Abstract
The importance of mitochondrial iron–sulfur cluster (ISC) biogenesis for human health has been well established, but the roles of some components of this critical pathway still remain uncharacterized in mammals. Among them is human heat shock cognate protein 20 (hHSC20), the putative human homolog of the specialized DnaJ type co-chaperones, which are crucial for bacterial and fungal ISC assembly. Here, we show that the human HSC20 protein can complement for its counterpart in yeast, Jac1p, and interacts with its proposed human partners, hISCU and hHSPA9. hHSC20 is expressed in various human tissues and localizes mainly to the mitochondria in HeLa cells. However, small amounts were also detected extra-mitochondrially. RNA interference-mediated depletion of hHSC20 specifically reduced the activities of both mitochondrial and cytosolic ISC-containing enzymes. The recovery of inactivated ISC enzymes was markedly delayed after an oxidative insult of hHSC20-deficient cells. Conversely, overexpression of hHSC20 substantially protected cells from oxidative insults. These results imply that hHSC20 is an integral component of the human ISC biosynthetic machinery that is particularly important in the assembly of ISCs under conditions of oxidative stress. A cysteine-rich N-terminal domain, which clearly distinguishes hHSC20 from the specialized DnaJ type III proteins of fungi and most bacteria, was found to be important for the integrity and function of the human co-chaperone.
INTRODUCTION
J-proteins (also referred to as DnaJ or Hsp40 proteins) constitute a highly diverse family of co-chaperone proteins that regulate the activity and specificity of their designated partners, members of the heat shock protein 70 (HSP70) class of molecular chaperones. Together, these chaperone/co-chaperone complexes contribute to the folding, assembly and maintenance of functional proteins within cells (1–7). The J-domain proteins are divided into three classes according to their structural organization. Type III proteins consist of an N-terminal J-domain that is essential for the transient interaction with its chaperone partner fused to a C-terminal substrate-binding domain. Unlike other J-proteins, Type III proteins do not act as molecular chaperones on their own and they tend to interact selectively with a limited subset of client proteins or even a single substrate (5,8). Recently, such highly specialized class III proteins were shown to be involved in the maturation of iron–sulfur proteins in bacteria and yeast (reviewed in 3,9,10).
Iron–sulfur clusters (ISCs) are ancient co-factors that remain among the most versatile of prosthetic groups employed by contemporary proteins. They play important roles in electron-transfer reactions and non-redox catalysis, acting as sensors and functional switches and serving as crucial structural elements (reviewed in 11–15). The in vivo assembly and transfer reactions of ISC biosynthesis require complex molecular machineries, and disturbances within these pathways are often associated with serious pathological consequences (16,17). A seminal work in bacteria has led to the discovery of three different systems involved in ISC maturation (reviewed in 18). All employ a cysteine desulfurase that provides activated sulfur moieties to scaffold proteins upon which the nascent clusters are pre-assembled prior to their transfer to specific target proteins. Other components appear to represent system-specific additions to this fundamental theme. For instance, a DnaJ type III co-chaperone, called Hsc20 (heat shock cognate protein 20 or HscB), and its HSP70-like partner, referred to as Hsc66 (or HscA), are recognized as crucial factors in the central biosynthetic route of ISC formation in many Gram-negative bacteria (3,9,18). Eukaryotes presumably inherited this ISC pathway from the proteobacterial precursor of contemporary mitochondria. In line with this assumption, a similar co-chaperone/chaperone couple, referred to as Jac1p (J-type accessory chaperone) and Ssq1p (Hsp70 homolog), was linked to mitochondrial ISC biogenesis in the yeast Saccharomyces cerevisiae (10,19–25). Both the bacterial and the fungal co-chaperone/chaperone systems appear to be highly specialized, and only the U-type scaffold proteins of the respective organisms have been identified as relevant substrates thus far (7,9,19,26–30).
Although it is now generally accepted that specialized chaperone/co-chaperone complexes are required for the ISC machineries of bacteria and yeast, their precise functions remain unclear. In vivo studies in S. cerevisiae suggest its primary role in efficient cluster transfer from scaffold protein to the apoprotein rather than a role in assembling the cluster on the scaffold (31,32). This notion is consistent with the in vitro experiments performed in bacterial systems, which demonstrated that transfer of a [2Fe–2S] cluster from the IscU scaffold protein to apoferredoxin was enhanced in the presence of HscA, HscB and ATP (33,34). However, the same system did not facilitate any transfer of a [4Fe–4S] cluster from IscU to apo-aconitase in vitro (35), although previous in vivo studies had indicated a direct role for HscA and HscB in the maturation of aconitase in Azotobacter vinelandii (36).
Over the last decade, it has become apparent that many components of the bacterial ISC machinery have been conserved in the human genome (reviewed in 17,37). However, an HSP70 chaperone specifically dedicated to ISC biogenesis has not been identified. Unlike S. cerevisiae and a small subset of related fungi, vertebrates apparently employ a multifunctional mitochondrial HSP70 (HSPA9), also known as mortalin/mthsp70/PBP74/GRP75 (38,39). HSPA9 activity has been implicated in many different processes, including the facilitation of protein folding and import, intracellular trafficking, stress response, hematopoiesis and control of cell proliferation and tumor progression (reviewed in 40,41). Recent studies in yeast led to the speculation that the multifunctional mitochondrial HSP70 of higher eukaryotes might function in ISC biogenesis (39), but there is, as yet, no direct experimental evidence that supports for a role of mammalian HSPA9 in ISC biogenesis.
The apparent absence of a chaperone that is solely dedicated to the ISC assembly in higher eukaryotes would appear to warrant even greater participation of a specialized co-chaperone to specify function. Genes that encode putative homologs of the dedicated DnaJ proteins from bacteria and yeast have been annotated in the genomes of many eukaryotes, including the human genome (2,42). On the basis of its high sequence similarity, the human co-chaperone hHSC20, also referred to as DNAJC20/HSCB, was predicted to be involved in ISC biogenesis (42). Although the crystal structure of the hHSC20 was solved recently (43), it still remains functionally uncharacterized. We have, therefore, performed functional analyses of the hHSC20 to determine its potential role in the ISC assembly.
RESULTS
The HSC20s of higher eukaryotes represent unusual DnaJ type III proteins with a cysteine-rich motif at the N-terminus
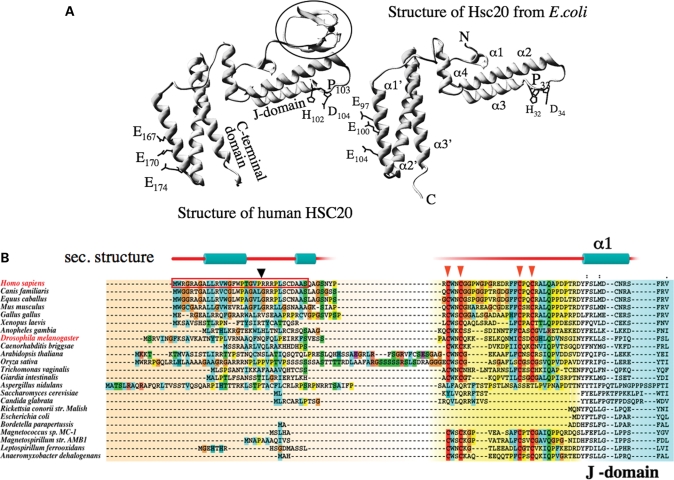
Only one gene that encodes a protein with high similarity to the general features of the specialized co-chaperones from Escherichia coli (HscB) and S. cerevisiae (Jac1p) is annotated in the human genome. On the basis of its primary sequence, the predicted hHSC20 shares ∼34 and 29% overall identity with its potential bacterial and fungal homologs, respectively (42). As revealed by the comparison of the recently solved structure of hHSC20 with the structure of HscB from E. coli (43,44), the high degree of sequence homology translates into a remarkable structural conservation of the J- and the C-terminal domains of the two proteins (Fig. 1A). These regions in both specialized co-chaperones from bacteria and yeast were found to be crucial for the interaction with the HSP70 chaperone partner and the respective scaffold proteins IscU/Isu (45–47). Furthermore, residues that were already implicated in these interactions in yeast (29,46) are apparently similarly positioned and oriented in the hHSC20 (Fig. 1A). However, the high degree of similarity is restricted to the C-terminal 2/3 of the human protein. The N-terminus of the hHSC20 is clearly different from the specialized DnaJ type III co-chaperones of fungi and most bacteria. Adjacent to an extended putative mitochondrial-targeting sequence (residues 1–35), the human protein contains an extra domain, which harbors two CXXC modules (C41/C44 and C58/C61; Fig. 1B). Their ability to coordinate a zinc ion in vitro results in zinc-finger-like structure (43). The high conservation of the CXXC modules in the predicted homologs of most eukaryotes and in proteins of a small number of specialized bacteria (see Fig. 1B and discussion) suggests an important, yet to be defined, function.
Figure 1.
The tertiary structures of hHSC20 and HscB from E. coli are highly similar, but hHSC20 contains a highly structured N-terminal extension. (A) The crystal structures of hHSC20 (BVO3) (43), and HscB from E. coli (1fpo) (44), suggest a remarkable structural similarity in both the J- and the C-terminal domains of the proteins. Amino acids believed to be directly involved in the interactions with the HSP70 partner, including the HPD motif of the J-domain, or the cluster of glutamates believed to be important for interaction with the scaffold protein, ISCU, are labeled and numbered in each structure. A unique N-terminal metal-binding domain, which is absent in the characterized co-chaperones from most bacteria and yeast, is highlighted in the human structure. (B) Multiple sequence alignments of the conserved and distinctive N-terminal regions of HSC20/HscB homologs from higher eukaryotes compared with fungal and bacterial species. The comparison of various species revealed that a twice-repeated metal-coordinating CXXC motif was highly conserved in the N-termini of HSC20s of higher eukaryotes, in the corresponding proteins of the anaerobic amitochondriata, G. intestinalis and T. vaginalis, and in some bacterial species that have unusually high iron demands. The evolutionary conservation of this intriguing domain in almost all eukaryotic representatives (except the fungal proteins) implies that this domain performs an important, yet undefined function. The conserved cysteines are highlighted by triangles. A putative mitochondrial leader sequence is boxed in the human protein; a predicted mitochondrial cleavage site is indicated by an arrowhead. Predicted secondary structure elements of the hHSC20 N-terminus are shown above the sequences.
Functional complementation
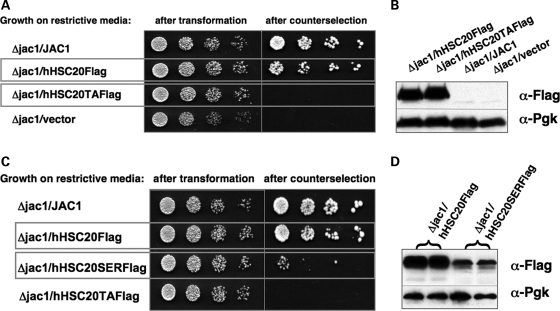
A yeast complementation assay was chosen in order to test whether hHSC20 could function in ISC biogenesis. To this end, a yeast strain with a chromosomal deletion of jac1, which is covered by plasmid-borne copies of the gene [courtesy of Dr A. Dancis (24)], was co-transformed with plasmids encoding the Flag-tagged versions of the human protein. As the dedicated co-chaperone of S. cerevisiae is critical for viability (23,24) (reviewed in 10), Jac1p-deficient cells can only grow in the presence of a functional substitute. After treatment with cycloheximide, which selects against vector-encoded copies of jac1, hHSC20WT was able to complement for Jac1p (Fig. 2A, compare left and right panels). The loss of leucine auxotrophy of hHSC20-expressing cells after counterselection clearly established the absence of the original covering plasmid (data not shown). To study the specificity of the complementation, a version of the human protein (hHSC20TA) was generated in which the HPD tripeptide of the J-domain, expected to be important for chaperone binding, was replaced by alanines. Equivalent mutations in Jac1p resulted in defective binding to its dedicated chaperone partner and led to a dramatic growth phenotype in vivo (23,24,29). As shown in Figure 2A, hHSC20TA could not rescue the viability of the Jac1p deletion strain even though the expression levels of the wild-type and mutant proteins were similar (Fig. 2B). These results clearly indicate that the severe growth defect of hHSC20TA-expressing cells was caused by altered protein function rather than differences in expression. Therefore, a functional J-domain appears to be critical for the biological activity of hHSC20. Taken together, these data strongly imply that hHSC20 represents the true ortholog of Jac1p.
Figure 2.
