Abstract
Twenty-two amino acid substitutions at seven conserved amino acid residues in the acetohydroxyacid synthase (AHAS) gene have been identified to date that confer target-site resistance to AHAS-inhibiting herbicides in biotypes of field-evolved resistant weed species. However, the effect of resistance mutations on AHAS functionality and plant growth has been investigated for only a very few mutations. This research investigates the effect of various AHAS resistance mutations in Lolium rigidum on AHAS functionality and plant growth. The enzyme kinetics of AHAS from five purified L. rigidum populations, each homozygous for the resistance mutations Pro-197-Ala, Pro-197-Arg, Pro-197-Gln, Pro-197-Ser or Trp-574-Leu, were characterized and the pleiotropic effect of three mutations on plant growth was assessed via relative growth rate analysis. All these resistance mutations endowed a herbicide-resistant AHAS and most resulted in higher extractable AHAS activity, with no-to-minor changes in AHAS kinetics. The Pro-197-Arg mutation slightly (but significantly) increased the Km for pyruvate and remarkably increased sensitivity to feedback inhibition by branched chain amino acids. Whereas the Pro-197-Ser and Trp-574-Leu mutations exhibited no significant effects on plant growth, the Pro-197-Arg mutation resulted in lower growth rates. It is clear that, at least in L. rigidum, these five AHAS resistance mutations have no major impact on AHAS functionality and hence probably no plant resistance costs. These results, in part, explain why so many Pro-197 AHAS resistance mutations in AHAS have evolved and why the Pro-197-Ser and the Trp-574-Leu AHAS resistance mutations are frequently found in many weed species.
Keywords: AHAS resistance mutation, enzyme kinetics, herbicide resistance, Lolium rigidum, relative growth rate
Introduction
Acetohydroxyacid synthase (AHAS, EC 2.2.1.6), also referred to as acetolactate synthase (ALS), is the first enzyme in the pathway for the biosynthesis of the branched chain amino acids valine, leucine, and isoleucine (reviewed by Duggleby and Pang, 2000). AHAS is the common target site of five AHAS-inhibiting herbicide chemistries, namely sulfonylurea (SU), imidazolinone (IMI), triazolopyrimidine, pyrimidinyl-thiobenzoates, and sulphonyl-aminocarbonyl-triazolinone (Saari et al., 1994, and references therein; Santel et al., 1999). Various AHAS-inhibiting herbicides (also referred to as AHAS herbicides) have been in widespread commercial use in world agriculture for nearly three decades. Global and persistent use of AHAS-inhibiting herbicides has consequently resulted in the rapid evolution of many AHAS herbicide-resistant weed populations. Worldwide, there are biotypes of 108 weed species with evolved AHAS herbicide resistance (Heap 2010, http://www.weedscience.com). In many cases, evolved resistance is target site-based, due to resistant plants having one or more specific resistance-endowing single point mutations in the target AHAS gene. A total of 22 resistance-endowing gene mutations at seven conserved amino acid residues in the AHAS gene have so far been identified in field-evolved resistant weed biotypes (reviewed by Tranel and Wright, 2002; Powles and Yu, 2010; also see Tranel et al., 2010, http://www.weedscience.org). To date, mutations at Pro-197 (especially the Pro-197-Ser) are most commonly reported and the Trp-574-Leu mutation is also frequently identified (Powles and Yu, 2010; Tranel et al., 2010, http://www.weedscience.org).
It is known that certain gene mutations endowing target site-based herbicide resistance have adverse pleiotropic effects on plant growth and fitness (reviewed in Vila-Aiub et al., 2009). A significant contributor to resistance costs associated with target site herbicide-resistance genes may arise if a resistance mutation causes change in enzyme functionality (e.g. impaired enzyme activity, reduced substrate affinity, altered feedback inhibition) resulting in insufficient or excessive product biosynthesis (Vila-Aiub et al., 2009). The individual impact of the known 22 resistance-endowing AHAS gene mutations on AHAS functionality and their concomitant effect on plant fitness remains unknown and needs empirical evaluation (Powles and Yu, 2010). Resistance-conferring amino acid substitutions are structural changes in AHAS that prevent or limit effective herbicide binding (McCourt et al., 2006; Duggleby et al., 2008). Therefore it is very likely that some resistance-conferring mutations would impair AHAS functionality. Indeed, for other herbicides, it is known that resistance mutations reduce enzyme activity (e.g. Healy-Fried et al., 2007, for EPSPS mutations; Yu et al., 2007b, for ACCase mutations), but the situation is complex for AHAS. Depending on plant species and the particular AHAS herbicide resistance-endowing amino acid substitution, there are studies showing reduced (Eberlein et al., 1997, 1999; Ashigh and Tardif, 2007), increased (Boutsalis et al., 1999; Purrington and Bergelson, 1999; Yu et al., 2003; 2007a) or unchanged (Boutsalis et al., 1999; Preston et al., 2006) AHAS activity. It is important to note that, of the 22 known AHAS resistance-conferring amino acid substitutions in field-evolved resistant weed species, only a few have been evaluated for their impact on both AHAS functionality and plant growth (Eberlein et al., 1997, 1999; Alcocer-Ruthling et al., 1992).
To conduct precise comparative studies on enzyme kinetics and their effects on the growth of resistant versus susceptible plants, it is necessary to have genetically well characterized plants. Here Lolium rigidum populations were generated with all plants individually homozygous for the resistance mutations Pro-197-Ala, Pro-197-Arg, Pro-197-Gln, Pro-197-Ser or Trp-574-Leu. The effect of these mutations on AHAS functionality and on plant growth was examined by determining AHAS kinetics and relative growth rate compared with the wild-type enzyme- and herbicide-susceptible populations. This is the first systematic study assessing the effect of various field-evolved AHAS resistance mutations on both AHAS kinetic properties and plant growth.
Materials and methods
Plant material
Information on the eight Lolium rigidum populations used in the AHAS kinetics study is provided in Table 1. Studies were conducted with the known AHAS herbicide-resistant L. rigidum populations WLR1 and RSG (Christopher et al., 1992; Yu et al., 2008). From these two resistant populations, five sub-populations were created that were each individually homozygous for the resistance mutations Pro-197-Ala, Pro-197-Arg, Pro-197-Gln, Pro-197-Ser, and Trp-574-Leu (Table 1). This was achieved by identifying (via sequencing and PCR-based marker analysis) at least six plants homozygous for each of these mutations, growing these homozygous plants to maturity, and allowing bulk-cross pollinating (in pollen-proof cages) to produce seed. Homozygosity of the progeny plants for the specific AHAS mutation in each purified population and the absence of other AHAS mutations was confirmed using PCR-based marker analysis (Yu et al., 2008). L. rigidum populations known to be susceptible to AHAS herbicides (VLR1) or possessing herbicide susceptible AHAS (WALR60, WALR70), (hereafter referred to as S1, S2, S3 or collectively as S) were used as wild type controls (Yu et al., 2009; Owen and Powles, 2010). For herbicide treatments and enzyme assays, seedlings of each population were grown in plastic trays containing potting mixture in a naturally illuminated and temperature controlled (25–30/20–25 °C, day/night) greenhouse with regular watering and fertilization. Two additional herbicide-susceptible L. rigidum populations (H3/6, referred as S4, and H4/6 as S5) collected from agricultural fields (Owen et al., 2007; M Owen and Q Yu, unpublished results) were used for plant growth analysis (see ‘Plant Growth Assessment’ below).
