Abstract
Primary open-angle glaucoma (POAG) is one of the three principal subtypes of glaucoma and among the leading cause of blindness worldwide. POAG is defined by cell death of the retinal ganglion cells (RGCs) and surrounding neuronal cells at higher or normal intraocular pressure (IOP). Coded by one of the three genes responsible for POAG, WD repeat-containing protein 36 (WDR36) has two domains with a similar folding. To address whether WDR36 is functionally important in the retina, we developed four transgenic mice strains overexpressing a wild-type (Wt) and three mutant variants of D606G, deletion of amino acids at positions 605–607 (Del605–607) and at 601–640 (Del601–640) equivalent to the location of the D658G mutation observed in POAG patients. A triple amino acid deletion of mouse Wdr36 at positions 605–607 corresponding to the deletion at positions 657–659 in humans developed progressive retinal degeneration at the peripheral retina with normal IOP. RGCs and connecting amacrine cell synapses were affected at the peripheral retina. Axon outgrowth rate of cultured RGC directly isolated from transgenic animal was significantly reduced by the Wdr36 mutation compared with Wt. Molecular modeling of wild and mutant mouse Wdr36 revealed that deletion at positions 605–607 removed three residues and a hydrogen bond, required to stabilize anti-parallel β-sheet of the 6th β-propeller in the second domain. We concluded that WDR36 plays an important functional role in the retina homeostasis and mutation to this gene can cause devastating retinal damage. These data will improve understanding of the functional property of WDR36 in the retina and provide a new animal model for glaucoma therapeutics.
INTRODUCTION
Glaucoma is characterized by progressive loss of retinal ganglion cells (RGCs), degeneration of axons in the optic nerve leading to visual field defects. Primary open-angle glaucoma (POAG) is one of the major causes of irreversible blindness leading to 12% of all global blindness and by the year 2020 over 11 million people are predicted to be affected (1,2). POAG is often associated with elevated intraocular pressure (IOP) >21 mmHg, which becomes a major contributory factor for typical glaucomatous changes, including optic nerve head cupping and visual field loss. At present, at least 24 different loci have been linked to various forms of glaucoma and over a decade, three genes, myocilin (MYOC), optineurin (OPTN) and WD repeat-containing protein 36 (WDR36), have been identified as monogenic genes associated with POAG (3–5). WDR36 was identified in the POAG loci GLC1G at 5q22.1 with segregation to POAG in the original paper (5). Hauser et al. (6) reported correlations of WDR36 variants and POAG severity, while others described disease-causing variants at equal frequency as the control (7–9). These mixed results suggest that WDR36 may act as a modifier gene for POAG or only be responsible for selective familial POAG as in MYOC and OPTN variants (6,8).
WDR36 is a 100 kDa protein, containing multiple guanine nucleotide-binding WD40 repeats (WDR), an AMP-dependent synthetase and ligase, and a cytochrome cd1-nitrite reductase-like WD40-associated domain (5). Previous studies have shown that the loss of WDR36 activated the p53 stress–response pathway that disrupts nucleolar morphology and rRNA processing, resulting in a decrease of the 18S rRNA mature form (10). Wdr36 knockout in zebrafish developed abnormal gut and smaller eye with progressive degeneration of the lens development. Homology modeling by Footz et al. (11) showed WDR36 containing 14 WDR, likely to fold in two connected seven-bladed β-propeller domains. WDR36 is ubiquitously expressed in all tissues and shares sequence homology with yeast Utp21. It is localized to the nucleoli and cytoplasm (10). Footz et al. further described that POAG-causing variants in WDR36 did not produce any significant defects in yeast viability or tRNA processing, but when combined with disruption of STI1, which synthetically interacts with UTP21, 5 of the 11 variants had increased or decreased cell viability due to reduced or elevated levels of pre-rRNA (11). These WDR36 variants included ones found in control populations, suggesting WDR36 as a modifier gene that requires another gene defect or unknown risk factor for the onset of POAG.
In this paper, we focused at the functionally sensitive site in the two 7-bladed β-propeller structure studied previously in humans and yeast. Overexpression of mutant Wdr36 produce significant and specific retinal damage in mice. Molecular modeling of wild-type (Wt) and mutant Wdr36 mice revealed explanation for the phenotypic results obtained in this study.
RESULTS
Construction of Wt and mutant Wdr36 transgenic mice
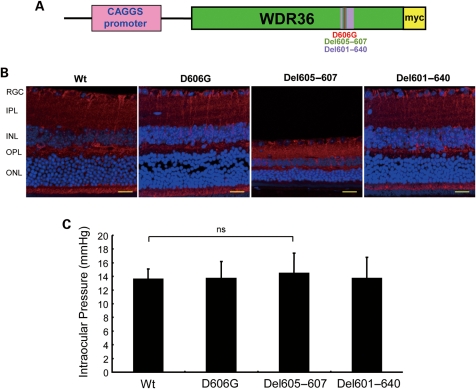
Four independent transgenic mice lines overexpressing Wt, D606G, deletion of amino acid 605–607 (Del605–607) located in seventh β-propeller in the second domain, and at positions 601–640 (Del601–640), a deletion of the entire seventh β-propeller of same domain, were developed independently in B7D2F1 (DBA/2J+C57BL/6J) mouse strain (Fig. 1A). The copy number of each mutant cDNA construct per mouse was approximately two to four, determined by TaqMan real-time polymerase chain reaction (PCR) assay (data not shown). The D606G is equivalent to the D658G mutation preferentially found in patients with POAG (5). Ubiquitous expression and distribution of Wt and mutant Wdr36 protein was visualized by immunostaining of myc-tag peptides fused to each Wdr36 construct (Fig. 1B).