Expression of WT hHSC20 efficiently complements for Jac1p in S. cerevisiae. (A) A JAC1 shuffle strain (24) was co-transformed with the following constructs: YCplac22-GPDprom-JAC1 (WT JAC1 ORF), YCplac22-GPDprom-HSC20-3xFlag (WT hHSC20 cDNA), YCplac22-GPDprom-hHSC20TA-3xFlag [hHSC20 mutant with an altered J-domain in which the critical residues H102, P103 and D104 were replaced by alanines (Triple A)] and YCplac22-GPDprom (empty vector). Transformants were analyzed by plating 5-fold dilutions of cells in a 5 µl volume on synthetic dropout (SD) medium without l-leucine (-Leu) and l-tryptophan (−Trp; left panel), and SD without Trp supplemented with cycloheximide to counterselect against the covering plasmid (right panel). Plates were incubated at 30°C for 4 days before photography. Despite the significant differences in the N-termini between the yeast and human protein, hHSC20 could rescue the Δjac1 deletion strain, clearly demonstrating that hHSC20 performs a function similar to that of its yeast counterpart. Conversely, a mutant version of the human protein with an altered J-domain (hHSC20TA) did not rescue. (B) Steady-state levels of hHSC20WT and hHSC20TA were very similar in transformed yeast. Expression levels of the WT hHSC20 and hHSC20TA were very similar, as revealed by western blotting of yeast total lysates from each of the transformed yeast strains in (A). (C) An hHSC20 mutant in which the conserved cysteines of the distinctive N-terminal domain are replaced by serines only poorly complements for Jac1p deficiency. The JAC1 shuffle strain was co-transformed with the following constructs: YCplac22-GPDprom-JAC1 (WT JAC1 ORF), YCplac22-GPDprom-HSC20-3xFlag (WT hHSC20 cDNA), YCplac22-GPDprom-hHSC20Ser-3xFlag (hHSC20 mutant with an altered N-terminal domain in which the four highly conserved cysteines indicated in the alignment in Fig. 1A were substituted by serines) and YCplac22-GPDprom-hHSC20TA-3xFlag (hHSC20 mutant with an altered J-domain). Transformants were analyzed as described in A. (D) Indication that the distinctive N-terminal domain of hHSC20 may be important for its stability. Steady-state levels of hHSC20WT and hHSC20Ser were strikingly different in transformants, as is shown in a western blot representative of three independent experiments.
In order to begin to understand the role of the N-terminal domain, a mutant was constructed (hHSC20Ser), in which the four conserved cysteines were replaced by serine in order to disrupt the putative zinc-binding center. Interestingly, hHSC20Ser showed a very poor complementation (Fig. 2C). As the cysteine-rich domain does not exist in the endogenous yeast protein, Jac1p, the absence of complementation was unexpected. It might be explained, at least in part, by the significantly lower steady-state levels of the full-length serine mutant compared with hHSC20WT and hHSC20TA (Fig. 2B and D). Longer exposure of the respective western blots revealed numerous bands of lower molecular weight, which were specifically labeled with the monoclonal α-Flag antibody (Supplementary Material, Fig. S1), possibly indicating that the hHSC20Ser was unusually prone to degradation. These results and the even weaker complementation obtained when the cysteines were replaced by alanines (Uhrigshardt et al., unpublished data) suggested that the N-terminal cysteine motifs are important, perhaps because they bind a metal and stabilize a specific conformation of hHSC20.
hHSC20 interacts with its predicted partners
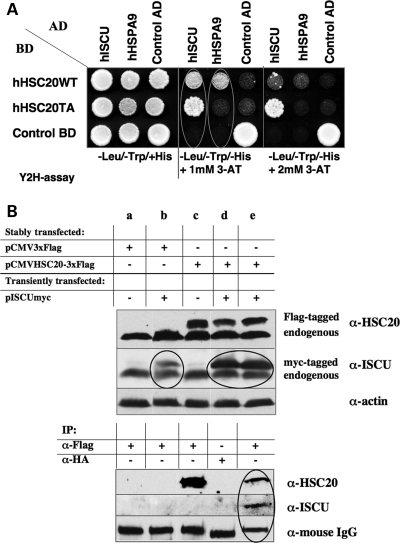
The results presented before demonstrated that hHSC20 was able to functionally replace Jac1p in S. cerevisiae, allowing a robust growth of Δjac1 cells. To further assess the potential role of hHSC20 in human ISC biogenesis, we tested whether hHSC20 could physically interact with its putative partners, the scaffold protein hISCU and hHSPA9 (mortalin), the only known mitochondrial HSP70 in mammalian cells. Previous studies on hISCU had clearly proved its involvement in human ISC biogenesis (48–50). In contrast, there are no direct data that link the multifunctional hHSPA9 to ISC assembly. Using yeast two-hybrid (Y2H) assays, we demonstrated that the wild-type hHSC20 interacted with its putative substrates, hISCU, and its predicted chaperone partner, hHSPA9 (Fig. 3A). Thus, in consistent with the results of the corresponding proteins in bacteria and yeast (30,51,52), the human co-chaperone can bind to either the substrate (ISCU scaffold) or the chaperone in the absence of the other partner. However, the individual interactions are apparently weak, as they were suppressed at a higher level of stringency (Fig. 3A, third panel). An hHSC20 mutant with an altered J-domain (hHSC20TA) did not bind to hHSPA9, whereas it could still bind to hISCU. Indeed, the strength of the interaction between hHSC20TA and the scaffold protein appeared to be even higher (Fig. 3A). These results correlated well with the complementation studies and demonstrated again the importance of a functional J-domain for an effective co-chaperone/chaperone interaction. However, despite an unaltered J-domain, hHSC20WT did not interact with an unrelated HSP70 (cytosolic hHSPA8;Supplementary Material, Fig. S3), indicating that additional specifying elements exist on either the co-chaperone or its dedicated partner, hHSPA9.
Figure 3.
hHSC20 interacts with its expected partners, the scaffold protein, hISCU, and the HSP70 protein, hHSPA9. (A) Y2H studies were performed to investigate the potential interactions of hHSC20 with its expected partners. hHSC20, hISCU and hHSPA9 were expressed as fusions to the Gal4-binding domain (BD) or Gal4 activation domain (AD), without the N-terminal mitochondrial-targeting sequences (MTS) to minimize the risk of unspecific interactions. Co-transformation with vectors encoding the known interactors Rabex-5 (control AD) and Rabaptin-5 (control BD) (91) provided positive and negative controls, respectively. The strains were plated on SD lacking l-leucine and l-tryptophan (−Leu/−Trp) with l-histidine (+His) as a control for loading and growth of the co-transformants. Specific interactions were monitored by activation of HIS3 gene transcription following plating on SD plates lacking l-histidine, l-leucine and l-tryptophan (-His/−Leu/−Trp), in the presence of increasing concentrations of the competitive inhibitor of the HIS3 protein, 3-amino-1,2,4-triazole (3-AT). All experiments were performed in triplicate. (B) hHSC20 interacts with hISCU in vivo. HeLa cells, either stably transfected with pCMVHSC20-3xFLAG or the empty vector, were co-transfected with pISCUmyc (50). Thirty-six hours post-transfection total cellular extracts were incubated with or without the indicated agarose bead-conjugated antibodies. The extract inputs (top panel, ∼5% of the total) and eluates from immunoprecipitates (bottom panel) were examined by western blotting using antibodies to endogenous hHSC20 and hISCU.
In an independent approach, we were also able to demonstrate a specific interaction of hHSC20 with its designated substrate, hISCU, in HeLa cells. An epitope-tagged version of hHSC20 (hHSC20-3xFlag) was stably expressed in HeLa cells (see Fig. 3B, upper panel, compare lanes a and b with lanes d and e). Co-transfection of the stable line with an hISCUmyc-encoding plasmid (48) resulted in ∼3–5 times higher steady-state levels of hISUmyc when compared with the vector controls (Fig. 3B, upper panel, compare lane b to lanes d and e). The mechanism by which hHSC20 expression increased the steady-state levels of hISCU is currently unknown, but it is tempting to speculate that co-expression of the co-chaperone and the scaffold hISCU stabilized the potential substrate. To evaluate the complex formation in vivo, immunoprecipitation was carried out with specific antibodies to the hHSC20 construct (α-Flag) and compared with unrelated antibodies (α-HA). As shown in Figure 3B, lower panel, only the use of the α-Flag beads led to co-immunoprecipitation of the epitope-tagged hHSC20 and hISCUmyc (lane e). No signal for the two proteins was detectable when the unrelated α-HA beads were employed (lane d). Although clearly specific, the low amounts of ISCUmyc captured suggest a weak or transient interaction between the scaffold protein and the co-chaperone. This assumption, which is supported by the results obtained in the Y2H assay (see above), might explain the apparent absence of co-precipitating endogenous hISCU, which is less abundant. Notably, significantly lower amounts of hHSC20-3xFlag were detectable in immunoprecipitations from cells that co-expressed hISCUmyc compared with mock-transfected controls (Fig. 3B, lanes c and e). Recent studies revealed that a conserved patch in the C-terminal domain of the specialized co-chaperones from E. coli and S. cerevisiae might represent the principal binding site for the respective scaffold protein (46,47). As the key residues are conserved and similarly oriented in the C-terminal domain of the hHSC20 (Fig. 1A), it is likely that the interaction with its potential substrate occupies a similar region on the human co-chaperone. hISCU binding could hinder the access of the α-Flag antibody to the epitope tag and thereby diminish the detection of hHSC20 in immunoprecipitations from co-expressing cells. hHSPA9 (mortalin) was not detected in co-immunoprecipitates derived from hHSC20/hISCUmyc co-expressing cells, using either α-myc or α-Flag antibodies (data not shown). However, interactions between HSP70 chaperone proteins and their co-chaperone partners and client proteins are known to be transient. Thus, consistent with the Y2H assays, which allow the detection of such weak and short-lived interactions, the human chaperone apparently does not form complexes with its partners that are sufficiently stable to withstand the conditions of immunoprecipitations.
hHSC20 localizes predominantly to the mitochondria and is widely expressed in human tissues
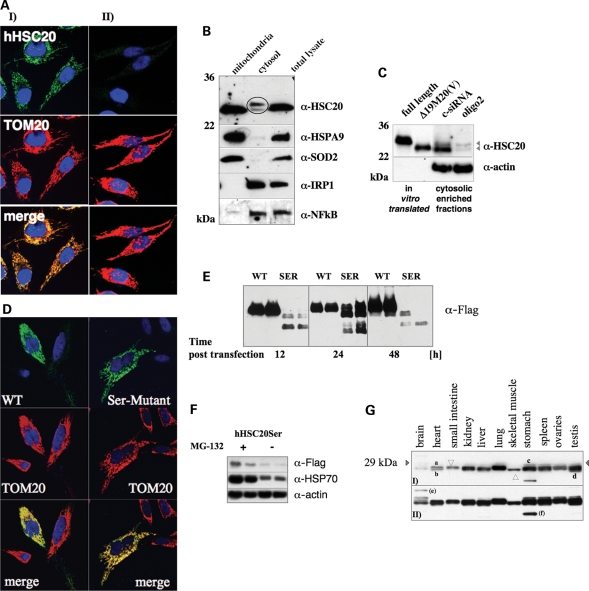
Examination of the sequence of hHSC20 revealed a putative mitochondrial leader sequence (residues 1–35) in the N-terminus of the human protein with a potential cleavage site between residues 21 and 22 (Fig. 1B). To address the question of whether hHSC20 indeed localizes to mitochondria, as already indicated by the yeast complementation experiments, and to gain a better insight into hHSC20 function in human cells, we assessed its intracellular localization in HeLa cells. We found that endogenous hHSC20 was mainly mitochondrial (Fig. 4A, panel I). Depletion of the hHSC20 by a gene-specific, small-interfering RNA (siRNA) abolished the hHSC20-derived signal, verifying the specificity of the anti-hHSC20 antibody (Fig. 4A, panel II).
Figure 4.
