Abstract
The aggregation of mutant polyglutamine (polyQ) proteins has sparked interest in the role of protein quality-control pathways in Huntington's disease (HD) and related polyQ disorders. Employing a novel knock-in HD mouse model, we provide in vivo evidence of early, sustained alterations of autophagy in response to mutant huntingtin (mhtt). The HdhQ200 knock-in model, derived from the selective breeding of HdhQ150 knock-in mice, manifests an accelerated and more robust phenotype than the parent line. Heterozygous HdhQ200 mice accumulate htt aggregates as cytoplasmic aggregation foci (AF) as early as 9 weeks of age and striatal neuronal intranuclear inclusions (NIIs) by 20 weeks. By 40 weeks, striatal AF are perinuclear and immunoreactive for ubiquitin and the autophagosome marker LC3. Striatal NIIs accumulate earlier in HdhQ200 mice than in HdhQ150 mice. The earlier appearance of aggregate pathology in HdhQ200 mice is paralleled by earlier and more rapidly progressive motor deficits: progressive imbalance and decreased motor coordination by 50 weeks, gait deficits by 60 weeks and gross motor impairment by 80 weeks of age. At 80 weeks, heterozygous HdhQ200 mice exhibit striatal and cortical astrogliosis and a ∼50% reduction in striatal dopamine receptor binding. Increased LC3-II protein expression, which is noted early and sustained throughout the disease course, is paralleled by increased expression of the autophagy-related protein, p62. Early and sustained expression of autophagy-related proteins in this genetically precise mouse model of HD suggests that the alteration of autophagic flux is an important and early component of the neuronal response to mhtt.
INTRODUCTION
Huntington's disease (HD) is a dominantly inherited neurodegenerative disorder marked by involuntary movements, cognitive impairment and psychiatric abnormalities as well as dysfunction and subsequent loss of medium spiny striatal neurons and neocortical neurons. HD is incurable and leads to inexorable decline and death. Polyglutamine (polyQ)-encoding expanded CAG repeats are the cause of HD and eight other neurodegenerative diseases (1). PolyQ expansions lead to abnormal conformations and altered functions of the mutant proteins, resulting in neurotoxicity. Unaffected individuals possess a polyQ repeat of 35 or fewer residues in the disease protein, huntingtin (htt), whereas fully penetrant disease results from polyQ expansions greater than 39 residues (2,3). Increasing polyQ length correlates with earlier onset of manifest disease. A common histopathologic hallmark of HD and other polyQ diseases is polyQ-containing protein deposits in the cytoplasm and nucleus (4,5).
Murine genetic models are versatile tools for studying pathogenic mechanisms and identifying therapeutic targets in HD. Knock-in models expressing a mutated (expanded) murine homologue of huntingtin in the appropriate genomic context are in some respects the most accurate models and exhibit features consistent with HD. Compared with transgenic models, however, existing HD knock-in models exhibit relatively normal life spans with later onset of behavioral abnormalities and neuropathologic features than do transgenic models (6,7). Experience with knock-in models shows that longer CAG/polyQ repeats generate more robust motor and pathological phenotypes (8–12). A knock-in model of HD with a more aggressive phenotype than available currently would be advantageous for the studies of pathogenesis and for therapeutic experiments. Here, we report a knock-in model derived from the HdhQ150 knock-in line (also previously named Hdh(CAG)150 and CHL2) (8) with ∼200 CAG/polyQ repeats (HdhQ200). HdhQ200 knock-in mice exhibit an accelerated behavioral and neuropathological phenotype compared with the parent HdhQ150 line.
The apparent pathogenic role of misfolded and aggregated polyQ proteins has sparked interest in defining the role of protein quality control pathways in polyQ diseases. One advantage of knock-in models is that they provide a normal physiological context in which to investigate protein quality-control pathway responses to mutant htt (mhtt) expression. Two primary protein quality-control pathways responding to abnormally folded proteins, the ubiquitin-proteasome pathway (UPS) and the autophagy-lysosomal degradation pathway, are major routes for mhtt clearance. The relative importance of these pathways in HD remains unknown. The possibility that UPS contributes to the neuronal response to mhtt expression was suggested by the early discovery that intranuclear htt-immunoreactive aggregates in HD brain tissue and in transgenic HD mice also contain ubiquitin and proteasome subunits (13,14) and that the impairment of proteasome activity increased mutant protein accumulation (15). Subsequent reports in polyQ diseases, however, yielded conflicting results with various studies indicating increased, diminished or unchanged UPS activity (16–20). A possible role for autophagy in HD was suggested by the studies of post-mortem HD brain indicating that cytoplasmic mhtt aggregates have a distribution similar to that of autophagosomes–lysosomes (21). The subsequent work indicated that autophagy is an important component of the cellular response to mhtt and other expanded polyQ proteins (22–30). Much of this work has been accomplished with in vitro and invertebrate model systems. Utilizing the HdhQ200 model, we now report an association of mhtt immunoreactive cytoplasmic aggregates with autophagosomes, and early and sustained induction of autophagy-associated proteins, suggesting that autophagy is indeed an important component of neuronal response to mhtt expression in vivo.
RESULTS
Expression analysis
We described previously that the HdhQ150 mRNA levels are reduced in mouse striatum to 66% of wild-type (WT) levels. The present results confirm our previous findings and show that the HdhQ200 mRNA levels do not significantly differ from the HdhQ150 mRNA levels (Supplementary Material, Fig. S1). Increase in repeat length from Q150 to longer lengths (Q200 to Q220) did not qualitatively alter intensities of these bands on western blots that were variable and always less intense than endogenous WT htt bands as previously shown for the Q150 protein (Supplementary Material, Fig. S1).
The HdhQ200 mutation causes a progressive decline in overall health and progressive motor abnormalities
HdhQ200 heterozygous mice exhibited progressive decline in overall health and appeared moribund by 80 weeks at which time animals exhibited diminished spontaneous movement, appeared thin, exhibited staggering gait and had poorly groomed fur. There were two deaths, 1 female and 1 male out of 10 animals, in the HdhQ200 heterozygous group at ∼75 weeks. No seizures were observed in any animal, either spontaneous or induced by handling.
HdhQ200 heterozygous female mice gain weight normally up to 40 weeks, at which point animals begin to show weight reduction. By 50 weeks, HdhQ200 heterozygous females exhibited a 10–22% weight decline compared with WT littermate controls from 50–80 weeks, respectively, P < 0.05 (Fig. 1A). This differs from the original HdhQ150 knock-in line in which weight loss was not observed in heterozygotes up to 100 weeks, though weight loss was observed in HdhQ150 homozygotes at 70 weeks (8). Weight is maintained in male HdhQ200 heterozygous mice from 10 to 80 weeks, P > 0.05 (Fig. 1B). Homozygous HdhQ200 mice gained weight normally up to 13 weeks (data not shown) but then exhibited weight loss between 20 and 40 weeks of age. Homozygous HdhQ200 mice exhibit an earlier and greater weight decrement, ∼30–40% weight reduction compared with WT controls, P < 0.05 (Fig. 1C, P < 0.05). HdhQ200 heterozygous mice also exhibit age-dependent reduced strength. Muscle strength was measured with a grip-strength meter, which assesses force exerted by forelimbs or hindlimbs of mice at various ages. HdhQ200 heterozygous mice exhibit progressive reduction in muscle strength in both the forelimbs and hindlimbs at 80 weeks, which was normal at 20, 30, 50 and 60 weeks (Fig. 1D). At 80 weeks, HdhQ200 heterozygous mice exhibited an ∼30% reduction in grip-strength of forelimbs and hindlimbs [Fig. 1D; forelimbs: WT 1.2 ± 0.07 (mean + SEM) newtons (N); heterozygous HdhQ200 mice 0.85 ± 0.08 N; hindlimbs: WT 0.86 ± 0.06 N; heterozygous HdhQ200 mice 0.57 ± 0.07 N; P < 0.05]. To assess gait impairments, we examined the gait pattern with the use of footprint analysis. HdhQ200 heterozygous mice exhibited a normal gait pattern consistent with WT littermates up to 60 weeks at which time HdhQ200 heterozygous mice showed the loss of gait pattern with progressive worsening to 80 weeks; the loss of gait pattern by this method was not apparent in Hdh150 heterozygous mice until 70 weeks. In contrast, WT littermates exhibited consistent and congruent overlaps of hindpaw over forepaw prints at all ages (Fig. 1E).
Figure 1.
HdhQ200 mutation causes age-related weight reduction, loss of grip-strength and gait impairment. (A) Heterozygous HdhQ200 female mice exhibit a significant weight loss as early as 50 weeks compared with WT littermate controls (n = WT-HdhQ200 animals as follows 10 weeks: 4–3; 20 weeks: 8–13; 40 weeks: 10–11; 50 weeks: 10–8; 60 weeks: 10–8; 70 weeks: 10–12; 80 weeks: 8–8) (B) whereas males maintain normal weight (n = WT/HdhQ200 animals as follows 10 weeks: 3–5; 20 weeks: 10–9; 40 weeks: 8–7; 50 weeks: 4–4; 60 weeks: 4–4; 70 weeks: 6–6; 80 weeks: 5–3). (C) Homozygous HdhQ200 mice exhibit an earlier and greater weight decrement compared with WT littermate controls seen as early as 20 weeks of age [(weight loss is equally robust in both genders) (n = WT-HdhQ200 animals as follows 20 weeks: 15–4; 20 weeks: 8–5; 40 weeks: 12–11)]. (D) Heterozygous HdhQ200 mice exhibit a significant reduction in grip-strength force at 80 weeks compared with WT controls in both forelimbs and hindlimbs. (E) Loss of gait pattern was observed at 60 weeks in heterozygous HdhQ200 mice with progressive worsening to 80 weeks, whereas WT littermates exhibit consistent and congruent overlaps of hindpaw over forepaw prints at all ages. Error bars represent a standard error of the mean (SEM) and asterisks denote significance at P < 0.05.