Table 1.
Information on the Lolium rigidum populations used and their growth response to the two AHAS inhibiting herbicides sulfometuron and imazapyr
| Population name | AHAS mutation | Original populations | % Survival |
|
| Sulfometuron | Imazapyr | |||
| (20 g ha−1) | (200 g ha−1) | |||
| Control populations | ||||
| S1 | Wild type | VLR1 | 0 | 0 |
| S2 | Wild type | WALR60 | 0 | 0 |
| S3 | Wild type | WALR70 | 0 | 0 |
| Purified resistant populations | ||||
| R-197-Ala | Pro-197-Ala | WLR1 | 100 | 0 |
| R-197-Arg | Pro-197-Arg | WLR1 | 100 | 0 |
| R-197-Gln | Pro-197-Gln | WLR1 | 100 | 0 |
| R-197-Ser | Pro-197-Ser | WLR1 | 100 | 0 |
| R-574-Leu | Trp-574-Leu | RSG | 100 | 100 |
Seedlings (90–100 per treatment) were treated at the 2–3-leaf stage and mortality was determined 21 d after treatment. R, resistant; S, susceptible.
Herbicide treatments
To confirm herbicide resistance and the cross-resistance pattern for plants with each individual AHAS resistance mutation, seedlings (90–100) of the eight populations used in this study were treated at the 2–3 leaf stage with the sulfonylurea (SU) herbicide sulfometuron (20 g ha−1) and the imidazolinone (IMI) herbicide imazapyr (200 g ha−1), respectively, using a spray cabinet with a moving-boom delivering 106 l ha−1 water at a pressure of 200 kPa through two flat fan nozzles. These AHAS herbicide rates are lethal to the S populations only.
AHAS in vitro assay
Earlier work reported much lower extractable AHAS activity in S compared to resistant (R) L. rigidum populations (Yu et al., 2004). Considerable optimization of the AHAS assay revealed that, specifically for the S populations, reducing the extraction time achieves higher AHAS activity and, especially, that the Sephadex G25 column desalting step could be eliminated without effects on AHAS activity (also see Singh et al., 1988, for the effect of ammonium sulphate concentration on AHAS activity), Km, and the sensitivity to herbicide and branched chain amino acid inhibition. This modified protocol improved extractable AHAS activity and reproducibility, especially for the susceptible L. rigidum plants.
At the 2–3 leaf stage, the above-ground leaf material (about 4 g) was harvested at soil level from each population (at least 20 seedlings per harvest), snap-frozen in liquid nitrogen and stored at –80 °C. The AHAS in vitro assay was conducted according to the method of Yu et al. (2004) with modifications. The frozen material was ground to a fine powder with a mortar and pestle in liquid nitrogen and homogenized in 3 vols of cold grinding buffer containing 0.1 M K2HPO4 (pH 7.5), 0.5 mM MgCl2, 0.5 mM thiamine pyrophosphate (TPP), 10 μM flavin adenine dinucleotide (FAD), 10 mM sodium pyruvate, 10% v/v glycerol, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulphonyl fluoride (PMSF) and 0.5% soluble PVP. The homogenate was filtered through two layers of Miracloth and centrifuged at 27 000 g for 15 min. About 6–7 ml supernatant was brought to 50% saturation with (NH4)2SO4 by drop-wise addition of an equal volume of 100% (NH4)2SO4, and the solution was allowed to stand on ice for 10 min with low-speed stirring. The protein was then precipitated at 27 000 g for 20 min. The pellet was redissolved in 4.5 ml reaction buffer containing 50 mM HEPES [N-(2-hydroxyethyl) piperazine-N′-(2-ethanesulphonic acid)], pH 7.5, 200 mM sodium pyruvate, 20 mM MgCl2, 2 mM TPP, and 20 μM FAD and was immediately used in the assay. The reaction mixture contained 100 μl enzyme extract and 100 μl of AHAS inhibitor solution (herbicide or amino acid prepared in water) and was incubated at 37 °C for 60 min. The reaction was stopped with 40 μl of 6 N H2SO4 and incubated at 60 °C for 15 min to convert acetolactate to acetoin. Then, 190 μl of creatine solution (0.55%) and 190 μl of α-naphthol solution (5.5% in 5 N NaOH) were added and the mixture incubated at 60 °C for 15 min. Enzyme activity was determined colorimetrically (530 nm) by measuring the amount of acetoin formed using commercial acetoin as a standard. The protein concentration of the crude extract was measured by the Bradford method. Reaction mixtures that were acidified (40 μl of 6 N H2SO4) prior to the addition of enzyme were used as standard background controls for non-enzymatic formation of acetoine.
As pyruvate decarboxylase (PDC) (Muhitch, 1988) and other multiple acetoin-forming enzymes (Forlani et al., 1999) in plant tissues may interfere with the assay, the contribution of the direct formation of acetoin (not via acidic conversion) by non-AHAS enzyme activities was determined using NaOH (40 μl 4 N NaOH) to terminate the reaction instead of 6 N H2SO4 (Tanaka, 2003; Pornprom et al., 2005; DL Shaner, personal communication). Pilot studies showed that other acetoin-forming, non-AHAS activity in shoot tissue of L. rigidum plants accounts for 15–30% of the total apparent AHAS activity measured using the standard acid control. Furthermore, non-AHAS activity (PDC) has been shown to contribute to reduced AHAS sensitivity to herbicide or branched chain amino acid feedback inhibition in maize kernels (Muhitch, 1988; DL Shaner personal communication). Therefore, it is suggested that background control for non-AHAS activities in the in vitro AHAS assay is necessary, at least for L. rigidum. In addition, as preliminary experiments demonstrated that AHAS activity in stem tissue is 2–3-fold higher than in the leaf blade, 3-week-old seedlings were always used and all above-ground material was pooled.
For the AHAS herbicide sensitivity test, the technical grade SU herbicide sulfometuron (dissolved in 20% acetone) was used at final concentrations of 0, 0.001, 0.01, 0.1, 1.0, 10, 100, and 500 μM. For Km (pyruvate) determination, pyruvate was omitted from the extraction and reaction buffers, and final concentrations of 0.39–100 mM were used in the reaction mixtures. Both the H2SO4 and NaOH controls were used for each pyruvate concentration.
