Abstract
Auxin response factors (ARFs) are key regulators of plant growth and development. Through interaction with auxin/indole acetic acid (Aux/IAA) proteins, they influence the expression of auxin response genes. An ARF gene family has been predicted in rice, but the functions of the individual structural domains of the OsARFs remain obscure. Bioinformatics was used to analyse the position of the DNA-binding domain (DBD), middle region (MR), and C-terminal dimerization domain (CTD) of OsARFs, and experimentally confirmed the presence of a classical monopartite nuclear localization signal (NLS) in the DBD. The DBD was shown to contribute to nuclear localization of OsARF proteins in addition to its known DNA-binding function. Interactions between 14 integrated OsARFs and 15 OsIAA proteins were tested using yeast two-hybrid assays. It was found that eight OsARF activators interacted with the 15 OsIAA proteins, while six OsARF repressors did not. The interactions between the MR+CTD or CTD of 10 OsARFs and 15 OsIAA proteins were also tested and the results were consistent with those of each intact OsARF, although some slight differences in interaction intensity were observed by α-galactosidase quantitative assays. The truncated CTD of OsARF11 did not interact with any OsIAA, implying that the CTD is required for ARF–IAA dimerization, and that the MR influences the interaction intensity in yeast. A subset of the interactions in yeast were also observed in tobacco plants using firefly luciferase complementation imaging assays, indicating that these interactions are specific in plants, and might have a special role in the auxin signalling response. This study provides new insight into the structure of OsARF proteins and ARF–Aux/IAA interactions.
Keywords: Firefly luciferase complementation imaging assay, nuclear localization signal, OsARF activators, OsARF repressors, OsIAA, yeast two-hybrid assay
Introduction
The auxin response factor (ARF) and auxin/indole acetic acid (Aux/IAA) protein families are required for transcriptional regulation of auxin response genes, and play a central role in auxin signalling and plant development (Guilfoyle et al., 1998, Guilfoyle and Hagen, 2001, 2007). A typical ARF contains a conserved N-terminal DNA-binding domain (DBD), a non-conserved middle region (MR), and a conserved C-terminal dimerization domain (CTD) (Guilfoyle et al., 1998, Guilfoyle and Hagen, 2001, 2007). The DBD of an ARF binds specifically to TGTCTC auxin response elements (AuxREs) in promoters, to regulate expression of auxin response genes (Ulmasov et al., 1997a, b). The CTD of ARF, which resembles domains III and IV of Aux/IAA proteins, is involved in homo- and heterointeraction (Ulmasov et al., 1997a). The MR, located between the DBD and CTD, determines whether the ARF functions as a transcriptional activator or repressor (Ulmasov et al., 1999a; Tiwari et al., 2003). The Aux/IAA family proteins lack DBDs, and constitute one class of three early auxin response genes. The other two families are Gretchen Hagen3 and small auxin up RNA (Abel and Theologis, 1996; Hagen and Guilfoyle, 2002). Aux/IAA genes encode short-lived nuclear proteins that generally contain four highly conserved domains, referred to as domains I, II, III, and IV (Abel et al., 1994; Hagen and Guilfoyle, 2002; Liscum and Reed, 2002). Domain I acts as a strong transcriptional repression domain (RD) and interacts with TOPLESS (TPL), which co-represses transcription of auxin response genes (Tiwari et al., 2004; Szemenyei et al., 2008); domain II is responsible for rapid degradation of Aux/IAA proteins (Worley et al., 2000; Ouellet et al., 2001); and domains III and IV mediate homo- and heterodimerization between Aux/IAA proteins and ARFs (Kim et al., 1997; Ulmasov et al., 1997a, b).
Extensive research suggests that auxin response specificity is regulated by interactions between ARF and Aux/IAA proteins. Yeast two-hybrid (YTH) assays and other physical assays have provided information on a number of ARF–Aux/IAA interactions and, in some cases, the interactions have been genetically documented. In Arabidopsis, ARF1 and MONOPTEROS (MP)/ARF5 have been shown to heterodimerize with AUXIN RESISTANT 3 (AXR3)/IAA17 in YTH assays (Ouellet et al., 2001). MP/ARF5 and NONPHOTOTROPIC (NPH4)/ARF7 interact with BODENLOS (BDL)/IAA12 in YTH and plant protoplast assays, and MP/ARF5 interacts genetically with BDL/IAA12 (Hamann et al., 2002; Hardtke et al., 2004; Weijers et al., 2006). Tatematsu et al. (2004) showed that the CTDs of MP/ARF5, NPH4/ARF7, and ARF8 heterodimerize with IAA1, IAA6, IAA13, and MASSUGU2 (MSG2)/IAA19 by YTH assays, and that NPH4/ARF7 also interacts with MSG2/IAA19 in in vitro pull-down assays. Fluorescence cross-correlation spectroscopy further demonstrated the interaction between ARF5/ARF7 and MSG2/IAA19 in HeLa cells (Muto et al., 2006). In another study, SOLITARY-ROOT (SLR4)/IAA14 interacted equally well with MP/ARF5, NPH4/ARF7, and ARF19 in YTH assays (Fukaki et al., 2005). CRANE/IAA18 also interacts with NPH4/ARF7 and ARF19 in YTH systems (Uehara et al., 2008). Tiwari et al. (2003) showed that MP/ARF5 interacts with IAA17 in plant protoplast assays, but ARF1 failed to interact with IAA17. Taken together, these results imply that in Arabidopsis, the ARF activators MP/ARF5, NPH4/ARF7, ARF8, and ARF19 can interact with a variety of Aux/IAAs by YTH assays, and other physical interaction assays. On the other hand, few interactions have been reported between ARF repressors and Aux/IAAs (Song et al., 2009), and whether ARF1 repressor interacts with AXR3/IAA17 is unclear from previous reports (Ouellet et al., 2001; Tiwari et al., 2001), making it important to investigate ARF–Aux/IAA interactions further.