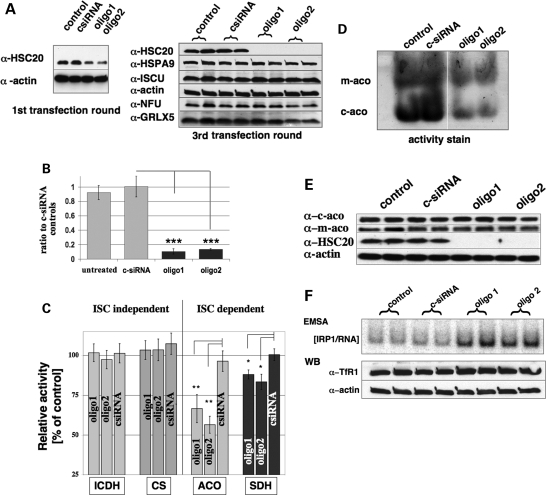
hHSC20 localizes predominantly to mitochondria. (A) Immunofluorescence studies revealed a predominantly mitochondrial localization of hHSC20 in HeLa cells. Cells were treated with an affinity purified antiserum against the hHSC20 and a monoclonal α-TOM20 antibody, and subsequently immunodecorated with α-rabbit Alexa488 and α-mouse Cy3 antibodies. (I) Top, hHSC20; middle, TOM20 (mitochondrial marker); bottom, merge of top and middle panels. (II) Immunolabeling was performed after two consecutive treatments with an hHSC20-specific siRNA (oligo2). (B) Subcellular fractionation demonstrated the existence of an extra-mitochondrial pool of hHSC20. HeLa cells were fractionated into cytosolic and mitochondrial fractions. Equal amounts of protein were subjected to SDS–PAGE and analyzed by immunoblotting. Antibodies against IRP1 and NFκB, cytosolic proteins, and hHSPA9, the mitochondrial HSP70 also known as mortalin, and SOD2, both components of the mitochondrial matrix, were used as controls. (C) Comparison of endogenous HSC20 species with in vitro translated versions of the protein and RNAi-treated cells confirms the presence of two cytosolic hHSC20 species. Plasmids encoding a full-length protein (HSC20FL) and a version of hHSC20 starting at position 20, Δ19M20, were used for in vitro expression. The resulting proteins were compared with the endogenous species detected by the hHSC20 antibody in cytosolic-enriched fractions from cells after two consecutive treatments with a non-specific control siRNA (c-siRNA) or an hHSC20-specific siRNA (oligo2). (D) Cysteine to serine mutations in the N-terminal domain of hHSC20 did not interfere with mitochondrial localization, as indicated by a similar distribution of the Ser-mutant protein. HeLa cells were transiently transfected with either pCMVHSC20WT-3xFLAG or pCMVHSCSer-3xFLAG. Twenty-four hours post-transfection, HeLa cells were treated with a monoclonal α-Flag antibody and a polyclonal α-TOM20 antibody, and subsequently immunodecorated with α-mouse Alexa488 and α-rabbit Cy3 antibodies. (Top) hHSC20WT-Flag (left) and hHSC20Ser-Flag (right); (middle) TOM20 (mitochondrial marker) and (bottom) merge of top and middle panels. (E) Cysteine to serine mutations in the N-terminal domain of hHSC20 resulted in dramatically reduced steady-state levels of the mutant protein. HeLa cells were transiently transfected with either pCMVHSC20WT-3xFLAG or pCMVHSCSer-3xFLAG and harvested at the indicated times post-transfection. Equal amounts of protein were subjected to SDS–PAGE and analyzed by immunoblotting using a monoclonal α-Flag antibody. A representative blot (n = 3) done in duplicates is shown. (F) Inhibition of proteasome does not result in substantial hHSC20Ser protection. Twelve hours after transfection with hHSC20Ser-Flag, HeLa cells were treated with the proteasomal inhibitor MG-132 (final concentration of 10 µg/ml). Cells were harvested 12 h post-treatment, and total lysates were subjected to western blotting. The robust induction of HSP72 clearly indicates proteasomal inhibition (92). A representative blot done in duplicates is shown. (G) hHSC20 protein is widely expressed in human tissues. Human multiple-tissue western blots were used to determine distribution. A commercially available membrane (Imgenex, 20 µg protein/lane) was probed with an affinity purified, antiserum against hHSC20. The expression of hHSC20 was essentially ubiquitous in tissues, with low levels found in brain and elevated levels present in lung. A (i) short and a (ii) longer exposure of the same blot are shown. Estimated molecular weights: (a) 27, (b) 26, (c) 29, (d) 25, (e) 34 and (f) 22 kDa.
The predominant mitochondrial localization of the hHSC20 was further supported by the subcellular fractionation experiments (Fig. 4B). However, two proteins of low abundance were consistently detected in the same fraction as the cytosolic proteins, iron regulatory protein 1 (IRP1) and NFκB. The apparent absence of mitochondrial markers [superoxide dismutase 2 (SOD2) and mHSP70] in these fractions argued against contamination from leaky mitochondria. The apparent estimated molecular weights of the two species of ∼27 and 26 kDa corresponded to the calculated molecular masses of the precursor (27.4 kDa) and a putative mature mitochondrial form after cleavage of the presequence at position 21 (25.2 kDa). Comparison with the corresponding in vitro translated versions of the hHSC20 supported this assignment (Fig. 4C), although the matches were not perfect. However, the clearly reduced intensities of both cytosolic bands after treatment with hHSC20-specific siRNAs supported the notion that the corresponding proteins indeed represented variants of hHSC20 (Fig. 4C, compare third and fourth lane). Taken together, and in agreement with an assumed function in the central human ISC assembly pathway, hHSC20 localized predominantly to the mitochondria in HeLa cells, but there was also evidence for a small extra-mitochondrial pool in mammalian cells.
To test for a potential impact of the cysteine-rich N-terminal domain of hHSC20 on the cellular localization of the protein, HeLa cells were transiently transfected with either an epitope-tagged wild-type version or a mutant in which the four conserved cysteines were mutagenized to serines (hHSC20Ser). As shown in Figure 4D, the localization pattern of both the proteins was indistinguishable from endogenous hHSC20, showing a strong overlap with the mitochondrial marker TOM20. However, despite the identical subcellular localization of the two proteins, the steady-state levels of wild-type and serine mutant proteins were found to be strikingly different (Fig. 4E). The hHSC20WT was expressed well 12 h after transfection, the expression of the serine mutant did not become unambiguously detectable until after 24 h and the protein was barely detectable at later time points (Fig. 4E). We did not observe diminished cell numbers in hHSC20Ser-transfected cultures, which argued against a dominant negative effect exerted by the mutant protein. The detection of α-Flag-sensitive bands of lower molecular weight in total lysates from hHSC20Ser-transfected cells implied that the degradation of the mutant protein was enhanced (Supplementary Material, Figs S1 and S3). Interestingly, only a small increase in the steady-state levels of full-length hHSC20Ser could be obtained upon the inhibition of the major proteolytic machinery of the cytosol, the proteasome (Fig. 4F). This result might suggest that the degradation of hHSC20Ser occurred mainly in the mitochondrial compartment. Overall, the observations in HeLa cells clearly resembled the results obtained in the yeast complementation studies, where the very low levels of the full-length hHSC20Ser-mutant protein, combined with increased presence of multiple lower molecular-weight species, suggested that the N-terminal cysteine motifs are needed for the integrity and function of hHSC20.
Finally, to evaluate the distribution hHSC20, we performed western blot analysis on various human tissues included on a commercially available blot. Although equal amounts of protein were loaded from each human tissue, β-actin loading appeared uneven, most likely because of the tissue-specific differences in abundance of this protein by itself (data not shown). Hence, we were limited to make qualitative rather than quantitative comparisons. Potential isoforms of the protein were detected in all tissues analyzed, with the highest abundance observed in extracts from the lung. However, in addition to these two major forms, with apparent molecular masses of ∼27 and 26 kDa, additional bands were also observed in the brain, heart, liver, stomach, spleen, ovary and testis, possibly revealing the tissue-specific modifications of hHSC20 (Fig. 4G).
Human HSC20 is involved in ISC biogenesis
The results presented thus far suggested a potential role of hHSC20 in human ISC biogenesis, and to further address an involvement of the protein in human ISC assembly, we employed RNA interference (RNAi) as a method to characterize the gene function. siRNAs corresponding to nucleotides 486–506 (oligo1) and 449–469 (oligo2) of the human HSC20 cDNA (NM 172002) were transfected into HeLa cells, and three rounds of transfection were performed in intervals of 3 days. Both siRNAs effectively reduced the hHSC20 protein levels to ∼30 ± 10% (n = 3) of the controls on day 3 of the first transfection. After three rounds of transfection, hHSC20 dropped below the levels which were detectable with our antibody. Like actin, which was chosen as an internal control, the amounts of its potential partners, hHSPA9 and hISCU, as well as the steady-state levels of two alternative carrier/scaffold proteins hGLRX5 (53) and hNFU (54) remained unchanged upon hHSC20 depletion (Fig. 5A). However, although the hHSC20 protein levels were below the limits of detection, the nature of siRNA-based silencing made it unlikely that hHSC20 synthesis had been completely abolished. Indeed, as shown by quantitative RT–PCR, after three rounds of transfection, ∼10% (oligo1) and 14% (oligo2) of the hHSC20 transcripts were detectable in cells treated with the specific oligos (Fig. 5B). Consistent with a role in ISC biogenesis, the depletion of hHSC20 was followed by a progressive decrease in the activities of the ISC-dependent enzymes, aconitase and succinate dehydrogenase (SDH). After 9 days of silencing, treatment with oligo1 and oligo2 resulted in moderate (SDH: ∼83 and 88%, and total aconitase: ∼57 and 67%), but significant reductions in the activities of the ISC-dependent enzymes. Notably, total aconitase activity (representing the combined activities of the mitochondrial and cytosolic isoenzymes) was affected earlier and more severely (Fig. 5C and Supplementary Material, Fig. S4). In contrast, the activities of isocitrate dehydrogenase (ICDH) and citrate synthase (CS), which do not require [Fe–S] clusters, were completely spared by hHSC20 knockdown (Fig. 5C), suggesting that the siRNA approach was gene-specific and relatively non-toxic.
Figure 5.
hHSC20 is involved in ISC biogenesis. (A) RNAi-based knockdown leads to undetectable levels of hHSC20 protein. HeLa cells were transfected with two hHSC20-specific siRNAs (oligo1 and oligo2) or control siRNA (c-siRNA) in single or multiple sequential transfections (for details see Materials and Methods). Although hHSC20 became undetectable after three transfections, the steady-state levels of actin (control), its potential partners hHSPA9 and hISCU, as well as of two alternate scaffold/carrier proteins, hGLRX5, and hNFU, remained essentially unaltered. Representative blots of at least three independent experiments done in duplicates are shown. (B) hHSC20 mRNA levels were significantly diminished after the third transfection round. Total RNA was prepared after the third transfection round and analyzed by qRT–PCR, using primers specific to the hHSC20 or the GAPDH genes, respectively. Error bars represent the SD of three independent experiments (***P < 0.001). (C) RNAi-based silencing of hHSC20 specifically reduced the activities of ISC-dependent enzymes. Activities of ISC-dependent enzymes, aconitase (Aco, representing the combined activities of mitochondrial and cytosolic isoenzymes) and SDH, and of two ISC-independent enzymes, ICDH and CS, were determined after the third transfection round (for details, see Methods and Materials). Activities of non-transfected controls were set to 100%. Results represent the means of at least three independent experiments, performed in duplicates ±SD. (D) hHSC20 depletion resulted in reduced activities of ISC-dependent enzymes in both the mitochondrial and the cytosolic compartment. Equal protein amounts were loaded per lane and subjected to native gel electro-phoresis. Aconitase activities were visualized by an in-gel assay (50). After the third transfection round, both mitochondrial and cytosolic aconitase activities were markedly reduced by treatment with oligo1 and oligo2, but not by treatment with c-siRNA (for details, see Materials and Methods). (E) Protein levels of cytosolic and mitochondrial aconitase were not altered by hHSC20 knockdown. Antibodies against mitochondrial and cytosolic aconitase revealed no significant decrease in the corresponding protein levels in hHSC20-depleted cells. β-Actin levels were used as control. A blot representative of three independent experiments is shown. (F) hHSC20 depletion activates IRP1 and stabilizes TfR1. IRE binding activities in the various cell lysates were increased, as determined by electro-mobility shift assay (EMSA) (82). The background of this assay is described in the text. Western blots revealed a slight increase in TfR1 protein levels in hHSC20-depleted HeLa cells.
General knockdown of different components of the ISC machinery in mammalian cells has been previously shown to affect both mitochondrial and cytoplasmic ISC proteins (50,55–57). Using a non-denaturing gel assay, we demonstrated that the depletion of hHSC20 also caused a significant loss of both mitochondrial and cytosolic aconitase activities, which were separated and assayed as described previously (50) (Fig. 5D). Western blot analyses of the two enzymes clearly showed that the substantially decreased activities could not be attributed to a significant reduction in the corresponding protein levels (Fig. 5E). The activities of both aconitase isoforms strictly depend on the presence of an intact cubane [4Fe–4S]2+ cluster (58). Both enzymes are susceptible to oxidant-induced inactivation (58–61). However, the interconversion of IRP1 from the cytoplasmic aconitase form that contains a [4Fe–4S]2+ cluster to the RNA-binding form requires the absence or removal of the entire cluster (58,62). As shown in Figure 5F, hHSC20 depletion resulted in a significant activation of IRP1, consistent with the impaired ISC assembly. This notion is further supported by a∼2-fold increase in the transferrin receptor 1 (TfR1) protein levels in the hHSC20-depleted cells. TfR1, an established target of IRP1 activation, is positively regulated through the IRP1-mediated stabilization of its transcript, which in turn leads to increased protein levels (62). Taken together, these results provide strong evidence for an integral function of hHSC20 in human ISC biogenesis, which has an impact on both mitochondrial and cytosolic ISC-containing proteins.
hHSC20 plays a critical role in assembly and/or repair of ISC under conditions of oxidative stress
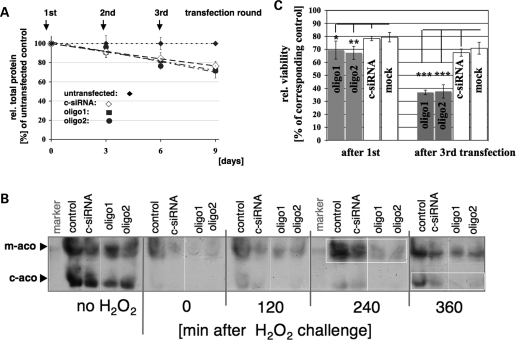
Owing to the vital functions of ISC-containing proteins within a mammalian cell (reviewed in 16,17), knockdown of components implicated in ISC biogenesis should lead to a significant impairment of cell growth and viability. Unexpectedly, treatment with oligo1 or oligo2 did not result in significant growth defects even after 9 days of silencing (Fig. 6A and Supplementary Material Fig. S5). As already reflected by the relatively moderate reduction in total aconitase, and in particular, SDH activities (Fig. 5C), at least in HeLa cells under normal growth conditions, very low hHSC20 levels were apparently sufficient to maintain the steady-state levels of ISC proteins.