Plant growth assessment
Populations used for growth analysis included three herbicide-sensitive populations, S1 (VLR1), S4 (H3/6), and S5 (H4/6), and resistant populations homozygous for Pro-197-Ser, Pro-197-Arg, or Trp-574-Leu AHAS-resistance mutations. Seeds of these populations were germinated on 0.6% agar (w/v) solidified water for 5 d at alternating 25/15 °C cycle with a 12 h photoperiod. Seventy-five uniform seedlings from each population were transplanted to single plastic pots containing potting soil (50% peatmoss and 50% river sand) and grown in a greenhouse at day/night temperatures of 20/15 °C with natural sunlight. Pots were watered and rearranged regularly to randomize environmental differences in the greenhouse. Above-ground biomass from about 25 individual plants of each population were harvested 14, 28, and 42 d after transplanting, oven-dried at 70 °C for 3 d and then weighed for dry biomass estimation.
Statistics
In vitro herbicide dose response and enzyme kinetics analysis:
The I50 value (herbicide concentration causing 50% inhibition of the enzyme activity) was estimated using non-linear regression analysis (Yu et al., 2007b). Km values were calculated using non-linear regression analysis by fitting the data to the Michaelis–Menten equation ν=VS/(Km+S), where S is the concentration of the substrate pyruvate, ν is the reaction velocity at any pyruvate concentration, and V is the maximal reaction velocity. Each assay contained two technical replicates and at least two to three independent enzyme extracts were used for each assay set. Data were subjected to analysis of variance using SAS Software (OnlineDoc® 9.1.3., Cary, NC., SAS Institute Inc. 2004). Means were separated using Fisher's protected least significant difference (LSD) test at the 5% level of probability.
Plant growth assessment:
The inclusion of various plant harvests enabled the estimation of plant relative growth rates (RGR) for the AHAS susceptible (S1, S4, and S5) and resistant (Pro-197-Ser, Pro-197-Arg, and Trp-574-Leu) populations. The inclusion of three susceptible populations for RGR estimation and comparison was planned to minimize the effect of genetic background differences between AHAS-susceptible and -resistant populations (Cousens et al., 1997; Vila-Aiub et al., 2009).
The unbiased formula proposed by Hoffmann and Poorter (2002) was used to determine RGR. The variance (V) of RGR was estimated according to Causton and Venus (1981):
where lnW2 is the mean of the ln-transformed plant weights at harvest time 2; lnW1 is the mean of the ln-transformed plant weights at harvest time 1. The degree of freedom associated with RGR is n–2, where n is the total number of plants in two harvest intervals. One-way analysis of variance (ANOVA) with Dunnett's post-test was performed to compare RGR estimates between AHAS S and R populations (GraphPad Prism 5.0, San Diego, California, USA).
Results
Herbicide treatment confirmed AHAS herbicide resistance of each purified L. rigidum population
Seedlings (90–100) of S and each purified R population were treated with the SU herbicide sulfometuron and with the IMI herbicide imazapyr. As expected, all S plants were killed (Table 1). All R plants with the various Pro-197 resistance mutations survived the SU herbicide sulfometuron but were killed by the IMI herbicide imazapyr, whereas the R plants with the Trp-574-Leu mutation survived both SU and IMI herbicides (Table 1). This confirms several studies which demonstrate that Pro-197 mutations confer resistance only to SU herbicides whereas the Trp-574-Leu confers resistance to both SU and IMI herbicides (Tranel et al., 2010, http://www.weedscience.org).
In vitro AHAS assays confirmed an herbicide-resistant AHAS enzyme
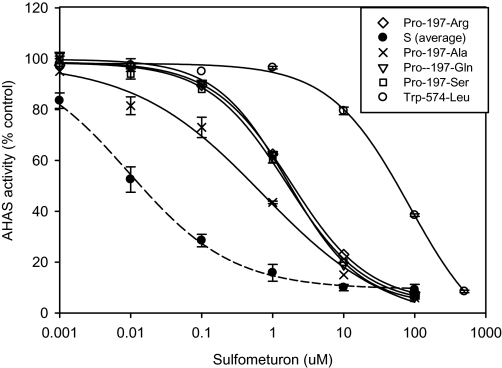
Herbicide sensitivity of the AHAS isolated from R and S plants was determined using the SU herbicide sulfometuron. As shown in Table 2 and Fig. 1, wild-type AHAS isolated from each of three S populations was, as expected, strongly inhibited by sulfometuron, with I50 values ranging from 0.005 μM to 0.09 μM (averaged 0.0075 μM). However, AHAS isolated from purified R populations homozygous for the various resistance mutations was clearly resistant, with I50 values 95 to >1333-fold greater than S populations. Thus, as expected, mutations at Pro-197 or Trp-574-Leu mutation confer high level resistance to sulfometuron.
Table 2.
AHAS I50 values of the resistant and susceptible L. rigidum populations for the SU herbicide sulfometuron
| Population | I50 (μM) | R/S (I50) ratio |
| S (average) | 0.0075 d | |
| R-197-Ala | 0.71 c | 95 |
| R-197-Arg | 1.81 a | 241 |
| R-197-Gln | 1.48 b | 197 |
| R-197-Ser | 1.57 b | 209 |
| R-574-Leu | >10 | >1333 |
I50 values from the three S populations were averaged. Means (n=2 or 3) with different letters are significantly different, P=0.05)
Fig. 1.
In vitro inhibition of AHAS activity of the susceptible (average of the three S populations, closed symbols, dotted line) and purified resistant Lolium rigidum populations (open symbols, solid lines) by sulfometuron. Data are means ±standard error (n=2 or 3).
Resistance mutations result in higher extractable AHAS activity
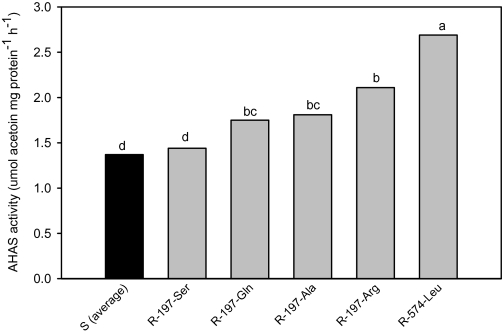
The total protein concentration in the reaction mixture was normalized to 340 μg for all samples, as this protein level catalyses a linear rate of acetoin formation within 60 min incubation at 37 °C, under the current experimental conditions. When AHAS activity was measured on an equal protein basis, AHAS extracted from the R populations (except for the R-197-Ser population) was found to display significantly higher AHAS activity than S populations (Fig. 2). For example, the R-197-Arg and R-574-Leu populations displayed, respectively, 40% and 55% higher AHAS activity than the S. These measurements were made with 3-week-old seedlings (Fig. 2) and 42-d-old plants (data not shown) and the R populations always displayed higher AHAS activity than the S control. By comparing AHAS activity of the five resistant populations with that of three S populations, it is clear that a significantly higher extractable AHAS activity is associated with most of these resistance mutations in these L. rigidum populations.
