Abstract
There is extensive evidence that increases in blood and tissue concentrations of steroid hormones and of insulin-like growth factor I (IGF-I) are associated with breast cancer risk. However, studies of common variation in genes involved in steroid hormone and IGF-I metabolism have yet to provide convincing evidence that such variants predict breast cancer risk. The Breast and Prostate Cancer Cohort Consortium (BPC3) is a collaboration of large US and European cohorts. We genotyped 1416 tagging single nucleotide polymorphisms (SNPs) in 37 steroid hormone metabolism genes and 24 IGF-I pathway genes in 6292 cases of breast cancer and 8135 controls, mostly Caucasian, postmenopausal women from the BPC3. We also imputed 3921 additional SNPs in the regions of interest. None of the SNPs tested was significantly associated with breast cancer risk, after correction for multiple comparisons. The results remained null when cases and controls were stratified by age at diagnosis/recruitment, advanced or nonadvanced disease, body mass index, with or without in situ cases; or restricted to Caucasians. Among 770 estrogen receptor-negative cases, an SNP located 3′ of growth hormone receptor (GHR) was marginally associated with increased risk after correction for multiple testing (Ptrend = 1.5 × 10−4). We found no significant overall associations between breast cancer and common germline variation in 61 genes involved in steroid hormone and IGF-I metabolism in this large, comprehensive study. Although previous studies have shown that variations in these genes can influence endogenous hormone levels, the magnitude of the effect of single SNPs does not appear to be sufficient to alter breast cancer risk.
INTRODUCTION
Extensive evidence from in vitro, animal and epidemiologic studies indicate that increases in blood and tissue concentrations of endogenous steroid hormones and insulin-like growth factor I (IGF-I) favor the development of breast cancer.
Bittner in 1947 was the first to propose, based on mouse mammary tumor models, that endogenous steroid hormones were a cause of breast cancer (1). Epidemiologic support for the hypothesis derived from the observations that most of the established risk factors for breast cancer influence endogenous hormone production (2,3). It is now well established that elevated circulating and urinary levels of estrogens and androgens predict increased risk of breast cancer in postmenopausal women (4–9). Some epidemiologic studies also support a role for endogenous estrogens and androgens in premenopausal breast cancer (10), and estrogen produced between puberty and first full-term pregnancy has long been proposed as a critical determinant of lifetime cancer risk (2,11). More recently, it has been reported that prolactin levels may also influence breast cancer risk (12–14), although these findings are much less conclusive than those for estrogens and androgens. Estrogens stimulate cell division in the breast (3) and can be converted to genotoxic and mutagenic metabolites (15). Androgens have been hypothesized to modify breast cancer risk directly, by stimulating or inhibiting the proliferation of breast cancer cells, or indirectly, by conversion to estrogens (8). Postmenopausal steroid hormone therapy significantly increases the incidence of breast cancer (16,17), whereas anti-estrogen agents that competitively bind to the estrogen receptor (ER), such as tamoxifen, can reduce the primary occurrence and spread of breast tumors (15).
IGF-I metabolism is central to the regulation of anabolic (growth) processes as a function of available energy and elementary substrates such as amino acids. IGF-I stimulates signal transduction pathways that enhance anabolic processes, inhibits apoptosis and stimulates cell proliferation in a wide variety of cell types, including breast epithelium, and enhances tumor growth (18–22). Prospective epidemiological studies have shown higher breast cancer risk in women with elevated serum IGF-I concentrations, measured either as absolute concentrations or relative to the levels of IGFBP-3, the main IGF-I-binding protein in plasma, although not all studies have observed this association (23–28). Recently, a pooled analysis of individual data from 17 prospective studies demonstrated a modest, positive association between circulating IGF-I and breast cancer risk in both pre- and postmenopausal women (29).
Heritability studies based on twins and families suggest that a sizable fraction of between-subject variance in circulating levels of some steroid hormones and IGF-I is under the influence of genetic determinants and familial environmental and lifestyle factors, with the magnitude of the effect ranging from 39 to 65% (30–37).
Over the past decade, the search for candidate susceptibility genes associated with breast cancer has logically focused on polymorphisms in genes known to be relevant for the synthesis, bioavailability and metabolism of steroid hormones and IGF-I. Although several common polymorphisms have been consistently associated with circulating hormone levels, none have been convincingly associated with breast cancer risk directly (38–49). Few reported associations have been replicated because most candidate gene studies, especially those published in the 1990s, were underpowered to detect modest associations and did not comprehensively capture variation across the entire gene region.
To address these limitations of earlier studies, the Breast and Prostate Cancer Cohort Consortium (BPC3) was formed to bring together large, well-established cohorts assembled in the USA and Europe that had DNA for genotyping, extensive baseline questionnaire data from cohort participants, and effective ascertainment of cancer outcomes during follow-up (50). Here, we present the associations with breast cancer risk of 1416 tagging polymorphisms in 37 steroid hormone metabolism and 24 IGF-I pathway genes in 6292 cases of breast cancer and 8135 controls from BPC3.
RESULTS
After exclusions, the analyses include 6292 breast cancer cases and 8135 controls from six cohorts. Table 1 shows summary characteristics of the study population. Cases and controls were predominantly Caucasian (78%) and postmenopausal at the time of enrollment/DNA collection (79%). The mean age at diagnosis was 63 [ranging from 57.1 years for European Prospective Investigation into Cancer and Nutrition (EPIC) to 70.1 years for Cancer Prevention Study II (CPS-II)].
Table 1.
Summary characteristics of study population
| CPS-II |
EPIC |
MEC |
NHS |
PLCO |
WHS |
All |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| Number | 472 | 490 | 1283 | 2297 | 1610 | 1954 | 1218 | 1685 | 1066 | 1089 | 643 | 620 | 6292 | 8135 |
| Age at diagnosis (mean, SD) | 70.1 (6.2) | – | 57.1 (8.6) | – | 65.1 (8.5) | – | 62.9 (7.3) | – | 66.3 (5.7) | – | 60.1 (7.5) | – | 63.1 (8.5) | – |
| Stage of breast cancer (%) | ||||||||||||||
| In situ | 22 | – | 8 | – | 1 | – | 17 | – | 16 | – | 0 | – | 10 | – |
| Localized | 62 | – | 49 | – | 73 | – | 52 | – | 37 | – | 67 | – | 57 | – |
| Advanced | 14 | – | 11 | – | 26 | – | 29 | – | 21 | – | 25 | – | 22 | – |
| Unknown | 2 | – | 32 | – | 0 | – | 2 | – | 26 | – | 6 | – | 12 | – |
| Estrogen receptor status (%) | ||||||||||||||
| Positive | 52 | – | 33 | – | 62 | – | 60 | – | 48 | – | 79 | – | 55 | – |
| Negative | 7 | – | 9 | – | 16 | – | 16 | – | 7 | – | 15 | – | 12 | – |
| Unknown | 41 | – | 58 | – | 21 | – | 24 | – | 45 | – | 6 | – | 33 | – |
| BMI (mean, SD) | 25.2 (4.6) | 25.5 (4.5) | 26.0 (4.4) | 26.2 (4.6) | 26.7 (5.6) | 27.0 (6.0) | 25.4 (4.7) | 25.5 (4.6) | 27.1 (5.2) | 27.0 (5.4) | 25.5 (4.4) | 26.1 (5.2) | 26.1 (5.0) | 26.3 (5.2) |
| Menopause | ||||||||||||||
| Premenopausal | 0 | 0 | 379 | 712 | 177 | 319 | 248 | 312 | 0 | 0 | 140 | 128 | 944 | 1471 |
| Perimenopausal/unknown | 0 | 0 | 133 | 221 | 39 | 33 | 126 | 145 | 10 | 9 | 99 | 120 | 407 | 528 |
| Postmenopausal | 472 | 490 | 771 | 1364 | 1394 | 1602 | 844 | 1228 | 1056 | 1080 | 404 | 372 | 4941 | 8135 |
| Ethnicity (%) | ||||||||||||||
| White | 98 | 99 | 100 | 100 | 25 | 22 | 94 | 94 | 91 | 90 | 95 | 95 | 77 | 78 |
| Hispanic | 1 | 0 | 0 | 0 | 21 | 20 | 0 | 0 | 1 | 1 | 1 | 1 | 6 | 5 |
| African American | 1 | 1 | 0 | 0 | 21 | 22 | 1 | 1 | 4 | 3 | 1 | 1 | 6 | 6 |
| Asian | 0 | 0 | 0 | 0 | 26 | 21 | 0 | 1 | 3 | 4 | 1 | 1 | 8 | 6 |
| Hawaiian | 0 | 0 | 0 | 0 | 7 | 15 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 4 |
| Other/unknown | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 | 0 | 1 | 2 | 2 | 1 | 1 |
Success rates and quality control
The percentage of SNPs selected for genotyping in cancer cases and controls that were successfully genotyped (based on the filtering methods described in the Materials and Methods section) ranged between 89.0 and 93.5%, calculated by cohort. Most failures were due to low call rates (<75% in a given cohort). Fewer than 1% of the SNPs were excluded from further analysis because of departure from Hardy–Weinberg equilibrium (P < 10−5). Allele frequencies were similar across the cohorts; only two SNPs showed Fst values higher than 2%. A total of 403 samples (2.72% of those where genotyping was attempted) were excluded due to call rate <75%. See Supplementary Material, Table S2 for more detail. A total of 1416 SNPs in the 61 candidate genes were successfully genotyped and used for statistical analyses.
