Abstract
The reduced expression of the Sp4 gene in Sp4 hypomorphic mice resulted in subtle vacuolization in the hippocampus as well as deficits in sensorimotor gating and contextual memory, putative endophenotypes for schizophrenia and other psychiatric disorders. In this study, we examined both spatial learning/memory and hippocampal long-term potentiation (LTP) of Sp4 hypomorphic mice. Impaired spatial learning/memory and markedly reduced LTP were found. To corroborate the functional studies, the expression of N-methyl-D-aspartate (NMDA) glutamate receptors was investigated with both western blot and immunohistochemical analyses. The reduced expression of the Sp4 gene decreased the level of the NR1 subunit of NMDA receptors in Sp4 hypomorphic mice. In human, SP4 gene was found to be deleted sporadically in schizophrenia patients, corroborating evidence that polymorphisms of human SP4 gene are associated with schizophrenia and other psychiatric disorders. Impaired NMDA neurotransmission has been implicated in several human psychiatric disorders. As yet, it remains unclear how mutations of candidate susceptibility genes for these disorders may contribute to the disruption of NMDA neurotransmission. Sp4 hypomorphic mice could therefore serve as a genetic model to investigate impaired NMDA functions resulting from loss-of-function mutations of human SP4 gene in schizophrenia and/or other psychiatric disorders. Furthermore, aberrant expression of additional genes, besides NMDAR1, likely also contributes to the behavioral abnormalities in Sp4 hypomorphic mice. Thus, further investigation of the Sp4 pathway may provide novel insights in our understanding of a variety of neuropsychiatric disorders.
INTRODUCTION
Sp4, a member of the Sp1 family of transcription factors, recognizes GC-rich sequences readily identified in the ‘CpG islands’ around the promoters of a variety of genes (1). In contrast to the ubiquitous expression pattern of the Sp1 gene, the Sp4 gene is expressed restrictively in the nervous system (2,3). The complete absence of the Sp4 gene impaired postnatal development of the hippocampal dentate gyrus by reducing cell proliferation, dendritic growth and dendritic arborization in Sp4 null mutant mice (4). The incomplete suppression of Sp4 expression, however, resulted in highly branched dendrites during the maturation of in vitro-cultured cerebellar granule neurons (5). The activation of glutamate receptors was reported to cause degradation of Sp4 proteins (6). The reduced expression of the Sp4 gene in Sp4 hypomorphic mice resulted in subtle vacuolization in the hippocampus as well as deficits in sensorimotor gating and memory, putative endophenotypes for schizophrenia and other psychiatric disorders (3,7–11). Here, we found that the reduced expression of Sp4 gene dramatically decreased the expression of NMDAR1, markedly reduced long-term potentiation (LTP) in hippocampal CA1 and impaired the formation of spatial memory.
Impaired N-methyl-D-aspartate (NMDA) neurotransmission has been suggested to cause prefrontal cognitive deficits in schizophrenia (12). This theory is based on the observation that acute administrations of non-competitive NMDA receptor antagonists, such as PCP and ketamine, induce schizophrenia-like symptoms in healthy people and worsen some symptoms in schizophrenia (13–15). Recently, patients with anti-NMDA-receptor encephalitis (anti-NMDAR1) were found to display many schizophrenia-like symptoms and/or loss of memory (16,17). Current antipsychotics have little effect on improving cognition in schizophrenia patients (18). These treatment-resistant cognitive deficits contribute substantially to functional disability (19).
Mouse hypoglutamatergic models for schizophrenia have been developed with the deletion of nmdar1 gene (20–22). These mutant mice displayed deficient sensorimotor gating and memory, and served as valuable genetic models for biological studies on schizophrenia. However, genetic evidence to suggest human NMDAR1 as a susceptibility gene for schizophrenia and other psychiatric disorders is lacking in either genome-wide association studies (GWAS) or copy-number variation (CNV) studies. Therefore, impaired NMDA functions are more likely indirect effects of the primary susceptibility genes in the pathogenesis of schizophrenia. During the examination of spatial learning/memory deficit in Sp4 hypomorphic mice, we found that NMDAR1 expression was remarkably reduced. Our analysis of the CNV database revealed that human SP4 gene was deleted sporadically in patients with schizophrenia. Our studies therefore suggested that Sp4 hypomorphic mouse represent a novel hypoglutamatergic model for psychiatric diseases, including schizophrenia.
RESULTS
We previously demonstrated that Sp4 hypomorphic mice displayed subtle vacuolization in hippocampus and deficits in sensorimotor gating and contextual memory. However, it was unknown what molecular mechanism underlies these phenotypes. To extend the characterization of hippocampus-dependent memory and understand the underlying molecular mechanisms, we focused our studies on the hippocampal CA1 region where the Sp4 gene is expressed abundantly (3).