Figure 1.
Development of Wdr36 transgenic mouse. (A) Schematic diagram of the mouse Wdr36 construct used in this study. Positions of mutations and deletions are shown. (B) Expression of each mutant Wdr36 was immunostained with anti-myc (red) and anti-Wdr36 (green) antibody. Image shown are co-localized. Scale bar, 20µm. (C) IOP was measured at 16 months during 10 am to noon using impact-rebound tonometer. This method gave normal range for all transgenic mice (n = 6).
IOP measurement for Wt and mutant mice
Elevated IOP is one of the major risk factors for glaucoma. We measured IOP in Wt and transgenic mice using noninvasive impact-rebound tonometer as described previously (12). The average IOP for Wt, D606G and Del601–640 was similar and in the normal range of approximately 13 mmHg at all examined ages (Fig. 1C). The Del605–607 mice gave a slight increase of IOP but statistically non-significant compared with other strains.
Histological comparison of Wt and mutant Wdr36 transgenic mice eye
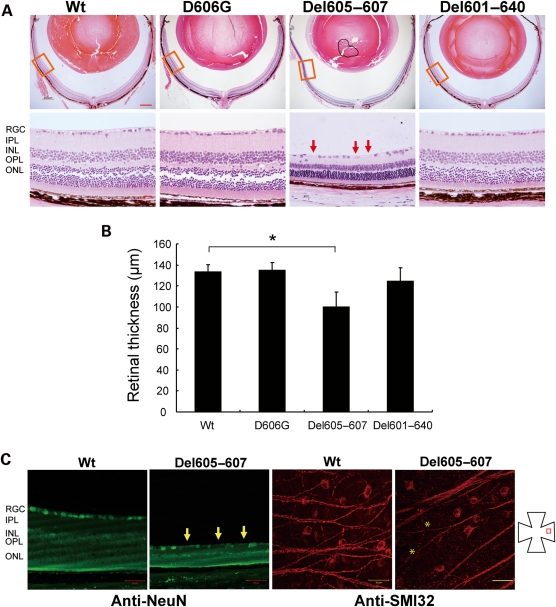
Hematoxylin–eosin (H&E) staining of the entire eye section showed no obvious abnormality for mice over expressing Wt, D606G and Del601–640. However, significant change was observed for Del605–607 mice exclusively in the peripheral retina at 16 months of age (Fig. 2A). Approximately 25% reduction in the peripheral retina thickness was observed for Del605–607 mice compared with the Wt mice (*P < 0.05; Fig. 2B). To maintain statistical quality, the measurements of retinal thickness were collected at the peripheral retina ∼1.0–1.2 mm from the optic nerve head. Within the same peripheral region, RGC loss was observed by immunostaining with anti-NeuN, a neuron-specific marker, and counted through the entire retina sections (Fig. 2C). RGC loss at the peripheral retina was placed horizontally and visualized by anti-SMI32 immunostaining of the whole-mount retina in Del605–607 mice in comparison with Wt mice (Fig. 2C). Cornea, lens and the anterior segment of Del605–607 mice eye were histologically normal even after 16 months.
Figure 2.
Comparison of retina morphology of Wt and mutant transgenic mice at the peripheral retina. (A) Hematoxylin–eosin staining of retina section is shown for 16-month-old transgenic mice. Scale bar, 200 µm (upper panel), 20 µm (lower panel). (B) Quantification of the retina thickness and RGC numbers of 16-month-old transgenic mice (n = 6). Significant thinning of the retina and loss of the RGCs (arrow) was specifically observed for Del605–607 mice (*P < 0.05). RGC numbers was counted in the NeuN-immunostained paraffin section. (C) Reduction of RGC number at the peripheral retina (asterisks) is shown by NeuN immunostaining (green, scale bar 50 µm) of paraffin section and SMI32 immunostaining (red, scale bar 100 µm) of flat mount retina for Wt and Del605–607 mice.