Main effects of genotyped SNPs
We calculated Meff values for each candidate gene separately and for the whole study (by adding the individual gene Meff values). The study-wide Meff was 1065. We therefore used a study-wide significance P-threshold of 0.05/1065 = 4.7 × 10−5.
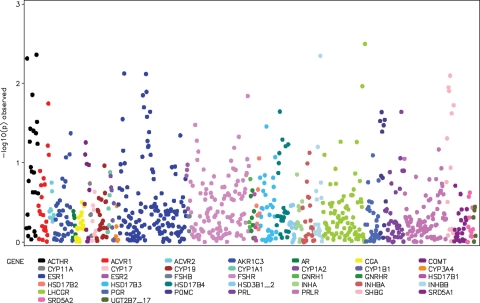
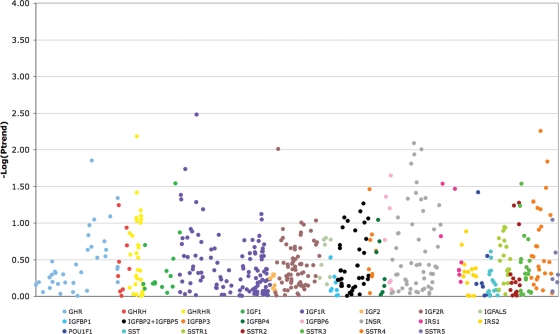
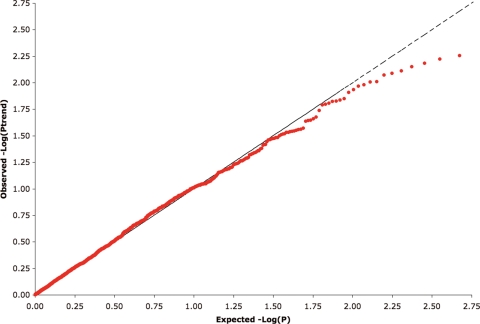
We observed no associations (Ptrend < 4.7 × 10−5 or P2df < 4.7 × 10−5) between any of the polymorphisms genotyped and overall breast cancer risk (Ptrend 0.003–0.996; P2df 0.001–0.999; Figs 1 and 2), nor did the distribution of observed P-values differ from that expected (Fig. 3). Tables 2 (steroid hormone-related genes) and 3 (IGF-I-related genes) show the minimum observed P-values of analyses performed on all subjects with invasive breast cancer. The results remained null when the sample was restricted to Caucasian women, or when analyses were stratified by: (a) age at diagnosis/recruitment (>55 years, ≤55 years); (b) advanced or nonadvanced disease (advanced disease being defined as regional or distant metastasis); (c) with or without inclusion of in situ cases (Supplementary Material, Tables S3 and S4).
Figure 1.
Manhattan plot showing associations of the 866 SNPs genotyped in steroid hormone-related genes with breast cancer risk. Each dot represents an SNP, and different colors indicate distinct genes. The y-axis shows the P-value for trend on a logarithmic scale.
Figure 2.
Manhattan plot showing associations of the 550 SNPs genotyped in IGF-I-related genes with breast cancer risk. Each dot represents an SNP, and different colors indicate distinct genes. The y-axis shows the P-value for trend on a logarithmic scale.
Figure 3.
Quantile–quantile plots for the test statistic (trend test). Red dots are the results of each typed SNP. Under the null hypothesis of no association at any locus, the points would be expected to follow the dotted line.
Table 2.
Associations between SNPs in steroid hormone-related genes and breast cancer risk
| Gene abbreviation | Gene name | Number of SNPs genotyped | Minimum observed P-value |
|
|---|---|---|---|---|
| Ptrenda | P2dfa | |||
| ACTHR (MC2R) | Adrenocorticotropic hormone receptor/melanocortin 2 receptor | 23 | 0.006 | 0.013 |
| ACVR1 | Activin A receptor, type I | 21 | 0.029 | 0.080 |
| ACVR2A | Activin A receptor, type IIA | 9 | 0.090 | 0.186 |
| AKR1C3 | Aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) | 36 | 0.096 | 0.117 |
| AR | Androgen receptor | 6 | 0.427 | 0.371 |
| CGA | Glycoprotein hormones, alpha polypeptide | 17 | 0.310 | 0.081 |
| COMT | Catechol-O-methyltransferase | 10 | 0.032 | 0.007 |
| CYP11A1 | Cytochrome P450, family 11, subfamily A, polypeptide 1 | 4 | 0.098 | 0.198 |
| CYP17A1 | Cytochrome P450, family 17, subfamily A, polypeptide 1 | 9 | 0.066 | 0.138 |
| CYP19A1 | Cytochrome P450, family 19, subfamily A, polypeptide 1 | 21 | 0.055 | 0.060 |
| CYP1A1 | Cytochrome P450, family 1, subfamily A, polypeptide 1 | 6 | 0.098 | 0.058 |
| CYP1A2 | Cytochrome P450, family 1, subfamily A, polypeptide 2 | 2 | 0.128 | 0.251 |
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 8 | 0.197 | 0.175 |
| CYP3A4 | Cytochrome P450, family 3, subfamily A, polypeptide 4 | 5 | 0.232 | 0.195 |
| ESR1 | Estrogen receptor 1 | 131 | 0.008 | 0.011 |
| ESR2 | Estrogen receptor 2 | 4 | 0.070 | 0.100 |
| FSHB | Follicle stimulating hormone, beta polypeptide | 3 | 0.337 | 0.588 |
| FSHR | Follicle stimulating hormone receptor | 114 | 0.011 | 0.026 |
| GNRH1 | Gonadotropin-releasing hormone 1 | 3 | 0.257 | 0.129 |
| GNRHR | Gonadotropin-releasing hormone receptor | 7 | 0.080 | 0.193 |
| HSD17B1 | Hydroxysteroid (17-beta) dehydrogenase 1 | 4 | 0.150 | 0.344 |
| HSD17B2 | Hydroxysteroid (17-beta) dehydrogenase 2 | 7 | 0.031 | 0.084 |
| HSD17B3 | Hydroxysteroid (17-beta) dehydrogenase 3 | 32 | 0.066 | 0.028 |
| HSD17B4 | Hydroxysteroid (17-beta) dehydrogenase 4 | 24 | 0.087 | 0.024 |
| HSD3B1_2 | Hydroxy-delta-5-steriod dehydrogenase, 3 beta- and steroid delta-isomerase 1 and 2 | 14 | 0.193 | 0.176 |
| INHA | Inhibin, alpha | 12 | 0.162 | 0.052 |
| INHBA | Inhibin, beta A | 23 | 0.033 | 0.057 |
| INHBB | Inhibin, beta B | 16 | 0.003 | 0.008 |
| LHCGR | Luteinizing hormone/choriogonadotropin receptor | 82 | 0.007 | 0.009 |
| PGR | Progesterone receptor | 28 | 0.090 | 0.069 |
| POMC | Proopiomelanocortin | 12 | 0.023 | 0.061 |
| PRL | Prolactin | 35 | 0.030 | 0.017 |
| PRLR | Prolactin receptor | 81 | 0.090 | 0.044 |
| SHBG | Sex hormone-binding globulin | 15 | 0.028 | 0.044 |
| SRD5A1 | Steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 25 | 0.159 | 0.293 |
| SRD5A2 | Steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) | 12 | 0.115 | 0.267 |
| UGT2B7/17 | UDP glucuronosyltransferase 2 family, polypeptide B7 and B17 | 5 | 0.423 | 0.277 |
All cancer cases are included, except in situ carcinomas and their controls. All associations supported by P ≤ 0.01 in at least one of the two tests are reported in bold.