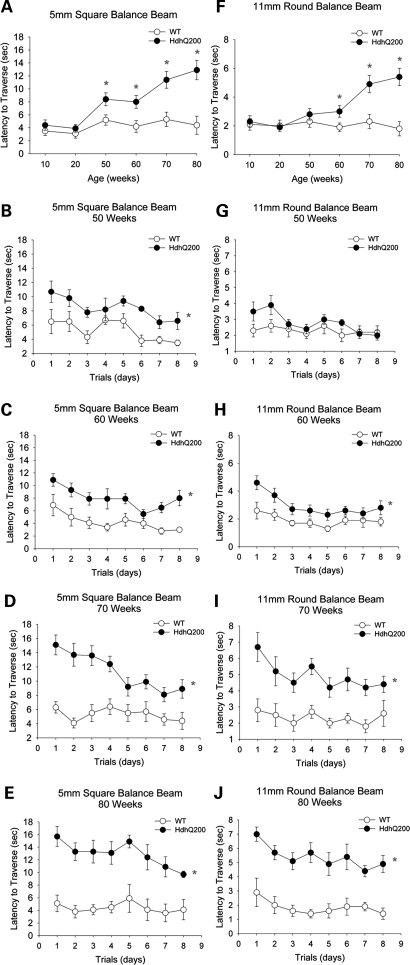
To further assess motor behavior deficits, we evaluated animals with a balance beam task (using 11 mm round and 5 mm square beams), rotarod and footprint analysis. As described previously in HdhQ150 mice, the balance beam test proved to be the most sensitive measure of motor impairment, detecting motor deficits early and even when the rotarod failed to show any deficits (8). Other groups have reported similar experience with the balance beam task in expanded polyQ model mice (31) (A. Lieberman, personal communication). Utilizing two different size beams expands the ability to gage and detect motor deficits.
HdhQ200 heterozygous mice exhibit progressive motor deficits beginning as early as 50 weeks on the 5-mm square beam (Fig. 2B–E) and 60 weeks on the larger 11-mm round beam (Fig. 2G–J), summarized in Fig. 2A and F, respectively. At 50 weeks, HdhQ200 heterozygous mice required approximately twice as much time as WT controls to traverse a 5-mm square beam [Fig. 2A; 5 mm square beam, F(1,18) = 17.3, P < 0.05 between-group effect; averaged time across trial days: WT, 5.2 ± 0.8 (mean ± SEM)s; heterozygous HdhQ200 mice, 8.4 ± 1.0 s]. These motor deficits progressively worsen to 80 weeks of age, where the average latency to traverse increases three times on the 5-mm square beam (Fig. 2D; 5 mm square beam, F(1,14) = 12, P < 0.05 between-group effect; WT, 4.4 ± 1.4 s; heterozygous HdhQ200 mice, 12.9 ± 1.5 s). Females and males were identical in balance beam performance (data not shown). In homozygous HdhQ200 mice, motor deficits were detected as early as 20 weeks of age. At 20 weeks, homozygous HdhQ200 animals required twice as much time to traverse the 5-mm balance beam as controls and three times as much time at 40 weeks of age [Supplementary Material, Fig. S2; 20 weeks: F(1,3) = 12.3, P < 0.05 between-group effect; 40 weeks: F(1,6) = 73.8, P < 0.05].
Figure 2.
HdhQ200 mutation causes progressive motor deficits. Heterozygous HdhQ200 mice were evaluated for ability to traverse a 5-mm square beam and a 11-mm round beam at 10, 20, 50, 60, 70 and 80 weeks. Motor deficits are progressive with age as summarized in (A and F). HdhQ200 heterozygous mice exhibit progressive motor deficits on both the 5-mm (B–E) and 11-mm balance beams (G–J). Motor deficits became apparent on the 5-mm beam at 50 weeks and on the 11-mm beam at 60 weeks of age. Error bars represent the SEM and asterisks denote significance at P < 0.05.
HdhQ200s have a markedly accelerated motor phenotype compared with the HdhQ150 line. In both heterozygous and homozygous HdhQ150 mice, motor deficits on the balance beam were not significant until 100 weeks (8). We also measured the latency to fall with the accelerating rotarod. With this test, however, heterozygous and homozygous HdhQ200 mice did not differ from WT littermate controls at any age (data not shown).
Striatal neuron dysfunction in HdhQ200 mice
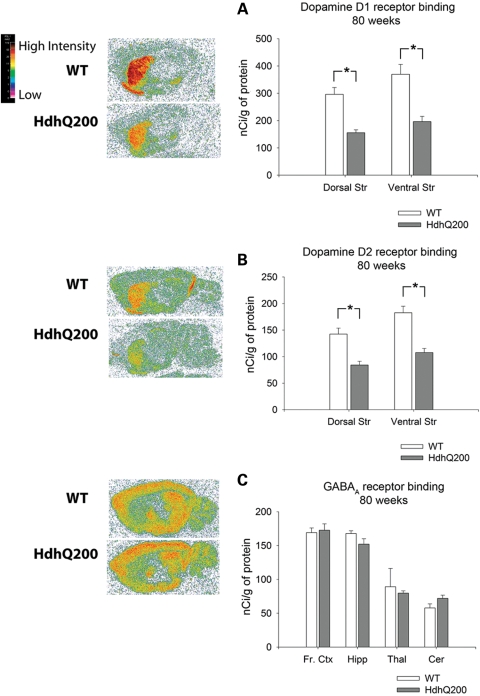
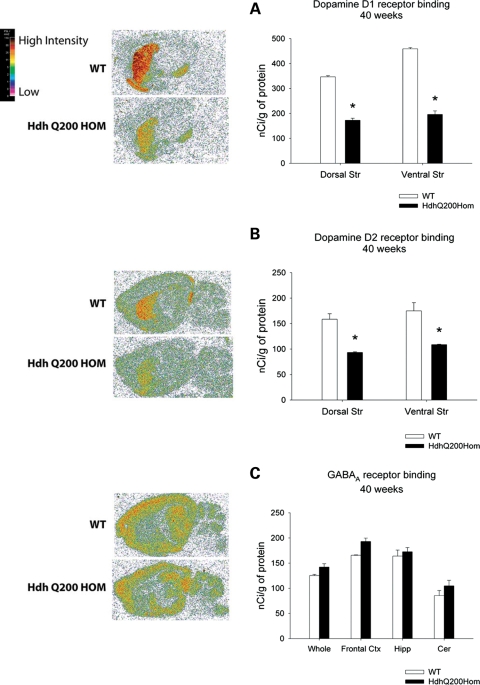
Imaging studies of HD patients show up to a 50% loss of striatal D1 and D2 receptor binding in presymptomatic subjects suggesting a prolonged period of striatal dysfunction prior to the onset of manifest disease (32). To examine striatal dysfunction, we performed quantitative receptor autoradiography at 80 weeks, when behavioral deficits were most pronounced, to assess the expression of dopamine D1 and D2 receptors, both of which are highly expressed on striatal dendrites. Eighty-week-old HdhQ200 heterozygous mice exhibited a ∼50% decrease in D1 receptor binding in both dorsal and ventral striata. Analysis of the ventral striatum focused primarily on the nucleus accumbens and ventral striatum proper. We excluded the olfactory tubercle as it is disproportionately large in rodents relative to humans. The dorsal striatum was considered the upper half of the striatal complex [Fig. 3A; D1 dorsal striatum (Dorsal Str): WT, 296.24 ± 25 (mean nCi/g ± SEM); heterozygous HdhQ200 mice, 155.8 ± 10.6, P < 0.001; ventral striatum (Ventral Str): WT, 369.8 ± 36; heterozygous HdhQ200 mice 196.4 ± 19, P < 0.01]. HdhQ200 heterozygous mice exhibited similar decreases in D2 receptor binding in both dorsal and ventral striata (Fig. 3B; D2 dorsal striatum: WT, 142.38 ± 11.2; heterozygous HdhQ200 mice, 84.2 ± 7.1, P < 0.001; ventral striatum: WT, 182.7 ± 12.2; heterozygous HdhQ200 mice, 107.7 ± 7.7, P < 0.001). To survey the integrity of extrastriatal regions, we assessed GABAA/benzodiazepine receptor binding as it is highly expressed in most brain regions outside the striatum. There were no differences in GABAA/benzodiazepine receptor binding in any brain region sampled [Fig. 3C; frontal cortex (Fr. Ctx): WT, 169.2 ± 6.7; heterozygous HdhQ200 mice, 172.8 ± 9.4; hippocampus (Hipp): WT, 167.9 ± 4; heterozygous HdhQ200 mice, 152.2 ± 8.0; thalamus (Thal): WT, 89.2 ± 27; heterozygous HdhQ200 mice, 79.7 ± 3.5; cerebellum (Cer): WT, 60 ± 6.2; heterozygous HdhQ200 mice, 70 ± 5.2; P > 0.05]. Homozygous HdhQ200 mice also exhibited the loss of striatal D1 and D2 receptor binding sites: 50% and 40% reductions in D1 and D2 receptors by 40 weeks of age, at which time the mice appear moribund (Fig. 4A and B; D1 Dorsal Str: WT, 346.7 ± 4.3; homozygous, 172.7±8.1, P < 0.001; Ventral Str: WT, 458.9 ± 5.0; homozygous, 196.1 ± 14.2; D2 Dorsal Str: WT, 158.5 ± 10.8; homozygous, 93.4 ± 1.3, P < 0.001; Ventral Str: WT, 175.0 ± 16.0; homozygous, 108.7 ± 0.8, P < 0.01). In contrast, previous results from our homozygous HdhQ150 mice show the normal levels of dopamine receptor binding at 40 weeks of age (8). Homozygous HdhQ200 mice exhibit no differences in extrastriatal regions measured with GABAA/benzodiazepine receptor binding (Fig. 4C). These results suggest relatively selective striatal neuron vulnerability in HdhQ200 mice.
Figure 3.
Heterozygous HdhQ200 mutation causes a loss of striatal dopamine D1 and D2 receptor expression. (A) The 80-week-old heterozygous HdhQ200 mice exhibit a 47% decrease in D1 receptor binding in both dorsal and ventral striata. (B) Heterozygous HdhQ200 mice exhibit similar decreases in D2 receptor binding in both dorsal and ventral striata. (C) Heterozygous HdhQ200 mice show no differences in GABAA/benzodiazepine receptor binding, which surveys the integrity of extrastriatal regions. Frontal cortex (Fr. Ctx), hippocampus (Hipp), thalamus (Thal) and cerebellum (Cer). Error bars represent the SEM, and asterisks denote P < 0.05.
Figure 4.
Earlier loss of striatal dopamine D1 and D2 receptor expression in homozygous HdhQ200 mice. Homozygous HdhQ200 mice exhibit earlier loss of striatal D1 and D2 receptor binding sites than heterozygous HdhQ200 mice: 50 and 40% reductions in D1 and D2 receptors by 40 weeks of age (A and B). There were no differences in extrastriatal regions measured with GABAA/benzodiazepine receptor binding (C). Whole brain (Whole), Frontal cortex (Frontal Ctx), hippocampus (Hipp) and cerebellum (Cer). Error bars represent the SEM, and asterisks denote significance at P < 0.05.