Histopathological evaluation of the retinal layers in Wdr36 Del605–607 mice
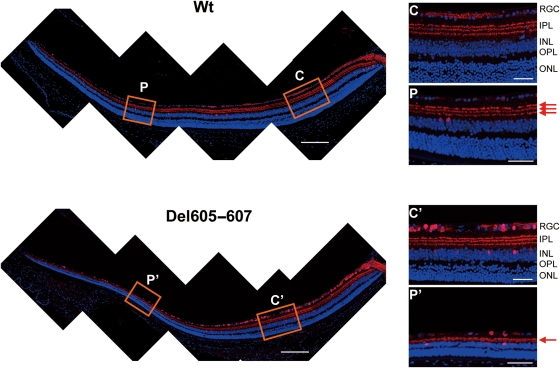
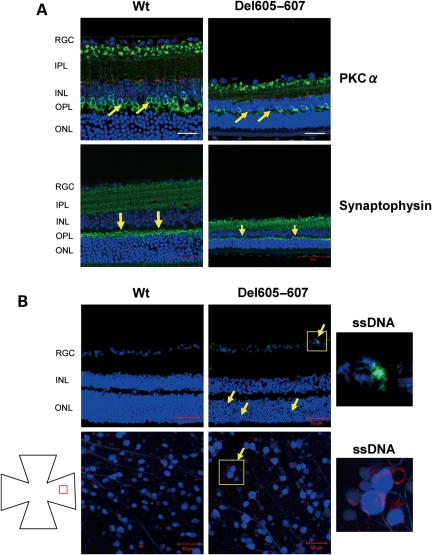
To determine which retinal cell components are vulnerable to the Del605–607 deletion, we performed immunohistochemical analysis using retinal cell-specific markers. In the inner plexiform layer (IPL), synapses of RGC and amacrine cells were visualized by calretinin immunostaining of retina sections of Wt and Del605–607 mice at 16 months (Fig. 3). A significant degeneration was observed in the peripheral retina in Del605–607 mice (Fig. 3, Panel P′) in contrast to the central retina of Wt and Del605–607 mice (Fig. 3, Panel C′). Loss of another type of amacrine cells and rod bipolar cells in the peripheral retina between the Del605–607 and Wt was detected by staining with antibodies against tyrosine hydroxylase (red, dopaminergic amacrine cell) and PKC α (green, rod bipolar cell) (Fig. 4A). Loss and/or abnormality of amacrine cells and bipolar cells was also observed in the Del605–607 mice. Expression of synaptophysin, a synaptic vesicle marker, was reduced in the outer plexiform layer (OPL) in Del605–607 mice compare with Wt mice (Fig. 4A).
Figure 3.
Disruption of synapses between RGC and amacrine cells in Del605–607 mice. Immunostaining of the retina sections with anti-calretinin antibody, a specific marker for RGCs and amacrine cells. Disruption of synapses between RGC and amacrine cells was specifically observed in peripheral (P) retina for Del605–607 mouse (P′) but not in the central (C) retina. Scale bar, 20 µm.
Figure 4.
Degeneration of outer plexiform layer (OPL) and outer nuclear layer at the peripheral retina of Del605–607 mouse. (A) Immunostaining of the retina sections with tyrosine hydroxylase (red) and PKC α (green), a specific maker for dopaminergic amacrine cells and rod bipolar cells, respectively (upper panel). Cell loss and size reduction of bipolar cells was observed. Scale bar, 20 µm. Immunostaining of the retina sections with synaptophysin, a specific marker for neuronal presynaptic vesicles (lower panel). Synapse disruption (arrow) was observed in the OPL of Del605–607 mouse peripheral retina. Scale bar, 50 µm. (B) Immunostaining of cryosection and flat mount retina with specific apoptosis marker, ssDNA. Apoptosis cells were observed in the all cell layers in the peripheral retina of Del605–607 mice. Scale bar, 50 µm.
Apoptosis assay by single-stranded DNA immunohistochemistry
Apoptotic cell death of RGC is one of the hallmarks of glaucoma pathogenesis. To visualize the apoptotic event in Wt and Del605–607 mice, we performed immunohistochemistry for single-stranded DNA (ssDNA), a detection marker for apoptosis-associated DNA damage. We detected ssDNA-positive cells (apoptotic cells) in the retinal ganglion cell layer and outer nuclear layer of the peripheral retina in Del605–607 mice (Fig. 4B, arrows). In the retinal flat mount, apoptotic RGCs were also observed in the peripheral region of Del605–607 mice (Fig. 4B).
Evaluation of Wt and Del605–607 primary RGC axon out growth
To evaluate the axon outgrowth of RGCs for Del605–607 mice, we first developed a double transgenic mouse by mating Del605–607 mice with Thy1 promoter-driven cyan fluorescence protein (Thy1-CFP) transgenic mouse, which enables visualization of RGC with CFP filter. Primary RGC culture of Del605–607/Thy1-CFP double transgenic mice was successfully established and the axon growth was monitored photographically (Fig. 5A). The RGC axon out growth rate of Del605–607/Thy1-CFP transgenic mice was reduced by ∼60% compared with those from Thy1-CFP mice alone at the fifth day of culture. This finding demonstrated that the Del605–607 mutation can directly influence the RGC axon outgrowth in vivo.
Figure 5.
Reduced axonal extension in primary cultured RGCs by Del605–607 mutation. (A) Primary cultured RGCs of Wt and Del605–607 mouse (upper panel). Trance of axonal extension are shown in lower panel. Shorted axons were observed. (B) Quantification of the related axon length with Image J software.
Molecular modeling of mouse Wdr36
In order to model a three-dimensional structure of mouse Wdr36, we thread a sequence of Wt Wdr36 through the database of known protein structures using the SMART 6 engine (13). The engine recognized eight WD-40 β-propeller repeats, each repeat a motif of 40 amino acid residues often terminating in Trp-Asp (W-D) dipeptide. The E-value was changed from 1.46E−01 to 1.43E+1 for these repeats. The most significant change was observed for the motif at 559–598 with an E-value of 2.2E−10. Modeling by PHYRE (14) suggests that a structure of mouse Wdr36 is homologous to yeast actin interacting protein 1 (PDB-file: 1nr0) as the best hit between 14 different protein structures with a very low PHYRE E-value. This structure is likely to fold in two 7-bladed β-propeller domains (total 14 WD-40 repeats) forming a circularized structure in positions from 1 through 642 of the mouse Wdr36 sequence. The rest of the amino acid sequence (residues 646–899) show similarity to the large fragment of Utp21 domain of unknown structure that starts at position 668 and ends at position 896 with the SMART E-value for the domain of 2.40E−39. This result confirmed that Wdr36 and yeast Utp21 are structurally similar and share two 7-bladed β-propeller domains as a common structural motif (10,11).