aP-values of tests where genotypes were coded either as counts of minor alleles (trend test) or as two indicator variables, one for heterozygotes and one for minor allele homozygotes (two degrees of freedom test).
Table 3.
Associations between SNPs in IGF-I-related genes and breast cancer risk
| Gene abbreviation | Gene name | Number of SNPs genotyped | Minimum observed P-value |
|
|---|---|---|---|---|
| Ptrenda | P2dfa | |||
| GHR | Growth hormone receptor | 35 | 0.014 | 0.027 |
| GHRH | Growth hormone-releasing hormone | 9 | 0.058 | 0.165 |
| GHRHR | Growth hormone-releasing hormone receptor | 29 | 0.007 | 0.0120 |
| IGF1 | Insulin-like growth factor 1 (somatomedin C) | 14 | 0.029 | 0.013 |
| IGF1R | Insulin-like growth factor 1 receptor | 118 | 0.003 | 0.006 |
| IGF2 | Insulin-like growth factor 2 (somatomedin A) | 9 | 0.503 | 0.056 |
| IGF2R | Insulin-like growth factor 2 receptor | 73 | 0.010 | 0.005 |
| IGFALS | Insulin-like growth factor-binding protein, acid labile subunit | 9 | 0.160 | 0.026 |
| IGFBP1 | Insulin-like growth factor-binding protein 1 | 8 | 0.294 | 0.114 |
| IGFBP2_5b | Insulin-like growth factor-binding protein 2 and Insulin-like growth factor-binding protein 5 | 37 | 0.054 | 0.025 |
| IGFBP3 | Insulin-like growth factor-binding protein 3 | 8 | 0.035 | 0.103 |
| IGFBP4 | Insulin-like growth factor-binding protein 4 | 7 | 0.091 | 0.226 |
| IGFBP6 | Insulin-like growth factor-binding protein 6 | 7 | 0.022 | 0.002 |
| INSR | Insulin receptor | 54 | 0.008 | 0.001 |
| IRS1 | Insulin receptor substrate 1 | 8 | 0.029 | 0.047 |
| IRS2 | Insulin receptor substrate 2 | 15 | 0.131 | 0.122 |
| POU1F1 | POU class 1 homeobox 1 | 7 | 0.038 | 0.089 |
| SST | Somatostatin | 16 | 0.245 | 0.509 |
| SSTR1 | Somatostatin receptor 1 | 19 | 0.113 | 0.041 |
| SSTR2 | Somatostatin receptor 2 | 15 | 0.053 | 0.076 |
| SSTR3 | Somatostatin receptor 3 | 19 | 0.029 | 0.048 |
| SSTR4 | Somatostatin receptor 4 | 27 | 0.006 | 0.010 |
| SSTR5 | Somatostatin receptor 5 | 7 | 0.091 | 0.239 |
All cancer cases are included, except in situ carcinomas and their controls. All associations supported by P ≤ 0.01 in at least one of the two tests are reported in bold.
aP-values of tests where genotypes were coded either as counts of minor alleles (trend test) or as two indicator variables, one for heterozygotes and one for minor allele homozygotes (two degrees of freedom test).
bThe genes for IGFBP2 and IGFBP5 are contiguous, and separated by only 7 kb; therefore, these two genes were considered as a single unit during SNP selection and further analyses.
When analyses were stratified by body mass index (BMI; <25, 25–30, ≥30 kg/m2), the test for heterogeneity for the trend test was of borderline statistical significance for one SNP, rs6744967 in LHCGR (P for heterogeneity = 0.0006). The stratified results suggested that the SNP was inversely associated with breast cancer among women with a BMI of >30. In this stratum, we observed odds ratio (OR) = 0.73, 99% CI: 0.57–0.93 for T/C versus T/T, and OR=0.63, 99% CI: 0.47–0.84 for C/C versus T/T (Supplementary Material, Table S4).
Data on ER status were available for 4193 cases (66.6% of the total). Of these, 3423 (81.6%) were ER positive and 770 (18.4%) were ER negative. Among the ER-negative cases, the minor allele of SNP rs719756 within GHR was associated with increased risk, and the association approached a statistical significance when taking into account multiple testing (Ptrend = 1.5 × 10−4). A test for heterogeneity for SNP rs719756 showed that the estimates of association in the ER-negative and -positive strata are different (P = 0.002). Among SNPs in steroid hormone-related genes, rs1543403 in ESR1 showed a differential association between ER-positive and -negative breast cancer cases [no association in the ER-positive stratum, in the ER-negative stratum Ptrend = 5.7 × 10−4; OR = 1.20 (0.90–1.60) for one copy of the allele and OR = 1.54 (1.11–2.12) for two copies]. No statistically significant associations were observed with ER-positive cases after correction for multiple testing. Detailed results are reported in Supplementary Material, Table S5.
Imputation of HapMap SNPs and their associations
We successfully imputed 3921 SNPs located in the gene regions from which we selected tagging single nucleotide polymorphisms (tagSNPs) (corresponding to 39.5% of SNPs present in database of single nucleotide polymorphisms in the same regions) for which the estimated correlation with genotyped SNPs was higher than 30%. However, none of these imputed SNPs were significantly associated with breast cancer risk when corrected for multiple testing (using a threshold of P = 4.7 × 10−5, Supplementary Material, Table S6).
Tests of pairwise interactions between SNPs
Of all 40 049 tests of statistical models including pairwise interactions tested, 43 showed a P-value of <10−3, with minimum observed P-values of 2.8 × 10−5 between SNPs rs7605927 and rs2164808 of POMC and 5.3 × 10−5 between SNPs rs1521479 and rs8038056 of IGF1R.
In order to further explore the possibility of association between breast cancer risk and SNPs in this gene, we reconstructed haplotypes in a region of IGF1R characterized by high linkage disequilibrium (LD) which encompasses rs1521479 and rs8038056. Statistical analysis of haplotypes did not show any significant association with breast cancer risk. Since none of the 12 genotyped SNPs in POMC approached statistical significance in the main effect models (Table 2 and Fig. 1), we attribute this finding to chance.