To date, no systematic or extensive investigations of potential interactions between ARF and Aux/IAA proteins have been reported in Arabidopsis or other plants. The potential for interaction between ARF and Aux/IAA proteins is high, because both ARF and Aux/IAA are encoded by multigene families in plants. For example, the Arabidopsis genome encodes 23 ARFs and 29 Aux/IAA proteins (Liscum and Reed, 2002; Guilfoyle and Hagen, 2007), and the rice genome encodes 25 ARFs and 31 Aux/IAA proteins (Jain et al., 2006; Wang et al., 2007; Song et al., 2008). To elucidate the function of the structural domains in OsARF and the relationship between OsARFs and OsIAAs, the structure of OsARF genes was analysed and classified in detail by bioinformatics, and the function of each domain experimentally confirmed. The position of the OsARF nuclear localization signal (NLS) was determined, and the interactions between 14 OsARFs and 15 OsIAA proteins were tested using YTH assays. The interactions between several OsARF–OsIAA pairs were further tested with a firefly luciferase complementation imaging (LCI) assay in tobacco.
Materials and methods
Sequence analysis and prediction of amino acid contents
Loci numbers of OsARF genes were obtained from http://plntfdb.bio.uni-potsdam.de/v2.0 (Oryza sativa subsp. japonica ARF family). Accession numbers of full-length OsIAA and OsARF sequences were from Jain et al. (2006) and Wang et al. (2007). The sequences of the OsARF and OsIAA genes were obtained from the knowledge-based Oryza molecular biological encyclopedia (KOME, http://www.cdna01.dna.affrc.go.jp/cDNA). The coding sequences of OsARFs were derived by prediction using The Institute for Genomic Research (TIGR: http://www.tigr.org). The software MEGA 4.0 was used for calculation of amino acid contents of the MR domain in OsARFs.
Plant materials and growth conditions
Nicotiana benthamiana plants were grown in vermiculite containing Murashige and Skoog salt nutritional liquid in a greenhouse (12 h/light, 25 °C; 12 h/dark, 18 °C). Six-week-old N. benthamiana plants were used for Agrobacterium-mediated transient expression.
Constructs for 35S:OsARF–sGFP fusion proteins
Candidate NLSs from OsARF proteins were predicted by the program ScanProsite (Gattiker et al., 2002) using classical NLS motifs: K(K/R)X(K/R) for monopartite NLSs and (K/R)(K/R)X10-12(K/R)3/5 for bipartite NLSs, where X indicates any amino acid. Full-length cDNA clones of OsARF were provided by the Rice Genome Resource Center (RGRC) in Japan. The open reading frame (ORF) of the OsARF19 gene, and elements I–VI of the OsARF18, 19, 22, and 25 genes (Supplementary Table S2 available at JXB online) were directly amplified from the cDNAs using the primers in Supplementary Table S3, and digested with BamHI/SalI, KpnI/SalI, XbaI/SalI, and SacI/BamHI, respectively. Individual gene fragments were cloned in-frame into the binary vector pCAMBIA1300 containing a 35S:sGFP (green fluorescent protein) cassette to create an OsARF–sGFP fusion protein. All 35S:OsARF–sGFP fusion constructs were transiently expressed in onion epidermal cells using the Bio-Rad biolistic PDS-1000/He system under 1100 psi (http://www.bio-rad.com/), carried out as described previously (Varagona et al., 1992). Fluorescence was imaged using a Carl Zeiss laser scanning system LSM510 (http://www.zeiss.com/).