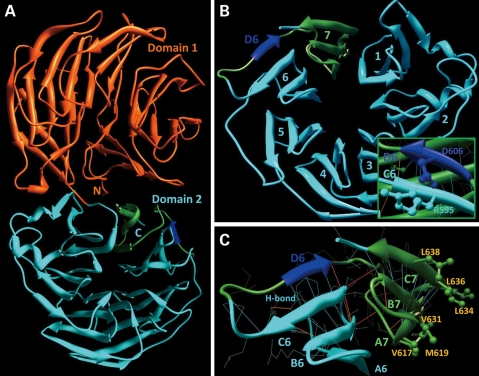
Further, the structural features of the Wdr36 are similar to that of Caenorhabditis elegans UNC-78/AIP1 described earlier (15). The WDR structure revealed two distinct domains (Fig. 6A). The individual domain is a seven-bladed β-propeller exhibiting a significant structural similarity (Fig. 6B). Each blade is arranged into four anti-parallel β-strands labeled from A to D starting from the center of the propeller. Each blade is stabilized by hydrogen bonds between main chain atoms of β-strands as exhibited in Figure 6C for blades 6 and 7 that were targeted by site-directed mutagenesis in this work. Interaction between D and C β-strands in blade 6 is supported by four main chain–main chain hydrogen bonds, including Ile605O–Thr596N (2.8 Å), Ile605N–Thr 596O (3.0 Å), Cys607O–Val594N (3.3 Å) and Cys607N–Val594O (2.8 Å), and by one side chain–side chain hydrogen bond Asp606OD2–Arg595NH1 (1.8 Å). The circular shape of Wdr36 is maintained by the closure mechanism similar to that of a seven-bladed β-propeller in which the immediate amino-terminus serves as an outer β-strand (D) for the blade 7 (15,16) and by hydrophobic interactions located at the surface of blade 7 residues Val617, Met619, Val631, Leu634, Ile636 and Leu638 (Fig. 6C) and at the surface of the blade 1 residues Leu329, Ile332, Tyr334, Ile342 and Phe361 (not shown).
Figure 6.
The WDR36 three-dimensional structure is obtained by homology modeling. The ribbon model with β-strands shown by arrows illustrates a proposed structure of WDR36 containing 14 WD40 repeats. N- and C-terminal domains with a seven-bladed β-propeller fold are shown by orange and cyan, respectively (A). In domain 2, individual blades with the WD40 structural motif are numbered from 1 to 7 (B). Similar to others, blade 6 is composed of an antiparallel β-sheet with the individual β-strands labeled from A6 to D6 (C). The individual β-strand D6 and the seventh blade of the domain 2 which correspond to mutations Del605–607 and Del601–640 are shown by blue and green, respectively. The Wt amino acid side chains that are important for the analysis of sequence variants are shown by ball and stick models. Hydrogen bonds are shown by cyan and orange lines. Interaction between residues R595 and D606 is revealed as an inset in (B). The missense mutation D606G cause the D606 side chain removal and breaks the single side chain–side chain hydrogen bond Asp606OD2–Arg595NH1. Amino acids are shown by a single letter code.
DISCUSSION
In this study, phenotypic features of mice overexpressing mutant Wdr36 were determined and characterization of RGC was performed in vitro and in vivo. Molecular modeling techniques were additionally applied to simulate mouse Wdr36 protein structure for calculating the structural change induced by three mutations that were introduced in transgenic mouse models. This is the first demonstration of an association between gene mutations in Wdr36 and changes in a phenotype observed in a mammalian system.
The design of the D606G transgene construct was based on the original paper by Monemi et al. (5). Our molecular modeling shows that mouse D606G mutant, equivalent to the human D658G mutation, is located in the seventh β-propeller of the second domain. This seventh β-propeller of the second domain is conserved for both amino acid sequence and molecular structure. Previous studies have predicted that the D658G variant was strongly associated with severe phenotype in glaucoma patients (5,8,11). To elucidate molecular functions in this region, we also developed a 3 amino acid deletion (Del605–607) containing D606 position at the seventh β-propeller of the second domain and a 40 amino acid deletion (Del 601–640), which deletes the entire seventh β-propeller of the second domain.
Two previous functional studies have demonstrated that WDR36 is involved in ribosome biogenesis similar to the yeast Utp21, which is a member of the small subunit processome complex responsible for maturation of 18S rRNA (10,11). Footz et al. has further demonstrated that expressing the equivalent human variants in Utp21 can cause significant reduction or elevation of the pre-rRNA level in the nucleoli. Among the 11 variants tested, Utp21 D621G equivalent to the human WDR36 D658G gave the lowest pre-rRNA level in yeast and the highest pre-rRNA level by 216% in the Sti1 null yeast strain (11). In human, all genomic sequence variants exclusive to glaucoma patients or variants with low or high incidence in control population did not correlate with functional studies in yeast. The authors of this paper concluded that human R529Q, I604V, D658G and M671V are likely to encode subtle defects in WDR36 that in concert with another susceptible gene(s) or with certain environmental factor(s) may trigger the onset of POAG.