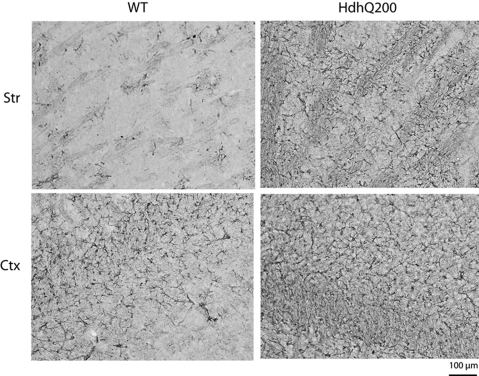
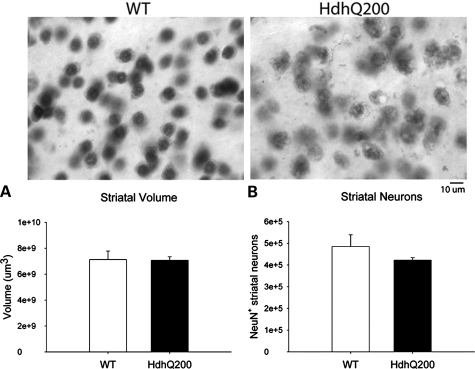
Reduction in striatal dopamine receptors was accompanied by increased GFAP immunostaining, a marker for astrogliosis, at 80 weeks (Fig. 5). This increase was higher in the striatum than cortex exhibiting a 2.4-fold increase and a 1.3-fold increase, respectively, compared with WT control regions (Supplementary Material, Fig. S3). Mildly increased GFAP immunostaining was apparent as early as 40 weeks and restricted to the striatum and cortex in HdhQ200 heterozygous mice (data not shown). Striatal dopamine receptor loss at 80 weeks, however, was not accompanied by striatal neuron loss. HdhQ200 heterozygous mice were examined for neuronal loss by unbiased stereology. Although striatal neurons of HdhQ200 heterozygous mice consistently presented differences in morphology and intensity of NeuN immunoreactivity compared with WT controls, they showed no significant decrease in striatal volume or striatal neuron number [Fig. 6A, striatal volume: WT, 7 130 665 000 ± 649 353 569.9 μm3; heterozygous HdhQ200 mice, 7 074 556 000 ± 272 643 375.9 μm3, P > 0.05; Fig. 6B, striatal neuron number: WT, 485 267 ± 54 147 N; heterozygous HdhQ200 mice, 422 492 ± 11 503 N (mean ± SEM); P > 0.05]. HdhQ200 striatal neurons consistently appeared to contain many (or larger) cytoplasmic vacuoles. Together, these results indicate striatal neuron dysfunction accompanied by astrogliosis in HdhQ200 knock-in mice.
Figure 5.
Heterozygous HdhQ200 mice exhibit increased striatal and cortical GFAP immunoreactivity. Increased GFAP immunoreactivity is most robust in striatal and cortical regions of heterozygous HdhQ200 mice at 80 weeks of age. Striatum (Str) and cortex (Ctx).
Figure 6.
Heterozygous HdhQ200 mice do not exhibit striatal neuron loss. Neurons were identified by NeuN immunoreactivity at 80 weeks. Heterozygous HdhQ200 mice show no significant decrease in striatal volume (A) or striatal neuron number (B).
HdhQ200 mice exhibit compartmentalization of heterogeneous aggregate species
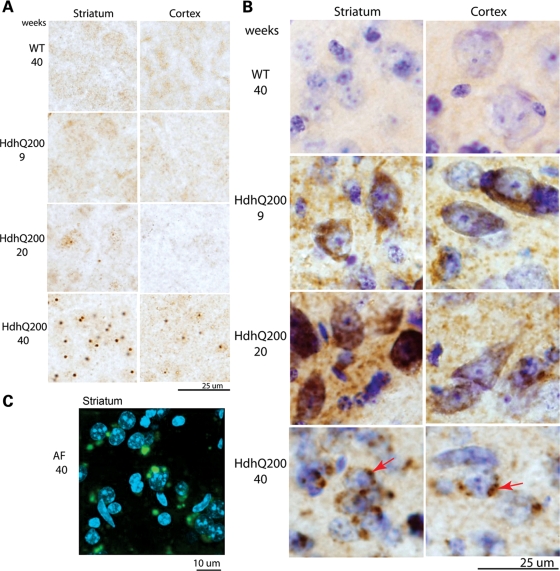
Neuronal intranuclear inclusions (NIIs) are a pathological feature of polyQ disease found in striata and cortices of HD post-mortem brains (14,33). NIIs were detected in HdhQ200 heterozygous mice with immunohistochemistry employing an N-terminal htt-specific antibody. NIIs were absent at 9 weeks, present minimally at 20 weeks and deposited robustly at 40 weeks. NIIs were largely restricted to the striatum and cortex with greater striatal abundance (Fig. 7A). NIIs were round and uniform with a diameter between 3 and 5 µm, consistent with NIIs described in HD post-mortem brains (14). NIIs were seen in HdhQ200 heterozygous mice 20 weeks earlier than in HdhQ150 mice (8) (Fig. 11 for a summary and comparison timeline of the HdhQ200 to the HdhQ150 knock-in mice).
Figure 7.
Heterozygous HdhQ200 mice exhibit different patterns of progressive accumulation of striatal and cortical NIIs and AF. (A) NIIs were labeled with an N-terminal htt antibody. NIIs are apparent as early as 20 weeks and become more robust and abundant by 40 weeks in both striatum and cortex. (B) HdhQ200 heterozygous mice exhibit progressive AF. AF were expressed diffusely in striatal neurons as early as 9 weeks and expressed robustly at 20 weeks. No AF are detected in WT mice at any age. At 40 weeks, AF appear distinctly punctate and perinuclear (denoted by arrows). (A and B) Heterogeneous populations of mhtt aggregates exist in different subcellular compartments, one cytoplasmic (AF) and the other nuclear (NIIs). (C) In heterozygous HdhQ200 striata at 40 weeks, perinuclear compartmentalization of punctuate AF was confirmed by confocal microscopy in which AF were labeled with Alexa Fluorophore 488 (green) and nuclei with DAPI (blue).
Figure 11.
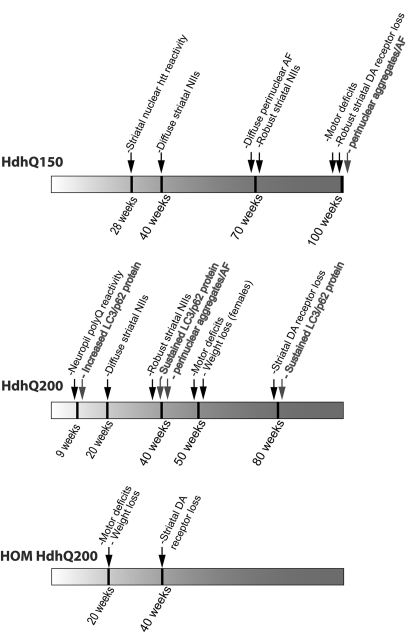
A pathological timeline summary. A pathological timeline summary and comparison of the HdhQ150 heterozygous mice, HdhQ200 heterozygous mice and HdhQ200 homozygous mice (HOM HdhQ200). The sequence of behavioral and pathologic changes is accelerated in HdhQ200 mice.
Cytoplasmic aggregates may precede NII formation in HD and are a neuropathologic feature of mouse models of HD (14,34). To further assess mhtt protein deposition, adjacent sections from 9-, 20- and 40-week-old HdhQ200 heterozygous and control mice were assayed for aggregation foci (AF), a sensitive histological technique used to detect polyQ aggregation competence in HD post-mortem samples and in rodent models of HD (35–37). In HdhQ200 heterozygous mice, AF are apparent as early as 9 weeks (Fig. 7B), when NIIs are not present, and are abundant by 20 weeks, when NIIs are barely detectable (Fig. 7A). By 40 weeks, when NIIs are expressed robustly, AF are clearly distinguishable from NIIs (Fig. 7B) and, like NIIs, are more abundant in striatum than cortex. By 40 weeks, AF become distinctively punctate and perinuclear in location. The perinuclear localization of AF was confirmed by confocal microscopy in 40-week-old HdhQ200 mice (Fig. 7C). These results indicate heterogeneous populations of mhtt aggregates in different subcellular compartments; one cytoplasmic (AF), and the other nuclear (NIIs), with differing accumulation kinetics.
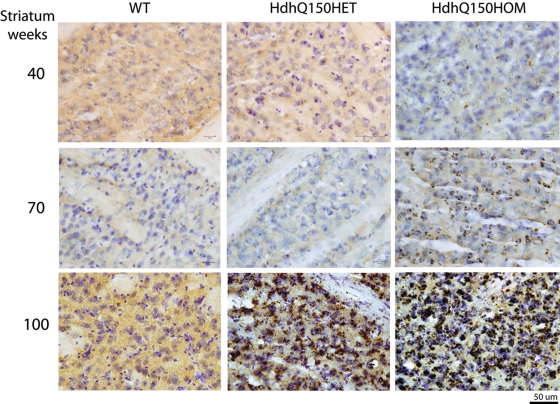
AF also accumulate in the HdhQ150 knock-in line but more slowly. Perinuclear AF are detected in homozygous HdhQ150 mouse brains at 70 and 100 weeks of age compared with 40 weeks in heterozygous HdhQ200 mice (Fig. 8).
Figure 8.
Heterozygous Hdh150 knock-in mice also accumulate AF but more gradually. AF are visible in heterozygotes at 70 weeks and homozygotes at 40 weeks and become distinctly perinuclear at 70 weeks in homozygotes. By 100 weeks, AF are prominent and robust in both heterozygotes and homozygotes.
Aggregated htt was also detected in younger HdhQ200 knock-in mice by Agarose Gel Electrophoresis for Resolving Aggregates (AGERA) using the polyQ-specific antibody, 1C2 (38). Soluble fractions of whole brain lysates from heterozygous HdhQ200 knock-in mice were run on 1.5% SDS agarose gels. High molecular weight species were evident in lysates from 2-, 9-, 20-, 40- and 80-week-old HdhQ200 heterozygous knock-in brains (Supplementary Material, Fig. S4).