YTH assays of OsARF–OsIAA pairs
YTH analysis was performed according to the instructions for the Matchmaker GAL4 Two-Hybrid System 3 (Clontech). The full-length cDNA clones of OsIAA and OsARF family members were provided by the RGRC. ORFs of OsIAA genes were cloned into pGBKT7, and transformed into yeast Y187. ORFs, CTDs, and MR+CTDs of OsARF were cloned into pGADT7 and transformed into yeast AH109. Yeast Y187 containing pGBKT7+ORFs of individual OsIAA genes were mated with AH109 containing pGADT7+ORFs, CTDs, or MR+CTDs of individual ARFs, according to the manufacturer's protocol. Plates containing the chromogenic substrate X-α-Gal (5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside) were used to detect protein interactions. Mated strains were spread on low-stringency SD–Leu/–Trp and high-stringency SD–Ade/–His/–Leu/–Trp/X-α-Gal. Strains from high-stringency plates were saved and re-cultured for α-galactosidase assays. The software Cluster 3.0 and Java tree view 1.1 were used to construct α-galactosidase activity graphs. The positions of MR domains in OsARFs are given in Supplementary Fig. S1 and Supplementary Table S1 at JXB online. Primers are listed in Supplementary Tables S4–S7. Interaction of pGBKT7-53 and pGADT7-T was used as a positive control, and non-interaction of pGBKT7-Lam and pGADT7-T was used as a negative control. p-Nitrophenyl-D-galactopyranoside (Sigma) solution was used as substrate for α-galactosidase quantitative assays (Supplementary Table S8). Absorbance was measured using a Shimadzu spectrophotometer (UV-2550).
Western blot analysis
Total proteins were extracted from yeast (strain AH109), which carried various OsARFs cloned into the pGADT7 expression vector [fused to amino acids 768–881 of the GAL4 activation domain (AD)], as described by the Yeast Protocols Handbook (Clontech, PT3024-1). A 30 μg aliquot of proteins was resolved by SDS–PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane that was used for subsequent western blot analysis according to standard procedures. Goat anti-haemagglutinin (HA) polyclonal antibodies conjugated to horseradish peroxidase (HRP; GenScript corporation, A00169) were used for the detection of OsARF fusion proteins containing the HA epitope tag (western blot detection kit, KGP1122, KeyGEN).
Agrobacterium-mediated transient expression in N. benthamiana leaves for LCI assays
The full-length cDNAs of OsARF 1 and OsARF16 were ligated into pCAMBIA1300-NLuc (2–416aa), and OsIAA1, 8, 12, and 13 were ligated into pCAMBIA1300-NLuc (2–416aa) or CLuc (398–550aa) (Chen, 2008). Constructs were transformed into Agrobacterium tumefaciens strain GV3101 for infiltrating N. benthamiana leaves. OsARFs and OsIAAs, amplified with the primers given in Supplementary Table S9 at JXB online, were cloned into NLuc or CLuc using KpnI/SalI, to generate fusion constructs. After 3 d, Agrobacterium-infiltrated areas on leaves were injected with 1 mM D-luciferin potassium salt for detecting luminescence. Fluorescence images were collected using a NightOWL LB 981 CCD camera (Berthold).
Results
Most OsARF proteins contain a DBD, MR, and CTD
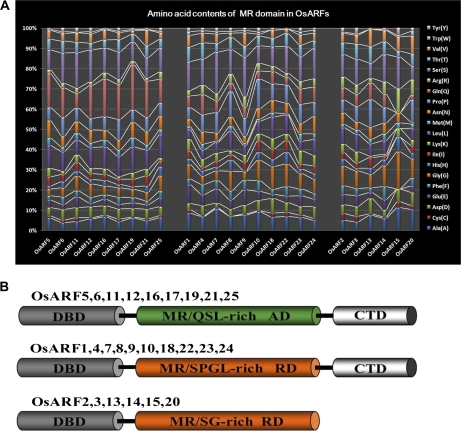
Domain positions in 25 OsARF proteins are shown in Supplementary Fig. S1 at JXB online. Amino acid contents of the MRs and classification of the OsARFs are given in Fig. 1A and B. All OsARFs contain a conserved, plant-specific B3-type DBD (except OsARF20 which contains two DBDs), which require additional N-terminal and C-terminal amino acids for efficient binding to TGTCTC AuxREs in vitro (Ulmasov et al., 1999b; Wang et al., 2007; Guilfoyle and Hagen, 2007). The 25 OsARF proteins can be divided into three groups based on the MR amino acid sequence and the presence or absence of CTDs. The OsARF family includes nine putative transcriptional activators, OsARF5, 6, 11, 12, 16, 17, 19, 21, and 25, all with an MR enriched in glutamine (Q), serine (S), and leucine (L). This type of MR has been shown to function as an AD in Arabidopsis ARFs (Ulmasov et al., 1999a; Tiwari et al., 2003). The other OsARFs are putative transcriptional repressors, because they contain MRs enriched in S, L, proline (P), and glycine (G), similar to RDs found in Arabidopsis ARF repressors (Ulmasov et al., 1999a; Tiwari et al., 2003). ARF2, 3, 13, 14, 15, and 20 do not contain a CTD, while the other 19 OsARFs do.
Fig. 1.
Analysis of amino acid contents and classification of OsARFs. (A) Amino acid contents of MR domains in putative OsARF proteins. The horizontal axis indicates OsARF, and the vertical axis indicates the corresponding amino acid contents. Coloured bars show the different amino acids. The positions of MR domains in OsARFs are shown in Supplementary Fig. S1 and Supplementary Table S1 at JXB online. (B) The modular structure of the OsARF family. DBD, DNA-binding domain; CTD, C-terminal dimerization domain; MR, middle region; RD, repression domain; AD, activation domain; Q, glutamine; S, serine; L, leucine; P, proline; G, glycine. (This figure is available in colour at JXB online.)