DISCUSSION
To date, our study provides the most comprehensive analysis of breast cancer risk in relation to common variation in genes associated with synthesis, bioavailability and metabolism of steroid hormones and IGF-I, both in terms of coverage of genes belonging to the two pathways and of SNPs belonging to each gene. In contrast to genome-wide association studies (GWAS), our approach included systematic polymorphism discovery through deep resequencing in and around candidate genes, imputation across all gene regions genotyped in the study, and examination of pairwise interactions for all genotyped SNPs. Our study provides evidence refuting the hypothesis that common genetic polymorphisms in the two pathways substantially affect breast cancer risk. The absence of an association with common variants is striking given the importance of circulating steroid hormone (4–9) and IGF-I levels (23–27,29), as well as postmenopausal steroid hormone use (16,17) in predicting breast cancer risk. Nevertheless, we believe that these null findings are convincing, given the size and comprehensive approach of our study, which included intensive SNP tagging and mathematical imputation of SNP variants identified through HapMap that were not directly genotyped.
Our study, including over 6000 breast cancer cases and 8000 controls, had >80% power to detect mono-allelic (‘main effect’) associations between variants in any of the genes studied [minor allele frequency (MAF) ≥ 0.05, OR ≥ 1.25; MAF ≥ 0.1, OR ≥ 1.2; or MAF ≥ 0.2, OR ≥ 1.15] and breast cancer risk at P < 0.0001. It is worth noting that the risks detectable in our study are of the same magnitude as those typically found in GWAS.
We did observe some significant P-values at conventional levels of statistical significance (e.g. P < 0.05, P < 0.01) that would have been classified as ‘positive’ in a single candidate gene study. We adopted a threshold of 4.7 × 10−5, to minimize false positives among the large number of SNPs tested. At this level of significance, no single SNP was associated with breast cancer risk; however, we cannot exclude that some of the SNP variants with small P-values are indeed associated with breast cancer risk at approximately the ORs we observed but that adjustment for the large number of SNPs tested obscured these associations. On balance, we prefer to be conservative and limit the chances of declaring false positives. We maintain that a major strength of our study is that with a coordinated approach and large sample size, the ORs and confidence intervals we observed substantially minimize the probability that we have missed common alleles with large effect sizes (e.g. OR ≥ 1.5) (51).
Our results do not support an association between breast cancer and a single SNP (rs3020314) in ESR1, which was recently reported from an analysis of >50 000 breast cancer cases among women of European descent in the Breast Cancer Association Consortium (BCAC) (52). In that study, carriers of the minor allele of rs3020314 had an OR of 1.05 (P = 0.004). The study by Dunning et al. included a subset of the NHS subjects used for the present work (955 cases and 1631 controls). In the BPC3, we did not genotype rs3020314. However, we genotyped two SNPs [rs3020377 (P for trend = 0.56) and rs3020394 (P for trend = 0.90)] that tag rs3020314, as well as 129 other SNPs in ESR1. Two of our SNPs in ESR1 (rs1361024 and rs1514347) have P-values for trend of 0.01. Six of our ESR1 SNPs have P-values for trend between 0.0004 and 0.001 among our advanced breast cancer cases, but the trends all suggest inverse associations for the minor allele. In our studies, ER status was known for a large fraction of the cases, which enabled us to analyze our data stratifying by ER status. Among women with ER-negative tumors, ESR1 rs1543403 was associated with a borderline significantly increased risk (Ptrend = 5.7 × 10−4). Since the BCAC includes most of the existing case–control studies of breast cancer, it may be difficult to find another independent set of data that can resolve these conflicting findings, especially given that the observed effect reported by the BCAC was small.
One SNP of GHR showed an association with ER-negative breast cancer that was close to significance when taking into account multiple testing (Ptrend = 1.5 × 10−4). Although this result is promising, the SNP is located 3.5 kb downstream from the end of the last exon of GHR and, therefore, its function is not obvious. Further studies, involving larger numbers of ER-negative cases, will show if this result can be replicated.
Besides testing for main effects, we assessed all possible models for pairwise interactions between SNPs within each gene. This approach tests all possible cis-acting interactions between SNPs at the same gene locus, at least for all pairs of SNPs within regions of high LD (haplotype blocks) where there is little uncertainty about the allelic phase (53). One interaction test between two SNPs in POMC showed a P-value of 2.8 × 10−5, and an interaction test between two SNPs at the 3′ end of IGF1R showed a P-value of 5.3 × 10−5. However, after correction using conservative Bonferroni adjustment, these no longer met the threshold for statistical significance (P < 1.2 × 10−6, 0.05/40 049 tests). Additional haplotype reconstruction and statistical testing in these gene regions did not provide further evidence for associations with breast cancer risk. Examination of the SNPs from our discovery, sequencing and genotyping sets in these two gene regions did not show any obvious potentially functional SNPs (splice junction or nonsynonymous SNPs) in high LD. These signals may have been, therefore, observed by chance.
We chose not to test for higher order interactions, as the overall sample size of our study, although very large, was insufficient to test very large number of complex interactions. For the same reasons, we elected not to conduct more complex haplotype analyses for all genes, which would require testing models incorporating all main effects plus interactions (first- and higher order) of the full sets of SNPs in regions of high LD (53).
Previous analyses of our genetic data have found variants of some of our candidate genes to be associated with circulating hormone levels (45,46,49,54). Other studies have reported similar findings, and we have assessed these genetic variants either directly or indirectly by tagging or imputation in the current analysis (55–58). In these studies, however, the genetic variation explained only a minor part of the between-subject variance in hormone levels. On the basis of the existing knowledge about the quantitative relationship between various hormones and breast cancer risk (6,29), the modest changes in circulating levels observed previously would have too small an effect on breast cancer risk to be detectable in our study, despite its power. Nevertheless, these SNPs can be considered to have high prior probabilities of association with breast cancer risk. When we calculated the false-positive report probabilities (59) for these SNPs, we found no evidence that these associations with breast cancer risk were noteworthy, even if we assigned them a high prior (data not shown). It is also possible that common variations in the genes associated with steroid hormone and IGF metabolism may modestly affect breast cancer risk, possibly at an earlier stage of life, but that this relationship is obscured in postmenopausal women by the strong influence through the lifespan of environmental factors, such as adiposity, diet, physical activity, and menstrual and reproductive events, on steroid hormone and IGF levels. However, our results remained null in analyses restricted to women aged <55 years or with a BMI of <25 kg/m2.
Our results are consistent with recent GWAS, which also have failed to identify any loci in steroid hormone or IGF-I metabolism genes associated importantly with breast cancer risk (60–65). Across a wide variety of cancers, loci identified by GWAS have usually been found in genes not previously hypothesized to be related to the disease, or in intergenic regions not known to contain specific genes (60–70). This suggests far greater complexity in the etiology of these diseases and in genetic regulation than previously anticipated, and that a narrow focus on what we already know about widely accepted etiologic pathways will have limited success identifying the heritable components of these diseases.
In conclusion, although the experimental and epidemiologic evidence relating steroid hormones and the IGF axis to breast cancer risk is strong, this comprehensive large-scale study found no statistically significant associations between breast cancer risk and polymorphisms in 61 candidate genes in these two pathways in a population of mostly Caucasian, postmenopausal women.
MATERIALS AND METHODS
Study population
The BPC3 has been described in detail elsewhere (50). Briefly, the consortium includes large, well-established cohorts assembled in the USA and Europe that have both DNA samples and extensive questionnaire information: the American Cancer Society CPS-II (71), the EPIC (72), the Harvard Nurses' Health Study (NHS) (73) and Women's Health Study (WHS) (74), the Prostate, Lung, Colorectal, Ovarian Cancer Screening Trial (PLCO) (75) and the Multiethnic Cohort (MEC) (76). With the exception of the MEC, most women in these cohorts are Caucasians of US and European descent. Cases were identified in each cohort by self-report with subsequent confirmation of the diagnosis from medical records or tumor registries, and/or linkage with population-based tumor registries (method of confirmation varied by cohort). Controls were matched to cases by ethnicity and age, and in some cohorts, additional criteria, such as country of residence in EPIC.