Figure 6.
hHSC20 depletion did not lead to significant growth defects under normal growth conditions, but the recovery of aconitase activity and viability was impaired after oxidative challenge. (A) HeLa cells were transfected and passaged as described in Materials and Methods. At the days indicated, the total protein concentration per well as a measure for cellular growth was determined. Values derived from untransfected cells were set at 100%, and mean values ± SD of three independent experiments, done in duplicates, are shown. Treatment with both hHSC20-specific siRNAs and the unspecific control siRNA had a very similar impact on cellular growth. These results suggested that the minor growth defect was caused by the transfection reagent rather than by hHSC20 depletion. (B) hHSC20 silencing led to impaired recovery of mitochondrial and cytosolic aconitase activities after challenge with hydrogen peroxide. Cells on day 3 after the first transfection were incubated for 1 h with 500 µm of H2O2. Cells were harvested at the times indicated and were subsequently analyzed for mitochondrial and cytosolic aconitase activities. Equal protein amounts were loaded. A representative of three similar independent experiments is shown. (C) hHSC20 depletion led to increased sensitivity to oxidative stress. To investigate the effects of hHSC20 depletion on cell viability in conditions of oxidative stress, HeLa cells were seeded into 24-well plates and treated with the siRNAs as described. Forty-eight hours after the first and the third transfection, cells were exposed to 500 µm H2O2 for 1 h. Cell viability was measured 24 h after treatment by the MTT assay (see Materials and Methods). Values were normalized on the basis of corresponding simultaneously plated cultures that were subjected only to siRNA or Lipofectamine (mock) treatment to reveal the toxicity specific to H2O2. Data represent the results of three experiments done in duplicates and are expressed as the mean percent of untransfected control ± SD. *P< 0.05; **P < 0.001.
To further unravel the role of hHSC20 in human ISC biogenesis, we evaluated the effects of depletion of the protein under conditions of oxidative stress. As shown in Figure 5A, the first round of siRNA-based knockdown reduced the hHSC20 levels to ∼30% of the controls. Although this degree of depletion did not lead to a significant reduction in aconitase activities in the absence of an oxidative challenge, treatment with hydrogen peroxide clearly resulted in a markedly delayed recovery of both aconitase isoforms in hHSC20-deficient cells (Fig. 6B). Consistent with earlier results (50,57,63), the mitochondrial isoenzyme was less affected by H2O2 treatment and regained its activity earlier and more readily than the cytosolic aconitase (Fig. 6B). We also investigated the effects of hHSC20 deficiency on cell viability after short-term exposure to hydrogen peroxide. A moderate, but nevertheless significant, negative impact of hHSC20 depletion on cell survival was already evident after the first siRNA treatment, and it became clearly enhanced after three rounds of silencing (Fig. 6C). These results were consistent with either an impaired in vivo repair/regeneration of damaged or destroyed ISCs or an increased sensitivity to H2O2 in hHSC20-deficient cells.
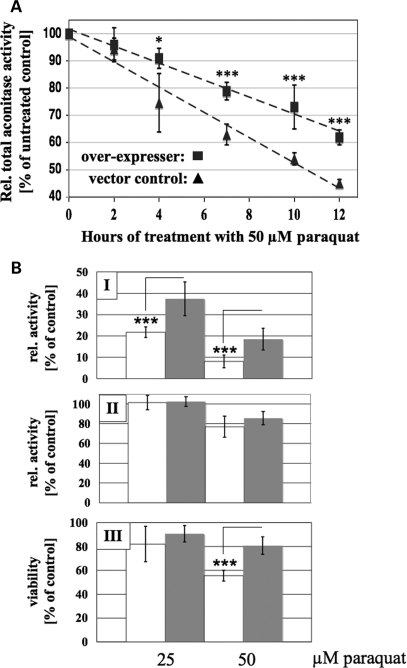
To explore the role of hHSC20 in response to the long-term exposure of oxidative stress, two HeLa cell lines, stably transfected with either hHSC20-3xFlag or empty vector, were exposed to the stable redox-cycling drug paraquat. As shown in Figure 7A, the decline in total aconitase activity in hHSC20-3xFlag-expressing cells was clearly delayed and less severe when compared with the controls. Conversely, the activity of the ISC-independent enzyme ICDH remained almost unaffected. Seventy-two hours of treatment with different paraquat concentrations (25 and 50 µm, respectively) resulted in a dramatic aconitase inactivation in both vector controls and hHSC20-3xFlag-expressing cells. However, in both cases, the loss of total aconitase activity was significantly more pronounced in the controls (Fig. 7B, top row). In contrast, even prolonged paraquat treatment resulted only in either none (25 µm) or a minor (50 µm) reduction in ICDH activities, which did not differ significantly in the two HeLa cell lines (Fig. 7B, middle row). These data indicate that slightly elevated levels of hHSC20, although not fully capable of preventing inactivation, clearly protected some ISC-containing proteins. Hence, the markedly higher resistance of hHSC20-3xFlag-expressing cells against paraquat-induced toxicity (Fig. 7B, bottom row) might be attributed to higher steady-state activity levels of critical ISC proteins.
Figure 7.
Overexpression of hHSC20 enhanced resistance to paraquat toxicity by delaying inactivation of aconitase and increasing viability. (A) Time course of aconitase inactivation upon exposure to paraquat. Cells, either stably transfected with pCMVHSC20WT-3xFLAG or empty vector, were seeded in the 12-well plates for 24 h before treatment with 50 µm paraquat. At the times indicated, cells were harvested and snap frozen. Enzymatic activities were determined as described in Materials and Methods. Activities of corresponding cultures, which were not treated with paraquat, were set to 100%. Although the overexpression of hHSC20 did not fully prevent inactivation, moderately elevated levels of hHSC20 clearly resulted in a significantly delayed inactivation of total aconitase activities (representing the combined activities of mitochondrial and cytosolic isoforms). The activities of the ISC-independent enzyme ICDH remained almost unaffected in both cell lines even after 12 h of paraquat treatment [91.3 ± 4.4% (hHSC20Flag), and 91.4 ± 6.7% (empty vector) of the ICDH activities in the corresponding controls]. Results represent the means of at least three independent experiments, performed in triplicates ± SD. Overexpresser, squares; vector control, triangles. (B) Overexpression of hHSC20 increases the resistance against paraquat toxicity. HeLa cells stably transfected with pCMVHSC20WT-3xFLAG, or empty vector were seeded in a 12 well and exposed to 0, 25 and 50 µm of paraquat. After 72 h, total aconitase (I) and ICDH (II) activities were measured. Cell viability was determined with the MTT assay (III). Activities/absorbances of corresponding cultures, which were not exposed to oxidative stress, were set to 100%. Consistent with a potential beneficial role of hHSC20 against the adverse effects of oxidative stress, we observed an increased resistance against paraquat-induced toxicity in cells that overexpressed hHSC20. All data represent the results of at least 3 independent experiments done in duplicates and are expressed as a mean percent of the corresponding untreated control ± SD. White bars, vector control; filled bars, pCMVHSC20WT-3xFLAG.
Long-term exposure to paraquat leads to markedly reduced hHSC20 protein levels
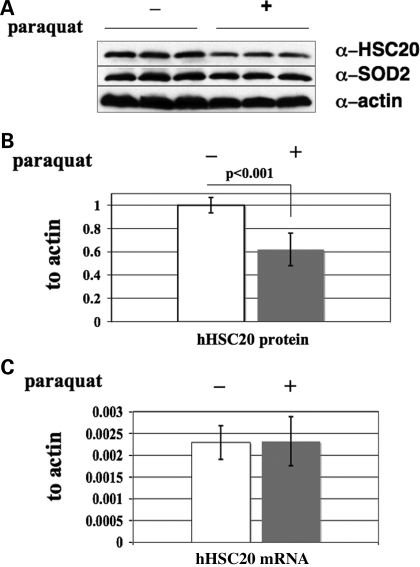
Thus far, our results implied a critical role of hHSC20 under conditions of stress. Therefore, we explored several potential regulatory mechanisms that might affect hHSC20 levels under conditions expected to alter the demand for ISC biosynthesis. To this end, HeLa cells were exposed to oxidative stress (50 µm paraquat) for 24–72 h. Although no obvious changes in endogenous hHSC20 protein abundance were detectable up to 48 h after treatment (data not shown), we observed a significant reduction in the hHSC20 steady-state levels (62 ± 16% of untreated controls; n = 9) in paraquat-treated cells after 72 h. In contrast, SOD2 and actin protein levels were not significantly affected (Fig. 8A and B). hHSC20 transcript levels were found to be surprisingly low per se (∼8- and 500-fold less abundant than the transcripts for hISCU and β-actin, respectively) and did not significantly change upon paraquat treatment (Fig. 8C). The striking discrepancies between the transcript and protein levels are consistent with possible regulation of hHSC20 at the post-transcriptional level and might either result from an increased rate of degradation or a decrease in stability under conditions of prolonged oxidative stress.
Figure 8.
Levels of hHSC20 diminish with long-term paraquat treatment, but transcription levels do not change. As revealed by western blotting, prolonged treatment with paraquat (72 h) leads to markedly decreased hHSC20 protein levels. (A) Representative western blot. β-Actin and SOD2 levels were used as controls. (B) Quantification of the reduction in hHSC20 protein levels in paraquat-treated cells. (C) However, as shown by qRT–PCR, hHSC20 transcript levels remain almost unchanged. β-Actin protein and transcript levels were used for normalization. Data represent the results of at least three independent experiments done at least in duplicates and are expressed as the mean percent of the corresponding untreated control ± SD. White bars, untreated, time-matched controls; Filled bars, treated with 50 µm paraquat for 72 h.
DISCUSSION
Among the 41 currently known J-domain proteins expressed in human cells (2), only DNAJC20/hHSC20 shares high homology to the specialized bacterial and yeast counterparts that are dedicated to ISC assembly. It was already predicted that hHSC20 would play an important role in human ISC biogenesis (42), but the functions of the human co-chaperone remained uncharacterized. When we expressed hHSC20 in a Jac1p-deficient yeast strain, the human protein clearly restored the severe growth defect, which accompanies the lack of the dedicated co-chaperone in S. cerevisiae (reviewed in 10) (24). These results strongly imply that hHSC20 represents the true ortholog of Jac1p and reveal a striking functional conservation of the two proteins through ∼1 billion years of evolution. Interaction of hHSC20 with its anticipated partner, the ISC scaffold protein hISCU in both Y2H and immunoprecipitation experiments performed in this study, further supported the role of the co-chaperone in human ISC biogenesis. Our Y2H studies also demonstrated a specific interaction between hHSC20 and hHSPA9 (mortalin), the only known mitochondrial resident HSP70 in human cells. hHSPA9 is more homologous to Ssc1p, the housekeeping mitochondrial chaperone of S. cerevisiae, than to Ssq1, the yeast chaperone dedicated to iron–sulfur biogenesis, and its role in human ISC biogenesis has not yet been established. However, recent in vitro studies demonstrated that peptide-binding domains of various multifunctional mitochondrial HSP70s specifically interacted with an ISCU-derived fragment (39). Furthermore, overexpression of the multifunctional Ssc1p corrected the negative impact of Ssq1 deficiency on ISC assembly (23,39). Thus, our observation that hHSC20 and hHSPA9 interact in vivo provides further support for an involvement of the multifunctional mitochondrial chaperones of higher eukaryotes in ISC biogenesis.
One of the most striking features of hHSC20 is the presence of a cysteine-rich domain in the extended N-terminus of the human protein. As revealed by the recently published crystal structure, metal co-ordination through the cysteine motifs leads to the formation of a zinc-finger-like domain in vitro (43). The apparent importance of the domain became obvious in the very weak rescue of Jac1p deficient yeast by an hHSC20 mutant, in which the four conserved cysteines of the N-terminus were replaced by serines. As the cysteine-rich domain does not exist in the endogenous yeast protein, the lack of complementation was unexpected. One explanation for the ineffective complementation by hHSC20Ser might be due to the clearly lower abundance of the mutant protein. Notably, markedly reduced the steady-state levels of hHSC20Ser compared with the wild-type protein were also evident upon transient expression in HeLa cells. Taken together, these results suggest an important role of the N-terminal domain for maintenance of the integrity and, consequently, the functionality of hHSC20. However, the high homology of hHSPA9, the presumed chaperone partner of hHSC20 (see above), with the multifunctional chaperone Ssc1p might offer yet another intriguing possibility. Ssc1p is present in yeast mitochondria at significantly higher levels than the specialized Ssq1, and the protein can function, albeit clearly less effectively, with Jac1p in the ISC assembly (24,39,46) (reviewed in 10). Thus, it is possible that the human co-chaperone functions together with the non-specific HSP70 and that the robustness of such an interaction might be influenced by the N-terminal zinc-finger-like domain of the hHSC20. In line with this assumption are recent findings on the role of two zinc-finger domains that are invariably present in class I J-proteins (2). One of these, Z2, was essential for functionality both in vitro and in vivo and likely represents the site of an additional interaction between class I co-chaperones with their HSP70 partners that facilitates conversion of the chaperone into its high-affinity binding state (reviewed in 3,64). Interestingly, the N-terminal domain of hHSC20 shares some structural similarity with the Z2 domain (65), and the presence of HSC20 homologs bearing a cysteine-rich N-terminal domain in higher eukaryotes correlates with the apparent lack of a chaperone specifically devoted to ISC biogenesis. Therefore, it is tempting to hypothesize that the zinc-finger-like domain of hHSC20 might be required to effect a more stringent interaction with a promiscuous HSP70 chaperone partner, which participates in multiple processes and interacts with different J-proteins as well as multiple client proteins. Notably, several hHSPA9-interacting proteins that contain zinc-finger-like domains are known to exist in the human mitochondria, including hTID1 (mitochondrial class I J-protein) and Hep1 (fractured J-protein), and these proteins could potentially compete with hHSC20 for binding (2,66). The presence of other proteins that compete to interact with hHSPA9 may help to explain why overexpression of hHSC20 protected cells from oxidative stress and diminished levels rendered cells more sensitive to oxidative stress. By binding to hHSPA9 and to hISCU, hHSC20 may direct the activities of a non-specific HSP70 protein into iron–sulfur biogenesis, which is particularly needed after ISCs are destabilized by oxidative stress. Interestingly, mutagenesis of the HPD motif in the J-domain of hHSC20, which is crucial for its interaction with hHSPA9, appeared to enhance the strength of the interaction between the co-chaperone and its substrate, hISCU. This observation is consistent with the possibility that hHSPA9 binding may force an hHSC20-hISCU complex to dissociate, which in turn might facilitate ISC-transfer from the scaffold protein to recipient target proteins.