A monopartite NLS in the DBD is responsible for nuclear localization of OsARFs
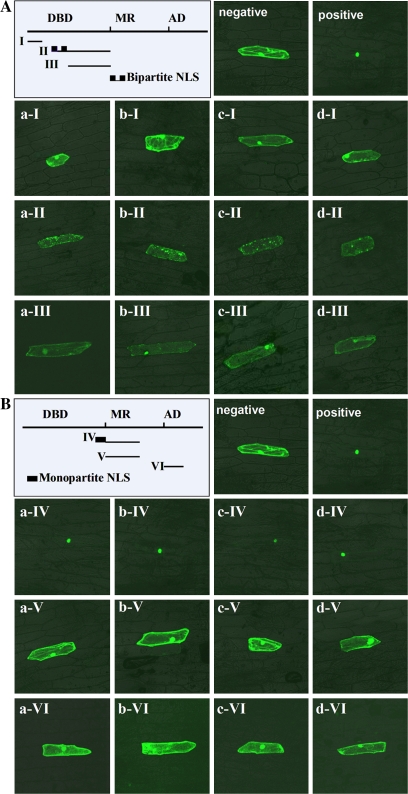
In OsARFs, the first NLS with a bipartite NLS structure is predicted to be in the middle of DBD, and the second, which resembles the monopartite NLS of simian virus 40, is predicted to be at the end of the DBD (Supplementary Table S2 at JXB online). The fluorescence of the 35S:OsARF19–sGFP fusion protein was observed only in the nucleus of onion epidermal cells, while the control 35S:sGFP fluorescence was observed throughout the entire cell (Fig. 2A, B, positive and negative control). Proteins fused to a bipartite NLS containing element II were detected throughout transformed cells, not only in the nucleus (Fig. 2A, aII–dII). Elements I, III, V, or VI fused to OsARF–sGFP caused expression throughout the entire cell, similar to 35S:sGFP (Fig. 2A, aI–dI and aIII–dIII; 2B, aV–dV and aVI–dVI). The fluorescence of sGFP fused to a monopartite NLS containing element IV accumulated exclusively in the nucleus (Fig. 2B, aIV–dIV). These data indicate that a monopartite NLS in the DBD had the capacity to direct nuclear localization of OsARF.
Fig. 2.
Analysis of the nuclear localization signal (NLS) in OsARFs. (A) Confirmation of the putative bipartite NLS of OsARF18, 19, 22, and 25. The line drawing on the left shows the probable sites of individual truncated elements in OsARF (details are given in Supplementary Table S2 at JXB online). Negative, 35S:GFP for negative control; positive, 35S:OsARF19–GFP for positive control; a–d represent the genes OsARF18/19/22/25, and I–III represent truncated elements of OsARFs. (B) Confirmation of the putative monopartite NLS of OsARF18, 19, 22. and 25. Negative and positive as A; a–d represent genes OsARF18/19/22/25, and IV–VI represent truncated elements of OsARFs. (This figure is available in colour at JXB online.)
Putative OsARF activators, but not repressors, interact with OsIAA proteins in YTH assay
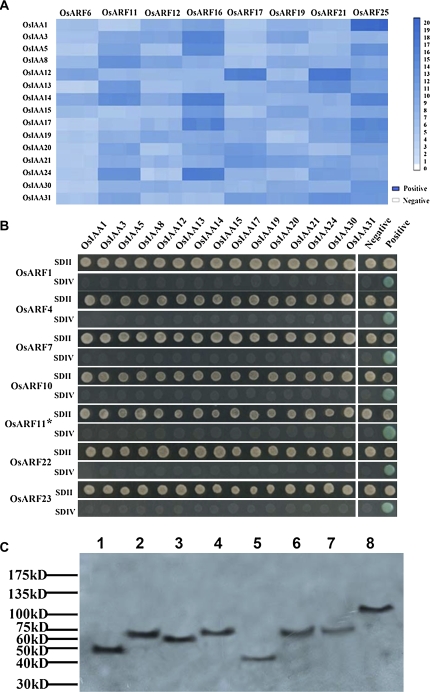
Each subfamily of OsARFs and IAA proteins was represented by 14 OsARFs and 15 IAA proteins. In previous reports, the CTD of most AtARFs (Hamann et al., 2002; Hardtke et al., 2004; Tatematsu et al., 2004; Fukaki et al., 2005; Weijers et al., 2006; Uehara et al., 2008) and the full length of some AtARFs (Ouellet et al., 2001) were used to test interactions with Aux/IAA proteins by YTH assay. Here, the full-length cDNAs of the OsARFs were tested for interaction with OsIAA proteins in a YTH system. As shown in Fig. 3A, the full-length cDNAs of putative OsARF6, 11, 12, 16, 17, 19, 21, and 25 activators interacted with the 15 OsIAA proteins, but interaction intensities varied (see α-galactosidase activity in Supplementary Table S8 at JXB online). The α-galactosidase activity for interaction between OsARF16/OsARF25 and the 15 OsIAA proteins was generally higher than activity for interactions between OsARF6 and the 15 OsIAA proteins. When yeast strain AH109 containing the putative OsARF11 activator with a truncated CTD (lacking 35 amino acids), or full-length cDNAs for putative OsARF1, 4, 7, 10, 22, and 23 repressors was pairwise mated with yeast Y187 containing the 15 OsIAA proteins, the mated yeast grew only on low-stringency plates, and not on high-stringency plates (Figs 3B, 4A). Furthermore, different results were observed for full-length OsARF11 and OsARF11 containing a truncated CTD (Fig. 3A, B). In contrast to the full-length protein, the truncated protein does not show interactions with these OsIAAs in yeast. To determine if the putative OsARF repressors and truncated OsARF11 activator were expressed at similar levels in the yeast strain AH109, western blotting was performed to detect these fusion proteins from pGADT7-OsARFs. OsARF1, 4, 7, 10, 22, 23, and the truncated OsARF11 were detected by western blotting (Fig. 3C). These results indicated that the full-length cDNAs of putative OsARF repressors did not interact with the 15 OsIAA proteins by YTH assay and the intact CTD is required for OsARF interaction with OsIAA proteins.