Informed consent was obtained from all subjects, and the project has been approved by the appropriate institutional review board for each cohort.
Gene and SNP selection
We selected all known genes involved in synthesis, bioavailability and metabolism of steroid hormones and IGF-I. We included only genes that are specific to steroid hormones and IGF-I, and we did not include genes involved in the downstream processing of signaling for either pathway. On the basis of the current knowledge of hormone physiology, we believe that our coverage of these two pathways is comprehensive.
A detailed description of SNP selection methodology is provided in Supplementary Material. Briefly, coding and evolutionarily conserved regions of selected genes were sequenced in a panel of 95 advanced breast cancer cases (15 from each of the five ethnic groups: African American, Latina, Japanese, Native Hawaiian and Caucasian) from the MEC and EPIC. A summary of the SNPs detected is reported in Supplementary Material, Table S1.
All SNPs identified by resequencing with MAF>5% in any of the five ethnic groups or >1% overall, as well as additional SNPs from HapMap (http://www.hapmap.org) were genotyped in multiethnic reference panels of up to 710 healthy subjects, using the Sequenom, TaqMan and Illumina platforms.
These genotyping data were then used to select tagging SNPs to genotype on cancer cases and controls from the cohorts. For genes in which TaqMan genotyping was planned (ACTHR/MC2R, AR, COMT, CYP11A, CYP17, CYP19A1, CYP1A1, CYP1A2, CYP1B1, ESR2, FSHB, GNRH1, GNRHR, HSD17B1, HSD17B2, PGR, POMC, IGF1, IGFBP1 and IGFBP3), a haplotype-tagging approach was used according to the method of Stram et al. (77), with criterion r2H ≥ 0.7. Tagging SNPs for the remaining 41 genes were selected later in the project and were genotyped using Illumina GoldenGate technology; all SNPs with MAF ≥0.05 in these genes were tagged with a minimum r2-value of ≥0.8. Haplotype tagging and pairwise tagging result in essentially the same sets of tagging SNPs over blocks of no or limited recombination, whereas the coverage with pairwise tagging is more complete in inter-block regions characterized by low LD. Although different tagging methods are not likely to select the same sets of tagging SNPs, they provide comparable power to detect associations (78).
The gene for growth hormone (GH1) was considered for this study, but it was dropped due to its high homology to four other genes that are physically clustered together with GH1 and share a very high sequence homology (79), making it virtually impossible to obtain reliable genotyping data from this region.
Genotyping
Genotyping in 6292 breast cancer cases and 8135 controls was performed in six laboratories (located at University of Southern California, National Cancer Institute, Harvard School of Public Health, Imperial College, International Agency for Research on Cancer and DKFZ). One hundred and seventy-two SNPs in 20 genes (listed in the subsection Gene and SNP selection) were genotyped by the 5′ nuclease (TaqMan) assay, using reagents by Applied Biosystems (Foster City, CA, USA). SNPs in the remaining 41 genes examined in the present analysis were genotyped by Illumina GoldenGate technology (San Diego, CA, USA). SNPs belonging to three genes, ACTHR, PGR and POMC, were genotyped on both platforms and did not show genotyping discrepancies. Samples of 30 HapMap CEU trios were genotyped in all laboratories to evaluate inter-lab reproducibility. For the 1536 SNPs included in the Illumina GoldenGate array, the concordance was 99.5% (before excluding failed SNPs or samples). Within each study, blinded duplicate samples (∼5%) were also included and concordance of these samples ranged from 97.2 to 99.9% across studies.
Data filtering and statistical analysis
Samples in which more than 25% of the SNPs failed were excluded from further analysis. Also excluded were poorly performing SNPs (those that failed on 25% or more samples in a given cohort), those that deviated significantly (P < 10−5) within a cohort from HWE genotype frequencies among European-ancestry controls, and those with MAF < 1% (within a cohort). Any SNP that was missing in more than three studies or exhibited large differences in European-ancestry allele frequencies across cohorts (fixation index Fst > 0.02) was excluded from further analysis.
We used the software MACH (http://www.sph.umich.edu/csg/abecasis/MaCH/index.html) to impute SNPs that were polymorphic in any of the HapMap reference panels using observed genotypes from the BPC3 subjects (Illumina or TaqMan) and phased haplotypes from HapMap samples (release #22) (80). Genotypes for European-ancestry subjects were imputed using the CEPH European (CEU) reference panel of HapMap; those for Japanese Americans were imputed using the combined Han Chinese and Japanese panels (CHB + JPT); those for remaining subjects (African Americans, Latinos, Native Hawaiians) were imputed using a ‘cosmopolitan’ panel of all HapMap samples (CEU + CHB + JPT + YRI) (81). Imputation was performed in analyses stratified by both study and ethnicity. We excluded SNPs for which the average correlation between the imputed and true underlying genotypes (estimated by comparing the variance of the estimated genotype scores with what would be expected if genotype scores were observed without error) was <30%; this threshold was chosen based on analyses of empirical data that maximized the number of accurately imputed genotypes while minimizing the proportion of excluded SNPs (82).
We analyzed the association between breast cancer risk and genotypes for each SNP with MAF >1% using unconditional logistic regression adjusted for four factors: age at diagnosis or selection as a control (5 year age intervals), study, ethnicity and (for the EPIC cohort) recruitment center. Genotypes were coded either as counts of minor alleles (trend test) or as two indicator variables, one for heterozygotes and one for minor allele homozygotes (two degrees of freedom test). We performed these analyses in all subjects, separately for each ethnicity. For the two largest ethnic groups (Caucasians and African Americans), we further performed statistical analyses separately for each study within ethnicity. We performed also analyses stratifying on four other variables: age at diagnosis (≤55, >55 years), advanced/nonadvanced disease (advanced disease was defined as having regional or distant metastasis), ER status, by BMI (<25, 25–30, ≥30 kg/m2) and by inclusion of in situ carcinoma cases.
In order to take into account the large number of tests performed in this project, we calculated for each gene the number of effective independent variables, Meff, by the use of the SNP Spectral Decomposition approach (83). We obtained a gene-wide Meff value for each gene and also a study-wide Meff value, by adding up the gene Meff's.
We analyzed all 40 049 possible SNP–SNP interaction models involving pairs of SNPs belonging to the same gene. We coded the SNPs by genotype category and we performed a likelihood ratio test of the model with all interaction terms against the reduced model that included only the main effects of the two SNPs.
Additional haplotype analyses in IGF1R were performed with the software ‘tagSNPs’ (77,84). This program calculates, for each individual, the expected numbers of copies (‘dosages’) of each of the haplotypes compatible with the individual's SNP genotypes. Association between haplotype dosages and breast cancer risk was tested with conditional logistic regression. This and all other regression analyses were conducted with SAS, v 9.1.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the US National Institutes of Health, National Cancer Institute (cooperative agreements U01-CA98233 to D.J.H., U01-CA98710 to M.J.T., U01-CA98216 to E.R. and R.K., and U01-CA98758 to B.E.H., and Intramural Research Program of NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics).