HdhQ200 mice exhibit early induction of autophagy markers
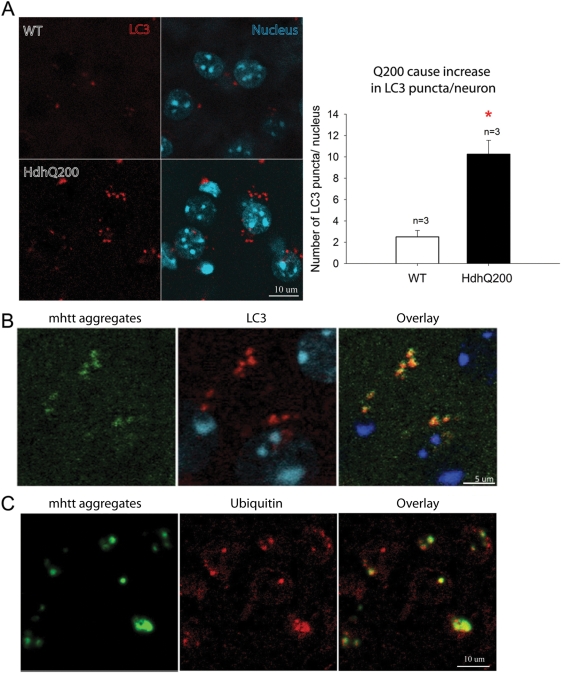
Increased autophagy markers and activity are described in brains of HD post-mortem patients and in murine genetic models of HD (21,39–41). Given the distinct perinuclear punctate pattern of AF, we sought to determine whether these aggregates might be associated with autophagy markers. Microtubule-associated protein 1 light chain 3, brain isoform B- phosphatidylethanolamine (LC3B hereafter called LC3) is a widely used marker for autophagy (40–42). Immunofluorescence labeling of LC3 in adjacent sections in the striatum of 40-week-old HdhQ200 heterozygous mice demonstrated a 4-fold increase in LC3 punctae per neuron compared with WT littermate controls (Fig. 9A). LC3 punctae co-localized with AF (Fig. 9B). Some abnormal protein aggregates are directed to the autophagy-lysosomal degradation pathway via ubiquitination (30). Consistent with this pathway, double labeling of AF and ubiquitin of adjacent sections demonstrated co-localization in HdhQ200 mice at 40 weeks (Fig. 9C).
Figure 9.
LC3 punctae and ubiquitin co-localize with mhtt aggregates. (A) Immunofluorescence labeling of LC3 (red) in striatum of 40-week-old HdhQ200 heterozygous mice exhibited a 4-fold increase in LC3 punctae per cell compared with WT littermate controls. (B) Double labeling of LC3 punctae (red) and AF (green) shows their co-localization (yellow) in HdhQ200 heterozygous striatum at 40 weeks. (C) Double labeling of AF (green) and ubiquitin (red) demonstrated co-localization of AF and ubiquitin (yellow) in heterozygous striatum at 40 weeks. Error bars represent the SEM, and asterisk denotes P < 0.05.
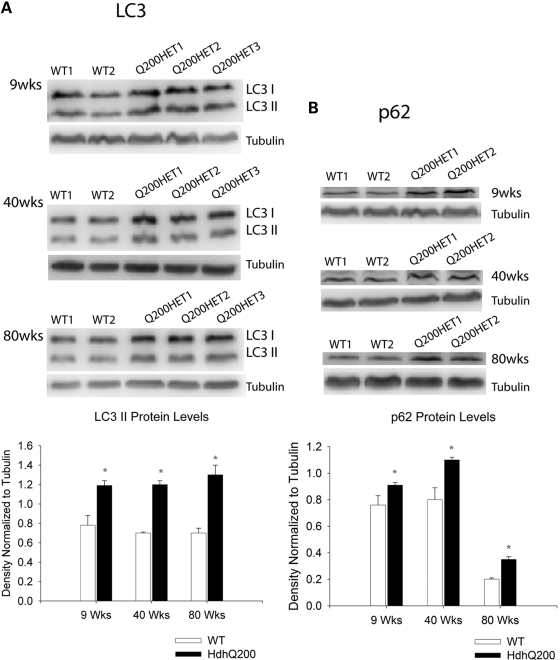
To address whether an autophagic response was induced at even earlier time points, LC3-II protein expression was assessed by western blotting. LC3-II is modified from its LC3-I cytosolic form to a more rapidly migrating, lipid-associated autophagosome membrane-bound form when autophagy is induced. LC3-II expression and localization is a widely used marker of autophagosome induction and is known to correlate with elevated levels of autophagic vesicles (42–44). LC3-II levels were increased significantly, by ∼40%, as early as 9 weeks in HdhQ200 mice compared with WT (Fig. 10A). This LC3-II increase was accompanied by a ∼20% increase in the level of p62 (Fig. 10B), a protein that facilitates selective delivery of misfolded, ubiquitinated proteins to the autophagosome (45).
Figure 10.
Heterozygous HdhQ200 elicits early and sustained response of autophagic markers. (A) Representative western blots of soluble fractions of total brain lysates showing significantly increased LC3-II with a ∼40% increase standardized to tubulin as early as 9 weeks and at 40 and 80 weeks of age in heterozygous HdhQ200 mice. (B) This LC3-II increase is accompanied by a ∼20% increase in p62 at all corresponding ages standardized to tubulin. Error bars represent the SEM, and asterisks denote significance at P < 0.05.
To extend our observations of an autophagic/lysosomal response, we employed electron microscopy to obtain ultrastructural evidence of an autophagy-lysosomal response. For this, we studied HdhQ150 homozygous mice at 70 weeks of age, when perinuclear, punctate AF are as abundant as in heterozygous HdhQ200 mice at 40 weeks of age. Electron micrographs of striatal neurons exhibited a marked increase in the number and size of electron-dense, single membrane-bound, cytoplasmic vesicles. These structural features are consistent with lysosomes and possibly autolysosomes, an intermediate autophagosome fusing with lysosomes to form a single membrane-bound vesicle (Supplementary Material, Fig. S5). During disease states, lysosomes often contain lipids and are non-uniform in structure, which may be suggestive of a protein aggregate containing lysosome. These lysosome-like structures in the HdhQ150 knock-in mice also increased in size, averaging 649 618 ± 7973 nm2 in total area, approximately a 6.9-fold increase compared with WT, which measured 94 555 ± 2704 nm2 in approximate area (arrows). Lysosome size has been shown to correlate with autophagy/lysosomal induction (46–49).
DISCUSSION
We provide in vivo evidence of early and sustained changes in autophagy in response to expanded polyQ htt protein expression in a novel knock-in mouse model of HD. The HdhQ200 knock-in model, generated by several rounds of selective breeding for further CAG expansions starting with HdhQ150 knock-in mice, manifests an accelerated and more robust version of the progressive disease phenotype reported in the parent line (8). The more aggressive phenotype of the HdhQ200 mice relative to the HdhQ150 mice is not due to greater levels of striatal/cortical HdhQ200 mRNA or protein. Figure 11 provides a summary and comparison of the time course of phenotypic features in HdhQ200 versus HdhQ150 knock-in mice. HdhQ200 mice accumulate polyQ protein aggregates (cytoplasmic AF) as early as 9 weeks of age and striatal NIIs as early as 20 weeks, whereas HdhQ150 heterozygous knock-in mice do not exhibit distinct AF until 70 weeks of age. Saturation of striatal neurons with NIIs occurs earlier in HdhQ200 mice, which also exhibit more neocortical NIIs than HdhQ150. This earlier appearance of aggregate pathology is paralleled by earlier and more rapidly progressive behavioral abnormalities. HdhQ200 mice show progressive motor impairments on the balance beam starting at 50 weeks, several months earlier than the age at which these abnormalities appear in the HdhQ150 mice. These motor deficits in HdhQ200 are followed by obvious and progressive gait deficits demonstrated by footprint analysis at 60 weeks. We did not find the widely used rotarod test useful in assessing motor deficits in this model. Severe motor deficits at 80 weeks are accompanied by striatal and cortical astrogliosis and a ∼50% reduction in striatal dopamine receptors. In contrast, HdhQ150 heterozygous mice exhibit smaller reductions in striatal dopamine receptor binding and at a later age (100 versus 80 weeks for heterozygous HdhQ150 and heterozygous HdhQ200 mice, respectively). It is likely that the behavioral abnormalities of HdhQ200 mice reflect neuronal dysfunction rather than overt neurodegeneration as stereological analysis did not reveal striatal neuron loss. NeuN staining of striatal neurons did reveal consistent observations of neurons containing many, often large cytoplasmic vacuoles, which may provide another marker for neuronal dysfunction prior to neuronal death. This inference is consistent with human imaging studies suggesting a prolonged period of striatal dysfunction prior to onset of manifest disease (32). Evidence supporting the idea that neuronal dysfunction, and not the loss of neurons per se, is likely to mediate important behavioral deficits is found in models of other polyQ diseases. One mouse model of SCA1, for example, develops ataxia as a result of neuronal dysfunction rather than cell loss (50). The important role of neuronal dysfunction is observed also in Alzheimer and Parkinson disease mouse models (51). Xia et al. (52) and Yamamoto et al. (53) demonstrated that eliminating expression of mutant proteins in SCA1 and in HD models, respectively, reversed motor dysfunction, offering further evidence for the importance of neuronal dysfunction. It is plausible that successful interventions for polyQ diseases will have to be targeted at neuronal dysfunction prior to neurodegeneration. The model reported here, with prominent neuronal dysfunction correlated with motor abnormalities, may be particularly useful for investigating mechanisms of neuronal dysfunction and testing interventions aimed at ameliorating neuronal dysfunction.
Homozygous HdhQ200 mice exhibit an even more accelerated phenotype with motor deficits commencing at 20 weeks accompanied by reduced weight, with 40–50% reduction in striatal D1 and D2 dopamine receptors at 40 weeks of age. Like the HdhQ150 model and several other knock-in lines (9–12), HdhQ200 mice are a model with good face and construct validity and with the added advantage of a more robust and aggressive phenotype than other reported HD knock-in lines.
This line is likely to be a useful addition to the stable of HD murine genetic models that include transgenic fragment models (R6/2 and derivatives, N-171), full-length transgenic models (YAC128 and BACHD) and several knock-in lines (7,54). Comparison of models can be confounded by background strain effects and differences in evaluation methods. A recent systematic analysis of several models provides useful information for comparing the HhdQ200 line with other models, though this analysis did not use the balance beam task, which we found particularly useful (54,55). The phenotype of heterozygous HdhQ200 mice begins later and progresses slowly than in R6/2 lines but like the R6/2 lines, exhibits deficits on more than one motor function test, including gait analysis and grip-strength. YAC128 and BACHD mice both exhibit rotarod deficits but the YAC128 deficits are apparently non-progressive. Neither the YAC128 nor the BACHD mice had marked gait analysis deficits with the BACHD mice exhibiting progressive deficits on a test of climbing behavior. Both YAC128 and BACHD mice exhibited a significant weight gain compared with WT controls, in contrast to R6/2 mice and female heterozygous HdhQ200 mice. The increased weight of these models may complicate interpretation of some motor tests, though this does not seem to be an issue with the rotarod test (54). For studies where neuronal loss would be a desired endpoint, the R6/2 lines, YAC128 lines or BACHD lines would be advantageous (56–58). Although the R6/2 model does exhibit striatal neuron loss, the profound behavioral abnormalities of this line seems out of proportion to the degree of neuronal loss, suggesting a large component of neuronal dysfunction, perhaps analogous in some respects to the HhdQ200 heterozygous mice.