These data suggest that in human WDR36 the D658G variant may be functionally the most influential mutation and contribute to the onset of the disease as the genetic modifier. In fact, WDR36 has been previously suggested to influence the severity of MYOC-based POAG (6). To verify the above-mentioned hypothesis, further investigation would be necessary. Such investigation is in progress by cross-mating Del605–607 mice with other glaucoma mice, such as the myocilin Y423H transgenic developed by Zhou et al. (17) to validate the POAG-like phenotype with RGC loss and increased IOP.
In spite of the fact that mutant Wdr36 was expressed in all tissues, it is surprising that defects were only observed in the retina. Previous studies by Skarie and Link (10) have shown loss of wdr36 function in zebrafish resulted in developmental defects including liver necrosis and absence of swim bladder inflation by 6 days post fertilization. Small head and eye was also reported with lens opacity and thickening of lens epithelium but relatively mild defect in the retina even after 6 months post fertilization. In contrast to zebrafish, transgenic mice with ubiquitous expression of the mutant Del605–607 revealed a progressive defect in the retina but not elsewhere including the anterior part of the eye even 16 months after birth. In gross anatomical examination, the size and structural abnormalities of head, eye and gut, which occurred in zebrafish, were not observed in mice. The vulnerability of the retina in the mouse model may suggest a differential role of Wdr36 in the two species, perhaps through interaction with different retina-specific molecules. On the other hand, Skarie and Link (10) also showed the involvement of the p53 pathway and subsequent activation of p21 on the effects of the loss of Wdr36, suggesting that the p53 pathway may also have influenced the phenotype of our transgenic mice.
Overall, the adult Wdr36 transgenic mice with abnormal phenotype serve as the potential POAG model for future use. To date, a number of animal models for glaucoma have been established (12,18–24). Interestingly, the phenotypes of human glaucoma mutation-based mouse models including myocilin Y423H (20,17), optineurin E50K (12) and Wdr36 Del605-607 (Figs 2–4) all involve loss of peripheral RGCs accompanied by peripheral retinal degeneration. Herein, we raise the question, what are the reasons for peripheral retinal degeneration? Previous reports have indicated that mouse models of glaucoma follow similar natural courses of peripheral retinal degeneration (12,17–20). These findings suggest that all three mouse models share a common signaling pathway for RGC death and beyond, although the function of each protein is distinct. Wdr36 transgenic mouse will be useful in combination with other models to identify molecules responsible for this common pathway of RGC death.
Prediction by homology structure of Wdr36 was used to analyze three mutant variants D606G, Del605–607 and Del601–640, respectively (Fig. 1A). The missense change of aspartic acid to glycine at position 606 interrupts the side chain–side chain hydrogen bond Asp606OD2–Arg595NH1 without affecting other hydrogen bonds that stabilize interaction between C6 and D6 β-strands (Fig. 6B, inset). This minor change suggests that the mutation will show either no effect or an insignificant structural change in the stability of the protein with age that could potentially appear at a very slow rate. In contrast, the mutation Del605–607 removes residues from positions 605 to 607 corresponding to the β-strand D6 (Fig. 6A–C, blue). This results in interruption of five hydrogen bonds in Wdr36 structure, and might affect the stability of the 6th blade of domain 2 (Fig. 6B). Although it is difficult to make a definite conclusion, our modeling predicts that the polypeptide chain might still adopt a β-propeller conformation. Thus, domain 2 will continue to adopt a seven-blade β-propeller structure, suggesting that protein–protein interactions that involve residues after position 607 might be significantly perturbed because all interacting residues will be shifted by three residues from their original positions. Further, this could exclude the mutant variant from the native pattern of protein–protein interactions, affecting biochemical pathways, and potentially leading to degenerative processes in the cell.
The Del601–640 construct removes β-strand D6 and seventh blade, shown by blue and green color, respectively, in Figure 6C. The deletion might dramatically affect a closure mechanism and hydrophobic interactions between blades 7 and 1 (Fig. 6C). As a result, this change significantly decreases protein stability by ‘unlocking’ the circular structure of the seventh-bladed β-propeller domain 2. The protein would definitely be expected to be misfolded, either forming insoluble aggregates or more likely be degraded in proteolytic pathways related to chaperone-mediated autophagy (25) or to the ubiquitin-proteasome system (26). If this hypothesis is correct, the mutant protein will be expressed but functionally inactive to influence the endogenous Wdr36.
Overall, the functional evidence in this study demonstrates an essential role for Wdr36 in mouse retina. These results will provide basis for future research to determine how WDR36 variants leads to POAG.