Supplementary Material
REFERENCES
- 1.Bittner J.J. The causes and control of mammary cancer in mice. Harvey Lect. 1947;42:221–246. [Google Scholar]
- 2.Henderson B.E. Estrogens as a cause of human cancer: The Richard and Hinda Rosenthal Foundation Award Lecture. Cancer Res. 1988;48:246–253. [PubMed] [Google Scholar]
- 3.Pike M.C., Spicer D.V., Dahmoush L., Press M.F. Estrogens, progestogens, normal breast cell proliferation, and breast cancer risk. Epidemiol. Rev. 1993;15:17–35. doi: 10.1093/oxfordjournals.epirev.a036102. [DOI] [PubMed] [Google Scholar]
- 4.Key T.J., Wang D.Y., Brown J.B., Hermon C., Allen D.S., Moore J.W., Bulbrook R.D., Fentiman I.S., Pike M.C. A prospective study of urinary oestrogen excretion and breast cancer risk. Br. J. Cancer. 1996;73:1615–1619. doi: 10.1038/bjc.1996.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hankinson S.E. Plasma sex steroid hormone levels and risk of breast cancer in postmenopausal women. J. Natl. Cancer. Inst. 1998;90:1292–1299. doi: 10.1093/jnci/90.17.1292. [DOI] [PubMed] [Google Scholar]
- 6.Key T., Appleby P., Barnes I., Reeves G. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J. Natl. Cancer Inst. 2002;94:606–616. doi: 10.1093/jnci/94.8.606. [DOI] [PubMed] [Google Scholar]
- 7.Missmer S.A., Eliassen A.H., Barbieri R.L., Hankinson S.E. Endogenous estrogen, androgen, and progesterone concentrations and breast cancer risk among postmenopausal women. J. Natl. Cancer Inst. 2004;96:1856–1865. doi: 10.1093/jnci/djh336. [DOI] [PubMed] [Google Scholar]
- 8.Kaaks R., Berrino F., Key T., Rinaldi S., Dossus L., Biessy C., Secreto G., Amiano P., Bingham S., Boeing H., et al. Serum sex steroids in premenopausal women and breast cancer risk within the European Prospective Investigation into Cancer and Nutrition (EPIC) J. Natl. Cancer Inst. 2005;97:755–765. doi: 10.1093/jnci/dji132. [DOI] [PubMed] [Google Scholar]
- 9.Zeleniuch-Jacquotte A., Gu Y., Shore R.E., Koenig K.L., Arslan A.A., Kato I., Rinaldi S., Kaaks R., Toniolo P. Postmenopausal levels of sex hormones and risk of breast carcinoma in situ: results of a prospective study. Int. J. Cancer. 2005;114:323–327. doi: 10.1002/ijc.20694. doi:10.1002/ijc.20694. [DOI] [PubMed] [Google Scholar]
- 10.Hankinson S.E. Circulating levels of sex steroids and prolactin in premenopausal women and risk of breast cancer. Adv. Exp. Med. Biol. 2008;617:161–169. doi: 10.1007/978-0-387-69080-3_15. doi:10.1007/978-0-387-69080-3_15. [DOI] [PubMed] [Google Scholar]
- 11.MacMahon B., Cole P. Endocrinology and epidemiology of breast cancer. Cancer. 1969;24:1146–1151. doi: 10.1002/1097-0142(196912)24:6<1146::aid-cncr2820240612>3.0.co;2-0. doi:10.1002/1097-0142(196912)24:6<1146::AID-CNCR2820240612>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Clevenger C.V., Furth P.A., Hankinson S.E., Schuler L.A. The role of prolactin in mammary carcinoma. Endocr. Rev. 2003;24:1–27. doi: 10.1210/er.2001-0036. doi:10.1210/er.2001-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tworoger S.S., Eliassen A.H., Sluss P., Hankinson S.E. A prospective study of plasma prolactin concentrations and risk of premenopausal and postmenopausal breast cancer. J. Clin. Oncol. 2007;25:1482–1488. doi: 10.1200/JCO.2006.07.6356. doi:10.1200/JCO.2006.07.6356. [DOI] [PubMed] [Google Scholar]
- 14.Lee S.A., Haiman C.A., Burtt N.P., Pooler L.C., Cheng I., Kolonel L.N., Pike M.C., Altshuler D., Hirschhorn J.N., Henderson B.E., Stram D.O. A comprehensive analysis of common genetic variation in prolactin (PRL) and PRL receptor (PRLR) genes in relation to plasma prolactin levels and breast cancer risk: the multiethnic cohort. BMC Med. Genet. 2007;8:72. doi: 10.1186/1471-2350-8-72. doi:10.1186/1471-2350-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yager J.D., Davidson N.E. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. doi:10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 16.Rossouw J.E., Anderson G.L., Prentice R.L., LaCroix A.Z., Kooperberg C., Stefanick M.L., Jackson R.D., Beresford S.A., Howard B.V., Johnson K.C., et al. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the Women's Health Initiative randomized controlled trial. JAMA. 2002;288:321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 17.Chlebowski R.T., Kuller L.H., Prentice R.L., Stefanick M.L., Manson J.E., Gass M., Aragaki A.K., Ockene J.K., Lane D.S., Sarto G.E., et al. Breast cancer after use of estrogen plus progestin in postmenopausal women. N. Engl. J. Med. 2009;360:573–587. doi: 10.1056/NEJMoa0807684. doi:10.1056/NEJMoa0807684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khandwala H.M., McCutcheon I.E., Flyvbjerg A., Friend K.E. The effects of insulin-like growth factors on tumorigenesis and neoplastic growth. Endocr. Rev. 2000;21:215–244. doi: 10.1210/edrv.21.3.0399. doi:10.1210/er.21.3.215. [DOI] [PubMed] [Google Scholar]
- 19.Hadsell D.L., Bonnette S.G. IGF and insulin action in the mammary gland: lessons from transgenic and knockout models. J. Mammary Gland Biol. Neoplasia. 2000;5:19–30. doi: 10.1023/a:1009559014703. doi:10.1023/A:1009559014703. [DOI] [PubMed] [Google Scholar]
- 20.Hursting S.D., Lavigne J.A., Berrigan D., Donehower L.A., Davis B.J., Phang J.M., Barrett J.C., Perkins S.N. Diet-gene interactions in p53-deficient mice: insulin-like growth factor-1 as a mechanistic target. J. Nutr. 2004;134:2482S–2486S. doi: 10.1093/jn/134.9.2482S. [DOI] [PubMed] [Google Scholar]
- 21.Yakar S., Leroith D., Brodt P. The role of the growth hormone/insulin-like growth factor axis in tumor growth and progression: lessons from animal models. Cytokine Growth Factor Rev. 2005;16:407–420. doi: 10.1016/j.cytogfr.2005.01.010. doi:10.1016/j.cytogfr.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. doi:10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 23.Hankinson S.E., Willett W.C., Colditz G.A., Hunter D.J., Michaud D.S., Deroo B., Rosner B., Speizer F.E., Pollak M. Circulating concentrations of insulin-like growth factor-I and risk of breast cancer. Lancet. 1998;351:1393–1396. doi: 10.1016/S0140-6736(97)10384-1. doi:10.1016/S0140-6736(97)10384-1. [DOI] [PubMed] [Google Scholar]
- 24.Kaaks R., Lundin E., Rinaldi S., Manjer J., Biessy C., Söderberg S., Lenner P., Janzon L., Riboli E., Berglund G., Hallmans G. Prospective study of IGF-I, IGF-binding proteins, and breast cancer risk, in northern and southern Sweden. Cancer Causes Control. 2002;13:307–316. doi: 10.1023/a:1015270324325. doi:10.1023/A:1015270324325. [DOI] [PubMed] [Google Scholar]
- 25.Renehan A.G., Zwahlen M., Minder C., O'Dwyer S.T., Shalet S.M., Egger M. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet. 2004;363:1346–1353. doi: 10.1016/S0140-6736(04)16044-3. doi:10.1016/S0140-6736(04)16044-3. [DOI] [PubMed] [Google Scholar]
- 26.Schernhammer E.S., Holly J.M., Pollak M.N., Hankinson S.E. Circulating levels of insulin-like growth factors, their binding proteins, and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2005;14:699–704. doi: 10.1158/1055-9965.EPI-04-0561. doi:10.1158/1055-9965.EPI-04-0561. [DOI] [PubMed] [Google Scholar]
- 27.Rinaldi S., Peeters P.H., Berrino F., Dossus L., Biessy C., Olsen A., Tjonneland A., Overvad K., Clavel-Chapelon F., Boutron-Ruault M.C., et al. IGF-I, IGFBP-3 and breast cancer risk in women: The European Prospective Investigation into Cancer and Nutrition (EPIC) Endocr. Relat. Cancer. 2006;13:593–605. doi: 10.1677/erc.1.01150. doi:10.1677/erc.1.01150. [DOI] [PubMed] [Google Scholar]