Fig. 2.
Extractable AHAS activity measured from partially purified enzyme extracts of the susceptible (average of the three S populations, black bar) and resistant (R) Lolium rigidum populations (grey bars) when plants were 18-d-old. Means (n=2 or 3) with different letters are significantly different (P=0.05).
Resistance mutations differ in their effects on AHAS Km (pyruvate)
The Km (pyruvate) values for the three S controls were very similar, ranging from 6.65 mM to 7.9 mM, in agreement with Km values reported for AHAS extracted from other plant species (Tanaka, 2003; Preston et al., 2006; also see the list by Duggleby and Pang, 2000). None of the five resistance mutations had a major impact on AHAS Km but each had different effects. The Pro-197-Ser and Trp-574-Leu did not change Km, the Pro-197-Ala and Pro-197-Gln populations significantly reduced (about 41%) Km values, wherease the Pro-197-Arg population significantly increased (by 31%) the Km value (Table 3), as compared to S populations. In addition, the AHAS Vmax values for R populations were generally higher than for the S populations and, especially the R-197-Arg, R-197-Gln, and R-197-Leu had significantly higher Vmax values (Table 3), confirming the results of higher AHAS specific activity for various resistance mutations, relative to the wild-type AHAS (Fig. 2).
Table 3.
AHAS Km (pyruvate) and Vmax values of the resistant and susceptible L. rigidum populations
| Population | Km (mM) | Vmax (μmol mg−1 protein h−1) |
| S (average) | 7.13 b | 1.11 c |
| R-197-Ala | 4.19 c | 1.86 bc |
| R-197-Arg | 10.36 a | 2.67 ab |
| R-197-Gln | 4.92 c | 2.87 ab |
| R-197-Ser | 7.14 b | 1.51 c |
| R-574-Leu | 6.46 b | 3.19 a |
Data from the three S populations were averaged. Means (n=2 or 3) with different letters are significantly different, (P=0.05).
Resistance mutations vary in their effects on feedback inhibition by branched chain amino acids
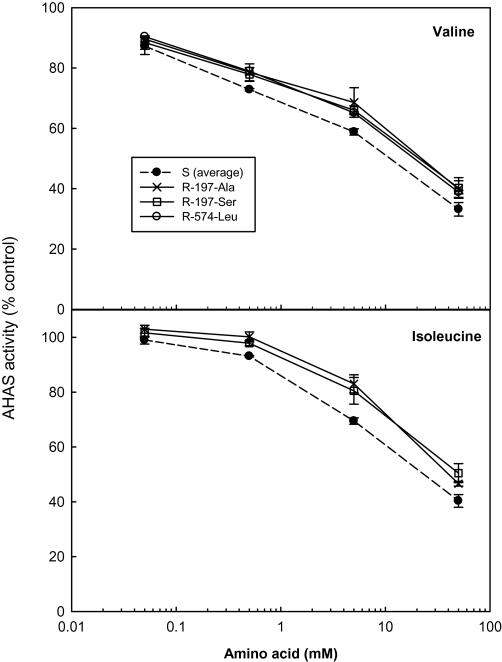
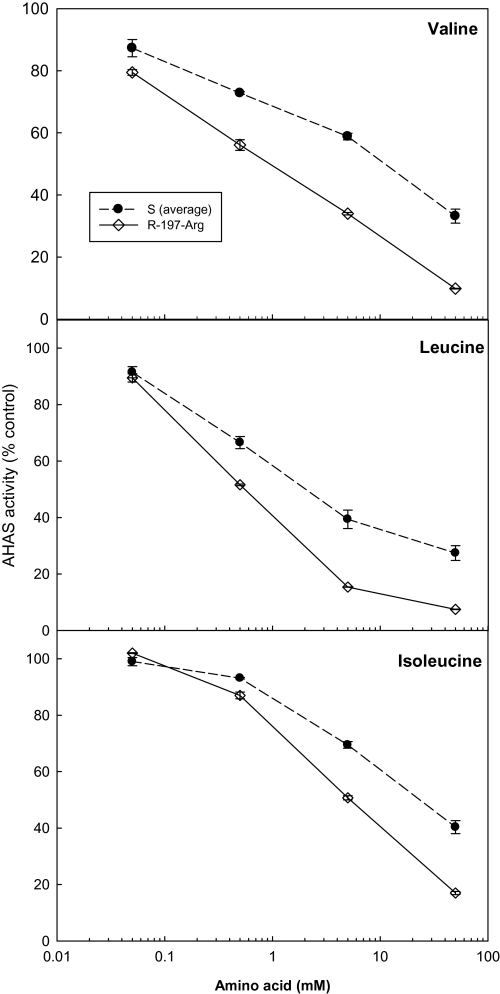
Several studies have established that AHAS from resistant plants show less feedback inhibition by branched chain amino acids (Eberlein et al., 1997, 1999; Preston et al., 2006; Ashigh and Tardif, 2007; Ashigh et al., 2009). In general, in the L. rigidum populations studied here, the single branched chain amino acids valine, leucine or isoleucine inhibited activity of both wild type and resistant AHAS at concentrations higher than 0.05 mM (Figs 3, 4), with leucine being the most effective inhibitor. AHAS activity of all three S populations was feedback inhibited to a similar extent by each of the three amino acids. However, sensitivity of resistant AHAS to amino acid inhibition was differentially modified by the specific resistance mutations, and accordingly, the resistance mutations can be divided into three groups (Table 4). The first group contains the Pro-197-Gln mutation with unchanged AHAS sensitivity to amino acid inhibition (data not shown). The second group contains the Pro-197-Ala, Pro-197-Ser, and Trp-574-Leu mutations, these mutations caused no-to-slight increase (5–10%) in AHAS sensitivity to amino acid inhibition (Fig. 3), depending on the specific amino acids. The third group contains the Pro-197-Arg mutation, which significantly increased (by up to 25%) AHAS sensitivity to amino acid inhibition (Fig. 4). This was also confirmed in another purified L. rigidum population (WALR50) homozygous for the same Pro-197-Arg mutation (data not shown).
Fig. 3.
Slightly decreased AHAS sensitivity to valine or isoleucine feedback inhibition for the resistant R-197-Ala, R-197-Ser, and R-574-Leu (open symbols, solid lines), as compared to that of the averaged S Lolium rigidum populations (closed symbols, dotted line). Data are means ±standard error (n=2 or 3).
Fig. 4.
Increased AHAS sensitivity to amino acid feedback inhibition for the R-197-Arg population (open symbols, solid line), as compared to that of the averaged S Lolium rigidum populations (closed symbols, dotted line). Data are means ±standard error (n=2 or 3).
Table 4.