MATERIALS AND METHODS
Development of transgenic mouse over expressing Wt and mutant Wdr36
Full-length Wdr36 cDNA was amplified from C57BL/6N mouse heart total RNA using DNA polymerase (PrimeSTAR HS DNA Polymerase, Takara, Tokyo, Japan) and cloned into pBluescript II KS(+) (Stratagene, La Jolla, CA, USA). D606G, and Del605–607, was introduced by site-directed mutagenesis (Quick Change XL Site Directed Mutagenesis Kit, Stratagene) in accordance with manufacturer's instructions. The primer sets used for mutagenesis of mouse Wdr36 D606G and Del605–607 are 5′-CTGGGTGCCTTATCGGCTGCTTTTTGTTGGAC-3′, 5′-GTCCAACAAAAAGCAGCCGATAAGGCACCCAG-3′ and 5′-ACCTTCCTTCTGGGTGCCTTTTTTTGTTGGACTCAGCGCC-3′, 5′-GGCGCTGAGTCCAACAAAAAAAGGCACCCAGAAGGAAGGT-3′ respectively. Del601–640 was constructed by amplification of mouse Wdr36 using primer set 5′-AATATTTCCCTCTATTCAGTTGT-3′ and 5′-AGGAAGGTCCCAAGTCCTAA-3′. Mutated cDNA was inserted into pCMV-Tag5 vector with chicken beta-actin promoter and CMV enhancer (pCAGGS) kindly provided by Dr Junichi Miyazaki (Osaka University). The Wt and mutant Wdr36 cDNA fragment was released from the pBroad2 vector using Pac I. The Wdr36 fragments were injected into pronuclear stage BDF1/C57BL6N embryos and transgenic mice were generated at PhoenixBio Co., Ltd (Utsunomiya, Japan). Offspring from 32 donor mice were screened for the transgene by isolating genomic DNA from tail biopsies followed by PCR. Primers used for PCR were: 5′-CAGAAACTCATCTCTGAAGAGGATCTGTAG-3′ and 5′-TTGTTCATGGCAGCCAGCATATGGCATATG-3′. All experimental data were obtained from 16-month-old Wt and mutant Wdr36 transgenic mice. All the experiments using mice were performed in accordance with the Association for Research in Vision and Ophthalmology statement for the Use of Animals in Vision Research.
Light microscopic histopathology of the optic nerve
After euthanized, mouse eyes were dissected and immersed in Davidson solution fixative overnight at 4°C. The eyes were embedded in paraffin and sectioned at 5 µm thickness along the vertical meridian through the optic nerve head. After deparaffinization and rehydration, sections were H&E stained.
Immunohistochemistry
The eyes were sectioned at 5 µm thickness along the vertical meridian through the optic nerve head. After deparaffinization and rehydration, the tissue sections were treated with Target Retrieval Solution (Dako Cytomation, Denmark). For cryosections, after perfusion with 4% paraformaldehyde (PFA), the eye balls were dissected and immersed in OCT compound. The frozen eyes were sectioned at 10 µm thickness. The sections were incubated with blocking solution for 1 h followed by overnight incubation with primary antibody against myc-tag (1:100 dilution; Abcam, Cambridge, MA, USA), NeuN (1:100 dilution; Millipore, Billerica, MA, USA), calretinin (1:500 dilution; Sigma, Sigma-Aldrich, St Louis, MO, USA), tyrosine hydroxylase (1:100 dilution; Millipore), PKC α (1:500 dilution; Millipore), synaptophysin (1:500 dilution; Abcam) or ssDNA (1:500 dilution; Immuno-Biological Laboratories, Gunma, Japan) in phosphate-buffered saline (PBS) containing 1% bovine serum albumin at 4°C. Slides were washed in PBS and then incubated with Alexa 488 or Alexa 568 (1:500 dilution; Invitrogen, Carlsbad, CA, USA) conjugated secondary antibody and with 4′,6′-diamidino-2-phenylindole (DAPI) for nuclear staining for 1 h at room temperature. The stained tissues were examined using confocal fluorescence laser microscopy (Radiance 2000, Bio-Rad Laboratories, Hercules, CA, USA). As negative control of the immunohistochemical staining, the sections were incubated with blocking solution without primary antibody (data not shown).
Whole-mount immunostaining
The whole-mount immunostaining was performed essentially as described (23,27). Anterior parts were dissected from enucleated eyes. The posterior parts were fixed in 4% PFA/PBS for 2h on ice and then incubated with the anti-SMI32 (1:200 dilution; Sternberger Monoclonals, Baltimore, MD, USA) and ssDNA antibody (1:500 dilution; Immuno-Biological Laboratories) for 7 days at 4°C. Slides were washed in PBS containing 0.1% Triton X-100 and then incubated with Alexa 488- or Alexa 568-conjugated secondary antibody (1:500 dilution; Invitrogen) and with DAPI for nuclear staining for 2 days at 4°C. The retinas were then mounted with Vectashield (Vector Laboratories, Burlingame, CA, USA) and evaluated on a confocal microscope.
Measurement of IOP
The average IOP for each genotype was determined. IOP was measured using an impact-rebound tonometer (Colonal Medical Supply, Franconia, NH, USA) for mice of each genotype as described previously (12). Using the rebound tonometer, we were able to measure IOP in awake and non-sedated mice of various ages. Measurement of IOP was always performed in the morning between 10 am to noon. The numbers of mice successfully assessed for each genotype were 8 Wt mice and 32 mutant transgenic mice at 16 months after birth.