We also demonstrate early neuronal cytoplasmic deposition of mhtt protein aggregates in the form of polyQ recruitment competent AF well before NIIs are present. This observation is consistent with the detection of cytoplasmic aggregates in HD brains; post-mortem brains with early grade disease contain more neuropil aggregates than NIIs (14,59). Although these earlier studies used immunohistochemistry to identify cytoplasmic aggregates, AF have been identified in HD post-mortem brain and the AF method is a highly sensitive histochemical method with which to detect aggregating polyQ proteins (as AF) in polyQ disease mouse models and HD post-mortem brain (36), where, in the latter case, these foci appear to correspond to precursors in the formation of neuropil aggregates, which, like NIIs, lose the ability to further recruit polyQ. Data from some mouse models of HD suggest the correlation of cytoplasmic aggregate accumulation with neuronal dysfunction and degeneration of neuronal processes (34,60). Collectively, human post-mortem analyses and murine experimental studies suggest that early pathologic events preceding perikaryal loss are associated with cytoplasmic expanded polyQ protein deposition (14,34,60). We do not wish to imply that cytoplasmic aggregates directly contribute to pathogenesis, though this remains a possibility; rather, we simply wish to emphasize that early cytoplasmic polyQ protein aggregates in neurons are a pathologic feature shared by human HD and HdhQ200 mice, further supporting the validity of this newest knock-in model.
The cellular distribution of AF in HdhQ200 mice evolves over time. Initially expressed diffusely throughout the cytoplasm, AF shift to a perinuclear localization by 40 weeks of age (Fig. 8A). Striatal perinuclear AF in striatal neurons co-localize with the autophagosome marker LC3 and the development of widespread AF is paralleled by increased expression in LC3 immunoreactive cytoplasmic punctae, suggesting increased expression of autophagosomes and association of AF with autophagosomes. The perinuclear localization of AF in 40-week-old mice and the absence of AF from the nucleus are consistent with an association with autophagosomes–lysosomes. Autophagosomes are transported to the perinuclear microtubule organizing center (MTOC) for fusion with lysosomes, and autophagosomes–lysosomes are excluded from the nucleus. An association of cytoplasmic mhtt inclusions with LC3 immunoreactivity was reported previously in 16-week-old transgenic R6/2 mice and in 1-year old homozygous HdhQ150 mice (61). Consistent with our morphologic results, western blotting indicated early and persistent elevation of LC3-II and another autophagy-associated protein, p62, in HdhQ200 heterozygous mouse brain. Increased autophagy should be associated with increased lysosome formation; indeed, we found ultrastructural evidence of increased lysosomes in HdhQ150 knock-in mouse striatal neurons. The specific types of aggregate species that compose AF, such as toxic monomers, oligomers or amyloid fibrils, are unknown and require further investigation. Taken together, our data indicate that cytoplasmic deposition of mhtt aggregates is an early event in this model and likely provokes a response by the autophagy system.
Macroautophagy (commonly referred to as autophagy) is a crucial cellular process for bulk degradation of organelles and long-lived proteins. Basal autophagy appears to be particularly important for protecting neurons from the toxic effects of misfolded proteins as murine knockouts of two key autophagy genes result in neurodegeneration with protein aggregate accumulations (62,63). In autophagy, cytoplasmic substrates are delivered via autophagosomes to lysosomes. Prior evidence suggests altered autophagy in HD. Sapp et al. (21) reported mhtt accumulation in neuronal perinuclear and cytoplasmic structures that resembled endosomal–lysosomal organelles in post-mortem HD brain. Similarly, autophagosome–lysosome-like structures have been reported in HD post-mortem brain by another group (40) and in lymphoblasts of HD subjects (39). A growing body of work in HD models suggests altered autophagy in response to mutant polyQ proteins and supports the view that increased autophagy is an important cytoprotective response to expanded repeat polyQ proteins (23,24,29,61,64,65). Post-translational modifications of mhtt, including acetylation and phosphorylation, appear particularly important in directing mhtt into the autophagic pathway (66–68). Autophagy has been implicated also in the cellular response to expanded polyQ proteins in models of spinobulbar muscular atrophy and SCA3 (23,69). Most studies indicating altered autophagy in polyQ diseases have been performed in cell-based or invertebrate models of HD and SBMA, though some work has been performed with transgenic mouse models. Our observations in a genetically precise and well-validated murine model of HD provide in vivo evidence that altered autophagy is likely an important cellular response to expanded repeat polyQ proteins.
Our data are consistent with an induction of autophagy. The protein LC3-II is uniquely associated with autophagosomes, and increased expression of LC3-II and LC3 immunoreactive vesicles is often interpreted as evidence of increased autophagic turnover. The protein p62 is another key actor in autophagic flux; this ubiquitin-binding protein is thought to mediate the entry of abnormal proteins, including mhtt, into autophagosomes by binding LC3-II (70). Increased p62 expression, however, is commonly interpreted as evidence of reduced autophagic flux (71–74). Jointly increased expression of LC3-II and p62 could reflect a downstream block in autophagic flux. Boland et al. (75), for example, inhibited lysosomal proteolysis and autophagosome-lysosomal fusion in cultured cortical neurons, increasing expression of LC3-II containing autophagic vesicles. Prior studies by Atwal et al. (76) and Shibata et al. (77) suggest that mhtt can impair autophagy. At present, differentiating increased autophagic flux from inhibition of autophagy downstream of autophagosome formation is only feasible with in vitro systems (44). Direct measurements of autophagic flux in vivo are needed to determine whether our findings in HdhQ200 mice result from induction of autophagy or a block in the pathway downstream of autophagosome formation. Our results are consistent with recent comprehensive work by Martinez-Vicente et al. (29) indicating that autophagic flux is increased in HD but functions inefficiently because of impaired ability to recognize and load cytoplasmic cargo into autophagosomes. The results of Martinez-Vicente et al. may reconcile our findings of increased expression of LC3-II and p62. Martinez-Vicente et al. also describe higher density of vacuolated structures within HD model neurons and HD lymphoblasts, a result consistent with our observation of increased vacuolization of striatal neurons in aged heterozygous HdhQ200 mice.
It is possible that elevated p62 expression could be driving a potentially protective cellular response. Komatsu et al. (78) demonstrated recently that elevated cellular p62, often associated with defective autophagy, stabilizes the levels of the protective stress response transcription factor Nrf2 by inhibiting Nrf2 ubiquitylation by the ubiquitin ligase Keap1.
Observations such as these have led to interest in manipulating autophagy pathways to promote clearance of expanded polyQ and other aggregation-prone proteins. Rubinsztein and colleagues have suggested mTOR-dependent and mTOR-independent interventions to increase autophagic flux and promote expanded repeat polyQ protein clearance (25–28,79). These strategies are attractive in view of the evidence that mhtt engages in a number of proximate mechanisms of pathogenesis, suggesting that interrupting only one proximate mechanism of pathogenesis will not be sufficient (3,80). It is possible, however, that increased autophagy might be detrimental to neuronal survival in HD (22). In a model of frontotemporal dementia due to dysfunctional ESCRT-III, Lee and Gao (81) demonstrated that dysregulated autophagy contributes to neurodegeneration and that a reduction in neuronal loss occurred when autophagy was inhibited. The apparent success, however, of interventions aimed at increasing autophagic flux in a variety of model systems of HD, and other polyQ diseases supports the safety of increasing autophagic flux for treatment of polyQ disorders.
The accelerated phenotype of the HdhQ200 mice is consistent with data from humans showing that larger polyQ expansions correlate with earlier onset of clinically manifest HD. Intergenerational repeat instability is a common feature of polyQ diseases with larger repeat expansions tending to cause earlier onset disease. Prior work with knock-in HD models indicated that longer repeats generate more robust motor and neuropathological phenotypes (8,10–12). Our work, together with prior experience in other knock-in models of HD, contrasts with interesting recent work in the widely used R6/2 transgenic model of HD. R6/2 mice express a transgene fragment that includes the human promoter region and exon 1 containing the expanded CAG/polyQ domain (82). The original R6/2 construct expressed ∼150 polyQ repeats, and R6/2 mice exhibited virtually complete mortality by 12–16 weeks of age. Subsequent increased polyQ repeat sizes in the range of 170–450 have been associated with a delayed onset of manifest disease and prolonged survival with a positive correlation between repeat size and survival. This is most marked with so-called R6/2x mice that exhibit very large repeat numbers, in excess of 335, and have a markedly attenuated phenotype (83,84). R6/2x mice exhibit reduced expanded polyQ protein expression compared with the parent R6/2 line, and the large polyQ repeat size may retard nuclear entry of expanded polyQ fragments. In their thoughtful analysis of R6/2 mice with very large repeats, Morton et al. note that mid-stage mice with very large repeats exhibit significant neurodegeneration, which is distinct from the relatively modest neuropathology of mid-stage R6/2 mice with ∼150 repeats. Morton et al. (84) suggest that amelioration of the phenotype in R6/2 mice with very long repeat lengths unmasks a slowly progressive phenotype with significant neurodegeneration that more closely resembles HD than the very aggressive phenotype of the parent R6/2 line. Further repeat length expansion in R6/2 mice lessens the R6/2 phenotype but perhaps by diminishing toxic effects of the transgene that are less relevant to HD.
In summary, we describe a new knock-in model of HD with good face and excellent construct validity and a sufficiently robust phenotype in heterozygous mice to permit efficient evaluation of disease mechanisms. Homozygous HdhQ200 mice have a particularly aggressive phenotype and may be useful in preclinical pharmacology experiments. Our observations in this model revealed the presence of early neuronal cytoplasmic aggregates of mhtt and suggest that altered autophagy is an early and sustained event in HD.
MATERIALS AND METHODS
Generation of the HdhQ200 knock-in line
HdhQ200 mice were produced by selective breeding that exploited germline repeat length instability in the HdhQ150 knock-in line described previously (85). Mice with large expansions were chosen for further breeding during backcrossing to C57BL/6J. After 7–8 generations of backcrossing, this strategy yielded the mice used in this study with ∼200 CAG repeats (200 ± 10 CAGs).
Mouse colony maintenance
All experiments were performed with the HdhQ200 knock-in murine model of HD maintained on a C57BL/6 genetic background. For all experiments, female and male heterozygous HdhQ200 knock-in mice were mated to produce homozygous, heterozygous and WT mice. Both male and female HdhQ200 mice and WT mice were used. All animals were housed in cages grouped by gender and provided with food and water ad libitum. Animals were housed in a specific pathogenic-free condition with a 12-h light/dark cycle maintained at 23°C. All procedures were conducted in strict compliance with the Guide for the Care and Use of Laboratory Animals as adopted by the NIH and approved by the Veterinary Medical Unit (VMU) at the Veterans Affairs Ann Arbor Health System.