Summary of sensitivity to feedback inhibition by branched chain amino acids of AHAS of purified resistant populations, relative to that of the S populations (refer to Figs 3–4)
| Population | Valine | Leucine | Isoleucine |
| Group I | |||
| R-197-Gln | No change | No change | No change |
| Group II | |||
| R-197-Ala | Slightly decreased | No change | Slightly decreased |
| R-197-Ser | Slightly decreased | No change | Slightly decreased |
| R-574-Leu | Slightly decreased | No change | No change |
| Group III | |||
| R-197-Arg | Increased | Increased | Increased |
Effects of AHAS resistance mutations on plant growth
Regardless of the population, plants showed RGR estimates that were significantly higher (P <0.0001) in the first growing period (1st–2nd harvest) than in the second growing period (2nd–3rd harvest) (Table 5). Comparisons of RGR estimates between S and R populations were then conducted within each growing period. In the first growing period, the three S populations showed a very similar RGR (averaged as 0.189±0.0023). Equally, resistant plants possessing the Pro-197-Arg, Pro-197-Ser, or Trp-574-Leu resistance mutation showed similar growth rates to individuals of the S populations during the first growing period (Table 5). During the second growth period (2nd–3rd harvest), there was some variability in RGR between the three S populations, with a mean RGR of 0.137±0.0044. As for the first growing period, the resistant plants with the Pro-197-Ser or Trp-574-Leu mutation had the same RGR as the S populations (Table 5). However, resistant plants with the Pro-197-Arg mutation exhibited a lower RGR (Table 5). Similarly, when observed over the whole plant growing period (28 d from 1st to 3rd harvest), the resistant plants with the Pro-197-Ser or Trp-574-Leu mutation had the same RGR as the S populations (Table 5). However, the resistant plants with the Pro-197-Arg mutation displayed a lower RGR than that of the S populations (Table 5). Thus, overall, these results indicate that, whereas the Pro-197-Ser or Trp-574-Leu mutation has no measureable effect on plant growth, the Pro-197-Arg may be associated with a subtle negative effect. Further detailed work, under competition, is required to confirm this observation.
Table 5.
Mean estimates of relative growth rates (RGR) associated with Lolium rigidum populations homozygous for different AHAS resistance alleles (R-197 and R-574).
| Population |
RGR (d−1) |
||
| 1st–2nd harvest | 2nd–3rd harvest | 1st–3rd harvests | |
| S (average) | 0.189 (0.0023) a | 0.137 (0.0024) a | 0.163 (0.0011) a |
| R-197-Arg | 0.190 (0.0029) a | 0.120 (0.00304) b | 0.154 (0.0016) b |
| R-197-Ser | 0.189 (0.0046) a | 0.130 (0.00295) a | 0.160 (0.0021) a |
| R-574-Leu | 0.195 (0.0029) a | 0.135 (0.00307) a | 0.165 (0.0016) a |
RGR estimates for three herbicide susceptible populations (S1, S4, and S5) were averaged. Three plant harvests were performed 14, 28, and 42 d after transplanting. Data are mean RGR (n=25–30) with standard error (in parenthesis) estimated for the first (1st–2nd harvest), second (2nd–3rd harvest), and whole (1st–3rd harvest) plant growing time intervals. Means with different letters indicate significant differences within each harvest period according to Dunnett's test (α=0.05).
Discussion
Resistance mutations and extractable AHAS activity
Several reports show that herbicide resistance-endowing AHAS gene mutations that limit effective herbicide binding to AHAS result in decreased AHAS activity (Eberlein et al., 1997, 1999; Ashigh and Tardif, 2007). However, in this L. rigidum study, conducted with plants homozygous for each of five resistance-endowing AHAS gene mutations, significantly higher extractable AHAS activity and Vmax for most R populations were consistently observed (Fig. 2; Table 3). This confirms previous observations with AHAS extracted from R versus S populations of Raphanus raphanistrum (Yu et al., 2003) and Hordeum leporinum (Yu et al., 2007a, b). Increased AHAS activity has been documented for various Pro-197 mutations or the Trp-574-Leu mutation in resistant weed biotypes, mutant cell lines, transgenic plants, and yeast (Chang and Duggleby, 1998; Boutsalis et al., 1999; Purrington and Bergelson, 1999; Duggleby et al., 2003).
It is not evident how some AHAS resistance mutations cause higher extractable AHAS activity. The higher activity observed for the resistance mutations is unlikely due to AHAS gene over-expression (Yu et al., 2003). There is no clear pattern to correlate the higher AHAS activity with the changes in enzyme kinetic properties (Tables 3, 4; Figs 3, 4). One of the possible explanations is that increased extractable AHAS activity is due to increased AHAS stability conferred by the specific resistance mutations. It is well known that plant AHAS is extremely labile and low in abundance, and therefore purification and characterization from plant tissues is difficult due to rapid loss of activity and sensitivity to amino acid feedback inhibition (Muhitch, 1988; Duggleby and Pang, 2000). It has been consistently observed in this and other studies (Yu et al., 2003, 2007a, b) that it is much easier to isolate AHAS from R plants with reproducible and higher activity levels, compared to AHAS isolated from S plants.
Plant AHAS, like most AHAS enzymes characterized to date, has a catalytic subunit and a regulatory subunit. The catalytic subunit, containing the cofactor thiamine diphosphate (ThDP; also referred as TPP) and the herbicide binding sites, is usually active alone. The regulatory subunit has no AHAS activity but can greatly stimulate the activity of the catalytic subunit and confers sensitivity to feedback inhibition by branched chain amino acids (Lee and Duggleby, 2001, 2002; reviewed by Duggleby et al., 2008). The increased extractable AHAS activity observed in R populations with AHAS resistance mutations may be due to improved cofactor binding/stability in the catalytic subunit, or increased stability of the regulatory subunit. Research by Kim et al. (2004) indicated that the existence of two disordered regions (i.e. a ‘mobile loop’ and a ‘C-terminal arm’) in the 3D crystal structure of the yeast AHAS catalytic subunit-herbicide complex is responsible for the binding/stabilization of the active dimer and of TPP. With resistant mutant A. thaliana AHAS expressed in E. coli, it was found that different resistance mutations do affect the binding of TPP and FAD (Chang and Duggleby, 1998).
In contrast to the results showing that AHAS herbicide-resistant L. rigidum can have significantly increased AHAS activity (Fig. 2), reduced or unchanged AHAS activity has been observed for a number of resistance mutations in some dicot weed species (Eberlein et al., 1997, 1999; Boutsalis et al., 1999; Preston et al., 2006; Ashigh and Tardif, 2007). Taken together, it follows that the difference in AHAS activity observed in R versus S biotypes is therefore likely to be related to specific resistance mutations in different plant species, frequency of resistance alleles in the resistant population, and the genetic background of the R and S populations under comparison. In the current study the effect of genetic background was largely eliminated by purifying different mutations from within one R population and by using three S populations as controls.