Deficient spatial learning/memory in Sp4 hypomorphic mice
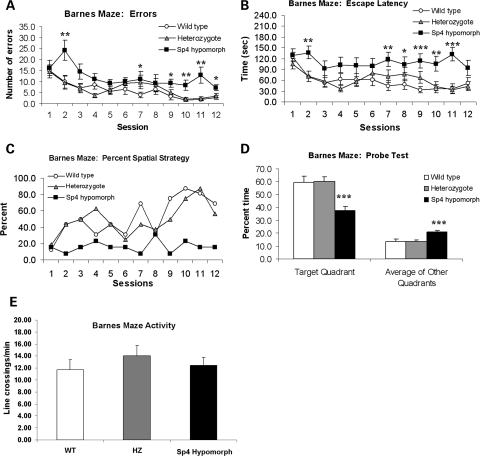
Hippocampal CA1 plays an essential role in the formation of spatial memory. To understand the role of Sp4 expression in CA1, we examined Sp4 hypomorphic mice in the Barnes maze test for spatial memory, chosen because these mice float and engage in thigmotaxis in the Morris water maze (unpublished data). The Barnes maze is essentially a land-based version of the Morris water maze. A mouse test cohort, consisting of sex- and age-matched siblings with the same genetic background, was first examined for their general health. No abnormalities were found (Supplementary Material). In the Barnes maze, their performance was measured as the number of errors, escape latency and strategy. The Sp4 hypomorphic mice made significantly more errors than either their wild-type or heterozygous siblings (F(2,42) = 13.5, P < 0.0001; Fig. 1A). Consistent with their higher number of errors, the Sp4 hypomorphic mice exhibited longer escape latencies than either wild-type or heterozygous mice (F(2,42) = 7.8, P < 0.005; Fig. 1B). Further analysis found that the Sp4 mutant mice spent most of their time randomly searching for the target hole with little improvement in performance, while wild-type and heterozygous mice gradually adopted a spatial strategy moving directly to the target hole during the 12 consecutive sessions (Fig. 1C). After the 12 sessions, a probe test was conducted with the target hole blocked off. The Sp4 hypomorphic mice spent significantly less time in the target quadrant than their sibling wild-type and heterozygous mice (F(2,42) = 10.0, P < 0.0005; Fig. 1D). To examine the potential contribution of non-cognitive activities, the locomotor activities were compared between wild-type and Sp4 hypomorphic mice in the Barnes maze. A 5 × 5 grid was superimposed onto the Barnes maze video data and line crossings (all four paws) on trial 2 were assessed. In order to control for differences in time on the maze, the crossings per minute were calculated. There was no genotypic difference in activity levels on the Barnes maze (F(2,39) = 0.600, ns; Fig. 1E). In addition, there were no differences between wild-type and Sp4 hypomorphic mice in head tracking in the optomotor test of vision, suggesting the absence of impaired visual ability in the Sp4 mutant mice. No differences were found either in anxiety-like behavior in the light/dark transfer test. Activity levels and exploration were assessed in the light/dark transfer test, the object exploration test, the social interaction test and in no case were there genotypic differences in these measures. Taken together, our results therefore suggest that Sp4 hypomorphic mice have a specific deficit in spatial learning/memory, which is unlikely contaminated by non-specific differences in behavioral output.
Figure 1.
Impaired spatial learning of Sp4 homozygous mice in the Barnes maze. A test cohort consisting of 6-month-old sibling mice with 16 wild-type (8 males and 8 females), 16 heterozygous (8 males and 8 females) and 13 Sp4 homozygous mice (7 males and 6 females) was used in the Barnes maze test. No sex effect was found. The combined data were therefore presented: (A) the number of times the mouse pokes its nose into, or hovers its head over a circular hole that does not contain the escape chamber; (B) the time between the initiation of the trial and the entrance of the mouse into the escape chamber; (C) the percent of mice using spatial cues to locate the location of the escape chamber, i.e. running directly to the escape chamber in each session; (D) after the 12 sessions, the escape chamber was removed and the time that each mouse spent in the target quadrant and in each other quadrant was recorded; (E) locomotor activities measured by line-crossing on trial 2. ANOVA was used for the statistical analysis, followed by post hoc Student's t-tests as appropriate (*P < 0.05, **P < 0.01, ***P < 0.001).
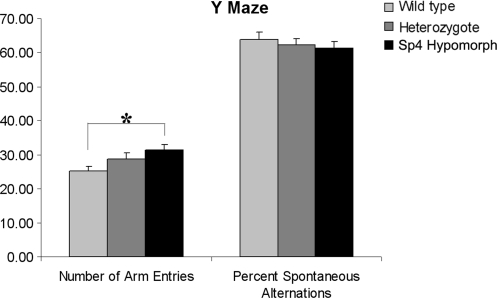
To further examine whether Sp4 hypomorphic mice may exhibit additional cognitive deficits, a Y-maze test was employed. Sp4 hypomorphic mice appeared somewhat more active than their wild-type littermates, exhibiting an increased number of entries (F(2,73) = 4.4, P < 0.05). The spatial alternation of the mutant mice was not, however, significantly different from that of their wild-type littermates in the Y-maze test (F(2,73) < 1, ns; Fig. 2), suggesting that short-term memory is relatively spared in the Sp4 hypomorphic mice.
Figure 2.
Spontaneous alternation in the Y-maze. A test cohort consisting of 32 wild-type, 26 heterozygous and 18 Sp4 homozygous mice was used in the Y-maze test. There was a significant increase in the number of arm entries made by the Sp4 homozygous mice in comparison with their wild-type littermates. Nevertheless, there was no significant difference in spontaneous alternation between different genotypes (*P < 0.05).