As expected from the presence of a predicted mitochondrial leader sequence, hHSC20 localized predominantly to the mitochondrial compartment, but a small extra-mitochondrial pool of hHSC20 was also detected in HeLa cells. The origin of the cytosolic fraction is currently unknown, but it is possible that the cytosolic formation of a zinc finger at the N-terminus could prevent efficient mitochondrial uptake, as was recently proposed for the small TIM proteins (67).
Like hHSC20, these components of the mitochondrial import machinery contain N-terminal cysteine motifs, which promote trapping of the TIM proteins in the intermembrane space (reviewed in 93). Hence, it is tempting to speculate that uptake of hHSC20 in the mitochondrial matrix could also be modulated through reversible zinc binding to its cysteine-rich N-terminal domain. Relative binding of zinc or other metals and relative levels of oxidative stress could contribute to a regulated and dynamic distribution of the protein between the cytosolic and mitochondrial compartments. Interestingly, extra-mitochondrial localization was also reported for AtHscB, the Jac1p ortholog of Arabidopsis thaliana (94), which, like the human protein, bears the N-terminal twin CXXC motifs.
Regardless of the underlying mechanism, the presence of the hHSC20 in the cytosolic compartment makes biological sense. Over the past years, an extra-mitochondrial localization has been demonstrated for many established and proposed components of the human mitochondrial ISC machinery (37,57,70–71,95). Among them are both predicted partners of hHSC20, the chaperone hHSPA9 (reviewed in 40,41) and the scaffold protein hISCU, for which an involvement in cytosolic ISC biogenesis has already been established (48–50,95). Thus, the cytosolic pool of the human co-chaperone might participate in cytosolic ISC biogenesis, which directly contributes to maintenance and assembly of extra-mitochondrial ISC proteins.
The progressive and significant reduction in activities of both mitochondrial and cytosolic ISC-dependent enzymes, as well as IRP1 activation, upon RNAi-mediated hHSC20 depletion, unequivocally established the role of the co-chaperone in human ISC assembly. However, even drastically reduced levels of hHSC20 had only a relatively moderate effect on ISC-dependent enzymes and resulted in a much milder phenotype when compared with similar studies in mammalian cells on other components of the central ISC pathway (50,55,56,72,73). Our results resemble previous in vivo studies of S. cerevisiae, which showed that despite the established essential nature of Jac1p, neither a marked reduction in protein levels nor expression of a Jac1p mutant with a reduced affinity for the scaffold Isu1p resulted in impaired viability or growth defects (23,28,46). However, the mild phenotype correlated with the presence of the specialized chaperone Ssq1p (28,30,46), and a severe growth defect was evident in cells that had to employ the multifunctional Ssc1p for ISC biogenesis (28,46).
Interestingly, and in clear contrast to recent studies in yeast (74), we did not observe any up-regulation of hISCU in hHSC20-depleted HeLa cells. Thus, the absence of a severe growth defect cannot be simply explained by a compensatory response mediated by elevated levels of the scaffold protein. In vitro studies with IscU from E. coli provided evidence that at least the bacterial scaffold protein was capable of catalyzing multiple cycles of ISC transfer to target proteins without the need of further assistance of a chaperone; however, the respective transfer rates were considered to be too slow to meet the in vivo demands (18,33). Notably, with hGLRX5, hNFU and hISA, three alternative carrier/scaffold proteins are present in HeLa cells that may act independent of a specialized co-chaperone/chaperone system (54,57,75). Although hGLRX5 and hNFU steady-state levels remained unaltered in hHSC20-depleted cells, it is possible that the presence of alternative scaffold systems mitigated the effects of hHSC20 loss. However, the lack of a severe growth phenotype resulting from even drastically reduced levels of hHSC20 is also consistent with the proposed catalytic role of the specialized co-chaperones (9). This notion is in agreement with the significantly lower abundance of hHSC20 compared with the scaffold hISCU in HeLa cells and might be further supported by the earlier and more pronounced effect of hHSC20 depletion on total aconitase activity than on SDH. As shown recently, with the exception of the notoriously unstable ISC of aconitase, the Fe–S co-factors of other ISC enzymes in mammalian mitochondria are surprisingly resistant to degradation (76). Thus, our findings might indicate that although the ISC machinery with severely reduced hHSC20 levels still has enough residual capacity to maintain biogenesis and transfer of stable ISC enzymes, maintenance of more vulnerable clusters with faster turnover is already compromised by silencing. Interestingly, after oxidative insults, even moderately reduced levels of hHSC20 led to a diminished recovery of total aconitase activities and significantly decreased cellular viability. On the other hand, moderate overexpression of hHSC20 clearly delayed aconitase inactivation and protected from paraquat-induced toxicity. Our results are consistent with studies of the bacterium A. vinelandii, in which the severe growth phenotype observed upon the depletion of HscAB was partially alleviated by growth under conditions of low oxygen tension (36). Thus, our data and results in other model systems indicate that the dedicated co-chaperone plays an important role in ISC biogenesis in conditions that require enhanced ISC biogenesis (e.g. oxidative stress). We suggest that hHSC20 may act as a pacemaker of the human ISC biogenesis that determines the speed and performance of the pathway, perhaps by supporting a faster and more efficient cluster transfer from the scaffold hISCU to recipient proteins or by recruiting the promiscuous hHSPA9 to participate in Fe–S biogenesis. Although even very small hHSC20 concentrations appear to be sufficient for cell viability under general cell culture conditions, it is possible that hHSC20 deficiency might eventually trigger more dramatic phenotypes in some mammalian tissues that are prone to higher rates of oxidative stress, such as the lung and the retina. Intriguingly, hhsc20 was one of the three candidate genes implicated as the cause of the new form of optic atrophy (77).
However, despite the proposed higher requirements of the human co-chaperone under conditions of oxidative stress, we did not observe an up-regulation of hHSC20 in paraquat-treated HeLa cells. On the contrary, prolonged exposure to oxidative stress led to markedly reduced steady-state levels of the hHSC20 protein. The fact that the transcript levels remained unaltered is consistent with a post-transcriptional mode of regulation for hHSC20. The underlying mechanism is currently under intense investigation, but the significantly reduced levels of the co-chaperone could be caused by an increased rate of degradation or a decrease in stability. The intriguing similarities with the results obtained with the serine mutant might imply an involvement of the N-terminal domain in the maintenance of the steady-state levels. Notably, the cysteine-rich region engaged in metal co-ordination represents the only part of hHSC20 that is devoid of a predicted secondary structure, and it is likely that the N-terminus of the protein unfolds in the absence of metal. Thus, the oxidative damage of the metal co-ordinating cysteines might lead to the loss of the metal ligand and subsequently result in an unfolded state that, like the serine mutant, is prone to degradation. ISCs are not only the vulnerable targets of oxidative stress, but their decay significantly contributes to the generation of intracellular reactive oxygen species. It is, therefore, possible that reduced hHSC20 availability in conditions of chronic oxidative stress is even beneficial for the cell. It might reduce ISC biogenesis and diminish the damage caused by disintegration of ISCs. As mentioned above, no obvious changes in the hHSC20 levels were observed up to 48 h treatment with paraquat. Hence, it is tempting to speculate that more subtle post-translational modifications in the cysteine-rich domain (for instance, glutathionylations/nitrosylations) might modulate the functionality of the human co-chaperone.
An additional hint for a potential regulatory role of the N-terminal domain might be gained from the presence of very similar motifs in the N-termini of the predicted hHSC20 homologs from a small number of specialized bacterial species. Besides having high needs for iron, these otherwise phylogenetically unrelated organisms might share an unusual susceptibility to oxidative stress. Like the magnetotatic species and Anaeromyxobacter dehalogenans, they either represent facultative anaerobes that exist at the oxic–anoxic transition zone between anaerobic and aerobic habitats (68,78) or like Leptospirillum ferrooxidans and Acidobacterium capsulatum thrive in aerobic, but acidic environments that have very high concentrations of redox active transition metals (69,79). Such conditions could repeatedly expose these bacteria to intermittent severe oxidative insults, which could have forced them to develop highly responsive ISC biogenesis machineries that rapidly accommodate to changes in environmental conditions. The cysteine-rich motif could have an important regulatory role in these settings. Interestingly, strikingly similar N-terminal cysteine-rich motifs exist in the putative HSC20 proteins of the amitochondriata, Giardia intestinalis and Trichomonas vaginalis, where the correlation with facultative anaerobic life-style points again in the direction of a regulatory, sensing or repair function. Like many pathogens, these organisms likely encounter oxidative stress and iron starvation when they infect hosts, and they may combat these two detrimental environmental conditions by enhancing their capacity for ISC biogenesis and function.
Our work demonstrates that hHSC20 is indeed an integral component of the human ISC biosynthetic machinery, which appears to be particularly important under conditions that demand an enhanced ISC biogenesis. Moreover, our finding that hHSC20 levels are reduced by a long-term exposure to paraquat might suggest that the decrease in iron–sulfur protein activities observed in chronic oxidative stress may result not only from direct damage to ISCs, but also from diminished rates of re-assembly and repair due to the degradation of hHSC20. We believe, although more work is needed to establish the details of the molecular mechanism of the degradation, that our work opens a new perspective to understanding the role of HSC20 in higher eukaryotes. As it appears that hHSC20 competes with other proteins for binding to hHSPA9, we suggest that the levels of hHSC20 may determine the rate of ISC biogenesis and that after its expected ATP-dependent release from hHSPA9, hHSC20 may enhance the delivery of the ISC of hISCU to a specific set of target proteins. Much effort will be devoted in the future to understand how specificity of cluster transfer is achieved, and further studies of hHSC20 may yield important clues.
MATERIALS AND METHODS
If not indicated otherwise, reagents were purchased from Sigma. Cell media and supplements were either from Gibco or Cellgro. Western blot reagents were obtained from Pierce and BioRad.
Constructs
pGEX-Jac1ΔN75, which encodes an N-terminally GST-tagged version of the hHSC20 (GST- Jac1ΔN75) starting at amino acid 76, was a gift by Dr W.H. Tong. Recloning in the EcoRI and NotI sites of pET41a (Novagen) resulted in pET41a Jac1ΔN75, which encodes an N-terminal GST-His-tagged version of the protein. Before cloning in the respective destination vectors, PCR-amplified transcripts were routinely subcloned in the vector pCR2.1-TOPO by TOPO TA cloning following the protocol of the manufacturer (Invitrogen). For in vitro transcription/translation, hHSC20-encoding sequences were amplified from the human Image clone 3505128 (ATCC, accession BC65569), using the primers GFH20 and GRH20, and H20m1xmn1 and H20revsal1, respectively. pTnTHSC20FL (coding for full-length protein) and pTnTΔ19M20HSC20 (coding for the mature protein without predicted mitochondrial leader sequence) were engineered by cloning the corresponding sequences in the EcoRI or EcoRI/SalI sites of the pTnT vector (Promega). For pCMVHSC20-3xFLAG, the gene was amplified from Image clone 3505128 with the primers NheH20F and H20salR. After subcloning in pCR2.1-TOPO, the transcript was excised with SalI and BamHI and inserted in the corresponding sites of the mammalian expression vector pCMV-3Tag-8 (Stratagene). For yeast complementation studies, epitope-tagged versions of hHSC20 were amplified from pCMVHSC20-3xFLAG using the primers NdeIhhsc20f and flagSacIrev. Jac1 was amplified from the plasmid SP64T-Jac1 (courtesy of Dr Andrew Dancis) using the primers NdeJac1f and Jac1SacIrev. After subcloning in pCR2.1-TOPO, the transcripts were excised with NdeI and SacI and integrated in the corresponding sites of YCplac22-GPDprom (a generous gift of Dr Andrew Dancis), which contains a 2 µm ori, the strong glyceraldehyde-3-phosphate dehydrogenase isoenzyme 3 (GPD) promoter and TRP1 as a marker. For Y2H studies, the Δ19M20HSC20 encoding sequence was excised with EcoRI and SalI from the respective pCR2.1 construct and recloned in the corresponding sites of pGBKT7 (Clontech), resulting in pGBKT7-ΔHSC20. Sequences encoding Δ27M28ISCU and Δ25M26HSPA9 were amplified from pXShISU2myc (courtesy of Wing-Hang Tong) (48,50) and the human Image clone 2964588 (ATCC, accession BC024034) using the primers fNdeIhISCUdel, hISCUxhorev, fNdelhHSPA9del and hHSPA9xhorev, respectively. After subcloning in pCR2.1-TOPO, the fragments were excised with NdeI and XhoI and recloned in the corresponding sites of pGADT7 (Clontech). Site-specific mutations were introduced into hhsc20-coding DNA sequences using the QuickChange protocol (Stratagene). For details on the used mutagenesis primers, see Supplementary Material Table S1. All constructs were sequenced to confirm their authenticity.