Resistance mutations and AHAS affinity for pyruvate (Km)
In this study, it was observed that specific resistance mutations had different (unchanged, reduced or increased) effects on the AHAS Km (Table 3). Unchanged Km values have been previously observed for a number of AHAS resistance mutations in some plant species (Mourad et al., 1995; Chang and Duggleby, 1998; Eberlein, et al., 1999; Boutsalis et al., 1999; Preston et al., 2006), and slightly increased or reduced Km have also been observed in other cases (Chang and Duggleby, 1998; Preston et al., 2006). The crystal structure of plant AHAS in complex with AHAS-inhibiting herbicides revealed that herbicides do not bind at the active site, but bind within the substrate-access channel, thereby blocking substrate access to the active site (McCourt et al., 2006). This confirms that SU and IMI herbicides are non- or un-competitive inhibitors of AHAS, and explains (i) why these resistance mutations do not drastically change the pyruvate binding, and (ii) how there can be so many different herbicide-resistance mutations in AHAS without a major adverse impact on substrate binding and thus functionality. The current results showing no major changes in Km by various AHAS resistance mutations (Table 3) are in line with the AHAS crystal structure analysis. However, the slightly increased Km for the Pro-197-Arg mutation (Table 3) may have a subtle negative effect on plant growth under stress conditions.
Resistance mutations and AHAS sensitivity to feedback inhibition by branched chain amino acids
Unlike most bacterial and fungal AHAS that is sensitive only to valine, a characteristic of plant AHAS is sensitivity to each of the three branched chain amino acids (Miflin and Cave, 1972; Rathinasabapathi and King, 1991; Southan and Copeland, 1996; Eberlein et al., 1997, 1999; Preston et al., 2006; reviewed by Duggleby et al., 2008). There have been only limited studies conducted with known AHAS mutations on AHAS sensitivity to branched chain amino acid feedback inhibition. Reduced sensitivity has been demonstrated for the Pro-197-His, Pro-197-Thr and a few other resistance mutations in weed species (Eberlein et al., 1997, 1999; Preston et al., 2006; Ashigh and Tardif, 2007; Ashigh et al., 2009). Unchanged sensitivity has been reported for the Pro-197-Ser and Trp-574-Leu mutation in Arabidopsis thaliana mutant lines (Mourad et al., 1995), and in transgenic tobacco (Hattori et al., 1995). The current study has revealed that the effect of resistance mutations on AHAS sensitivity to amino acid feedback inhibition is related to both the specific mutation and amino acid (Figs 3, 4; Table 4). Interestingly, it was found that the Pro-197-Arg mutation significantly increased AHAS sensitivity to amino acid inhibition (Fig. 4). It is known that the AHAS sensitivity to branched chain amino acid inhibition is conferred by the regulatory subunits (Lee and Duggleby, 2001; reviewed by Duggley et al., 2008). However, the pathway is complex for the transmission of inhibitory signals within the regulatory subunits and across to the catalytic subunits (Lee and Duggleby, 2002). Further determination of the 3-D structure of the plant AHAS regulatory subunits, and catalytic plus regulatory subunits, could greatly assist in understanding the cross-talk between the subunits, and, consequently, how herbicide resistance mutations in the catalytic subunit affect the enzyme activity and its sensitivity to feedback inhibition.
The consequence of altered AHAS sensitivity to branched chain amino acid feedback inhibition is perhaps a change in amino acid pool concentrations. However, since the change of sensitivity is only evident at the higher amino acid concentrations (>0.05 mM), the physiological relevance of this impact in plants is not clear (reviewed by Tranel and Wright, 2002). Subtle changes observed for the Pro-197-Arg mutation (increased Km and sensitivity to amino acid feedback inhibition) indicate that this mutation has the potential to express a resistance cost (see below) (Vila-Aiub et al., 2009).
Pleiotropic effects of AHAS resistance mutations on plant growth
Target site herbicide resistance mutations can cause biochemical and physiological alterations that compromise plant survival and reproduction (i.e. resistance cost) (reviewed in Vila-Aiub et al., 2009). The basis for this assumption is that structural enzymatic changes caused by herbicide resistance mutations may result in either subtle or drastic modifications of substrate and/or inhibitor binding leading to insufficient (impaired) activity, imbalance (feedback inhibition) or excess (higher activity) of enzyme end-product biosynthesis. Some of the kinetics changes in herbicide target enzymes may involve additional energy costs with a negative effect on plant growth (Purrington and Bergelson, 1999). In order to understand the biochemical basis of resistance costs associated with various target site AHAS herbicide resistance mutations, the effect of the Pro-197-Ser, Pro-197-Arg or Trp-574-Leu mutation on L. rigidum RGR was evaluated. RGR is a useful eco-physiological parameter to denote the expression of herbicide resistance costs as variations in RGR are often positively correlated with variations in plant competitive and establishment ability (Grime and Hunt, 1975; Vila-Aiub et al., 2005).
It has been argued that increased AHAS activity may be associated with a herbicide resistance cost due to higher carbon investments and/or toxic effects of excessive amino acids synthesis (Vila-Aiub et al., 2009). The growth analysis has revealed that the higher extractable AHAS activity caused by the Pro-197-Ser and Trp-574-Leu mutations has no adverse effect on L. rigidum vegetative growth (Table 5). This finding contradicts the significant resistance cost associated with higher AHAS activity derived from the Pro-197-Ser mutation in laboratory-generated Arabidopsis thaliana transgenic lines (Purrington and Bergelson, 1999). In relation to the Trp-574-Leu AHAS resistance mutation, a strong pleiotropic effect on plant growth has been associated with this mutation in field-evolved resistant Amaranthus powellii populations (Tardif et al., 2006); however, the effect of the Trp-574-Leu mutation on AHAS enzyme functionality has not been reported and remains to be elucidated in this species. It is evident from the current results that higher extractable AHAS activity associated with the Pro-197-Ser and Trp-574-Leu mutations during early vegetative growth in L. rigidum does not necessarily involve any significant metabolic drain or toxicity with apparent adverse effects on growth. Alternatively, the higher measurable in vitro AHAS activity in the resistant mutants may not correlate with higher in vivo AHAS activity but may be associated with increased enzyme stability with negligible effects on plant growth. It remains to be established whether higher in vitro AHAS activity (or stability) originated from specific AHAS resistance mutations in L. rigidum show an associated resistance cost at different plant developmental stages such as reproduction and seed germination.
An interesting outcome of the growth analysis was that plants possessing the Pro-197-Arg resistance mutation showed lower RGR when compared to herbicide susceptible individuals during the slower plant growing period (Table 5). Of the AHAS resistance mutations evaluated in this study, the Pro-197-Arg mutation is the only one leading to increased AHAS activity (Fig. 3), Km value (Table 3), and sensitivity to valine, leucine, and isoleucine feedback inhibition (Fig. 4). The combination of a higher Km and increased feedback sensitivity may cause temporal impaired synthesis of branched chain amino acids during active plant growth periods, thus contributing to the observed subtle plant growth reduction. Further experiments are required to determine the biological significance of this finding on overall plant fitness.