Impaired LTP in the hippocampal CA1
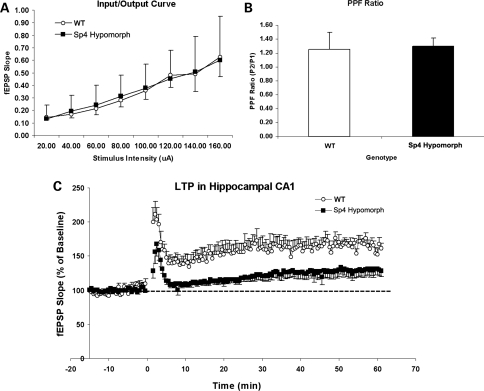
LTP in the hippocampal CA1 has been studied extensively for its role in the formation of spatial memory (23,24). Because the mouse Sp4 gene is expressed abundantly in the CA1 region (3), we examined whether reduced Sp4 expression might impair the formation of LTP in the CA1 region of the hippocampus in Sp4 mutant mice. Six pairs of sibling wild-type and Sp4 hypomorphic mice were used for electrophysiological recording in the hippocampal CA1. Field potentials were evoked by stimulation of Schaffer collateral/commissural afferents and recorded in the stratum radiatum of CA1. To examine synaptic transmission, different stimulation intensities were applied, and the resultant field potentials were recorded. There was no difference in their input/output ratio, suggesting that Sp4 hypomorphic mice had normal synaptic neurotransmission (Fig. 3A). Paired-pulse facilitation (PPF) was also examined and no difference was found, indicating the absence of pre-synaptic abnormalities in the Sp4 hypomorphic mice (Fig. 3B). Nevertheless, LTP was markedly impaired in the hippocampal CA1 of the Sp4 hypomorphic mice in comparison with the wild-type mice, when a high-frequency train of stimuli was delivered to the afferent fibers (Fig. 3C). NMDA receptors have been shown to be essential for the generation of LTP in the hippocampal CA1 region and in the formation of spatial memory (23). These data therefore prompted the hypothesis that the Sp4 gene plays an important role in the NMDA receptor-mediated signaling pathway in the hippocampal CA1 region.
Figure 3.
Electrophysiological recording of mouse hippocampal slices from adult wild-type and Sp4 mutant mice. (A) Input/output curves were constructed by plotting evoked field excitatory postsynaptic potentials (EPSPs) against different intensities of stimuli. There was no difference in the slope of I/O curve between the wild-type (n = 6) and Sp4 mice (n = 6). (B) PPF at inter-stimulus interval of 50 ms was quantified as the ratio (P2/P1) of the slope of the fEPSPs evoked by the second stimulus (P2) versus the first stimulus (P1). There was no difference in the PPF ratios between the wild-type (n = 6) and Sp4 mice (n = 6). (C) LTP was induced by high-frequency stimulation (4 × 100 Hz) of Schaffer collateral/commissural afferents at time = 0 and quantified by plotting the slope of fEPSPs against different time points after the high-frequency stimulation (Student's t-test, P < 0.001).
Decreased expression of NMDAR1 (NR1) protein
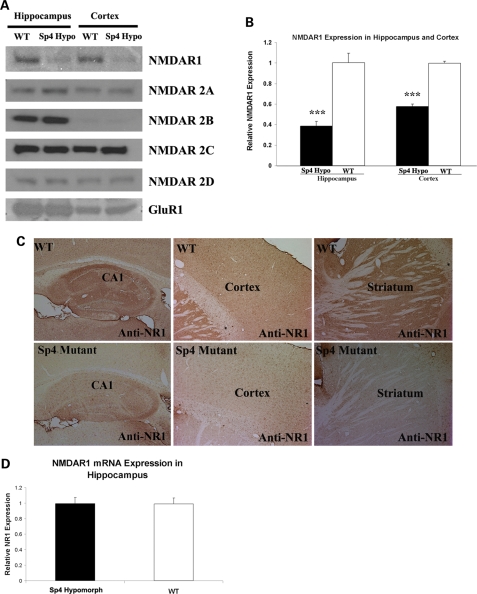
As our first step to understand the role of the Sp4 gene in the NMDA receptor-signaling pathway, we examined whether Sp4 might regulate the expression of NMDA receptors. NMDA receptors are tetrameric proteins consisting of two core NR1 subunits and two NR2 subunits coming from different NR2 family genes NR2A, NR2B, NR2C and NR2D (25,26). The expression of the NR1 protein in both the hippocampus and cortex of Sp4 hypomorphic mice was decreased to about 40–50% of the level in wild-type mouse brain (Fig. 4A and B). Nevertheless, no significant difference was found in the expression of four NR2 subunits (2A, 2B, 2C, 2D) (Fig. 4A). Due to the absence of abnormal synaptic transmission in our electrophysiological recordings, we did not analyze the expression of all non-NMDA receptor subunits except GluR1 in the Sp4 hypomorphic mice. As expected, we did not observe differential expression of GluR1 between wild-type and Sp4 hypomorphic mice (Fig. 4A). To assess the regional distribution of the down-regulation of NR1 expression in brain, we conducted immunohistochemical staining on mouse brain sections. As expected, a striking reduction of NR1 was confirmed in both the hippocampus and cortex of Sp4 hypomorphic mice in comparison with wild-type sibling mice (Fig. 4C). Moreover, it appeared that the expression of NR1 was decreased across the whole brain. The Sp family of transcription factors had been suggested to regulate the Nmdar1 gene expression by binding to the GC-rich region in the promoter (27,28). We therefore conducted quantitative RT–PCR assays to examine whether the level of the Nmdar1 mRNA expression was decreased in the hippocampus of the Sp4 hypomorphic mice. No difference was found in Nmdar1 mRNA expression between wild-type and Sp4 hypomorphic mice (Fig. 4D). The differential expression of NMDAR1 proteins could result from the expression of different Nmdar1 mRNA isoforms by alternative splicing and/or different 5′- and 3′-untranslated regions (UTRs) between wild-type and Sp4 hypomorphic mice. We therefore examined all possible alternative splicing and both 5′- and 3′-UTRs of NMDAR1 mRNA between wild-type and Sp4 hypomorphic mice by using RT–PCR but did not observe any differences (Supplementary Material, Fig. S1).
Figure 4.