Protein expression in bacteria and affinity purification of the hHSC20 antiserum
Escherichia coli BL21-CodonPlus (DE3)-Rosetta (Novagen) cells transformed with pET41a Jac1ΔN75 were cultured in LB medium containing 50 µg/ml of ampicillin at 37°C. After reaching an A600 of 1.2, cultures were diluted 1:1 in 2× LB and shifted to 30°C. Isopropyl-1-thio-d-galactopyranoside was added to the final concentration of 0.5 mm. After 4 h, cells were harvested by centrifugation (4000g, 20 min), resuspended in lysis buffer (50 mm Tris–HCl, pH 8.0, containing 100 mm NaCl, 2 mm EDTA, 1 mm dithiothreitol (DTT), 10 µg/ml of leupeptin, 1 mm phenylmethylsulfonyl fluoride, 0.5% Triton X-100) and ruptured by sonication. Jac1ΔN75 was purified from inclusion bodies by affinity chromatography using nickel-nitrilotriacetic acid superflow gel under denaturing conditions as recommended by the manufacturer (Qiagen, http://www1.qiagen.com/literature/handbooks protocol 18). The eluate was concentrated and subjected to preparative SDS–PAGE. The protein was electro-eluted from the gel (Centrilutor, Millipore), using a buffer containing 25 mm Na2CO3, pH 8.3, and 0.5% NP-40. GST-His-Jac1ΔN75 was coupled to CnBr-activated Sepharose 4FF (Amersham) according to the recommendations of the manufactures. A polyclonal antiserum to GST-Jac1ΔN75 (4432C, a gift of Dr W.H. Tong) was purified by incubating the antiserum with GST-His-Jac1ΔN75 conjugated to sepharose (4 h, 4°C). After three washes with 1× PBS, the purified antiserum was eluted in 1 ml increments with 0.1 m glycine, pH 2.4. 30 µl of 3 m Tris–HCl, pH 8.8, and 20 µl of 5 m NaCl were immediately added to protein-containing fractions.
Yeast complementation assays
Co-transformation of the yeast shuffle strain for JAC1 [MATa, ura3-52, lys2-801(amber), ade2-101(ochre), trp1-63, his3-200, leu2-1, jac1:HIS3 (pRS318-JAC1)] (24) with various constructs was carried out as follows: a 500 µl of aliquot of a 5 ml overnight culture (grown at 30°C) was spun down for 5 s in a sterile Eppendorf tube. The supernatant was discarded. The pellet was washed with sterile water and spun down again. After removal of the supernatant, 125 µl of EZ-transformation reagent (Q-BIO gene), 2 µl of carrier DNA and ∼100–200 ng of the desired construct were added to the cells and thoroughly mixed by aspirating. After incubation at 42°C for at least 1 h, the mixture was spun briefly again. The cellular material was transferred to selective plates (−Leu/−Trp). After 2 days at 30°C, single colonies were picked and resuspended in ultrapure water. The OD600nm was adjusted to 0.05; 5 µl increments of 5-fold consecutive dilutions were plated on −Leu/−Trp plates with 10 µg/ml of cycloheximide (for further details, see Fig. 2A and C).
Y2H analysis
For interaction studies, the Matchmaker 3 two-hybrid system (Clontech) was employed. Co-transformation of the AH109 reporter strain with pGADT7- and pGBKT7-based constructs was performed as described above. Plating of transformants and the positive and the negative experimental controls are described in the legend of Figure 3A.
Human cell culture, transfection and RNAi
HeLa cells were obtained from ATCC. If not indicated otherwise, cells were grown in the Dulbecco's modified Eagle's medium with 1 g/l of glucose, supplemented with 10% (v/v) fetal bovine serum, 100 U/ml of penicillin and 100 µg/ml of streptomycin at 37°C and 5% CO2. Fugene 6 (Roche) was used for transient and stable transfections of plasmids according to the recommendations of the manufactures. Selection for stably transfected clones was carried out in the 6-well plates in the presence of 750 µg/ml of HygromycinB (Invitrogen). Fourteen days after transfection, individual clones were transferred into 24 wells and analyzed for expression. Positive clones were maintained in the presence of 500 µg/ml of HygromycinB. For RNAi studies, cells were typically seeded in 12-well or 10-cm plates 24 h prior transfection, such that they reached ∼50% confluency at the time of transfection. Cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol for HeLa cells. The final concentration of siRNAs in the wells/plates was 6.3 pmol/µl. Routinely, three consecutive rounds of transfection were performed in intervals of 3 days. Cells were typically harvested and split 48 h post-transfection. If not indicated otherwise, retransfection and/or experimental analysis were carried out 72 h post-transfection. siRNAs corresponding to nucleotides 486–506 (oligo1) and to nucleotides 449–469 (oligo2) of the hHSC20 cDNA (NM 172002) and a negative control siRNA, which has no homology with any mammalian sequence, were purchased from Qiagen.
Immunoprecipation
HeLa cells stably transfected with hHSC20Flag, or empty vector, were transiently co-transfected with pXShISU2myc (courtesy to Wing-Hang Tong). After 36 h, cells from one 10 cm dish were harvested, washed with 1× PBS and lysed in 100 µl of ice-cold lysis buffer [50 mm Tris–HCl, pH 7.4, 40 mm KCl, 2.5 mm MgCl2, 5% (v/v) glycerol, 0.5% (v/v) NP-40, protease inhibitors (EDTA free, Roche) with 2 mm ADP]. Supernatants from a 10 min centrifugation at 17 000g were diluted 10-fold in lysis buffer w/o NP-40, and incubated at 4°C with 25 µl of α-Flag (M2), α-HA or α-myc-conjugated agarose beads (Sigma). After 2 h, the beads were washed three times with 1 ml of lysis buffer with 0.05% NP-40. Bound protein was eluted with SDS–PAGE sample buffer and subjected to western blot analysis. Immunoprecipitated proteins were probed with specific antibodies against hHSC20, ISCU (48) and hHSPA9 (Abcam, 1:1000).
In vitro transcription/translation
The plasmids pTnTHSC20FL, or pTnTΔ19M20HSC20, which encoded the full-length precursor or a version starting at position 20 of the hHSC20 were used in a transcription/translation system as described (80). Translation reactions were analyzed by SDS–PAGE followed by autoradiography and/or immunoblotting.
Immunofluorescence and confocal microscopy
HeLa cells, seeded on coverslips, were generally immunostained ∼24–36 h after transient transfection. Cells were washed twice in PBS, fixed in 4% paraformaldehyde for 15 min at room temperature (RT), followed by five washes with PBS. Permeabilization was carried out with 0.1% (v/v) Triton X-100 in PBS for 10 min, followed by two washes with PBS. Blocking was conducted in a 10% (v/v) solution of BlokHen (AvesLabs) in PBS for 1 h. After two washes with 1× PBS, cells were immunostained in with the primary antibodies: α-HSC20 (1:100, 4 h), mouse monoclonal α-TOM20 (Abcam, 1:500, 2 h), α-Flag M2 (Sigma, 1:500, 2 h) and rabbit polyclonal α-TOM20 (Santa Cruz, 1:500, 2 h). After three rinses with 1× PBS, the cells were successively labeled with the relevant secondary antibodies Alexa488-conjugated α-mouse (Molecular Probes, 1:1000, 1 h) and Cy3-conjugated α-rabbit (Jackson Labs, 1:1000, 1 h). Slides were mounted in Fluoromount-G (SouthernBiotech). Immunofluorescence was imaged with a confocal microscope system (LSM 510 META; Zeiss). For imaging green fluorescence, the 488 nm line of an argon/krypton laser was used, and emitted light was collected between 500 and 540 nm; for red fluorescence, the 543 nm line of an HeNe laser was used with a 488/543 dichroic mirror, and its emission was collected with a 560 nm long-pass filter.
Immunoblotting
Typically, 25–30 µg of protein/lane were subjected to SDS–PAGE and the proteins transferred to nitrocellulose membranes. The membranes were blocked with 5% non-fat milk (BioRad) in 1× PBS containing 0.1% Triton X-100 and probed with the relevant antibodies followed by horseradish peroxidase-coupled secondary antibodies (Amersham, 1:5000). Detection was performed using SuperSignal substrate (Pierce). A commercially available membrane (INSTA-human tissue blot, Imgenex), on which 20 µg/lane of various human tissue extracts had been electro-transferred, was probed with an affinity purified, polyclonal antiserum against the human HSC20 (1:1000) overnight. Other antibodies used only for western blot were: α-actin (Sigma, 1:5000), α-mitochondrial aconitase (81) (1:1000), α-Pgk1p (Molecular Probes, 1:1000), α-HSP70 (Abcam 2787, 1:1000), α-NFU (1:1000) (75) and α-GLRX5 (1:1000) (53).
Preparation of mitochondria-enriched fractions and cytosolic extracts
Cell fractionation was performed at 4°C using a mitochondrial/cytosol fractionation kit (BioVision). Briefly, cells were homogenized by 50 passes in a chilled glass homogenator in a 100-fold excess of cytosol extraction buffer (kit component) and 0.005% (v/v) digitonin. The homogenate was subjected to low-speed centrifugation (700g). This step was carried out twice. Pellets were discarded and the supernatant was spun for 15 min at 10 000g, resulting in a mitochondrial-enriched pellet and a cytosolic supernatant. The cytosolic extracts were centrifuged again for 5 min at 10 000g and then at 100 000g for 1 h to remove any particulate material. The mitochondrial pellet was washed twice in ∼50-fold excess of cytosolic extraction buffer without digitonin and lysed in 50 mm Tris–HCl, pH 7.4, 100 mm NaCl, 1% (v/v) Triton X-100 and protease inhibitors. The different fractions were analyzed by the standard western blotting with antibodies against hHSC20 (1:1000) and compartment-specific proteins. Mitochondria: Monoclonal α-mitochondrial HSP70 (HSPA9/mortalin) (Abcam, 1:1000) and monoclonal α-SOD2 (Abcam16956, 1:1000). Cytosol: α-NFκB (Santa Cruz; 1:500) and α-IRP1 (82) (1:5000).
Enzymatic assays
Aconitase, ICDH, CS and SDH activities were measured according to the published methods (59,83,84) with modifications. All enzymatic assays were carried out on the 96-well plates. A Multiscan MCC plate reader (Fisher) was used for data acquisition. For aconitase, ICDH and CS, cells were washed once with 1× PBS and lysed in 40 µl of ice-cold L1-buffer [50 mm Tris–HCl at pH 8.0, containing 1% (v/v) NP-40 (Fluka)] and EDTA-free protease inhibitor.
Total aconitase activity was determined by following the reduction in NADP+ at 340 nm in the subsequent ICDH reaction. Ten microliters of the lysate were added to 50 µl of R1-buffer [50 mm Tris-HCl, pH 8.0, with 50 mm NaCl, 5 mm MgCl2, 0.5 mm NADP+, 0.01 U ICDH (NADP+-dependent) from porcine heart (Sigma)]. The reaction was started by the addition of 40 µl of R2-buffer (R1 w/o NADP+ and ICDH, with 2.5 mm cis-aconitate). Activity was expressed as ΔE (340 nm)/Δt and was normalized for protein concentration.
ICDH activity was determined by following the reduction in NADP+ at 340 nm (see above). Five microliters of lysate were added to 45 µl of R3-buffer (R1 w/o ICDH). The reaction was initiated by the addition of 50 µl of R4-buffer (R3 w/o NADP+, with 5 mm isocitrate). Activity was expressed as described above.
CS activity was monitored by following the formation of 5-mercapto-2-nitrobenzoic acid at 412 nm. Five microliters of lysate were added to 45 µl of R5-buffer (100 mm Tris–HCl, pH 7.5, with 2.5 mm oxaloacetate). The reaction started by the addition of 45 µl of R6-buffer (100 mm Tris–HCl, with 0.2 mm DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)], 0.25 mm acetyl-CoA). Activity was defined as ΔE (412 nm)/Δt and normalized for protein concentration.
