4 cells stably expressing LC3-GFP reporter at 40 nM final concentration using reverse transfection. HiPerfect reagent (Qiagen) was diluted 1:20 in DMEM and 8 uL/well of the mixture was aliquoted into 384 well plates using WellMate liquid handling unit (Matrix). 2 uL of 1 uM arrayed siRNA pools/well were added robotically using Velocity11 Bravo. After 30 minutes incubation 500 cells/well in 40 uL full media were added using WellMate. Cells were incubated for 72h under standard culture conditions, counterstained with 0.5 μM Hoechst 33342 (Invitrogen) for 1h and fixed with 30 uL of 8% paraformaldehyde for 30 minutes. Cells were washed 3 times with PBS using Velocity11 ELX405 Plate Washer (Bio-Tek).
For the secondary screen we used deconvolved library in which the 4 components of each siRNA pool were separated into individual wells. The cells were transfected and treated as for primary screen except that siRNAs were used at 30 nM final concentration and HiPerfect was diluted 1:30 in OptiMEM (Invitrogen). The secondary screen transfections were done in 2 rounds, for a total of 5 replica plates, using a 1:1 mixture of LC3-GFP and FYVE-dsRed or Lamp1-RFP cells. All tertiary characterization screens were performed in either triplicate or duplicate. Each assay plate included 10–12 wells of non-targeting siRNA as well as mTOR, ATG5, PLK1 and/or Vps34 siRNA controls. To evaluate the contribution of inhibition of lysosomal degradation to the induction of autophagy, where indicated E64d was added at 10 μg/mL for the last 8 hours before fixation.
For follow up assays cells were transfected in 96-, 24- or 6-well plates using reverse transfection with 1 μL to 6 μL HiPerfect/mL, 10–30 nM final siRNA concentration and cells at 5 × 104 to 2 × 105 cells/mL, depending on the cell type. Cells were harvested after 48–72 hours after transfection.
Imaging and image quantification
Cells were imaged on an automated CellWoRx microscope (Applied Precision) at 10× magnification and 350 nm (Hoechst), 488 nm (LC3-GFP) and 550 nm (Lamp1-RFP and FYVE-dsRed) wavelengths. All images were quantified using VHSscan and VHSview image analysis software (Cellomics). Total cell number, total LC3-GFP intensity/cell as well as number, area and intensity of LC3-GFP positive autophagosomes/cell were scored. Dead and mitotic cells were excluded from analysis based on nuclear intensity. The final autophagy score for every well was obtained by multiplying total autophagosome intensity/cell times number of autophagosomes/cell and dividing by average cells intensity. This formula was empirically determined to most accurately measure LC3-GFP translocation from cytosol into autophagosomes as reflected by consistently significant z-scores and p-values when using siRNAs against mTOR and Atg5 controls under the assay conditions and to give the highest z′ score (0.52, where z′=1−(3SDmTOR+3SDAtg5)/(AvemTOR−AveAtg5) (Zhang et al., 1999)). FYVE-dsRed and Lamp1-RFP scores were obtained in a similar manner, except that in the case of Lamp1-RFP, which measures total accumulation of the reporter rather than its translocation, division by the average cell intensity was omitted.
In-cell-western assay
H4 cells were cultured in 384-well plates, fixed and counterstained as described for the LC3-GFP assay. Following imaging the cells were permeabilized in PBS + 0.2% Tx-100 (PBST) and stained with Alexa-680 NHS-ester at 20 ng/mL for 15 minutes. Cells were washed with PBST, incubated for 30 minutes in blocking buffer (1:1 LiCOR Blocking Buffer:PBST), and overnight in rabbit-anti-rpS6 phospho-235/236 (Cell Signaling Technologies), or mouse-anti-KDEL (Stressgen) antibody at 1:1000 in blocking buffer. Cells were washed in PBST and stained with an IRDye-800-conjugated secondary antibody (LiCOR) at 1:1000. The plates were scanned on the Aerius infrared imaging system (LiCOR). The intensity of both, the rpS6 phospho-235/236 or KDEL staining, and of NHS-ester staining were integrated, and the normalized phospho-S6 or KDEL score was calculated for each well by dividing phospho-rpS6 or KDEL intensity by NHS-ester intensity (Hoffman et al., 2010).
Statistical analysis
All screen data was normalized by conversion to logarithmic scale. For primary screen z-scores were calculated based on plate median (controls excluded) and Median Absolute Deviation (MAD), with z-score = (cell score − median plate score)/(plate MAD × 1.4826) (Chung et al., 2008). The screen hits were selected based on the median z-score of the 3 replica-plates with cut-offs set at z-score > 1.7 or < −1.9 (p < 0.02). The same method was used for the rpS6 and KDEL secondary screens except the assays were performed in duplicate. For LC3-GFP, FYVE-dsRed and Lamp1-RFP secondary screens z-scores were calculated based on non-targeting siRNA control (n=10–12) mean and standard deviation. For confirmation of hits in the LC3-GFP assay we required that at least 2 out of 4 individual siRNA oligonucleotides for each gene had median z-scores > 1.5 or < −1.5 based on 5 replica plates and were consistent with the primary screen z-score (p < 0.01). In all other secondary assays z-scores > 1.5 and < −1.5 were also considered significant. The final z-scores for confirmed genes were calculated based on average z-scores of all wells for oligonucleotides positive in the secondary LC3-GFP assay.