Reduced expression of NMDAR1 subunit in Sp4 homozygous mice. (A) Western blot analysis of protein lysates from both hippocampus and cortex in adult mice. (B) Quantification of NMDAR1 proteins in both hippocampus and cortex. A reduced expression of NMDAR1 proteins was observed in the Sp4 mutant mice (n = 5) in comparison with wild-type mice (n = 5). (C) Immunohistochemical staining of NMDAR1 expression in frozen sections of mouse brains. NMDAR1 expression was reduced in the hippocampus, cortex and striatum of the Sp4 hypomorphic mice. (D) Quantitative RT–PCR analysis of NMDAR1 mRNA expression in both wild-type (n = 6) and the Sp4 mutant mice (n = 6). No significant difference was observed in the expression of NMDAR1 mRNA in the hippocampus.
SP4 deletion in sporadic patients with schizophrenia
Despite mounting evidence for impaired NMDA functions in schizophrenia, genes encoding NMDA receptors do not display significant genetic associations with the disease. Considering that Sp4 regulates NMDAR1 expression, we would expect that the reduced expression of human SP4 gene might decrease the expression of human NMDAR1, which could contribute to the susceptibility for developing schizophrenia. With the advancement of technologies, large-scale genome-wide CNV scan has been conducted in thousands of patients and controls. After analyzing the data from 3391 schizophrenic patients, SP4 deletion was found in two sporadic patients (29) (http://pngu.mgh.harvard.edu/isc/isc-r1.cnv.bed.). No deletion or duplication was found in 3181 controls. The two SP4 deletions, with the sizes of 108 and 120 kb, respectively, excised the first three exons as well as the 5′-flanking regulatory region of SP4 gene (Fig. 5). The deletions therefore constitute lose-of-function mutations of the SP4 gene. The two nearest genes, SP8 and DNAH11, were located 557 and 85 kb away from the deletions, respectively, suggesting SP4 as the only physically affected gene.
Figure 5.
SP4 deletion in schizophrenia. Human SP4 gene was analyzed in the CNV database from International Schizophrenia Consortium (ISC) (http://pngu.mgh.harvard.edu/isc/isc-r1.cnv.bed.). Two sporadic patients carried SP4 deletion, and no controls had any SP4 mutations.
DISCUSSION
Sp4 transcription factors, restrictively expressed in neuronal cells, play important roles in hippocampal development and neurogenesis (4,5). In this study, we found that Sp4 hypomorphic mice displayed impaired spatial learning/memory, mimicking a cognitive deficit seen in schizophrenia (30–32). In addition to a spatial learning/memory deficit in the Barnes maze, Sp4 hypomorphic mice also displayed learning/memory deficits in both contextual fear conditioning and passive avoidance tests (3). Sp4 hypomorphic mice did not display a deficit in short-term memory, however, since they performed normally in the Y-maze task. The impaired hippocampus-dependent memories in these different memory paradigms are consistent with hippocampal abnormalities in Sp4 hypomorphic mice. However, there is a discrepancy in anxiety-related behaviors from two different tests. We did not observe increased anxiety-like behavior of Sp4 hypomorphic mice in light/dark transfer test in contrast to our previous report of increased anxiety-like behavior suggested in open field tests (3). Such inconsistencies between different tests for anxiety have been reported in other mutant mice, and attributed to different sets of overlapping genes for multidimensionality of anxiety (33–36).
Reduced NMDAR1 expression
Our further molecular studies revealed that the reduced expression of Sp4 remarkably decreased the expression of NMDAR1 throughout mouse brain, including hippocampus.
An impaired NMDA neurotransmission, supported by molecular and electrophysiological observations in Sp4 hypomorphic mice, provided a plausible molecular mechanism for the demonstrated deficits in both memory (23,37) and sensorimotor gating (20,21,38). The reduced expression of NMDAR1 was unlikely caused by localized subtle vacuolization in the hippocampus of adult Sp4 hypomorphic mice, because the reduction of NMDAR1 expression was also found in the cortex where no structural abnormalities were found. It will be interesting to determine whether decreased NMDAR1 expression could be the primary defect for the subtle vacuolization, as suggested by Olney et al. (39). It was surprising that we did not detect a significant difference of NMDAR1 RNA expression between wild-type and Sp4 hypomorphic mice. There are several possibilities for this disparity. First, our RNA expression analysis was conducted in the RNA samples prepared from the whole hippocampus. Sp4 may regulate NMDAR1 RNA expression only in a small number of specific types of neurons (e.g. interneurons), which may subsequently result in down-regulation of NMDAR1 (e.g. NMDAR1 turnover) in a large number of pyramidal neurons via abnormal NMDA neurotransmission networking. Second, since mouse Sp4 gene expression starts around E9 during embryogenesis (2), subtle developmental abnormalities (although not detected yet) cannot be ruled out during the development of Sp4 hypomorphic mice, which might indirectly contribute to down-regulation of NMDAR1 protein expression. Third, the reduced expression of Sp4 gene causes aberrant expression of many downstream target genes, which somehow results in down-regulation of NMDAR1 proteins. Fourth, Sp4 transcription factors may be involved in post-transcriptional regulation of NMDAR1 expression, such as Y-box proteins which function as both DNA- and RNA-binding proteins (40). These possible molecular mechanisms merit further investigation in future studies.