SDH activity was evaluated based on the succinate-dependent reduction in iodonitrotetrazolium chloride (INT). After washing with 1× PBS, 50 µl of L2-buffer (50 mm Tris–HCl, pH 7.5, containing 7.5 mm KCN, 20 mm sodium azide, 0.075% (w/v) digitonin, 15 g/l of CremophorREL and EDTA-free protease inhibitor were added to each well, and the cells were resuspended by aspiration. After 10 min pre-incubation at RT, 50 µl of R7-buffer (50 mm Tris–HCl, pH 7.5, with 4 mm INT, 15 g/l CremophorREL, with or without 50 mm Na2-succinate) were added to each well. After incubation at 37° C for 60 min, the samples were resuspended, and subsequently the plates were centrifuged (1000g, 2 min). Fifty microliters of the respective supernatants were transferred to a new well, and the absorbance at 492 nm was determined. Activity was defined as E(492 nm) sample with substrate –E(492 nm) sample without substrate and was normalized for protein concentration.
Aconitase gel assay
For in-gel aconitase assays, cells were lysed in buffer containing 20 mm Tris–HCl, pH 7.5, 40 mm KCl, 2 mm Na3-citrate, 1 mm DTT, 0.6 mm MnCl2, 1% (v/v) Triton X-100 and EDTA-free proteinase inhibitor. Assays were performed as previously described (50).
Electro-mobility assay
Gel retardation assays were performed as described (82).
Assay for cell viability
Cell viability was evaluated by MTT [3-(4,5-dimehylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] reduction (85). After specific treatment, cells were incubated at 37°C in 1× HANKS solution, which contained 5 mm glucose and 0.5 mg/ml of MTT. After 30 min, the mixture was taken off and cells were lysed in acidified isopropanol. After 30 min at 4°C, formazan formation was determined in the 96-well plates at 540 nm using a Multiscan MCC plate reader (Fisher).
qRT–PCR analysis
Total RNA from HeLa was isolated and purified by means of the RNAeasy Mini kit and RNase-free DNase set (QIAGEN) following the manufacturer's instructions. A total RNA and random primers of 0.5–1.0 µg were used for reverse transcription reactions (High-Capacity cDNA RT kit, Applied Biosystems, #4368814). qRT–PCR analysis was performed in triplicates on AlfPrizm6000 (Applied Biosystems). Gene-specific primers (Supplementary Material, Table S1) were developed using the GeneRunner program. Five microliters of diluted cDNA (5-fold in ultrapure water) and 62.5 nm forward and reverse primers were used in a final volume of 25 µl using a 2× RT–PCR master mix (Platinum SYBR Green qPCR SuperMix-UDG with ROX, Invitrogen, #11744). Thermal cycling conditions were as follows: 50°C for 2 min (incubation for the AmpErase UNG) preceding the first denaturation step at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1.5 min.
Miscellaneous
Protein concentrations were determined by the Micro BCA kit (Pierce). SDS–PAGE was carried out according to Laemmli (86). Primary structures were compared with respective sequences in the available databases by BLAST search (87). For the prediction of the putative mitochondrial-targeting sequence and cleavage site, the algorithms TargetP and SignalP were employed (88). Sequences were aligned by using the program CLUSTAL X (version 1.64b) (89). The PSIPRED Protein Structure Prediction Server (http://bioinf4.cs.ucl.ac.uk:3000/psipred/) was used for the secondary prediction. Crystal structures were visualized with Swiss-PDB viewer (90).
Data are expressed as the mean and standard deviation (SD). Statistical analysis was performed using the Student's t-test. P-values of <0.05 were considered statistically significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Supplementary Material
ACKNOWLEDGEMENTS
To my father Horst Dieter Uhrigshardt (11 February 1934 – 1 March 2009) in loving memory (H.U.). All authors like to thank: Dr Andrew Dancis for the generous gift of the jac1 shuffle strain and valuable advice. Drs Yogikala Prabhu and George Patterson for their appreciated help with the Y2H studies and the confocal microscope. Drs Ramanujam Hegde and Fanis Missirlis for reagents and insightful discussions of the manuscript. Drs Wing H. Tong, Hong Ye and Rafael Mattera for reagents. Mr Falin Patel for technical assistance. Finally, all former and current colleagues in the laboratory for support and advice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Walsh P., Bursac D., Law Y.C., Cyr D., Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. doi:10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu X.B., Shao Y.M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. doi:10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genevaux P., Georgopoulos C., Kelley W.L. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol. Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. doi:10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 4.Cheetham M.E., Caplan A.J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. doi:10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan C.Y., Lee S., Cyr D.M. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. doi:10.1379/1466-1268(2003)008<0309:MFROHF>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hennessy F., Nicoll W.S., Zimmermann R., Cheetham M.E., Blatch G.L. Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. doi:10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig E.A., Huang P., Aron R., Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. doi:10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- 8.Kelley W.L. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. doi:10.1016/S0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 9.Vickery L.E., Cupp-Vickery J.R. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron–sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol. 2007;42:95–111. doi: 10.1080/10409230701322298. doi:10.1080/10409230701322298. [DOI] [PubMed] [Google Scholar]
- 10.Craig E.A., Marszalek J. A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell. Mol. Life Sci. 2002;59:1658–1665. doi: 10.1007/PL00012493. doi:10.1007/PL00012493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouault T.A., Klausner R.D. Iron–sulfur clusters as biosensors of oxidants and iron. Trends Biochem. Sci. 1996;21:174–177. [PubMed] [Google Scholar]
- 12.Johnson M.K. Iron–sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol. 1998;2:173–181. doi: 10.1016/s1367-5931(98)80058-6. doi:10.1016/S1367-5931(98)80058-6. [DOI] [PubMed] [Google Scholar]
- 13.Beinert H. Iron–sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 14.Kiley P.J., Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 2003;6:181–185. doi: 10.1016/s1369-5274(03)00039-0. doi:10.1016/S1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 15.Imlay J.A. Iron–sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. doi:10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 16.Lill R., Muhlenhoff U. Maturation of iron–sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. doi:10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 17.Rouault T.A., Tong W.H. Iron–sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. doi:10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson D.C., Dean D.R., Smith A.D., Johnson M.K. Structure, function, and formation of biological iron–sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. doi:10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 19.Lill R., Muhlenhoff U. Iron–sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu. Rev. Cell. Dev. Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. doi:10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 20.Lill R., Dutkiewicz R., Elsasser H.P., Hausmann A., Netz D.J., Pierik A.J., Stehling O., Urzica E., Muhlenhoff U. Mechanisms of iron–sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes. Biochim. Biophys. Acta. 2006;1763:652–667. doi: 10.1016/j.bbamcr.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Strain J., Lorenz C.R., Bode J., Garland S., Smolen G.A., Ta D.T., Vickery L.E., Culotta V.C. Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron–sulfur cluster assembly. J. Biol. Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. doi:10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- 22.Schilke B., Voisine C., Beinert H., Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. doi:10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voisine C., Cheng Y.C., Ohlson M., Schilke B., Hoff K., Beinert H., Marszalek J., Craig E.A. Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2001;98:1483–1488. doi: 10.1073/pnas.98.4.1483. doi:10.1073/pnas.98.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim R., Saxena S., Gordon D.M., Pain D., Dancis A. J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe-S cluster proteins. J. Biol. Chem. 2001;276:17524–17532. doi: 10.1074/jbc.M010695200. doi:10.1074/jbc.M010695200. [DOI] [PubMed] [Google Scholar]
- 25.Lutz T., Westermann B., Neupert W., Herrmann J.M. The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J. Mol. Biol. 2001;307:815–825. doi: 10.1006/jmbi.2001.4527. doi:10.1006/jmbi.2001.4527. [DOI] [PubMed] [Google Scholar]
- 26.Hoff K.G., Cupp-Vickery J.R., Vickery L.E. Contributions of the LPPVK motif of the iron–sulfur template protein IscU to interactions with the Hsc66–Hsc20 chaperone system. J. Biol. Chem. 2003;278:37582–37589. doi: 10.1074/jbc.M305292200. doi:10.1074/jbc.M305292200. [DOI] [PubMed] [Google Scholar]
- 27.Tokumoto U., Nomura S., Minami Y., Mihara H., Kato S., Kurihara T., Esaki N., Kanazawa H., Matsubara H., Takahashi Y. Network of protein–protein interactions among iron–sulfur cluster assembly proteins in Escherichia coli. J. Biochem. 2002;131:713–719. doi: 10.1093/oxfordjournals.jbchem.a003156. [DOI] [PubMed] [Google Scholar]
- 28.Knieszner H., Schilke B., Dutkiewicz R., D'Silva P., Cheng S., Ohlson M., Craig E.A., Marszalek J. Compensation for a defective interaction of the hsp70 ssq1 with the mitochondrial Fe-S cluster scaffold isu. J. Biol. Chem. 2005;280:28966–28972. doi: 10.1074/jbc.M503031200. doi:10.1074/jbc.M503031200. [DOI] [PubMed] [Google Scholar]
- 29.Dutkiewicz R., Schilke B., Cheng S., Knieszner H., Craig E.A., Marszalek J. Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. J. Biol. Chem. 2004;279:29167–29174. doi: 10.1074/jbc.M402947200. doi:10.1074/jbc.M402947200. [DOI] [PubMed] [Google Scholar]
- 30.Dutkiewicz R., Schilke B., Knieszner H., Walter W., Craig E.A., Marszalek J. Ssq1, a mitochondrial Hsp70 involved in iron–sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J. Biol. Chem. 2003;278:29719–29727. doi: 10.1074/jbc.M303527200. doi:10.1074/jbc.M303527200. [DOI] [PubMed] [Google Scholar]
- 31.Muhlenhoff U., Gerber J., Richhardt N., Lill R. Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. doi:10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutkiewicz R., Marszalek J., Schilke B., Craig E.A., Lill R., Muehlenhoff U. The Hsp70 chaperone Ssq1p is dispensable for iron–sulfur cluster formation on the scaffold protein Isu1p. J. Biol. Chem. 2006;281:7801–7808. doi: 10.1074/jbc.M513301200. doi:10.1074/jbc.M513301200. [DOI] [PubMed] [Google Scholar]
- 33.Bonomi F., Iametti S., Ta D., Vickery L.E. Multiple turnover transfer of [2Fe2S] clusters by the iron–sulfur cluster assembly scaffold proteins IscU and IscA. J. Biol. Chem. 2005;280:29513–29518. doi: 10.1074/jbc.M504344200. doi:10.1074/jbc.M504344200. [DOI] [PubMed] [Google Scholar]
- 34.Chandramouli K., Johnson M.K. HscA and HscB stimulate [2Fe–2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. doi:10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unciuleac M.C., Chandramouli K., Naik S., Mayer S., Huynh B.H., Johnson M.K., Dean D.R. In vitro activation of apo-aconitase using a [4Fe–4S] cluster-loaded form of the IscU [Fe–S] cluster scaffolding protein. Biochemistry. 2007;46:6812–6821. doi: 10.1021/bi6026665. doi:10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- 36.Johnson D.C., Unciuleac M.C., Dean D.R. Controlled expression and functional analysis of iron–sulfur cluster biosynthetic components within Azotobacter vinelandii. J. Bacteriol. 2006;188:7551–7561. doi: 10.1128/JB.00596-06. doi:10.1128/JB.00596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouault T.A., Tong W.H. Opinion: iron–sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell. Biol. 2005;6:345–351. doi: 10.1038/nrm1620. doi:10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- 38.Huynen M.A., Snel B., Bork P., Gibson T.J. The phylogenetic distribution of frataxin indicates a role in iron–sulfur cluster protein assembly. Hum. Mol. Genet. 2001;10:2463–2468. doi: 10.1093/hmg/10.21.2463. doi:10.1093/hmg/10.21.2463. [DOI] [PubMed] [Google Scholar]
- 39.Schilke B., Williams B., Knieszner H., Pukszta S., D'Silva P., Craig E.A., Marszalek J. Evolution of mitochondrial chaperones utilized in Fe-S cluster biogenesis. Curr. Biol. 2006;16:1660–1665. doi: 10.1016/j.cub.2006.06.069. doi:10.1016/j.cub.2006.06.069. [DOI] [PubMed] [Google Scholar]
- 40.Wadhwa R., Taira K., Kaul S.C. An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: what, when, and where? Cell Stress Chaperones. 2002;7:309–316. doi: 10.1379/1466-1268(2002)007<0309:ahfcmm>2.0.co;2. doi:10.1379/1466-1268(2002)007<0309:AHFCMM>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deocaris C.C., Kaul S.C., Wadhwa R. On the brotherhood of the mitochondrial chaperones mortalin and heat shock protein 60. Cell Stress Chaperones. 2006;11:116–128. doi: 10.1379/CSC-144R.1. doi:10.1379/CSC-144R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun G., Gargus J.J., Ta D.T., Vickery L.E. Identification of a novel candidate gene in the iron–sulfur pathway implicated in ataxia-susceptibility: human gene encoding HscB, a J-type co-chaperone. J. Hum. Genet. 2003;48:415–419. doi: 10.1007/s10038-003-0048-9. doi:10.1007/s10038-003-0048-9. [DOI] [PubMed] [Google Scholar]