- 28.Schernhammer E.S., Holly J.M., Hunter D.J., Pollak M.N., Hankinson S.E. Insulin-like growth factor-I, its binding proteins (IGFBP-1 and IGFBP-3), and growth hormone and breast cancer risk in The Nurses Health Study II. Endocr. Relat. Cancer. 2006;13:583–592. doi: 10.1677/erc.1.01149. doi:10.1677/erc.1.01149. [DOI] [PubMed] [Google Scholar]
- 29.Endogenous Hormones and Breast Cancer Collaborative Group. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11:530–542. doi: 10.1016/S1470-2045(10)70095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong Y., Pedersen N.L., Brismar K., Hall K., de Faire U. Quantitative genetic analyses of insulin-like growth factor I (IGF-I), IGF-binding protein-1, and insulin levels in middle-aged and elderly twins. J. Clin. Endocrinol. Metab. 1996;81:1791–1797. doi: 10.1210/jcem.81.5.8626837. doi:10.1210/jc.81.5.1791. [DOI] [PubMed] [Google Scholar]
- 31.Harrela M., Koistinen H., Kaprio J., Lehtovirta M., Tuomilehto J., Eriksson J., Toivanen L., Koskenvuo M., Leinonen P., Koistinen R., et al. Genetic and environmental components of interindividual variation in circulating levels of IGF-I, IGF-II, IGFBP-1 and IGFBP-3. J. Clin. Invest. 1996;98:2612–2615. doi: 10.1172/JCI119081. doi:10.1172/JCI119081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verhaeghe J., Loos R., Vlietinck R., Herck E.V., van Bree R., Schutter A.M. C-peptide, insulin-like growth factors I and II, and insulin-like growth factor binding protein-1 in cord serum of twins: genetic versus environmental regulation. Am. J. Obstet. Gynecol. 1996;175:1180–1188. doi: 10.1016/s0002-9378(96)70025-x. doi:10.1016/S0002-9378(96)70025-X. [DOI] [PubMed] [Google Scholar]
- 33.Hall K., Hilding A., Thoren M. Determinants of circulating insulin-like growth factor-I. J. Endocrinol. Invest. 1999;22:48–57. [PubMed] [Google Scholar]
- 34.An P., Rice T., Gagnon J., Hong Y., Leon A.S., Skinner J.S., Wilmore J.H., Bouchard C., Rao D.C. Race differences in the pattern of familial aggregation for dehydroepiandrosterone sulfate and its responsiveness to training in the HERITAGE Family Study. Metabolism. 2001;50:916–920. doi: 10.1053/meta.2001.24926. doi:10.1053/meta.2001.24926. [DOI] [PubMed] [Google Scholar]
- 35.Meikle A.W., Stringham J.D., Woodward M.G., Bishop D.T. Heritability of variation of plasma cortisol levels. Metabolism. 1988;37:514–517. doi: 10.1016/0026-0495(88)90164-3. doi:10.1016/0026-0495(88)90164-3. [DOI] [PubMed] [Google Scholar]
- 36.Rotter J.I., Wong F.L., Lifrak E.T., Parker L.N. A genetic component to the variation of dehydroepiandrosterone sulfate. Metabolism. 1985;34:731–736. doi: 10.1016/0026-0495(85)90023-x. doi:10.1016/0026-0495(85)90023-X. [DOI] [PubMed] [Google Scholar]
- 37.Franks S., Webber L.J., Goh M., Valentine A., White D.M., Conway G.S., Wiltshire S., McCarthy M.I. Ovarian morphology is a marker of heritable biochemical traits in sisters with polycystic ovaries. J. Clin. Endocrinol. Metab. 2008;93:3396–3402. doi: 10.1210/jc.2008-0369. doi:10.1210/jc.2008-0369. [DOI] [PubMed] [Google Scholar]
- 38.Dunning A.M. A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 1999;8:843–854. [PubMed] [Google Scholar]
- 39.Coughlin S.S., Piper M. Genetic polymorphisms and risk of breast cancer. Cancer Epidemiol. Biomarkers Prev. 1999;8:1023–1032. [PubMed] [Google Scholar]
- 40.Lohmueller K.E., Pearce C.L., Pike M., Lander E.S., Hirschhorn J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. doi:10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 41.Pharoah P.D., Tyrer J., Dunning A.M., Easton D.F., Ponder B.A. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:e42. doi: 10.1371/journal.pgen.0030042. doi:10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong L.M., Potter J.D., White E., Ulrich C.M., Cardon L.R., Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299:2423–2436. doi: 10.1001/jama.299.20.2423. doi:10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mavaddat N., Dunning A.M., Ponder B.A., Easton D.F., Pharoah P.D. Common genetic variation in candidate genes and susceptibility to subtypes of breast cancer. Cancer Epidemiol. Biomarkers Prev. 2009;18:255–259. doi: 10.1158/1055-9965.EPI-08-0704. doi:10.1158/1055-9965.EPI-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fletcher O., Gibson L., Johnson N., Altmann D.R., Holly J.M., Ashworth A., Peto J., dos Santos Silva I. Polymorphisms and circulating levels in the insulin-like growth factor system and risk of breast cancer: a systematic review. Cancer Epidemiol. Biomarkers Prev. 2005;14:2–19. [PubMed] [Google Scholar]
- 45.Canzian F., McKay J.D., Cleveland R.J., Dossus L., Biessy C., Boillot C., Rinaldi S., Llewellyn M., Chajès V., Clavel-Chapelon F., et al. Genetic variation in the growth hormone synthesis pathway in relation to circulating insulin-like growth factor-I, insulin-like growth factor binding protein-3, and breast cancer risk: results from the European prospective investigation into cancer and nutrition study. Cancer Epidemiol. Biomarkers Prev. 2005;14:2316–2325. doi: 10.1158/1055-9965.EPI-04-0874. doi:10.1158/1055-9965.EPI-04-0874. [DOI] [PubMed] [Google Scholar]
- 46.Canzian F., McKay J.D., Cleveland R.J., Dossus L., Biessy C., Rinaldi S., Landi S., Boillot C., Monnier S., Chajès V., et al. Polymorphisms of genes coding for insulin-like growth factor 1 and its major binding proteins, circulating levels of IGF-I and IGFBP-3 and breast cancer risk: results from the EPIC study. Br. J. Cancer. 2006;94:299–307. doi: 10.1038/sj.bjc.6602936. doi:10.1038/sj.bjc.6602936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng I., Penney K.L., Stram D.O., Le Marchand L., Giorgi E., Haiman C.A., Kolonel L.N., Pike M., Hirschhorn J., Henderson B.E., et al. Haplotype-based association studies of IGFBP1 and IGFBP3 with prostate and breast cancer risk: the multiethnic cohort. Cancer Epidemiol. Biomarkers Prev. 2006;15:1993–1997. doi: 10.1158/1055-9965.EPI-06-0361. doi:10.1158/1055-9965.EPI-06-0361. [DOI] [PubMed] [Google Scholar]
- 48.Wagner K., Hemminki K., Försti A. The GH1/IGF-1 axis polymorphisms and their impact on breast cancer development. Breast Cancer Res. Treat. 2007;104:233–248. doi: 10.1007/s10549-006-9411-9. doi:10.1007/s10549-006-9411-9. [DOI] [PubMed] [Google Scholar]
- 49.Patel A.V., Cheng I., Canzian F., Le Marchand L., Thun M.J., Berg C.D., Buring J., Calle E.E., Chanock S., Clavel-Chapelon F., et al. Breast and Prostate Cancer Cohort Consortium. IGF-1, IGFBP-1 and IGFBP-3 polymorphisms predict circulating IGF levels but not breast cancer risk: findings from the Breast and Prostate Cancer Cohort Consortium (BPC3) PLoS One. 2008;3:e2578. doi: 10.1371/journal.pone.0002578. doi:10.1371/journal.pone.0002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunter D.J., Riboli E., Haiman C.A., Albanes D., Altshuler D., Chanock S.J., Hayes R.B., Henderson B.E., Kaaks R., Stram D.O., et al. A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat. Rev. Cancer. 2005;5:977–985. doi: 10.1038/nrc1754. doi:10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 51.Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol. Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- 52.Dunning A.M., Healey C.S., Baynes C., Maia A.T., Scollen S., Vega A., Rodríguez R., Barbosa-Morais N.L., Ponder B.A., Low Y.L., et al. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum. Mol. Genet. 2009;18:1131–1139. doi: 10.1093/hmg/ddn429. doi:10.1093/hmg/ddn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cordell H.J., Clayton D.G. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am. J. Hum. Genet. 2002;70:124–141. doi: 10.1086/338007. doi:10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haiman C.A., Dossus L., Setiawan V.W., Stram D.O., Dunning A.M., Thomas G., Thun M.J., Albanes D., Altshuler D., Ardanaz E., et al. Genetic variation at the CYP19A1 locus predicts circulating estrogen levels but not breast cancer risk in postmenopausal women. Cancer Res. 2007;67:1893–1897. doi: 10.1158/0008-5472.CAN-06-4123. doi:10.1158/0008-5472.CAN-06-4123. [DOI] [PubMed] [Google Scholar]