SP4 as a susceptibility gene for psychiatric disorders
Hippocampal abnormalities, deficient sensorimotor gating and impairments in learning/memory have been studied extensively as putative endophenotypes for schizophrenia and other psychiatric disorders. It should be noted, however, that none of these endophenotypes are specific for any particular psychiatric disorder. In this study, we found that human SP4 gene was deleted in sporadic schizophrenia patients. Sp4 hypomorphic mice therefore recapitulated several endophenotypes of schizophrenia, where impaired NMDA neurotransmission has been implicated. There are likely more downstream effectors other than NMDAR1 that contribute to the phenotypes in Sp4 hypomorphic mice. However, the Sp4 hypomorphic mouse could represent a valuable model for us to understand hypoglutamatergic function, one of the key features of human schizophrenia and other psychiatric disorders (13). Although SP4 deletions are rare, subtle alterations of SP4 gene may contribute to modest risk in the general population of schizophrenia and/or bipolar disorder as suggested in our genetic association studies (41). Recently, GWAS identified SP4 as a leading candidate susceptibility gene for major depressive disorder (42,43) which can share the same genetic mutations with schizophrenia and bipolar disorder (44,45).
Recently, an aberrant over-expression of SP4 transcription factors was found in the cytoplasm of degenerating neurons in the post-mortem brains of Alzheimer's disease patients (46). It is not surprising that abnormal expression of SP4 may also contribute to Alzheimer's disease, since NMDA receptors have been suggested to play important roles in Alzheimer's disease (47). Therefore, we anticipate that the SP4 molecular pathway could play an important role for a range of psychiatric and neurological diseases characterized by abnormal NMDA neurotransmission.
MATERIALS AND METHODS
Mouse strains and breeding
The Sp4 hypomorphic mice were generated as described (3) and maintained in both S129 and Black Swiss backgrounds. The Sp4 test cohort was generated by breeding the heterozygous Sp4 mice between the two genetic backgrounds. Therefore, all the test adult mice (about 6 months old) have common parenting and the same genetic mixed S129/Black Swiss background as the F1 generation. PCR was used for Sp4 mouse genotyping as previously described (3). Mice were housed in a climate-controlled animal colony with a reversed day/night cycle. Food (Harlan Teklab, Madison, WI, USA) and water were available ad libitum, except during behavioral testing. All behavioral testing procedures were approved by the UCSD and The Scripps Research Institute Institutional Animal Care and Use Committee prior to the onset of the experiments. Mice were maintained in American Association for Accreditation of Laboratory Animal Care approved animal facilities at the local Veteran's Administration Hospital. This facility meets all Federal and State requirements for animal care.
Barnes maze
In the Barnes maze test, an opaque Plexiglas disk 75 cm in diameter was used. Twenty holes, 5 cm in diameter, were located 5 cm from the perimeter, and a black Plexiglas escape box (19 × 8 × 7 cm) was placed under one of the holes. Distinct spatial cues were located all around the maze and were kept constant throughout the study. On the first day of testing, a training session was performed, which consisted of placing the mouse in the escape box and leaving it there for 1 min. One min later, the first session was started. At the beginning of each session, the mouse was placed in the middle of the maze in a 10 cm high cylindrical black start chamber. After 10 s, the start chamber was removed, a buzzer (80 dB) and a light (400 lux) were turned on, and the mouse was set free to explore the maze. The session ended when the mouse entered the escape tunnel or after 3 min elapsed. When the mouse entered the escape tunnel, the buzzer was turned off and the mouse was allowed to remain in the dark for 1 min. When the mouse did not enter the tunnel by itself, it was gently put in the escape box for 1 min. The tunnel was always located underneath the same hole (stable within the spatial environment), which was determined randomly for each mouse. The platform and escape box were cleaned with 70% ethanol between mice. Mice were tested once a day for 12 days for the acquisition portion of the study. For the 13th test (probe test), the escape tunnel was removed and the mouse was allowed to explore the maze freely for 3 min. The time spent in each quadrant was determined and the percent time spent in the target quadrant (the one originally containing the escape box) was compared with the average percent time in the other three quadrants. This is a direct test of spatial memory because there is no potential for local cues to be used in the mouse's behavioral decision. ANOVA was used for the statistical analysis, followed by post hoc Student's t-tests as appropriate (*P < 0.05, **P < 0.01, ***P < 0.001).
Y-maze
A single 5 min test was performed in which each mouse was placed in the center of the Y. Arm entries were recorded by video camera, and the total number of arm entries and the order of entries were determined. Spontaneous alternations were defined as consecutive triplets of different arm choices. ANOVA was used for the statistical analysis.
Hippocampal slice recording
Mice were anesthetized with isoflurane, and their brains were rapidly removed into ice-cold artificial cerebrospinal fluid (ACSF) gassed with 95% O2/5% CO2. Transverse hippocampal slices (400 µM thick) were cut on a Leica VT 1000S (McBain Instruments, Chatsworth, CA, USA). Fresh slices were incubated in chamber with cabogenated ACSF and recovered at room temperature for at least 1.5 h before they were transferred to recording chamber. The ACSF was composed of 125 mm NaCl, 25 mm NaHCO3, 3.75 mm KCl, 2 mm CaCl2, 1.2 mm MgCl2, 1.25 mm NaH2PO4 and 10 mm d-glucose. For electrophysiological recordings, slices were transferred to a submerged recording chamber, maintained at room temperature and perfused continuously with ACSF at a rate of 2–3 ml/min. Field potentials were recorded with extracellular recording electrodes (2–3 MΩ) filled with ACSF and placed in stratum radiatum of CA1. Field potentials were evoked by monophasic stimulation (duration, 200 µs) of Schaffer collateral/commissural afferents with concentric bipolar tungsten stimulating electrode (Frederick Haer Company, Bowdoinham, ME, USA). Stable baseline responses were collected every 30 s using a stimulation intensity (10–30 µA) yielding 50–60% of the maximal responses. A Multiclamp-700B amplifier and Digidata 1322A (Molecular Devices, Union City, CA, USA) were used to record and store data with pCLAMP 9.2 software (Molecular Devices), respectively. LTP was induced by stimulating afferent fibers at high-frequency stimulation (HFS: 4 × 100 Hz).