In summary, AHAS herbicide-resistant L. rigidum populations, each individually homozygous for the Pro-197-Ala, Pro-197-Arg, Pro-197-Gln, Pro-197-Ser or Trp-574-Leu resistance mutation, were generated. The effect of these mutations on AHAS functionality and on plant growth was investigated by determining AHAS kinetics and by assessing plant relative growth rates at the vegetative stage, compared with the wild-type enzyme and various herbicide-susceptible populations. Each of these resistance mutations endowed AHAS resistant to the sulfonylurea herbicide sulfometuron (up to >1000-fold), but with some effects on AHAS kinetics. Nearly all the resistance mutations resulted in higher extractable AHAS activity (both specific activity and Vmax), and whilst most of them caused no-to-minor changes in AHAS kinetics, the Pro-197-Arg mutation slightly (but significantly) increased the Km for pyruvate and markedly increased sensitivity to feedback inhibition by branched chain amino acids. Compared with the other resistance mutations examined, the Pro-197-Ser and Trp-574-Leu mutations displayed the least affected enzyme kinetics. Correspondingly, the Pro-197-Ser and Trp-574-Leu mutations did not show an adverse impact on plant productivity, while the Pro-197-Arg mutation showed a transient negative effect on plant growth. The undisturbed AHAS kinetics and unaffected vegetative growth for the Pro-197-Ser and Trp-574-Leu mutations help explain, in part, why these two resistance mutations often evolve in field populations of many weed species under AHAS herbicide selection.
Acknowledgments
This research is partially funded by the Australian Research Council and the Grains Research and Development Corporation of Australia. Drs D Goggin, T Gaines, and D Shaner are particularly acknowledged for critical and helpful comments on the manuscript.
References
- Alcocer-Ruthling M, Thill DC, Mallorysmith C. Monitoring the occurrence of sulphonylurea-resistant prickly lettuce (Lactuca serriola) Weed Technology. 1992;6:437–440. [Google Scholar]
- Ashigh J, Corbett CAL, Smith PJ, Laplante J, Tardif FJ. Characterization and diagnostic tests of resistance to acetohydroxyacid synthase inhibitors due to Asp376Glu substitution in Amaranthus powellii. Pesticide Biochemistry and Physiology. 2009;95:38–46. [Google Scholar]
- Ashigh J, Tardif F. An Ala205Val substitution in acetohydroxyacid synthase of Eastern black nightshade (Solanum ptychanthum) reduces sensitivity to herbicides and feedback inhibition. Weed Science. 2007;55:558–565. [Google Scholar]
- Boutsalis P, Karotam J, Powles SB. Molecular basis of resistance to acetolactate synthase-inhibiting herbicides in Sisymbrium orientale and Brassica tournefortii. Pesticide Science. 1999;55:507–516. [Google Scholar]
- Causton DR, Venus JC. The biometry of plant growth. London, UK: Edward Arnold; 1981. [Google Scholar]
- Chang AK, Duggleby RG. Herbicide-resistant forms of Arabidopsis thaliana acetohydroxyacid synthase: characterization of the catalytic properties and sensitivity to inhibitors of four defined mutants. Biochemical Journal. 1998;333:765–777. doi: 10.1042/bj3330765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher JT, Powles S, Holtum JAM. Resistance to acetolactate synthase-inhibiting herbicides in annual ryegrass (Lolium rigidum) involves at least two mechanisms. Plant Physiology. 1992;100:1909–1913. doi: 10.1104/pp.100.4.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousens RD, Gill GS, Speijers EJ. Comment: number of sample populations required to determine the effects of herbicide resistance on plant growth and fitness. Weed Research. 1997;37:1–4. [Google Scholar]
- Duggleby RG, McCourt JA, Guddat LW. Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiology and Biochemistry. 2008;46:309–324. doi: 10.1016/j.plaphy.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Duggleby RG, Pang SS. Acetohydroxyacid synthase. Journal of Biochemistry and Molecular Biolology. 2000;33:1–36. [Google Scholar]
- Duggleby RG, Pang SS, Yu H, Guddat LW. Systematic characterization of mutations in yeast acetohydroxyacid synthase, interpretation of herbicide-resistance data. European Journal of Biochemistry. 2003;270:2895–2904. doi: 10.1046/j.1432-1033.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- Eberlein CV, Guttieri MJ, Berger PH, Fellman JK, Mallory-Smith CA, Thill DC, Baerg RJ, Belknap WR. Physiological consequence of mutation for ALS-inhibitor resistance. Weed Science. 1999;47:383–392. [Google Scholar]
- Eberlein CV, Guttieri MJ, Mallory-Smith CA, Thill DC, Baerg RJ. Altered acetolactate synthase activity in ALS-inhibitor resistant prickly lettuce (Lactuca serriola) Weed Science. 1997;45:212–217. [Google Scholar]
- Forlani G, Mantelli M, Nielsen E. Biochemical evidence for multiple acetoin-forming enzymes in cultured plant cells. Phytochemistry. 1999;50:255–262. [Google Scholar]
- Grime JP, Hunt R. Relative growth-rate: its range and adaptive significance in a local flora. The Journal of Ecology. 1975;63:393–422. [Google Scholar]
- Hattori J, Brown D, Mourad G, Labbe H, Ouellet T, Sunohara G, Rutledge R, King J, Miki B. An acetohydroxy acid synthase mutant reveals a single site involved in multiple herbicides resistance. Molecular and General Genetics. 1995;246:419–425. doi: 10.1007/BF00290445. [DOI] [PubMed] [Google Scholar]
- Healy-Fried ML, Funke T, Priestman MA, Han H, Schönburnn E. Structural basis of glyphosate tolerance resulting from mutations of Pro101 in E. coli EPSP synthase. Journal of Biological Chemistry. 2007;282:32949–32955. doi: 10.1074/jbc.M705624200. [DOI] [PubMed] [Google Scholar]
- Hoffmann WA, Poorter H. Avoiding bias in calculations of relative growth rate. Annals of Botany. 2002;80:37–42. doi: 10.1093/aob/mcf140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KimBeak JDG, Kim YT, Choi JD, Yoon MY. Effects of deletions at the C-terminus of tobacco acetohydroxyacid synthase on the enzyme activity and cofactor binding. Biochemical Journal. 2004;384:59–68. doi: 10.1042/BJ20040427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Duggleby RG. Identification of the regulatory subunit of Arabidopsis thaliana acetohydroxyacid synthase and reconstitution with its catalytic subunit. Biochemistry. 2001;40:6836–6844. doi: 10.1021/bi002775q. [DOI] [PubMed] [Google Scholar]