Primary culture of retinal ganglion cells
To evaluate axon outgrowth of primary RGC, Del605–607 mice were mated with Thy1-CFP transgenic mouse (28–30) [B6.Cg-Transgenic (Thy1-CFP) 23Jrs/J] obtained from Jackson Laboratory (Bar Harbor, ME, USA) to develop Del605–607/Thy1-CFP double transgenic mice. The double transgenic mice at postnatal 5 days were euthanized and the eye globes enucleated to dissect anterior parts from the eye. The retinas were dissociated by SUMITOMO nerve-cell culture system/dissociation solutions (Sumitomo Bakelite, Tokyo, Japan) and cultured in SUMITOMO culture medium for 7 days on poly-l-lysine-coated culture plates.
Molecular modeling of mouse Wdr36
The sequence of Mus musculus WD repeat domain 36 isoform 1, WDR36 (NP_001103485.1), was exported to protein homology/analogy recognition engine Phyre version 0.2 (http://www.sbg.bio.ic.ac.uk/~phyre/) where C. elegans homologue of yeast actin interacting protein 1 (UNC-78/AIP1) (PDB file: 1nr0) was identified as the top-score structural homolog hit and the structure of two consecutive WD40 domains was generated. Two structural domains in WDR36 protein sequence were confirmed with the program REPRO (31). Amino acid sequences of mice Wdr36 and C. elegans UNC-78/AIP1 were aligned with PROMALS3D (http://prodata.swmed.edu/promals3d). Individual WD40 repeat motifs and Utp21 C-terminal domain were localized using the modular architecture research tool SMART 6 available at http://smart.embl-heidelberg.de. The Wdr36-1nr0 alignment exported to Look version 3.5.2 to automatically generate homology models of mutant proteins with deletions Del605–607 and Del601–640 by the automatic segment matching method (32) followed by 500 cycles of energy minimization. The missense mutation Asp606Gly (D606G) was generated and refined by self-consistent ensemble optimization (33), which applies the statistical mechanical mean-force approximation iteratively to achieve the global energy minimum structure.
Statistical analysis
All data were expressed as the mean ± standard deviation (SD). Statistical differences were analyzed by the ANOVA or Student's t-test. *P < 0.05 was considered statistically significant.
FUNDING
This research was supported in part by the grants to T.I. by the Japan Ministry of Health, Labour, and Welfare, the Japan Ministry of Education, Culture, Sports, Science and Technology and Grant-in-Aid for JSPS fellows for Z.-L.C. Funding to pay the Open Access Charge was provided by the Ministry of Health, Labour, and Welfare of Japan and the Ministry of Education, Culture, Sports, Science and Technology of Japan.
ACKNOWLEDGEMENTS
The authors thank Dr S. Zigler Jr for critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Quigley H.A. Number of people with glaucoma worldwide. Br. J. Ophthalmol. 1996;80:389–393. doi: 10.1136/bjo.80.5.389. doi:10.1136/bjo.80.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley H.A., Broman A.T. The number of people with glaucoma world wide in 2010 and 2020. Br. J. Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. doi:10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone E.M., Fingert J.H., Alward W.L., Nguyen T.D., Polansky J.R., Sunden S.L., Nishimura D., Clark A.F., Nystuen A., Nichols B.E., et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. doi:10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 4.Rezaie T., Child A., Hitchings R., Brice G., Miller L., Coca-Prados M., Heon E., Krupin T., Ritch R., Kreutzer D., et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. doi:10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 5.Monemi S., Spaeth G., DaSilva A., Popinchalk S., Ilitchev E., Liebmann J., Ritch R., Heon E., Crick R.P., Child A., et al. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum. Mol. Genet. 2005;14:725–733. doi: 10.1093/hmg/ddi068. doi:10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 6.Hauser M.A., Allingham R.R., Linkroum K., Wang J., LaRocque-Abramson K., Figueiredo D., Santiago-Turla C., del Bono E.A., Haines J.L., Pericak-Vance M.A., et al. Distribution of WDR36 DNA sequence variants in patients with primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 2006;47:2542–2546. doi: 10.1167/iovs.05-1476. doi:10.1167/iovs.05-1476. [DOI] [PubMed] [Google Scholar]
- 7.Weisschuh N., Wolf C., Wissinger B., Gramer E. Variations in the WDR36 gene in German patients with normal tension glaucoma. Mol. Vis. 2007;13:724–729. [PMC free article] [PubMed] [Google Scholar]
- 8.Pasutto F., Mardin C.Y., Michels-Rautenstrauss K., Weber B.H., Sticht H., Chavarria-Soley G., Rautenstrauss B., Kruse F., Reis A. Profiling of WDR36 missense variants in German patients with glaucoma. Invest. Ophthalmol. Vis. Sci. 2008;49:270–274. doi: 10.1167/iovs.07-0500. doi:10.1167/iovs.07-0500. [DOI] [PubMed] [Google Scholar]
- 9.Miyazawa A., Fuse N., Mengkegale M., Ryu M., Seimiya M., Wada Y., Nishida K. Association between primary open-angle glaucoma and WDR36 DNA sequence variants in Japanese. Mol. Vis. 2007;13:1912–1919. [PubMed] [Google Scholar]
- 10.Skarie J.M., Link B.A. The primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Hum. Mol. Genet. 2008;17:2474–2485. doi: 10.1093/hmg/ddn147. doi:10.1093/hmg/ddn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Footz T.K., Johnson J.L., Dubois S., Boivin N., Raymond V., Walter M.A. Glaucoma-associated WDR36 variants encode functional defects in a yeast model system. Hum. Mol. Genet. 2009;18:1276–1287. doi: 10.1093/hmg/ddp027. doi:10.1093/hmg/ddp027. [DOI] [PubMed] [Google Scholar]