Western blot and immunochemical staining
Mice were anesthetized with carbon dioxide, and the brains were quickly taken out and put on ice. Hippocampi were then dissected out on ice. Both hippocampi and cortex were homogenized by using Dounce Homogenizer in 1X Passive Lysis Buffer (Promega) supplemented with Protease Inhibitor Cocktail (Sigma). The protein concentration was measured with Bradford Assay Kit (Pierce). Fifty microgram of total proteins were loaded on SDS–PAGE gel (PAGEgel), and blotted to PVDF membrane (Bio-Rad) after separation with electrophoresis. The concentration of primary antibodies for western blot was generally used as recommended by the manufacturers. In the studies, the following antibodies were used: mouse monoclonal antibody anti-NMDAR1 (BD Biosciences); Rabbit polyclonal anti-NMDAR2A IgG (abcam); Rabbit polyclonal anti-NMDAR2B IgG (abcam); Rabbit polyclonal anti-NMDAR2C IgG (abcam); Rabbit polyclonal anti-NMDAR2D IgG (abcam); Rabbit polyclonal anti-GluR1 IgG (abcam). The secondary antibodies, HRP-conjugated anti-mouse IgG and HRP-conjugated anti-rabbit IgG, were obtained from Amersham and Sigma, respectively. The Image J, free software for image quantification from NIH (http://rsbweb.nih.gov/ij/), was used for the quantification of NMDAR1 protein expression. For mouse immunohistochemical staining, frozen sections (20 µm) were cut from mouse brains fixed by transcardial perfusion of 4% formaldehyde buffered in PBS and cryo-protected. Mouse monoclonal antibody anti-NMDAR1 was used as primary antibody with 1:100 dilutions. The VECTASTAIN elite ABC kit (Vector Laboratories) was used for signal amplification, and DAB (Roche Diagnostics) was used for color reaction.
SUPPLEMENTARY MATERIAL
FUNDING
These studies were supported by grants from the National Institutes of Health MH073991 (M.A.G.) and MH081037 (X.Z.), National Institute of Neurological Disorders and Stroke P30NS057096 (Stuart Lipton) and by the Veterans Affairs VISN 22 Mental Illness Research, Education, and Clinical Center.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Pamela Sklar for the CNV database, and Dr Jared W. Young for advice and review of this manuscript.
Conflict of Interest statement. J.R.K. is a founder and holds equity in Psynomics, Inc. The terms of this arrangement have been reviewed and approved by UCSD in accordance with its conflict of interest policies.
REFERENCES
- 1.Heisler L.E., Torti D., Boutros P.C., Watson J., Chan C., Winegarden N., Takahashi M., Yau P., Huang T.H., Farnham P.J., et al. CpG Island microarray probe sequences derived from a physical library are representative of CpG Islands annotated on the human genome. Nucleic Acids Res. 2005;33:2952–2961. doi: 10.1093/nar/gki582. doi:10.1093/nar/gki582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Supp D.M., Witte D.P., Branford W.W., Smith E.P., Potter S.S. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev. Biol. 1996;176:284–299. doi: 10.1006/dbio.1996.0134. doi:10.1006/dbio.1996.0134. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X., Long J.M., Geyer M.A., Masliah E., Kelsoe J.R., Wynshaw-Boris A., Chien K.R. Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Mol. Psychiatry. 2005;10:393–406. doi: 10.1038/sj.mp.4001621. doi:10.1038/sj.mp.4001621. [DOI] [PubMed] [Google Scholar]
- 4.Zhou X., Qyang Y., Kelsoe J.R., Masliah E., Geyer M.A. Impaired postnatal development of hippocampal dentate gyrus in Sp4 null mutant mice. Genes. Brain Behav. 2007;6:269–276. doi: 10.1111/j.1601-183X.2006.00256.x. doi:10.1111/j.1601-183X.2006.00256.x. [DOI] [PubMed] [Google Scholar]
- 5.Ramos B., Gaudilliere B., Bonni A., Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proc. Natl Acad. Sci. USA. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. doi:10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao X., Yang S.H., Simpkins J.W., Barger S.W. Glutamate receptor activation evokes calpain-mediated degradation of Sp3 and Sp4, the prominent Sp-family transcription factors in neurons. J. Neurochem. 2007;100:1300–1314. doi: 10.1111/j.1471-4159.2006.04297.x. doi:10.1111/j.1471-4159.2006.04297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braff D.L., Geyer M.A., Swerdlow N.R. Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology. 2001;156:234–258. doi: 10.1007/s002130100810. doi:10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- 8.Geyer M.A., Krebs-Thomson K., Braff D.L., Swerdlow N.R. Pharmacological studies of prepulse inhibition models of sensorimotor gating deficits in schizophrenia: a decade in review. Psychopharmacology. 2001;156:117–154. doi: 10.1007/s002130100811. doi:10.1007/s002130100811. [DOI] [PubMed] [Google Scholar]
- 9.Dempster E.L., Toulopoulou T., McDonald C., Bramon E., Walshe M., Wickham H., Sham P.C., Murray R.M., Collier D.A. Episodic memory performance predicted by the 2bp deletion in exon 6 of the ‘alpha 7-like’ nicotinic receptor subunit gene. Am. J. Psychiatry. 2006;163:1832–1834. doi: 10.1176/ajp.2006.163.10.1832. doi:10.1176/appi.ajp.163.10.1832. [DOI] [PubMed] [Google Scholar]