Genotyping
All genotyping was performed at Laragen (Laragen Inc., Los Angeles, CA, USA). Tail biopsies were sent to Laragen Inc. for genotyping and CAG sizing to confirm CAG stability and consistency of the CAG repeat number. The HdhQ200 mutant allele ranged from 190 to 210 CAG repeats.
Expression analysis
QRT–PCR was performed by the ΔΔCt method to β-actin control using Applied Biosystems reagents as previously described in Dixon et al. (86). All mice were between 14 and 27 weeks of age and were homozygotes for Hdh alleles. For western blots, tissues from the cortex and striatum of WT, heterozygous Q150 and heterozygous Q200–Q220 mice between 12 and 23 weeks of age were homogenized in a lysis buffer [50 mm Tris–HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 0.1 mm phenylmethylsulfonyl fluoride and a 10-µg/ml concentration of each of aprotinin, leupeptin and pepstatin] with a glass/glass Dounce homogenizer, prior to being sonicated on ice and spun at 2000g for 10 min at 4°C. Protein concentration of the supernatant was determined using the bicinchoninic acid assay method, and cellular lysates were diluted to a final concentration of 1 mg/ml in 2× reducing stop buffer [0.25 m Tris–HCl (pH 7.5), 2% SDS, 25 mm dithiothreitol, 5 mm EGTA, 5 mm EDTA, 10% glycerol and 0.01% bromophenol blue as tracking dye] and incubated in a boiling water bath for 5 min. Proteins were separated by SDS–PAGE on 3–16% gradient gel then transferred to nitrocellulose. Membranes were probed with the indicated primary antibodies [mouse anti-polyQ expansion 1C2 (1:5000 dilution; Millipore, Billerica, MA, USA), and mouse anti-htt 2166 (1:2000 dilution; Millipore)] followed by incubation with horseradish peroxidase–conjugated secondary antibodies according to the standard protocols. Blots were developed using peroxidase substrate chemiluminescence.
Behavioral examination
To assess motor deficits, animals were tested on a standard battery of motor tests as described previously (8). These included the balance beam test with 11 mm round and 5 mm square beams, rotarod, footprint analysis and grip-strength meter analysis (San Diego Instruments, San Diego, CA, USA). To observe progressive motor decline, HdhQ200 and WT control animals were assessed at six time points: 10, 20, 50, 60, 70 and 80 weeks of age. If not otherwise mentioned, six animals per group were used at 10- and 20-week time points; 10 animals per group for remaining time points; however, two animals died at around 75 weeks, resulting in an N of 8 for the HdhQ200 heterozygous group for the 80-week time point. Animals at 80 weeks were euthanized for pathologic examination, as HdhQ200 animals appeared moribund at this time point. Homozygous mice were assessed similarly at 13, 20, 30 and 40 weeks of age, with 4–5 animals analyzed per group. All behavioral evaluations were videotaped and performed with the examiner blind to genotype. Brains were collected on the last day of testing.
All animals were euthanized according to the national guidelines. Euthanasia was carried out by decapitation. This method was approved by the VMU.
Immunohistochemistry, immunofluorescence and stereology
Immunohistochemistry, stereology and receptor autoradiography analyses were performed as described previously (8). Briefly, one fresh frozen hemisphere was prepared for autoradiographic receptor binding analyses, with the corresponding hemisphere fixed in 4% paraformaldehyde for 24 h, cryoprotected in 20% sucrose in 0.1 m phosphate buffer for an additional 24 h at 4°C, frozen and stored at −80°C for later stereological analysis and immunohistochemistry. All studies were coded and performed blind to the genotype. Free-floating 40-µm sections were processed with primary antisera against the neuronal antigen NeuN (Millipore; 1:100 dilution), GFAP (Dako, Produktionsvej 42aDK-2600 Glostrup, Denmark; 1:1000 dilution), htt N-terminal (N-18) antibody (Santa Cruz Biotechnology, Inc.; 1:250 dilution) and LC3B (Novus Biologicals, Littleton CO, USA; 1:1000 dilution). Detection of immunoreactivity was performed using the Vectastain Elite kit (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer's protocol, and sections were developed in ImmPACT DAB (Vector Laboratories) for visualization. To control for background staining, sections were processed in the absence of primary antisera. Sections were mounted on Superfrost slides (Fisher Scientific, Pittsburgh, PA, USA) and, once dried, were dehydrated in graded alcohols and xylene. Coverslips were affixed with DPX (Electron Microscopy Sciences, Hatfield, PA, USA). Quantification of reactive astrogliosis was quantified using NIH ImageJ software. Particle number was quantified with the ‘analyze particles’ function in threshold and measure functions with the scale standardized to the scale bar and set globally.
Unbiased stereological counts of neurons were obtained from the striatum of animals at 80 weeks of age using the StereoInvestigator software (Micro-Brightfield, Colchester, VT, USA). The optical fractionator method was used to generate an estimate of neuronal number with positively stained NeuN immunoreactive cells counted in an unbiased selection of serial sections in a defined volume of the striatum. Striatal volume was reconstructed by the StereoInvestigator software. Serially cut sagittal tissue sections (every fourth section) were analyzed for one entire hemisphere of animals in each genotype cohort (n = 5 per group).
Quantitative receptor autoradiography
Fresh frozen hemispheres were cut serially in the parasagittal plane on a cryostat in 15 µm sections at −12°C, thaw mounted on Superfrost slides, and stored at −80°C (n = 5 animals per group). Autoradiographic studies were performed as described previously (8). Briefly, assays for D1 and D2 dopamine receptors were performed as follows: slides with sections were incubated in 25 mm Tris, 100 mm NaCl, 1 mm MgCl2, 0.001% ascorbic acid, 1 µm pargyline, pH 7.2 with radioactive ligand 0.55 nm [3H]SCH23390 (specific activity 89 Ci/mmol; Amersham, UK) for D1 and 0.75 nm [3H]Spiperone (specific activity 96 Ci/mmol; Amersham) with 100 nm mianserin for D2 for 2.5 h at room temperature. Non-specific binding was determined in the presence of 1 µM cis-flupenthixol for D1and 50 µm dopamine for D2, respectively. For GABA-A/benzodiazepine receptors, slides were prewashed in 50 mm Tris-citrate buffer, pH 7.2, at 4°C for 30 min, air-dried and incubated in 50 mm Tris-citrate buffer, pH 7.2, with 5 nm [3H]flunitrazepam (specific activity 85 Ci/mmol; Perkin Elmer, Waltham, MA, USA) for 1 h at 4°C. Non-specific binding was determined in a ligand buffer with 2 µm clonazepam.
Slides and 14C standards (ARC, St Louis, MO, USA) are arrayed in a standard Fujifilm BAS cassette (Fujifilm Bas cassette 2025), and an autoradiogram is generated by direct apposition of the tissue to the emulsion side of a tritium imaging plate (BAS-TR2025; Fuji Photo Film, Japan) for 8 h.
Autoradiograms were analyzed by quantitative densitometry using an MCID-M2 image analysis system (Interfocus Ltd, England). Optical density (OD) measurements were made in each section in which the structure was visible, sampling as large an area as possible. These parasagittal sections sampled the entire medial–lateral extent of the striatum. Tissue radioactivity was determined by comparison of ODs with a calibration curve obtained from the co-expressed standards. Specific binding was determined by subtracting non-specific binding from the total binding.
Aggregation foci
Synthetic peptide bPEGQ30, prepared at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT, USA; http://info.med.yale.edu/wmkeck), composed of 30 glutamine residues flanked by two lysyl residues at each end to enhance solubility and, attached at the N-terminus, a glutamine residue with a polyethylene glycol moiety linked at the ϒ-amide position, conjugated to biotin (polyQbiotin). For polyQbiotin detection of AF, hemispheres from paraformaldehyde-fixed preparations were sectioned at 40 µm and processed as described previously (36). Serially cut parasagittal tissue sections (every fourth section) were processed for one entire hemisphere of animals in each genotype cohort (n = 3 per group). Briefly, free-floating sections were subjected to two washes in 0.1 m PBS (pH 7.4) for 10 min each, followed by incubation in 1% NaBH4 in 0.1 m PBS for 30 min, followed by two washes in PBS for 10 min, three washes in PBSTx (PBS plus 0.4% TX-100) each at 10, 30 and 10 min. Sections were incubated in 50 nm biotinylated polyQ peptide (bPEGQ30 50 nm) in PBSTx for 18 h, then subjected to four 15 min washes in PBSTx, followed by 60 min incubation in an avidin-biotinylated enzyme complex (1:400 dilution ABC Elite kit; Vector Laboratories). Sections were washed in PBSTx twice for 15 min followed by incubation in 1:100 dilution biotinylated tyramide; 1:50 000 dilution H2O2 30% stock for 15 min. Floating sections were developed using ImmPACT DAB following the manufacturer's instructions for visualization. All washes and incubations were at room temperature. Sections were mounted, air-dried and dehydrated in graded alcohols and xylene, and coverslips affixed with DPX.
Confocal microscopy
For confocal and double-label immunofluorescence, free-floating sagittal sections from HdhQ200 mice were incubated with alexa fluorophore goat anti-rabbit (1:1000 Invitrogen, Carlsbad, CA, USA) and streptavidin 488 tyramide (1:1000, Invitrogen) in 0.1 m Tris–HCL buffer, pH 7.4, for 2 h at room temperature in darkness. Sections were then washed, mounted and coverslips affixed with ProLong Gold with DAPI (Invitrogen).
Striatal sections were examined and scanned in a Zeiss LSM 510 microscope (Thornwood, NY, USA) equipped with an argon/Hene1 mixed gas laser. Zeiss LSM version 4.2 Image Browser software was used to establish the optimal conditions for collecting images. Stacks of confocal images were captured with a 60× PlanApochromat objective (numerical aperture 1.4) with 2× zoom function. A series of images at 1 µm intervals throughout the entire 30 µm section were collected for analysis. Sections sampled represented regions throughout the entire medial–lateral extent of the striatum. Images are presented here as single optical images or projections of optical image stacks. Laser power was set at 10%, optical sections were scanned at increments of 1 µm, which correlates with the resolution value at z-plane, scanning was at 500 lps. Each optical section was the result of three scans followed by the Kalman filtering, and the size of the image was 512 × 512 pixels. The 594-labeled brain sections were studied in the same manner except that the laser power was set at 100% (n = 3 per group). For total counts of LC3 punctae per nucleus, the number of punctae per nucleus were averaged over counting 10–15 random field views through 1 µm thick sections over a total of three total sections per animal. All slides were coded and performed blind to the genotype.