- 12.Chi Z.L., Akahori M., Obazawa M., Minami M., Noda T., Nakaya N., Tomarev S., Kawase K., Yamamoto T., Noda S., et al. Overexpression of optineurin E50K disrupts Rab8 interaction and leads to a progressive retinal degeneration in mice. Hum. Mol. Genet. 2010;19:2606–2615. doi: 10.1093/hmg/ddq146. doi:10.1093/hmg/ddq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letunic I., Doerks T., Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–232. doi: 10.1093/nar/gkn808. doi:10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley L.A., Sternberg M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. doi:10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 15.Mohri K., Vorobiev S., Fedorov A.A., Almo S.C., Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. doi:10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- 16.Sondek J., Bohm A., Lambright D.G., Hamm H.E., Sigler P.B. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. doi:10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y., Grinchuk O., Tomarev S.I. Transgenic mice expressing the Tyr437His mutant of human myocilin protein develop glaucoma. Invest. Ophthalmol. Vis. Sci. 2008;49:1932–1939. doi: 10.1167/iovs.07-1339. doi:10.1167/iovs.07-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada T., Harada C., Nakamura K., Quah H.M., Okumura A., Namekata K., Saeki T., Aihara M., Yoshida H., Mitani A., et al. The potential role of glutamate transporters in the pathogenesis of normal tension glaucoma. J. Clin. Invest. 2007;117:1763–1770. doi: 10.1172/JCI30178. doi:10.1172/JCI30178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John S.W., Smith R.S., Savinova O.V., Hawes N.L., Chang B., Turnbull D., Davisson M., Roderick T.H., Heckenlively J.R. Essential iris atrophy, pigment dispersion, and glaucoma in DBA/2J mice. Invest Ophthalmol Vis Sci. 1998;39:951–962. [PubMed] [Google Scholar]
- 20.Senatorov V., Malyukova I., Fariss R., Wawrousek E.F., Swaminathan S., Sharan S.K., Tomarev S. Expression of mutated mouse myocilin induces open-angle glaucoma in transgenic mice. J. Neurosci. 2006;26:11903–11914. doi: 10.1523/JNEUROSCI.3020-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang B., Smith R.S., Hawes N.L., Anderson M.G., Zabaleta A., Savinova O., Roderick T.H., Heckenlively J.R., Davisson M.T., John S.W. Interacting loci cause severe iris atrophy and glaucoma in DBA/2J mice. Nat. Genet. 1999;21:405–409. doi: 10.1038/7741. [DOI] [PubMed] [Google Scholar]
- 22.Anderson M.G., Smith R.S., Hawes N.L., Zabaleta A., Chang B., Wiggs J.L., John S.W. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat. Genet. 2002;30:81–85. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- 23.Jakobs T.C., Libby R.T., Ben Y., John S.W., Masland R.H. Retinal ganglion cell degeneration is topological but not cell type specific in DBA/2J mice. J. Cell. Biol. 2005;171:313–325. doi: 10.1083/jcb.200506099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoilov I., Akarsu A.N., Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum. Mol. Genet. 1997;6:641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 25.Kaushik S., Cuervo A.M. Chaperone-mediated autophagy. Methods. Mol. Biol. 2008;445:227–244. doi: 10.1007/978-1-59745-157-4_15. doi:10.1007/978-1-59745-157-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg A.L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. doi:10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 27.Howell G.R., Libby R.T., Jakobs T.C., Smith R.S., Phalan F.C., Barter J.W., Barbay J.M., Marchant J.K., Mahesh N., Porciatti V., et al. Axons of retinal ganglion cells are insulted in the optic nerve early in DBA/2J glaucoma. J. Cell. Biol. 2007;179:1523–1537. doi: 10.1083/jcb.200706181. doi:10.1083/jcb.200706181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng G., Mellor R.H., Bernstein M., Keller-Peck C., Nguyen Q.T., Wallace M., Nerbonne J.M., Lichtman J.W., Sanes J.R. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. doi:10.1016/S0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 29.Vidal M., Morris R., Grosveld F., Spanopoulou E. Tissue-specific control elements of the Thy-1 gene. EMBO. J. 1990;9:833–840. doi: 10.1002/j.1460-2075.1990.tb08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raymond I.D., Vila A., Huynh U.C., Brecha N.C. Cyan fluorescent protein expression in ganglion and amacrine cells in a thy1-CFP transgenic mouse retina. Mol. Vis. 2008;14:1559–1574. [PMC free article] [PubMed] [Google Scholar]
- 31.George R.A., Heringa J. The REPRO server: finding protein internal sequence repeats through the web. Trends. Biochem. Sci. 2000;25:515–517. doi: 10.1016/s0968-0004(00)01643-1. [DOI] [PubMed] [Google Scholar]
- 32.Levitt M. Accurate modeling of protein conformation by automatic segment matching. J. Mol. Biol. 1992;226:507–533. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 33.Lee C. Predicting protein mutant energetics by self-consistent ensemble optimization. J. Mol. Biol. 1994;236:918–939. doi: 10.1006/jmbi.1994.1198. [DOI] [PubMed] [Google Scholar]