Fig. 3.
OsARF–OsIAA protein interactions by YTH assays. (A) Eight full-length OsARF activators interact with 15 OsIAA proteins. α-Galactosidase activity assays were used to evaluate the intensity of interactions between OsARF and OsIAA proteins. The intensity of the blue colour in the panels indicates the relative α-galactosidase activity (quantitative data are given in Supplementary Table S8 at JXB online). The colour standard is on the right. Positive, interaction of pGBKT7-53 with pGADT7-T as the positive control (activity value: 22). Negative, interaction of pGBKT7-Lam with pGADT7-T as the negative control (activity value: 0). (B) Full-length OsARF repressors fail to interact with any of the 15 OsIAA proteins. OsARF11* shows truncated OsARF11, which lacks 35 amino acids in its C-terminus. Yeast colonies were grown on SDII, synthetic dropout (SD)–Leu/–Trp; or SDIV, SD–Ade/–His/–Leu/–Trp/ X-α-Gal plates. Positive and negative controls are the same as in A. (C) Western blotting analysis of six full-length OsARF repressors and the OsARF11 activator with a truncated C-terminal region. Lane 1, positive control, mutiple tag cell lysate (52KD, Genscript); lanes 2–8, lysates from the yeast strain AH109 containing full-length OsARF1, 4, 7, 10, 22, 23, and OsARF11 lacking 35 C-terminal amino acids with Gal4-AD fusion proteins, respectively. Numbers at the right denote the protein ladder of molecular weights of the standards (GeneDireX Inc, PM-22) in kilodaltons. (This figure is available in colour at JXB online.)
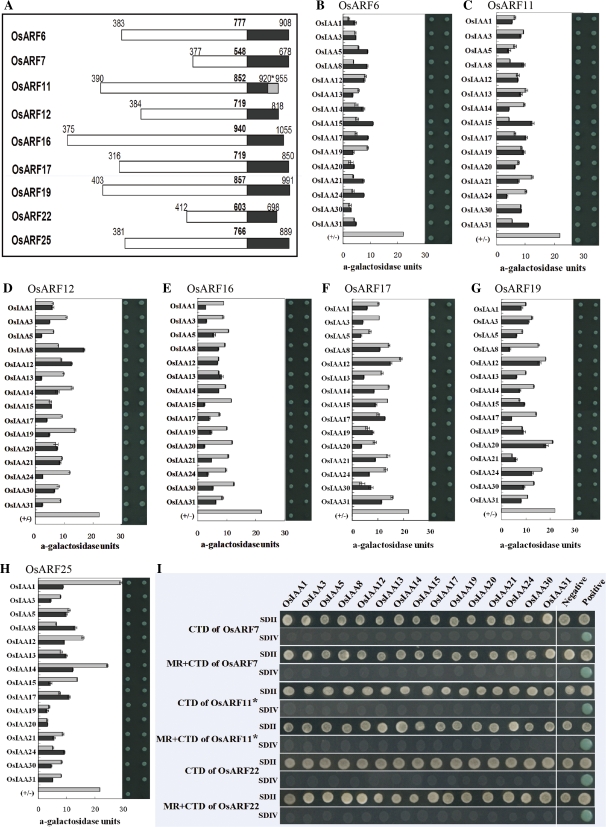
Fig. 4.