- 43.Bitto E., Bingman C.A., Bittova L., Kondrashov D.A., Bannen R.M., Fox B.G., Markley J.L., Phillips G.N.J. Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain. J. Biol. Chem. 2008;283:30184–30192. doi: 10.1074/jbc.M804746200. doi:10.1074/jbc.M804746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cupp-Vickery J.R., Vickery L.E. Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli. J. Mol. Biol. 2000;304:835–845. doi: 10.1006/jmbi.2000.4252. doi:10.1006/jmbi.2000.4252. [DOI] [PubMed] [Google Scholar]
- 45.Hoff K.G., Ta D.T., Tapley T.L., Silberg J.J., Vickery L.E. Hsc66 substrate specificity is directed toward a discrete region of the iron–sulfur cluster template protein IscU. J. Biol. Chem. 2002;277:27353–27359. doi: 10.1074/jbc.M202814200. doi:10.1074/jbc.M202814200. [DOI] [PubMed] [Google Scholar]
- 46.Andrew A.J., Dutkiewicz R., Knieszner H., Craig E.A., Marszalek J. Characterization of the interaction between the J-protein Jac1p and the scaffold for Fe-S cluster biogenesis, Isu1p. J. Biol. Chem. 2006;281:14580–14587. doi: 10.1074/jbc.M600842200. doi:10.1074/jbc.M600842200. [DOI] [PubMed] [Google Scholar]
- 47.Fuzery A.K., Tonelli M., Ta D.T., Cornilescu G., Vickery L.E., Markley J.L. Solution structure of the iron–sulfur cluster cochaperone HscB and its binding surface for the iron–sulfur assembly scaffold protein IscU. Biochemistry. 2008;47:9394–9404. doi: 10.1021/bi800502r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong W.H., Rouault T. Distinct iron–sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 2000;19:5692–5700. doi: 10.1093/emboj/19.21.5692. doi:10.1093/emboj/19.21.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li K., Tong W.H., Hughes R.M., Rouault T.A. Roles of the mammalian cytosolic cysteine desulfurase, ISCS, and scaffold protein, ISCU, in iron–sulfur cluster assembly. J. Biol. Chem. 2006;281:12344–12351. doi: 10.1074/jbc.M600582200. doi:10.1074/jbc.M600582200. [DOI] [PubMed] [Google Scholar]
- 50.Tong W.H., Rouault T.A. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron–sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. doi:10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Hoff K.G., Silberg J.J., Vickery L.E. Interaction of the iron–sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:7790–7795. doi: 10.1073/pnas.130201997. doi:10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silberg J.J., Vickery L.E. Kinetic characterization of the ATPase cycle of the molecular chaperone Hsc66 from Escherichia coli. J. Biol. Chem. 2000;275:7779–7786. doi: 10.1074/jbc.275.11.7779. doi:10.1074/jbc.275.11.7779. [DOI] [PubMed] [Google Scholar]
- 53.Ye H., Jeong S.Y., Ghosh M.C., Kovtunovych G., Silvestri L., Ortillo D., Uchida N., Tisdale J., Camaschella C., Rouault T.A. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J. Clin. Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye H., Rouault T.A. Human iron–sulfur cluster assembly, cellular iron homeostasis and disease. Biochemistry. 2010;49:4945–4956. doi: 10.1021/bi1004798. doi:10.1021/bi1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stehling O., Elsasser H.P., Bruckel B., Muhlenhoff U., Lill R. Iron–sulfur protein maturation in human cells: evidence for a function of frataxin. Hum. Mol. Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. doi:10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 56.Fosset C., Chauveau M.J., Guillon B., Canal F., Drapier J.C., Bouton C. RNA silencing of mitochondrial m-Nfs1 reduces Fe-S enzyme activity both in mitochondria and cytosol of mammalian cells. J. Biol. Chem. 2006;281:25398–25406. doi: 10.1074/jbc.M602979200. doi:10.1074/jbc.M602979200. [DOI] [PubMed] [Google Scholar]
- 57.Song D., Tu Z., Lee F.S. Human ISCA1 interacts with IOP1/NARFL and functions in both cytosolic and mitochondrial iron–sulfur protein biogenesis. J. Biol. Chem. 2009;284:35297–35307. doi: 10.1074/jbc.M109.040014. doi:10.1074/jbc.M109.040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beinert H., Kennedy M.C., Stout D.C. Aconitase as iron–sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 1996;96:2335–2373. doi: 10.1021/cr950040z. doi:10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 59.Kennedy M.C., Emptage M.H., Dreyer J.L., Beinert H. The role of iron in the activation-inactivation of aconitase. J. Biol. Chem. 1983;258:11098–11105. [PubMed] [Google Scholar]
- 60.Brown N.M., Kennedy M.C., Antholine W.E., Eisenstein R.S., Walden W.E. Detection of a [3Fe–4S] cluster intermediate of cytosolic aconitase in yeast expressing iron regulatory protein 1. Insights into the mechanism of Fe-S cluster cycling. J. Biol. Chem. 2002;277:7246–7254. doi: 10.1074/jbc.M110282200. doi:10.1074/jbc.M110282200. [DOI] [PubMed] [Google Scholar]
- 61.Jang S., Imlay J.A. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron–sulfur enzymes. J. Biol. Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. doi:10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rouault T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006;2:406–414. doi: 10.1038/nchembio807. doi:10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 63.Starzynski R.R., Lipinski P., Drapier J.C., Diet A., Smuda E., Bartlomiejczyk T., Gralak M.A., Kruszewski M. Down-regulation of iron regulatory protein 1 activities and expression in superoxide dismutase 1 knock-out mice is not associated with alterations in iron metabolism. J. Biol. Chem. 2005;280:4207–4212. doi: 10.1074/jbc.M411055200. doi:10.1074/jbc.M411055200. [DOI] [PubMed] [Google Scholar]
- 64.Linke K., Wolfram T., Bussemer J., Jakob U. The roles of the two zinc binding sites in DnaJ. J. Biol. Chem. 2003;278:44457–44466. doi: 10.1074/jbc.M307491200. doi:10.1074/jbc.M307491200. [DOI] [PubMed] [Google Scholar]
- 65.Martinez-Yamout M., Legge G.B., Zhang O., Wright P.E., Dyson H.J. Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J. Mol. Biol. 2000;300:805–818. doi: 10.1006/jmbi.2000.3923. doi:10.1006/jmbi.2000.3923. [DOI] [PubMed] [Google Scholar]
- 66.Zhai P., Stanworth C., Liu S., Silberg J.J. The human escort protein Hep binds to the ATPase domain of mitochondrial hsp70 and regulates ATP hydrolysis. J. Biol. Chem. 2008;283:26098–26106. doi: 10.1074/jbc.M803475200. doi:10.1074/jbc.M803475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morgan B., Ang S.K., Yan G., Lu H. Zinc can play chaperone-like and inhibitor roles during import of mitochondrial small Tim proteins. J. Biol. Chem. 2009;284:6818–6825. doi: 10.1074/jbc.M808691200. doi:10.1074/jbc.M808691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanford R.A., Cole J.R., Tiedje J.M. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 2002;68:893–900. doi: 10.1128/AEM.68.2.893-900.2002. doi:10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parro V., Moreno-Paz M. Gene function analysis in environmental isolates: the nif regulon of the strict iron oxidizing bacterium Leptospirillum ferrooxidans. Proc. Natl Acad. Sci. USA. 2003;100:7883–7888. doi: 10.1073/pnas.1230487100. doi:10.1073/pnas.1230487100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Land T., Rouault T.A. Targeting of a human iron–sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell. 1998;2:807–815. doi: 10.1016/s1097-2765(00)80295-6. doi:10.1016/S1097-2765(00)80295-6. [DOI] [PubMed] [Google Scholar]
- 71.Condo I., Ventura N., Malisan F., Tomassini B., Testi R. A pool of extramitochondrial frataxin that promotes cell survival. J. Biol. Chem. 2006;281:16750–16756. doi: 10.1074/jbc.M511960200. doi:10.1074/jbc.M511960200. [DOI] [PubMed] [Google Scholar]
- 72.Biederbick A., Stehling O., Rosser R., Niggemeyer B., Nakai Y., Elsasser H.P., Lill R. Role of human mitochondrial Nfs1 in cytosolic iron–sulfur protein biogenesis and iron regulation. Mol. Cell. Biol. 2006;26:5675–5687. doi: 10.1128/MCB.00112-06. doi:10.1128/MCB.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cavadini P., Biasiotto G., Poli M., Levi S., Verardi R., Zanella I., Derosas M., Ingrassia R., Corrado M., Arosio P. RNA silencing of the mitochondrial ABCB7 transporter in HeLa cells causes an iron-deficient phenotype with mitochondrial iron overload. Blood. 2007;109:3552–3559. doi: 10.1182/blood-2006-08-041632. doi:10.1182/blood-2006-08-041632. [DOI] [PubMed] [Google Scholar]
- 74.Andrew A.J., Song J.Y., Schilke B., Craig E.A. Posttranslational regulation of the scaffold for Fe-S cluster biogenesis, Isu. Mol. Biol. Cell. 2008;19:5259–5266. doi: 10.1091/mbc.E08-06-0622. doi:10.1091/mbc.E08-06-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tong W.H., Jameson G.N., Huynh B.H., Rouault T.A. Subcellular compartmentalization of human Nfu, an iron–sulfur cluster scaffold protein, and its ability to assemble a [4Fe–4S] cluster. Proc. Natl Acad. Sci. USA. 2003;100:9762–9767. doi: 10.1073/pnas.1732541100. doi:10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pearce L.L., Martinez-Bosch S., Manzano E.L., Winnica D.E., Epperly M.W., Peterson J. The resistance of electron-transport chain Fe–S clusters to oxidative damage during the reaction of peroxynitrite with mitochondrial complex II and rat-heart pericardium. Nitric Oxide. 2009;20:135–142. doi: 10.1016/j.niox.2008.12.001. doi:10.1016/j.niox.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barbet F., Hakiki S., Orssaud C., Gerber S., Perrault I., Hanein S., Ducroq D., Dufier J.L., Munnich A., Kaplan J., Rozet J.M. A third locus for dominant optic atrophy on chromosome 22q. J. Med. Genet. 2005;42 doi: 10.1136/jmg.2004.025502. electronic letter e1. doi:10.1136/jmg.2004.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bazylinski D.A., Frankel R.B. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. doi:10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 79.Hiraishi A., Kishimoto N., Kosako Y., Wakao N., Tano T. Phylogenetic position of the menaquinone-containing acidophilic chemo-organotroph Acidobacterium capsulatum. FEMS Microbiol. Lett. 1995;132:91–94. doi: 10.1111/j.1574-6968.1995.tb07816.x. doi:10.1111/j.1574-6968.1995.tb07816.x. [DOI] [PubMed] [Google Scholar]
- 80.Fons R.D., Bogert B.A., Hegde R.S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell. Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. doi:10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim H.Y., LaVaute T., Iwai K., Klausner R.D., Rouault T.A. Identification of a conserved and functional iron-responsive element in the 5′UTR of mammalian mitochondrial aconitase. J. Biol. Chem. 1996;271:24226–24230. doi: 10.1074/jbc.271.39.24226. [DOI] [PubMed] [Google Scholar]
- 82.Meyron-Holtz E.G., Ghosh M.C., Iwai K., LaVaute T., Brazzolotto X., Berger U.V., Land W., Ollivierre-Wilson H., Grinberg A., Love P., Rouault T.A. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. doi:10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parvin R., Atkinson D.E. Purification and some properties of yeast citrate synthase. Arch. Biochem. Biophys. 1968;128:528–533. doi: 10.1016/0003-9861(68)90060-x. doi:10.1016/0003-9861(68)90060-X. [DOI] [PubMed] [Google Scholar]
- 84.Munujos P., Coll-Canti J., Gonzalez-Sastre F., Gella F.J. Assay of succinate dehydrogenase activity by a colorimetric-continuous method using iodonitrotetrazolium chloride as electron acceptor. Anal. Biochem. 1993;212:506–509. doi: 10.1006/abio.1993.1360. doi:10.1006/abio.1993.1360. [DOI] [PubMed] [Google Scholar]
- 85.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. doi:10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 86.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. doi:10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 87.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. doi:10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 89.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. doi:10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. doi:10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 91.Mattera R., Tsai Y.C., Weissman A.M., Bonifacino J.S. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc-finger domain. J. Biol. Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. doi:10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- 92.Meriin A.B., Gabai V.L., Yaglom J., Shifrin V.I., Sherman M.Y. Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J. Biol. Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. doi:10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- 93.Bihlmaier K., Mesecke N., Kloeppel C., Herrmann J.M. The disulfide relay of the intermembrane space of mitochondria: an oxygen-sensing system? Ann. N Y Acad. Sci. 2008;1147:293–302. doi: 10.1196/annals.1427.005. [DOI] [PubMed] [Google Scholar]
- 94.Xu X.M., Lin H., Latijnhouwers M., Moller S.G. Dual localized AtHscB involved in iron sulfur protein biogenesis in Arabidopsis. PLoS One. 2009;4:e7662. doi: 10.1371/journal.pone.0007662. doi:10.1371/journal.pone.0007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Acquaviva F., De Biase I., Nezi L., Ruggiero G., Tatangelo F., Pisano C., Monticelli A., Garbi C., Acquaviva A.M., Cocozza S. Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2. J. Cell. Sci. 2005;118:3917–3924. doi: 10.1242/jcs.02516. doi:10.1242/jcs.02516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.