- Lee YT, Duggleby RG. Regulatory interaction in Arabidopsis thaliana acetohydroxyacid synthase. FEBS Letters. 2002;512:180–184. doi: 10.1016/s0014-5793(02)02253-6. [DOI] [PubMed] [Google Scholar]
- McCourt JA, Panf SS, King-Scott J, Guddat LW, Duggleby RG. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid aynthase. Proceedings of The National Academy of Sciences, USA. 2006;103:569–573. doi: 10.1073/pnas.0508701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin BJ, Cave PR. The control of leucine, isoleucine, and valine biosynthesis in a range of higher plants. Journal of Experimental Botany. 1972;23:511–516. [Google Scholar]
- Mourad G, Williams D, King J. A double mutant allele, csr1-4, of Arabidopsis thaliana encodes an acetolactate synthase with altered kinetics. Planta. 1995;196:64–68. doi: 10.1007/BF00193218. [DOI] [PubMed] [Google Scholar]
- Muhitch M. Acetolactate synthase activity in developing maize (Zea mays L.) kernels. Plant Physiolology. 1988;86:23–27. doi: 10.1104/pp.86.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, Powles SB. Glyphosate-resistant rigid ryegrass (Lolidum rigidum) populations in the Western Australian grainbelt. Weed Technology. 2010;24:44–49. [Google Scholar]
- Owen MJ, Walsh MJ, Llewellyn RS, Powles SB. Widespread occurrence of multiple herbicide resistance in Western Australian annual ryegrass (Lolium rigidum) populations. Australian Journal of Agricultural Research. 2007;58:711–718. [Google Scholar]
- Pornprom T, Usui K, Ishizuka K. Growth inhibition and acetolactate synthase activity of soybean seedlings and suspension-cultured cells treated with bensulphuron-methyl. Weed Biology and Management. 2005;5:150–153. [Google Scholar]
- Powles SB, Yu Q. Evolution in action: plants resistant to herbicides Annual Review of Plant Biology. 2010;61:317–347. doi: 10.1146/annurev-arplant-042809-112119. [DOI] [PubMed] [Google Scholar]
- Preston C, Stone LM, Rieger MA, Baker J. Multiple effects of a naturally occurring proline to threonine substitution within acetolactate synthase in two herbicide-resistant populations of Lactuca serriola. Pesticide Biochemistry and Physiology. 2006;84:227–235. [Google Scholar]
- Purrington CB, Bergelson J. Exploring the physiological basis of costs of herbicide resistance in Arabidopsis thaliana. The American Naturalist. 1999;154:S82–S91. doi: 10.1086/303285. [DOI] [PubMed] [Google Scholar]
- Rathinasabapathi B, King J. Herbicide resistance in Datura innoxia: kinetic characterization of acetolactate synthase from wild-type and sulphonylurea-resistant cell variants. Plant Physiology. 1991;96:255–261. doi: 10.1104/pp.96.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari LL, Cotterman JC, Thill DC. Resistance to acetolactate synthase inhibiting herbicide. In: Powles SB, Holtum JAM, editors. Herbicide resistance in plants, biology and biochemistry. Boca Raton, FL: Lewis Publishers; 1994. pp. 83–139. [Google Scholar]
- Santel HJ, Bowden BA, Sorensen VM, Mueller KH. Flucarbazone-sodium: a new herbicide for the control of wild oat and green foxtail in wheat. In: Proceedings of the Brighton Crop Protection Conference - Weeds, 23. 1999 [Google Scholar]
- Singh BK, Stidham MA, Shaner DL. Assay of acetohydroxyacid synthase. Analytical Biochemistry. 1988;171:173–179. doi: 10.1016/0003-2697(88)90139-x. [DOI] [PubMed] [Google Scholar]
- Southan MD, Copeland L. Physical and kinetic properties of acetohydroxyacid synthase from wheat leaves. Physiologia Plantarum. 1996;98:824–832. [Google Scholar]
- Tanaka Y. Properties of acetolactate synthase from sulphonylurea-resistant Scirpus juncoides Roxb. Var. ohwianus T. Koyama. Pestcide Biochemistry and Physiology. 2003;77:147–153. [Google Scholar]
- Tardif FJ, Rajcan I, Costea M. A mutation in the herbicide target site acetohydroxyacid synthase produces morphological and structural alterations and reduces fitness in Amaranthus powellii. New Phytologist. 2006;169:251–264. doi: 10.1111/j.1469-8137.2005.01596.x. [DOI] [PubMed] [Google Scholar]
- Tranel PJ, Wright TR. Resistance of weeds to ALS inhibiting herbicides: what have we learned? Weed Science. 2002;50:700–712. [Google Scholar]
- Vila-Aiub MM, Neve P, Powles SB. Resistance cost of a cytochrome P450 herbicide metabolism mechanism but not an ACCase target site mutation in a multiple resistant Lolium rigidum population. New Phytologist. 2005;167:787–796. doi: 10.1111/j.1469-8137.2005.01465.x. [DOI] [PubMed] [Google Scholar]
- Vila- Aiub MM, Neve P, Powles SB. Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytologist. 2009;184:751–767. doi: 10.1111/j.1469-8137.2009.03055.x. [DOI] [PubMed] [Google Scholar]
- Yu Q, Abdallah I, Han HP, Owen M, Powles SB. Distinct non-target-site mechanisms endow resistance to glyphosate, ACCase and ALS-inhibiting herbicides in multiple herbicide-resistant Lolium rigidum populations. Planta. 2009;230:713–723. doi: 10.1007/s00425-009-0981-8. [DOI] [PubMed] [Google Scholar]
- Yu Q, Collavo A, Zheng MQ, Owen M, Sattin M, Powles SB. Diversity of acetyl-coenzyme A carboxylase mutations in resistant Lolium populations: evaluation using clethodim. Plant Physiology. 2007b;145:547–558. doi: 10.1104/pp.107.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Friesen LJS, Zhang XQ, Powles SB. Tolerance to acetolactate synthase and acetyl-coenzyme A carboxylase herbicides in Vulpia bromoides is conferred by multiple resistance mechanisms. Pesticide Biochemistry and Physiology. 2004;78:21–30. [Google Scholar]
- Yu Q, Han H, Powles SB. Mutations of the ALS gene endowing resistance to ALS-inhibiting herbicides in Lolium rigidum populations. Pest Management Science. 2008;64:1229–1236. doi: 10.1002/ps.1624. [DOI] [PubMed] [Google Scholar]
- Yu Q, Nelson JK, Zheng MQ, Jackson M, Powles SB. Molecular characterisation of resistance to ALS-inhibiting herbicides in Hordeum leporinum biotypes. Pest Management Science. 2007a;63:918–927. doi: 10.1002/ps.1429. [DOI] [PubMed] [Google Scholar]
- Yu Q, Zhang XQ, Hashem A, Walsh MJ, Powles SB. ALS gene proline (197) mutations confer ALS herbicide resistance in eight separated wild radish (Raphanus raphanistrum) populations. Weed Science. 2003;51:831–838. [Google Scholar]