Interactions between CTDs or MR+CTDs of OsARFs and OsIAA proteins in YTH assays. (A) Positions of the MRs or CTDs of OsARFs for interaction studies. White, MRs; black, CTDs. Bold numbers are the last amino acid residue of the MR. Numbers on the left indicate the start positions of MRs, and numbers on the right indicate the CTD end based on the entire OsARF. The 955 in the grey box indicates the C-terminus for the entire OsARF11, and 920* shows the end of truncated OsARF11. (B–H) Interactions between CTDs or MR+CTDs of putative OsARF activators and the 15 OsIAA proteins. The left of each panel indicates α-galactosidase activity and the right of each panel shows yeast colonies grown on SDIV plates (SDII plates are not shown). Grey bars show the α-galactosidase activity of OsARF CTD–OsIAA protein interactions; black bars show α-galactosidase activity of OsARF MR+CTD–OsIAA protein interactions. (I) CTDs and MR+CTDs of an OsARF11 activator with a CTD truncation, and CTDs and MR+CTDs of OsARF7 and 22 repressors fail to interact with any of the 15 OsIAA proteins. Yeast colonies were grown on SDII, synthetic dropout (SD)–Leu/–Trp; or SDIV, SD–Ade/–His/–Leu/–Trp/X-α-Gal plates. Positive and negative controls are the same as in Fig. 3A. (This figure is available in colour at JXB online.)
Interactions between the CTD, MR+CTD, and full-length OsARFs, and OsIAA proteins were similar
To determine if the ARF MR has an effect on ARF–Aux/IAA binding, interactions between the CTD or MR+CTD of nine OsARFs and the 15 OsIAA proteins were tested using YTH assays. The positions of the MR or CTD of nine OsARF proteins used in the assays are shown in Fig. 4A, and Supplementary Fig. S1 at JXB online. The results indicated that the CTD or the MR+CTD of putative OsARF6, 11, 12, 16, 17, 19, and 25 activators interacted with the 15 OsIAA proteins (Fig. 4B–H). The α-galactosidase activities for interactions between most of the OsARF MR+CTDs and the OsIAA proteins were lower than for interactions between constructs with only the CTDs of OsARFs and OsIAA (Fig. 4B–H). The MR of OsARFs seemed, in general, to reduce the intensity of interactions. The CTD or MR+CTD from putative OsARF7 repressors or OsARF22 failed to interact with any of the 15 OsIAA proteins (Fig. 4I). In addition, the truncated CTD or MR+truncated CTD of the putative OsARF11 activator failed to interact with any of the 15 OsIAA proteins (Fig. 4I).
Interactions between OsARFs and OsIAA proteins in tobacco are partially consistent with results in yeast
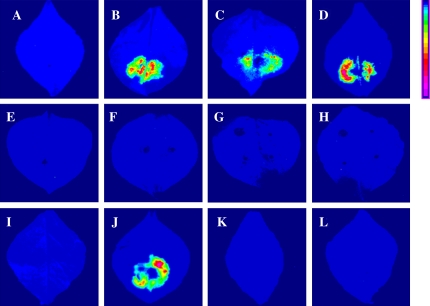
The firefly LCI assay is a simple, highly sensitive tool for testing interacting proteins in plants, using transient or stable transgenic expression (Chen et al., 2008). To understand the OsARF–OsIAAs interactions in plants, several combinations of OsARF1/16 and OsIAA1/8/12/13 were verified by LCI. Homodimers of OsIAA 1 or OsIAA12, and the positive control SGT1a-NLuc/CLuc-RAR1 resulted in strong luciferase complementation (Fig. 5B–D). The heterodimers of OsARF1 and OsIAA1/8/12/13, like the negative control, showed no visible fluorescence in tobacco leaves (Fig. 5A, E–H). A heterodimer of OsARF16 and OsIAA8 showed intense fluorescence, while OsARF16 and OsIAA1/OsIAA12/OsIAA13 did not (Fig. 5I–L). These results suggested that the interaction between OsARF and OsIAA proteins in plants may be more specific and finely controlled than in yeast.
Fig. 5.
Firefly luciferase complementation imaging assay of OsARF–OsIAA protein interactions in N. benthamiana leaves. Leaves were co-infiltrated with Agrobacterium containing the following vector pairs: (A) negative control, OsARF16-NLuc/CLuc; (B) positive control, SGT1a-NLuc/CLuc-RAR1; (C) OsIAA1-NLuc/OsIAA1-CLuc; (D) OsIAA12-NLuc/OsIAA12-CLuc; (E) OsARF1- NLuc/OsIAA1-CLuc; (F) OsARF1-NLuc/OsIAA8-CLuc; (G) OsARF1-NLuc/ OsIAA12-CLuc; (H) OsARF1-NLuc/OsIAA13-CLuc; (I) OsARF16-NLuc/OsIAA1- CLuc; (J) OsARF16-NLuc/OsIAA8-CLuc; (K) OsARF16-NLuc/OsIAA12-CLuc; (L) OsARF16-NLuc/OsIAA13-CLuc. Pseudocolour bar, right, shows the range of luminescence intensity from weak to strong. Images were collected 3 d after infiltration. The results were tested independently in triplicate. (This figure is available in colour at JXB online.)
Discussion
ARF transcription factors are categorized as transcriptional activators or repressors based upon the amino acid composition of the MR that is found between the DBD and the CTD in most ARF proteins. The ARF MR can function as an AD or an RD when fused to a heterologous DBD, and targeted to non-auxin response gene promoters (Tiwari et al., 2003). However, whether the Q, S, and L residues in Arabidopsis ARF activators or the S, L, P, and G residues in Arabidopsis ARF repressors are important for conferring activation and repression is unknown. Rice OsARF5, 6, 11, 12, 16, 17, 19, 21, and 25 (class IIa ARFs), which contain a high abundance of Q, S, and L in their MRs, resemble the Arabidopsis ARF activators ARF5, 6, 7, 8, and 19 (Fig. 1B). Thus, the class IIa OsARF subfamily might function as transcriptional activators for auxin response genes, while the other OsARFs containing MRs rich in S, L, P, and G might function as transcriptional repressors, analogous to the corresponding Arabidopsis ARFs.