- 10.Robles O., Blaxton T., Adami H., Arango C., Thaker G., Gold J. Nonverbal delayed recognition in the relatives of schizophrenia patients with or without schizophrenia spectrum. Biol. Psychiatry. 2008;63:498–504. doi: 10.1016/j.biopsych.2007.05.016. doi:10.1016/j.biopsych.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daumas S., Halley H., Lassalle J.M. Disruption of hippocampal CA3 network: effects on episodic-like memory processing in C57BL/6J mice. Eur. J. Neurosci. 2004;20:597–600. doi: 10.1111/j.1460-9568.2004.03484.x. doi:10.1111/j.1460-9568.2004.03484.x. [DOI] [PubMed] [Google Scholar]
- 12.Olney J.W., Newcomer J.W., Farber N.B. NMDA receptor hypofunction model of schizophrenia. J. Psychiatr. Res. 1999;33:523–533. doi: 10.1016/s0022-3956(99)00029-1. doi:10.1016/S0022-3956(99)00029-1. [DOI] [PubMed] [Google Scholar]
- 13.Krystal J.H., Karper L.P., Seibyl J.P., Freeman G.K., Delaney R., Bremner J.D., Heninger G.R., Bowers M.B., Jr, Charney D.S. Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch. Gen. Psychiatry. 1994;51:199–214. doi: 10.1001/archpsyc.1994.03950030035004. [DOI] [PubMed] [Google Scholar]
- 14.Lahti A.C., Holcomb H.H., Medoff D.R., Tamminga C.A. Ketamine activates psychosis and alters limbic blood flow in schizophrenia. Neuroreport. 1995;6:869–872. doi: 10.1097/00001756-199504190-00011. doi:10.1097/00001756-199504190-00011. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra A.K., Pinals D.A., Adler C.M., Elman I., Clifton A., Pickar D., Breier A. Ketamine-induced exacerbation of psychotic symptoms and cognitive impairment in neuroleptic-free schizophrenics. Neuropsychopharmacology. 1997;17:141–150. doi: 10.1016/S0893-133X(97)00036-5. [DOI] [PubMed] [Google Scholar]
- 16.Dalmau J., Gleichman A.J., Hughes E.G., Rossi J.E., Peng X., Lai M., Dessain S.K., Rosenfeld M.R., Balice-Gordon R., Lynch D.R. Anti-NMDA-receptor encephalitis: case series and analysis of the effects of antibodies. Lancet Neurol. 2008;7:1091–1098. doi: 10.1016/S1474-4422(08)70224-2. doi:10.1016/S1474-4422(08)70224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iizuka T., Sakai F., Ide T., Monzen T., Yoshii S., Iigaya M., Suzuki K., Lynch D.R., Suzuki N., Hata T., et al. Anti-NMDA receptor encephalitis in Japan: long-term outcome without tumor removal. Neurology. 2008;70:504–511. doi: 10.1212/01.wnl.0000278388.90370.c3. doi:10.1212/01.wnl.0000278388.90370.c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manschreck T.C., Boshes R.A. The CATIE schizophrenia trial: results, impact, controversy. Harv. Rev. Psychiatry. 2007;15:245–258. doi: 10.1080/10673220701679838. doi:10.1080/10673220701679838. [DOI] [PubMed] [Google Scholar]
- 19.Green M.F. What are the functional consequences of neurocognitive deficits in schizophrenia? Am. J. Psychiatry. 1996;153:321–330. doi: 10.1176/ajp.153.3.321. [DOI] [PubMed] [Google Scholar]
- 20.Mohn A.R., Gainetdinov R.R., Caron M.G., Koller B.H. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98:427–436. doi: 10.1016/s0092-8674(00)81972-8. doi:10.1016/S0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- 21.Duncan G.E., Moy S.S., Perez A., Eddy D.M., Zinzow W.M., Lieberman J.A., Snouwaert J.N., Koller B.H. Deficits in sensorimotor gating and tests of social behavior in a genetic model of reduced NMDA receptor function. Behav. Brain Res. 2004;153:507–519. doi: 10.1016/j.bbr.2004.01.008. doi:10.1016/j.bbr.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Belforte J.E., Zsiros V., Sklar E.R., Jiang Z., Yu G., Li Y., Quinlan E.M., Nakazawa K. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat. Neurosci. 2010;13:76–83. doi: 10.1038/nn.2447. doi:10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsien J.Z., Huerta P.T., Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. doi:10.1016/S0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 24.Serrano P., Friedman E.L., Kenney J., Taubenfeld S.M., Zimmerman J.M., Hanna J., Alberini C., Kelley A.E., Maren S., Rudy J.W., et al. PKMzeta maintains spatial, instrumental, and classically conditioned long-term memories. PLoS Biol. 2008;6:2698–2706. doi: 10.1371/journal.pbio.0060318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furukawa H., Singh S.K., Mancusso R., Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. doi:10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 26.Stephenson F.A., Cousins S.L., Kenny A.V. Assembly and forward trafficking of NMDA receptors (Review) Mol. Membr. Biol. 2008;25:311–320. doi: 10.1080/09687680801971367. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto S., Sherman K., Bai G., Lipton S.A. Effect of the ubiquitous transcription factors, SP1 and MAZ, on NMDA receptor subunit type 1 (NR1) expression during neuronal differentiation. Brain Res. Mol. Brain Res. 2002;107:89–96. doi: 10.1016/s0169-328x(02)00440-0. doi:10.1016/S0169-328X(02)00440-0. [DOI] [PubMed] [Google Scholar]
- 28.Liu A., Hoffman P.W., Lu W., Bai G. NF-kappaB site interacts with Sp factors and up-regulates the NR1 promoter during neuronal differentiation. J. Biol. Chem. 2004;279:17449–17458. doi: 10.1074/jbc.M311267200. doi:10.1074/jbc.M311267200. [DOI] [PubMed] [Google Scholar]