Transmission electron microscopy
Animals were deeply anesthetized and intracardially perfused with 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 mol/l Sorensen's buffer, pH 7.4 (n = 3 per group). Coronal striatal sections of 1-mm thick were collected and further fixed overnight at 4°C. Samples are post-fixed in 1% osmium tetroxide in 0.1 mmol/l cacodylate buffer. Samples were rinsed in distilled water and treated with 1% aqueous uranyl acetate overnight, dehydrated in ascending series of alcohol dilutions to 100%, followed by acetone. Samples are infiltrated with propylene oxide, embedded in Epon and sectioned. Semi-thin sections were stained with toluidine blue for tissue identification. Selected regions of interest were ultrathin sectioned at 70 nm in thickness and post-stained with uranyl acetate and lead citrate. Images were examined with a Philips CM 100 transmission electron microscope at 60 kV and digitally imaged using a Hamamatsu ORCA-HR camera (Hamamatsu City, Shizuoka, Japan). The area of electron-dense lysosome-like structure was quantified using NIH ImageJ software. Striatal sections of electron-dense lysosome-like structures at a direct magnification of ×13 500 were traced individually using the threshold and measure functions with the scale standardized to the scale bar and set globally.
Brain lysate, western blotting and AGERA
The following antibodies were used: mouse anti-polyQ expansion 1C2 (1:5000; Millipore), rabbit polyclonal anti-LC3B (1:1000; Novus Biologicals), rabbit polyclonal anti-p62 (1:1000; American Research Products, Belmont, MA, USA), mouse anti-α-tubulin (1:20 000; Sigma-Aldrich, St Louis, MO, USA) and peroxidase-conjugated, goat anti-rabbit, goat anti-mouse secondary antibodies (1:10 000; Jackson Immunoresearch, Westgrove, PA, USA). Primary antibodies are diluted in Tween-TBST with 5% milk and incubated with membranes overnight at 4°C. Secondary antibodies were incubated with membranes for 1 h (n = 3 per group).
Flash-frozen brains from HdhQ200 knock-in mice were homogenized in NETN buffer [50 mm Tris–HCL, pH 7.5, 150 mm NaCl, 0.5% (v/v) Nonidet P-40] with phosphatases: 25 mm NAF, Na3VO4, glycerophosphate, and 4× protease inhibitors (Sigmafast protease inhibitor tablets, Sigma-Aldrich), sonicated and microcentrifuged at 14 000g at 4°C for 20 min. For western blotting and agarose gel electrophoresis for resolving aggregates (AGERA), supernatants were supplemented with 1% final SDS and 100 mm DTT, boiled and loaded on 10% SDS–PAGE gels and transferred to polyvinylidene difluoride (PVDF) membrane in 5 mm Tris protein running buffer (38). For AGERA, samples were resolved on 1.5% w/v agarose in TAE buffer with 0.1% SDS and run in 5 mm Tris protein running buffer. For AGERA, Agarose-SDS gels are transferred to PVDF membrane using a semi-dry apparatus in a discontinuous buffer system [12 mm Tris base, 8 mm CAPS, 0.1% SDS cathode buffer; 12 mm Tris base, 8 mm CAPS, 15% methanol anode buffer (38)]. Membranes are washed in Tween-TBS, blocked in 5% milk in Tween-TBS, followed by chemiluminescence (Western Lightning, Perkin Elmer) and imaged using VersaDoc 5000 MP (Bio-Rad, Hercules, CA, USA) and quantified using Quantity One (Bio-Rad). Background was subtracted equally among lanes, and bands were standardized to tubulin.
Statistical analyses
All studies were performed blind to genotype. Comparisons of different groups (between genotypes) were performed on experiments requiring non-repeated trials (weight, immunohistochemistry, immunofluorescence, quantitative receptor autoradiography, stereology, biochemistry, grip-strength meter analysis and quantitative analysis with ImageJ) using an one-way ANOVA or two-tailed independent t-tests. Two-way repeated-measures ANOVA with repeated measures with a between-subject factor for genotype and repeated measures for training were performed on experiments for behavioral tests requiring repeated trials (11 mm round and 5 mm square beams, and rotarod). There were no interactions observed between genotype and training. All data are presented as the mean ± SEM. A critical P < 0.05 was used for statistical significance in all analyses. The Huynh–Feldt adjustment was used in correcting for the violation of sphericity when necessary to adjust non-uniform variance across days or groups. SPSS statistical software was used.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a VA Merit Review grant (to R.L.A.) and the National Institutes of Health (R21 NS059537 to R.L.A., R21 NS059647 to P.J.D., RO1 NS038712 to H.L.P. and T32 NS007222 to M.Y.H.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Erin Katz for her technical assistance. We thank Dr Tom Morrow for use of his MCID system, Dr Kirk Frey for use of his stereology apparatus, Dr Sokol Todi for assistance with western blotting and Dr Maria do Carmo Costa for assistance with intracardial perfusion.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Williams A.J., Paulson H.L. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. doi:10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warby S., Graham R., Hayden M.R. Huntington disease. 2008. www.genetests.org .
- 3.Imarisio S., Carmichael J., Korolchuk V., Chen C.W., Saiki S., Rose C., Krishna G., Davies J.E., Ttofi E., Underwood B.R., et al. Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412:191–209. doi: 10.1042/BJ20071619. doi:10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 4.Orr H.T., Zoghbi H.Y. Reversing neurodegeneration: a promise unfolds. Cell. 2000;101:1–4. doi: 10.1016/S0092-8674(00)80617-0. doi:10.1016/S0092-8674(00)80617-0. [DOI] [PubMed] [Google Scholar]
- 5.Paulson H.L., Perez M.K., Trottier Y., Trojanowski J.Q., Subramony S.H., Das S.S., Vig P., Mandel J.L., Fischbeck K.H., Pittman R.N. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. doi:10.1016/S0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 6.Ferrante R.J. Mouse models of Huntington's disease and methodological considerations for therapeutic trials. Biochim. Biophys. Acta. 2009;1792:506–520. doi: 10.1016/j.bbadis.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heng M.Y., Detloff P.J., Albin R.L. Rodent genetic models of Huntington disease. Neurobiol. Dis. 2008;32:1–9. doi: 10.1016/j.nbd.2008.06.005. doi:10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Heng M.Y., Tallaksen-Greene S.J., Detloff P.J., Albin R.L. Longitudinal evaluation of the Hdh(CAG)150 knock-in murine model of Huntington's disease. J. Neurosci. 2007;27:8989–8998. doi: 10.1523/JNEUROSCI.1830-07.2007. doi:10.1523/JNEUROSCI.1830-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickey M.A., Kosmalska A., Enayati J., Cohen R., Zeitlin S., Levine M.S., Chesselet M.F. Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington's disease mice. Neuroscience. 2008;157:280–295. doi: 10.1016/j.neuroscience.2008.08.041. doi:10.1016/j.neuroscience.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menalled L.B., Sison J.D., Dragatsis I., Zeitlin S., Chesselet M.F. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J. Comp. Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. doi:10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 11.Shelbourne P.F., Killeen N., Hevner R.F., Johnston H.M., Tecott L., Lewandoski M., Ennis M., Ramirez L., Li Z., Iannicola C., et al. A Huntington's disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Hum. Mol. Genet. 1999;8:763–774. doi: 10.1093/hmg/8.5.763. doi:10.1093/hmg/8.5.763. [DOI] [PubMed] [Google Scholar]
- 12.Wheeler V.C., Gutekunst C.A., Vrbanac V., Lebel L.A., Schilling G., Hersch S., Friedlander R.M., Gusella J.F., Vonsattel J.P., Borchelt D.R., et al. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum. Mol. Genet. 2002;11:633–640. doi: 10.1093/hmg/11.6.633. doi:10.1093/hmg/11.6.633. [DOI] [PubMed] [Google Scholar]
- 13.Davies S.W., Turmaine M., Cozens B.A., DiFiglia M., Sharp A.H., Ross C.A., Scherzinger E., Wanker E.E., Mangiarini L., Bates G.P. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. doi:10.1016/S0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 14.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. doi:10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 15.Chai Y., Koppenhafer S.L., Shoesmith S.J., Perez M.K., Paulson H.L. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. doi:10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 16.Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. doi:10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 17.Maynard C.J., Bottcher C., Ortega Z., Smith R., Florea B.I., Diaz-Hernandez M., Brundin P., Overkleeft H.S., Li J.Y., Lucas J.J., et al. Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc. Natl Acad. Sci. USA. 2009;106:13986–13991. doi: 10.1073/pnas.0906463106. doi:10.1073/pnas.0906463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowman A.B., Yoo S.Y., Dantuma N.P., Zoghbi H.Y. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 2005;14:679–691. doi: 10.1093/hmg/ddi064. doi:10.1093/hmg/ddi064. [DOI] [PubMed] [Google Scholar]
- 19.Davies J.E., Sarkar S., Rubinsztein D.C. The ubiquitin proteasome system in Huntington's disease and the spinocerebellar ataxias. BMC Biochem. 2007;8((Suppl. 1)):S2. doi: 10.1186/1471-2091-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortega Z., Diaz-Hernandez M., Maynard C.J., Hernandez F., Dantuma N.P., Lucas J.J. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J. Neurosci. 2010;30:3675–3688. doi: 10.1523/JNEUROSCI.5673-09.2010. doi:10.1523/JNEUROSCI.5673-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapp E., Schwarz C., Chase K., Bhide P.G., Young A.B., Penney J., Vonsattel J.P., Aronin N., DiFiglia M. Huntingtin localization in brains of normal and Huntington's disease patients. Ann. Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. doi:10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 22.Kegel K.B., Kim M., Sapp E., McIntyre C., Castano J.G., Aronin N., DiFiglia M. Huntingtin expression stimulates endosomal–lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montie H.L., Cho M.S., Holder L., Liu Y., Tsvetkov A.S., Finkbeiner S., Merry D.E. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2009;18:1937–1950. doi: 10.1093/hmg/ddp115. doi:10.1093/hmg/ddp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey U.B., Batlevi Y., Baehrecke E.H., Taylor J.P. HDAC6 at the intersection of autophagy, the ubiquitin-proteasome system and neurodegeneration. Autophagy. 2007;3:643–645. doi: 10.4161/auto.5050. [DOI] [PubMed] [Google Scholar]
- 25.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. doi:10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 26.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. doi:10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar S., Davies J.E., Huang Z., Tunnacliffe A., Rubinsztein D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. doi:10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 28.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H., Jahreiss L., Fleming A., Pask D., Goldsmith P., et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. doi:10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E., Harris S., Sulzer D., et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. doi:10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey U.B., Nie Z., Batlevi Y., McCray B.A., Ritson G.P., Nedelsky N.B., Schwartz S.L., DiProspero N.A., Knight M.A., Schuldiner O., et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 31.Liu J., Tang T.S., Tu H., Nelson O., Herndon E., Huynh D.P., Pulst S.M., Bezprozvanny I. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. J. Neurosci. 2009;29:9148–9162. doi: 10.1523/JNEUROSCI.0660-09.2009. doi:10.1523/JNEUROSCI.0660-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weeks R.A., Piccini P., Harding A.E., Brooks D.J. Striatal D1 and D2 dopamine receptor loss in asymptomatic mutation carriers of Huntington's disease. Ann. Neurol. 1996;40:49–54. doi: 10.1002/ana.410400110. doi:10.1002/ana.410400110. [DOI] [PubMed] [Google Scholar]