Developmental phenotypes and alterations in gene expression have been examined in a number of loss-of-function arf mutants in Arabidopsis (Guilfoyle and Hagen, 2007). While most Arabidopsis arf mutants do not show dramatic phenotypes or changes in auxin-responsive gene expression, loss-of-function arf5 and arf7 mutants display strong down-regulation of a number of auxin response genes (Harper et al., 2000; Mattsson et al., 2003; Okushima et al., 2005; Wang et al., 2005; Wilmoth et al., 2005). Based on global gene expression profiling with Arabidopsis arf7 arf19 double mutants, most auxin response genes seemed to be activated by ARF7, but a small number of genes appeared to be repressed, suggesting that ARF7 may function as both an activator and a repressor (Okushima et al., 2005). However, the microarray analysis could not distinguish if the genes were direct or indirect targets of ARFs (Guilfoyle and Hagen, 2007). More in-depth experiments will be required to determine if the biased amino acid sequences in ARF MRs are responsible for activation or repression function, and whether ADs and RDs can function in a single ARF.
The NLS of the ARF protein in Arabidopsis was predicted to be PQRNKRPR (Ulmasov et al., 1997), but had not yet been experimentally confirmed. By bioinformatic prediction, the ARF proteins in rice have two NLSs, a non-canonical bipartite NLS close to the middle of the DBD, and a monopartite NLS in the end of the DBD. Although no classical bipartite NLS was found in OsARF proteins, the C-terminal region of the B3 domain contains basic amino acids, and is considered to be a putative bipartite NLS, because recently a non-canonical bipartite NLS, with a spacer sequence of >20 amino acids, was identified in humans (Romanelli et al., 2002) and viruses (Gomez Corredor et al., 2009). Hence, the predicted non-canonical bipartite NLS was also studied to determine if it can direct the gene product into the nucleus. However, the studies confirmed that a monopartite NLS was efficient to direct OsARF18/19/22/25–GFP fusion proteins into the nucleus, but a bipartite NLS was not (Fig. 2A, B). Even though the results were unlike the AUX/IAA gene family, which has a bipartite NLS, they were consistent with previous predictions in AtARF (Ulmasov et al., 1997; Thakur et al., 2005). The data demonstrated that the DBD contributed the function of nuclear localization for the OsARF proteins.
Studies with Arabidopsis have shown that the CTD of Aux/IAA proteins and ARFs represents a dimerization domains that facilitates interactions between ARF and Aux/IAA proteins (Kim et al., 1997; Ulmasov et al., 1997; Guilfoyle and Hagen, 2007). To date, however, only a small number of Arabidopsis ARFs and Aux/IAA proteins have been tested for their ability to interact with each other. The limited information suggests, however, that Arabidopsis ARF activators interact strongly with other ARF activators and Aux/IAA repressors, but interact poorly with ARF repressors (Guilfoyle and Hagen, 2007). In contrast, Arabidopsis ARF repressors appear to interact strongly with other ARF repressors, but weakly with ARF activators and Aux/IAA repressors. To assess interactions between ARF and Aux/IAA proteins more thoroughly, quantitative YTH assays were used to test the interactions between OsARF and OsIAA proteins, including seven full-length OsARF activators, one OsARF activator with a CTD truncation, seven full-length OsARF repressors, and 15 OsIAA proteins. Quantitative interaction studies with full-length OsARFs indicated that all full-length tested OsARF activators interacted with all tested OsIAA proteins, while an OsARF activator lacking an intact CTD failed to interact with any of the OsIAA proteins. In marked contrast, none of the OsARF repressors displayed detectable interactions with the panel of OsIAA proteins. These results confirmed the suggestions in Arabidosis above.
A further assessment was made of whether removal of the OsARF DBD or MR affected the capacity of OsARFs to interact with OsIAA proteins. In general, ARFs with truncated DBDs and MRs behaved like the full-length ARFs in their capacity to interact with OsIAA proteins. Direct comparison of interactions between the CTD and MR+CTD suggested, however, that the presence of an activator MR reduced the interaction of the OsARF CTD with OsIAA proteins. These results suggested that interactions between ARFs and Aux/IAA proteins may differ, depending on whether full-length ARFs or truncated ARFs are used to assess interaction with IAA proteins. This might explain why Arabidopsis repressor ARFs have been observed to interact with Aux/IAA proteins in some cases, but not in others (Ulmasov et al., 1997b; Tiwari et al., 2003; Ouellet et al., 2006; Guilfoyle and Hagen, 2007). The data suggest that ARF–Aux/IAA interactions are restricted but not random.