- 55.Dunning A.M., Dowsett M., Healey C.S., Tee L., Luben R.N., Folkerd E., Novik K.L., Kelemen L., Ogata S., Pharoah P.D., et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J. Natl. Cancer Inst. 2004;96:936–945. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- 56.Johansson M., McKay J.D., Wiklund F., Rinaldi S., Verheus M., van Gils C.H., Hallmans G., Bälter K., Adami H.O., Grönberg H., et al. Implications for prostate cancer of insulin-like growth factor-I (IGF-I) genetic variation and circulating IGF-I levels. J. Clin. Endocrinol. Metab. 2007;92:4820–4826. doi: 10.1210/jc.2007-0887. doi:10.1210/jc.2007-0887. [DOI] [PubMed] [Google Scholar]
- 57.Thompson D.J., Healey C.S., Baynes C., Kalmyrzaev B., Ahmed S., Dowsett M., Folkerd E., Luben R.N., Cox D., Ballinger D., et al. Identification of common variants in the SHBG gene affecting sex hormone-binding globulin levels and breast cancer risk in postmenopausal women. Cancer Epidemiol. Biomarkers Prev. 2008;17:3490–3498. doi: 10.1158/1055-9965.EPI-08-0734. doi:10.1158/1055-9965.EPI-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johansson M., McKay J.D., Wiklund F., Rinaldi S., Hallmans G., Bälter K., Adami H.O., Grönberg H., Stattin P., Kaaks R. Genetic variation in the SST gene and its receptors in relation to circulating levels of insulin-like growth factor-I, IGFBP3, and prostate cancer risk. Cancer Epidemiol. Biomarkers Prev. 2009;18:1644–1650. doi: 10.1158/1055-9965.EPI-08-0893. doi:10.1158/1055-9965.EPI-08-0893. [DOI] [PubMed] [Google Scholar]
- 59.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. doi:10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A., et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. doi:10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 63.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K., et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. doi:10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 64.Thomas G., Jacobs K.B., Kraft P., Yeager M., Wacholder S., Cox D.G., Hankinson S.E., Hutchinson A., Wang Z., Yu K., et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. doi:10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. doi:10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Freedman M.L., Haiman C.A., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. doi:10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gudmundsson J., Sulem P., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. doi:10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 68.Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. doi:10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. doi:10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 70.Ghoussaini M., Song H., Koessler T., Al Olama A.A., Kote-Jarai Z., Driver K.E., Pooley K.A., Ramus S.J., Kjaer S.K., Hogdall E., et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calle E.E., Rodriguez C., Jacobs E.J., Almon M.L., Chao A., McCullough M.L., Feigelson H.S., Thun M.J. The American Cancer Society Nutrition Cohort: rationale, study design and baseline characteristics. Cancer. 2002;94:2490–2501. doi: 10.1002/cncr.101970. doi:10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 72.Riboli E., Hunt K.J., Slimani N., Ferrari P., Norat T., Fahey M., Charrondiere U.R., Hemon B., Casagrande C., Vignat J., et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5:1113–1124. doi: 10.1079/PHN2002394. doi:10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 73.Colditz G.A., Hankinson S.E. The Nurses' Health Study: lifestyle and health among women. Nat. Rev. Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. doi:10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 74.Rexrode K., Lee I., Cook N., Hennekens C., Buring J. Baseline characteristics of participants in the Women's Health Study. J. Women's Health Gend. Based Med. 2000;9:19–27. doi: 10.1089/152460900318911. doi:10.1089/152460900318911. [DOI] [PubMed] [Google Scholar]
- 75.Hayes R.B., Reding D., Kopp W., Subar A.F., Bhat N., Rothman N., Caporaso N., Ziegler R.G., Johnson C.C., Weissfeld J.L., et al. Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial Project Team. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control. Clin. Trials. 2000;21:349S–355S. doi: 10.1016/s0197-2456(00)00101-x. doi:10.1016/S0197-2456(00)00101-X. [DOI] [PubMed] [Google Scholar]
- 76.Kolonel L., Henderson B., Hankin J., Nomura A., Wilkens L., Pike M., Stram D., Monroe K., Earle M., Nagamine F. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stram D.O., Haiman C.A., Hirschhorn J.N., Altshuler D., Kolonel L.N., Henderson B.E., Pike M.C. Choosing haplotype-tagging SNPs based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum. Hered. 2003;55:27–36. doi: 10.1159/000071807. doi:10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- 78.Goode E.L., Fridley B.L., Sun Z., Atkinson E.J., Nord A.S., McDonnell S.K., Jarvik G.P., de Andrade M., Slager S.L. Comparison of tagging single-nucleotide polymorphism methods in association analyses. BMC Proc. 2007;1((Suppl. 1)):S6. doi: 10.1186/1753-6561-1-s1-s6. doi:10.1186/1753-6561-1-s1-s6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krawczak M., Chuzhanova N.A., Cooper D.N. Evolution of the proximal promoter region of the mammalian growth hormone gene. Gene. 1999;237:143–151. doi: 10.1016/s0378-1119(99)00313-3. doi:10.1016/S0378-1119(99)00313-3. [DOI] [PubMed] [Google Scholar]
- 80.Marchini J., Howie B. Comparing algorithms for genotype imputation. Am. J. Hum. Genet. 2008;83:535–539. doi: 10.1016/j.ajhg.2008.09.007. doi:10.1016/j.ajhg.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Bakker P.I., Burtt N.P., Graham R.R., Guiducci C., Yelensky R., Drake J.A., Bersaglieri T., Penney K.L., Butler J., Young S., et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat. Genet. 2006;38:1298–1303. doi: 10.1038/ng1899. doi:10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- 82.Sanna S., Jackson A.U., Nagaraja R., Willer C.J., Chen W.M., Bonnycastle L.L., Shen H., Timpson N., Lettre G., Usala G., et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008;40:198–203. doi: 10.1038/ng.74. doi:10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao X., Starmer J., Martin E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. doi:10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 84.Stram D.O., Leigh Pearce C., Bretsky P., Freedman M., Hirschhorn J.N., Altshuler D., Kolonel L.N., Henderson B.E., Thomas D.C. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case–control study of unrelated individuals. Hum. Hered. 2003;55:179–190. doi: 10.1159/000073202. doi:10.1159/000073202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.