- 29.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. doi:10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzo L.D., Jean-Marie, Van Der Linden M., Grangé D., Rohmer J.-G. Impairment of memory for spatial context in schizophrenia. Neuropsychology. 1996;10:376–384. doi:10.1037/0894-4105.10.3.376. [Google Scholar]
- 31.Greenwood T.A., Braff D.L., Light G.A., Cadenhead K.S., Calkins M.E., Dobie D.J., Freedman R., Green M.F., Gur R.E., Gur R.C., et al. Initial heritability analyses of endophenotypic measures for schizophrenia: the consortium on the genetics of schizophrenia. Arch. Gen. Psychiatry. 2007;64:1242–1250. doi: 10.1001/archpsyc.64.11.1242. doi:10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefebvre A.A., Cellard C., Tremblay S., Achim A., Rouleau N., Maziade M., Roy M.A. Familiarity and recollection processes in patients with recent-onset schizophrenia and their unaffected parents. Psychiatry Res. 2009;175:15–21. doi: 10.1016/j.psychres.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turri M.G., Datta S.R., DeFries J., Henderson N.D., Flint J. QTL analysis identifies multiple behavioral dimensions in ethological tests of anxiety in laboratory mice. Curr. Biol. 2001;11:725–734. doi: 10.1016/s0960-9822(01)00206-8. doi:10.1016/S0960-9822(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 34.Tsujimura A., Matsuki M., Takao K., Yamanishi K., Miyakawa T., Hashimoto-Gotoh T. Mice lacking the kf-1 gene exhibit increased anxiety- but not despair-like behavior. Front. Behav. Neurosci. 2008;2:4. doi: 10.3389/neuro.08.004.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmes A., Kinney J.W., Wrenn C.C., Li Q., Yang R.J., Ma L., Vishwanath J., Saavedra M.C., Innerfield C.E., Jacoby A.S., et al. Galanin GAL-R1 receptor null mutant mice display increased anxiety-like behavior specific to the elevated plus-maze. Neuropsychopharmacology. 2003;28:1031–1044. doi: 10.1038/sj.npp.1300164. [DOI] [PubMed] [Google Scholar]
- 36.Ramos A., Mormede P. Stress and emotionality: a multidimensional and genetic approach. Neurosci.Biobehav. Rev. 1998;22:33–57. doi: 10.1016/s0149-7634(97)00001-8. doi:10.1016/S0149-7634(97)00001-8. [DOI] [PubMed] [Google Scholar]
- 37.Tonegawa S., McHugh T.J. Molecular and Circuit Mechanisms for Hippocampal Learning. Berlin: Springer; 2008. [Google Scholar]
- 38.Bakshi V.P., Geyer M.A. Multiple limbic regions mediate the disruption of prepulse inhibition produced in rats by the noncompetitive NMDA antagonist dizocilpine. J. Neurosci. 1998;18:8394–8401. doi: 10.1523/JNEUROSCI.18-20-08394.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olney J.W., Labruyere J., Price M.T. Pathological changes induced in cerebrocortical neurons by phencyclidine and related drugs. Science. 1989;244:1360–1362. doi: 10.1126/science.2660263. doi:10.1126/science.2660263. [DOI] [PubMed] [Google Scholar]
- 40.Yang J., Medvedev S., Reddi P.P., Schultz R.M., Hecht N.B. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc. Natl Acad. Sci. USA. 2005;102:1513–1518. doi: 10.1073/pnas.0404685102. doi:10.1073/pnas.0404685102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X., Tang W., Greenwood T.A., Guo S., He L., Geyer M.A., Kelsoe J.R. Transcription factor SP4 is a susceptibility gene for bipolar disorder. PLoS ONE. 2009;4:e5196. doi: 10.1371/journal.pone.0005196. doi:10.1371/journal.pone.0005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyn S.I., Shi J., Kraft J.B., Potash J.B., Knowles J.A., Weissman M.M., Garriock H.A., Yokoyama J.S., McGrath P.J., Peters E.J., et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol. Psychiatry. 2009 doi: 10.1038/mp.2009.125. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi J., Potash J.B., Knowles J.A., Weissman M.M., Coryell W., Scheftner W.A., Lawson W.B., Depaulo J.R., Jr, Gejman P.V., Sanders A.R., et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol. Psychiatry. 2009 doi: 10.1038/mp.2009.124. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blackwood D.H., Fordyce A., Walker M.T., St Clair D.M., Porteous D.J., Muir W.J. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am. J. Hum. Genet. 2001;69:428–433. doi: 10.1086/321969. doi:10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millar J.K., Christie S., Anderson S., Lawson D., Hsiao-Wei Loh D., Devon R.S., Arveiler B., Muir W.J., Blackwood D.H., Porteous D.J. Genomic structure and localisation within a linkage hotspot of Disrupted In Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol. Psychiatry. 2001;6:173–178. doi: 10.1038/sj.mp.4000784. doi:10.1038/sj.mp.4000784. [DOI] [PubMed] [Google Scholar]
- 46.Boutillier S., Lannes B., Buee L., Delacourte A., Rouaux C., Mohr M., Bellocq J.P., Sellal F., Larmet Y., Boutillier A.L., et al. Sp3 and sp4 transcription factor levels are increased in brains of patients with Alzheimer's disease. Neurodegenerative Dis. 2007;4:413–423. doi: 10.1159/000107701. [DOI] [PubMed] [Google Scholar]
- 47.Lau C.G., Zukin R.S. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat. Rev. Neurosci. 2007;8:413–426. doi: 10.1038/nrn2153. doi:10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.