The correlation analysis between LC3-GFP and other secondary assays was performed based on individual assay well quadrant analysis: for each well a score of +1 was assigned if z-scores for both features were > 1.5 or both were < −1.5; a score of −1 if one z-score was > 1.5 while the other was < −1.5; a score of 0 if either z-score failed to reach the cut-off. The individual well scores were than summed up for each gene for all oligonucleotides considered significant in the LC3-GFP secondary assay and divided by the total number of wells assayed for these oligonucleotides. A correlation between features was considered to be positive if the final score was ≥ 0.5, negative if it was ≤ −0.5.
For identification of genes which knock-down was able to induce autophagy in the presence of E64d a double criteria was used: 1) Knock-down of the gene in the presence of E64d was able to induce autophagy above the levels observed with non-targeting siRNA + E64d by at least 1.5 s.d.; 2) Treatment with E64d was able to increase levels of autophagy following knock-down of the gene by at least 1.5 s.d.
Relative viability was calculated by dividing number of cells in each well based on Hoechst imaging by the average cell number in the plate. The reported viability for each hit gene reflects average viability of all wells for oligonucleotides positive in the secondary LC3-GFP assay.
Unless otherwise indicated, all remaining p values were calculated from a 2-tailed student t-test with equal variance. All error bars are standard error.
Western analysis
Cells were lysed in Lammeli sample buffer, resolved on 12% SDS-PAGE and transferred to PVDF membrane following standard protocols. The following antibodies were used: phospho-Foxo3a and Atg5 (Sigma) at 1:500; LC3 (Novus), p62 (Pharmigen), phospho-S6K (Thr389), phospho-Akt (Ser473), Stat3, phospho-Stat3 (Tyr705) and mTOR (all Cell Signaling) at 1:1000; phospho-S6 (Ser235/236) (Cell Signaling) and phospho-ERK 1/2 (Sigma) at 1:2000; tubulin (Sigma) at 1:5000. Blots were quantified using NIH ImageJ64 software.
Semi-quantitative RT-PCR
Total RNA was prepared using RNeasy mini kit (Qiagen). 1.25 μg RNA was used for cDNA synthesis using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) with oligo dT primers. Following primers were used: FGFR1 CTCCGGCCTCTATGCTTGCGTAAC and TGCGGCTGCGGGTCACTGTA, Stat3 ACCCCGGCTTGGCGCTGTCTCT and ACGGCTGCTGTGGGGTGGTTGG, CLCF1 TGGGCCTGGCGGATGGGATTATTA and TGGGTCAGCCGCAGTTTGTCATTG, β actin GACCTGACAGACTACCTCAT and AGACAGCACTGTGTTGGCTA.
Bioinformatics analysis
Enrichment analyses: genes were classified into functional categories: biological process, molecular function (PANTHER classification system (Mi et al., 2005)), canonical pathways (MSigDB (Subramanian et al., 2005)) and transcription factor binding sites (MSigDB and TRANSFAC v7.4 (www.gene-regulation.com)). Genes for which no annotations could be assigned were excluded. To assess the statistical enrichment of these categories for the hit genes relative to their representation in the global set of genes examined, p-values were computed using the hyper geometric probability distribution. Categories with p<0.05 are considered enriched. Similar enrichment results were obtained independently of the details of the hit selection protocol.
Protein interaction network: The network was constructed by iteratively connecting interacting proteins, with data extracted from genome-wide interactome screens (Ho et al., 2002; Ito et al., 2001), databases: HPRD (Mishra et al., 2006), MINT (Chatr-aryamontri et al., 2007), REACTOME (Joshi-Tope et al., 2005) and curated literature entries. The network uses graph theoretic representations, which abstract components (gene products) as nodes and relationships (interactions) between components as edges, implemented in the Perl programming language.
Supplementary Material
Acknowledgments
We thank Drs. Noboru Mizushima and Nathaniel Gray for reagents, Caroline Yi for critical reading of the manuscript and the members of the Yuan lab and the ICCB-Longwood for help during this work. This work was supported in part by NIH grants R37 AG012859 and PO1 AG027916 to JY, AI062773 and DK043351 to RJX, R01 GM051405 and R21 NS059428 to JB and NSF grant DMS-0706989 to JL. GH is supported by fellowships from the Helen Hay Whitney Foundation and the LAM Foundation, AN by the Crohn’s and Colitis Foundation of America, JE by CCIB Development Fund.
Footnotes
Author contributions: MML and JY conceived and planned the project; MML performed the screen, imaging and biochemical analysis; GH and JB developed and performed S6 in-cell-western assay; AN, JE and RJX performed bioinformatics analysis; WZ, BFP and EH assisted with biochemical analysis; XL and JL performed statistical analysis. The manuscript was written by MML and JY and commented on by all the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen. 2008;13:149–158. doi: 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hoffman GR, Moerke NJ, Hsia M, Shamu CE, Blenis J. A High-Throughput, Cell-Based Screening Method for siRNA and Small Molecule Inhibitors of mTORC1 Signaling Using the In Cell Western Technique. Assay Drug Dev Technol. 2010 doi: 10.1089/adt.2009.0213. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Krex D, Mohr B, Hauses M, Ehninger G, Schackert HK, Schackert G. Identification of uncommon chromosomal aberrations in the neuroglioma cell line H4 by spectral karyotyping. J Neurooncol. 2001;52:119–128. doi: 10.1023/a:1010680920087. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007a;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007b;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- Sakaki K, Wu J, Kaufman RJ. Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem. 2008;283:15370–15380. doi: 10.1074/jbc.M710209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassa A, Roux MP, Attaix D, Bechet DM. Class III phosphoinositide 3-kinase--Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003;376:577–586. doi: 10.1042/BJ20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.