- 33.Maat-Schieman M.L., Dorsman J.C., Smoor M.A., Siesling S., Van Duinen S.G., Verschuuren J.J., den Dunnen J.T., Van Ommen G.J., Roos R.A. Distribution of inclusions in neuronal nuclei and dystrophic neurites in Huntington disease brain. J. Neuropathol. Exp. Neurol. 1999;58:129–137. doi: 10.1097/00005072-199902000-00003. doi:10.1097/00005072-199902000-00003. [DOI] [PubMed] [Google Scholar]
- 34.Wang C.E., Zhou H., McGuire J.R., Cerullo V., Lee B., Li S.H., Li X.J. Suppression of neuropil aggregates and neurological symptoms by an intracellular antibody implicates the cytoplasmic toxicity of mutant huntingtin. J. Cell Biol. 2008;181:803–816. doi: 10.1083/jcb.200710158. doi:10.1083/jcb.200710158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen H.P., Kobbe P., Rahne H., Worpel T., Jager B., Stephan M., Pabst R., Holzmann C., Riess O., Korr H., et al. Behavioral abnormalities precede neuropathological markers in rats transgenic for Huntington's disease. Hum. Mol. Genet. 2006;15:3177–3194. doi: 10.1093/hmg/ddl394. doi:10.1093/hmg/ddl394. [DOI] [PubMed] [Google Scholar]
- 36.Osmand A.P., Berthelier V., Wetzel R. Imaging polyglutamine deposits in brain tissue. Methods Enzymol. 2006;412:106–122. doi: 10.1016/S0076-6879(06)12008-X. [DOI] [PubMed] [Google Scholar]
- 37.Slow E.J., Graham R.K., Osmand A.P., Devon R.S., Lu G., Deng Y., Pearson J., Vaid K., Bissada N., Wetzel R., et al. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc. Natl Acad. Sci. USA. 2005;102:11402–11407. doi: 10.1073/pnas.0503634102. doi:10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss A., Klein C., Woodman B., Sathasivam K., Bibel M., Regulier E., Bates G.P., Paganetti P. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington's disease. J. Neurochem. 2008;104:846–858. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- 39.Nagata E., Sawa A., Ross C.A., Snyder S.H. Autophagosome-like vacuole formation in Huntington's disease lymphoblasts. Neuroreport. 2004;15:1325–1328. doi: 10.1097/01.wnr.0000127073.66692.8f. [DOI] [PubMed] [Google Scholar]
- 40.Rudnicki D.D., Pletnikova O., Vonsattel J.P., Ross C.A., Margolis R.L. A comparison of Huntington disease and Huntington disease-like 2 neuropathology. J. Neuropathol. Exp. Neurol. 2008;67:366–374. doi: 10.1097/NEN.0b013e31816b4aee. doi:10.1097/NEN.0b013e31816b4aee. [DOI] [PubMed] [Google Scholar]
- 41.Petersen A., Larsen K.E., Behr G.G., Romero N., Przedborski S., Brundin P., Sulzer D. Expanded CAG repeats in exon 1 of the Huntington's disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum. Mol. Genet. 2001;10:1243–1254. doi: 10.1093/hmg/10.12.1243. doi:10.1093/hmg/10.12.1243. [DOI] [PubMed] [Google Scholar]
- 42.Tanida I., Ueno T., Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J. Biol. Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. doi:10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 43.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D.J. In search of an ‘autophagomometer. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. doi:10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 45.Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. doi:10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. doi:10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bains M., Florez-McClure M.L., Heidenreich K.A. Insulin-like growth factor-I prevents the accumulation of autophagic vesicles and cell death in Purkinje neurons by increasing the rate of autophagosome-to-lysosome fusion and degradation. J. Biol. Chem. 2009;284:20398–20407. doi: 10.1074/jbc.M109.011791. doi:10.1074/jbc.M109.011791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie Z., Klionsky D.J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. doi:10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 49.Xie Z., Nair U., Klionsky D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. doi:10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark H.B., Burright E.N., Yunis W.S., Larson S., Wilcox C., Hartman B., Matilla A., Zoghbi H.Y., Orr H.T. Purkinje cell expression of a mutant allele of SCA1 in transgenic mice leads to disparate effects on motor behaviors, followed by a progressive cerebellar dysfunction and histological alterations. J. Neurosci. 1997;17:7385–7395. doi: 10.1523/JNEUROSCI.17-19-07385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selkoe D.J. Alzheimer's disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. doi:10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 52.Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T., Paulson H.L., Yang L., Kotin R.M., Davidson B.L. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. doi:10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto A., Lucas J.J., Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. doi:10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 54.Menalled L.B., El-Khodor B.F., Patry M., Suárez-Fariñas M., Orenstein S.J., Zahasky B., Leahy C., Wheeler V., Yang X.W., MacDonald M., et al. Systematic behavioral evaluation of Huntington's disease transgenic and knock-in mouse models. Neurobiol. Dis. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. doi:10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crawley J.N. What's Wrong with My Mouse? New York: Whiley-Liss; 2000. [Google Scholar]
- 56.Stack E.C., Kubilus J.K., Smith K., Cormier K., Del Signore S.J., Guelin E., Ryu H., Hersch S.M., Ferrante R.J. Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington's disease transgenic mice. J. Comp. Neurol. 2005;490:354–370. doi: 10.1002/cne.20680. doi:10.1002/cne.20680. [DOI] [PubMed] [Google Scholar]
- 57.Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.Z., et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. doi:10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 58.Gray M., Shirasaki D.I., Cepeda C., André V.M., Wilburn B., Lu X.H., Tao J., Yamazaki I., Li S.H., Sun Y.E., et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. doi:10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutekunst C.A., Li S.H., Yi H., Mulroy J.S., Kuemmerle S., Jones R., Rye D., Ferrante R.J., Hersch S.M., Li X.J. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J. Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Z.X., Li S.H., Evans J., Pillarisetti A., Li H., Li X.J. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease. J. Neurosci. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwata A., Christianson J.C., Bucci M., Ellerby L.M., Nukina N., Forno L.S., Kopito R.R. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl Acad. Sci. USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. doi:10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. doi:10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 63.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. doi:10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 64.Sarkar S., Rubinsztein D.C. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;275:4263–4270. doi: 10.1111/j.1742-4658.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 65.Qin Z.H., Wang Y., Kegel K.B., Kazantsev A., Apostol B.L., Thompson L.M., Yoder J., Aronin N., DiFiglia M. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum. Mol. Genet. 2003;12:3231–3244. doi: 10.1093/hmg/ddg346. doi:10.1093/hmg/ddg346. [DOI] [PubMed] [Google Scholar]
- 66.Gu X., Greiner E.R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J.S., Thompson L.M., Wetzel R., Yang X.W. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. doi:10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeong H., Then F., Melia T.J., Jr, Mazzulli J.R., Cui L., Savas J.N., Voisine C., Paganetti P., Tanese N., Hart A.C., et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. doi:10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson L.M., Aiken C.T., Kaltenbach L.S., Agrawal N., Illes K., Khoshnan A., Martinez-Vincente M., Arrasate M., O'Rourke J.G., Khashwji H., et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell. Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. doi:10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menzies F.M., Huebener J., Renna M., Bonin M., Riess O., Rubinsztein D.C. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. doi:10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. doi:10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 71.Bjorkoy G., Lamark T., Pankiv S., Overvatn A., Brech A., Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. doi:10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 72.Marino G., Ugalde A.P., Salvador-Montoliu N., Varela I., Quiros P.M., Cadinanos J., van der Pluijm I., Freije J.M., Lopez-Otin C. Premature aging in mice activates a systemic metabolic response involving autophagy induction. Hum. Mol. Genet. 2008;17:2196–2211. doi: 10.1093/hmg/ddn120. doi:10.1093/hmg/ddn120. [DOI] [PubMed] [Google Scholar]
- 73.Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 74.Tasdemir E., Maiuri M.C., Galluzzi L., Vitale I., Djavaheri-Mergny M., D'Amelio M., Criollo A., Morselli E., Zhu C., Harper F., et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. doi:10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boland B., Kumar A., Lee S., Platt F.M., Wegiel J., Yu W.H., Nixon R.A. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. doi:10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. doi:10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 77.Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. doi:10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 78.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. doi:10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 79.Winslow A.R., Rubinsztein D.C. Autophagy in neurodegeneration and development. Biochim. Biophys. Acta. 2008;1782:723–729. doi: 10.1016/j.bbadis.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aronin N., Kim M., Laforet G., DiFiglia M. Are there multiple pathways in the pathogenesis of Huntington's disease? Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999;354:995–1003. doi: 10.1098/rstb.1999.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee J.A., Gao F.B. Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J. Neurosci. 2009;29:8506–8511. doi: 10.1523/JNEUROSCI.0924-09.2009. doi:10.1523/JNEUROSCI.0924-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. doi:10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 83.Dragatsis I., Goldowitz D., Del Mar N., Deng Y.P., Meade C.A., Liu L., Sun Z., Dietrich P., Yue J., Reiner A. CAG repeat lengths > or =335 attenuate the phenotype in the R6/2 Huntington's disease transgenic mouse. Neurobiol. Dis. 2009;33:315–330. doi: 10.1016/j.nbd.2008.10.009. doi:10.1016/j.nbd.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morton A.J., Glynn D., Leavens W., Zheng Z., Faull R.L., Skepper J.N., Wight J.M. Paradoxical delay in the onset of disease caused by super-long CAG repeat expansions in R6/2 mice. Neurobiol. Dis. 2009;33:331–341. doi: 10.1016/j.nbd.2008.11.015. doi:10.1016/j.nbd.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 85.Lin C.H., Tallaksen-Greene S., Chien W.M., Cearley J.A., Jackson W.S., Crouse A.B., Ren S., Li X.J., Albin R.L., Detloff P.J. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. doi:10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 86.Dixon K.T., Cearley J.A., Hunter J.M., Detloff P.J. Mouse Huntington's disease homolog mRNA levels: variation and allele effects. Gene Expr. 2004;11:221–231. doi: 10.3727/000000003783992234. doi:10.3727/000000003783992234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.