The YTH method is widely used in protein–protein interaction studies because of its suitability for large-scale screening. The YTH studies suggested that interactions between OsARF activators and OsIAA proteins are strong, and that interactions between OsARF repressors and OsIAA protein are weak, at best. These results are consistent with current models for auxin-regulated gene expression in Arabidopsis (Guilfoyle and Hagen, 2007). Those results provide clues about OsARF–OsIAA interactions, though they do not necessarily reflect the interactions in plants. The YTH system is prone to false positives in a heterologous system (Weijers et al., 2005; Chen et al., 2008). The interaction of two proteins often occurs in the presence of additional proteins or cellular factors; therefore, the YTH results might not accurately reflect the entire spectrum of ARF–Aux/IAA interactions that occur under natural conditions in plants. Similarly, false negatives might also emerge from YTH assays.
To understand the interactions between OsARFs and OsIAAs in plant, several interactions between OsARF–OsIAA pairs in tobacco were also analysed. Strong LCI was observed in only two homodimers, and in one heterodimer. Testing more OsARF–OsIAA pairs using the LCI assay would provide useful information about their interactions. Natural ARF–Aux/IAA interactions in plants are probably driven by a much lower concentration of ARF and Aux/IAA proteins than those assessed by YTH assays, and are probably regulated by differential expression patterns for the members of the ARF and Aux/IAA families.
In conclusion, confirmation of the NLS indicated that the DBD of OsARFs has nuclear localization function. The ability of OsARF activators and OsIAA to interact was greater than for OsARF repressors and OsIAA in yeast. The application of LCI facilitated research on ARF–Aux/IAA in plants.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The position of each domain of 25 OsARF proteins determined with http://www.ebi.ac.uk/Tools/t-coffee/. DBD, DNA-binding domain; MR, middle region; CTD, C-terminal dimerization domain.
Figure S2. Region of the nuclear localization signal (NLS).
Table S1. The sites of the MR domain in putative OsARF proteins.
Table S2. Sites of elements I–V of OsARFs for the nuclear localization signal.
Table S3. Primer sequences of OsARF elements for the nuclear localization signal.
Table S4. Primer sequences of 15 OsIAA genes for yeast-two hybrid analysis.
Table S5. Primer sequences of 15 OsARF genes for yeast two-hybrid analysis.
Table S6. Primer sequences of the CTD of OsARFs for yeast two-hybrid analysis.
Table S7. Primer sequences of the MR of OsARFs for yeast two-hybrid analysis.
Table S8. α-Galactosidase activity (mmol ml−1 min−1) in triplicate.
Table S9. Primer sequences of OsARF/OsIAA for firefly luciferase complementation imaging assay.
Supplementary Material
Acknowledgments
We thank the Laboratory of JianMing Zhou (National Institute of Biological Sciences, China) for providing the CAMBIA1300-NLuc/CLuc vector. This research is supported by the National Natural Science Foundation of China (grant no. 30770213), the National High Technology Research and Development Program of China (863 Program) (grant no. 2007AA10Z188), the Project sponsored by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (grant no. 20080435), and the Natural Science Foundation of Zhejiang province, China (grant no. Y3080111). We gratefully acknowledge the RGRC (Rice Genome Resource Center) in Japan for providing the full-length cDNA clones of OsIAA and OsARF gene families. We would like to thank Professors XueMei Chen and YueZhi Tao for their assistance in editing this manuscript.
References
- Abel S, Oeller PW, Theologis A. Early auxin-induced genes encode short-lived nuclear proteins. Proceedings of the National Academy of Sciences, USA. 1994;91:326–330. doi: 10.1073/pnas.91.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Theologis A. Early genes and auxin action. Plant Physiology. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiology. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. The Plant Journal. 2005;44:382–395. doi: 10.1111/j.1365-313X.2005.02537.x. [DOI] [PubMed] [Google Scholar]
- Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Applied Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- Gomez Corredor A, Archambault D. The bovine immunodeficiency virus Rev protein: identification of a novel lentiviral bipartite nuclear localization signal harbouring an atypical spacer sequence. Journal of Virology. 2009;83:12842–12853. doi: 10.1128/JVI.01613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Journal of Plant Growth Regulation. 2001;10:281–291. [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Current Opinion in Plant Biology. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Ulmasov T, Hagen G. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cellular and Molecular Life Sciences. 1998;54:619–627. doi: 10.1007/s000180050190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ. Auxin-responsive gene expression: genes, promoters, and regulatory factors. Plant Molecular Biology. 2002;15:533–543. [PubMed] [Google Scholar]
- Hamann T, Benkova E, Baurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes and Development. 2004;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. The Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa) Functional and Integrative Genomics. 2006;6:47–59. doi: 10.1007/s10142-005-0005-0. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. Protein–protein interactions among the Aux/IAA proteins. Proceedings of the National Academy of Sciences, USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Dai X, Zhao Y. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiology. 2006;140:899–908. doi: 10.1104/pp.105.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology. 2002;49:387–400. [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiology. 2003;131:1327–1339. doi: 10.1104/pp.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Nagao I, Demura T, Fukuda H, Kinjo M, Yamamoto KT. Fluorescence cross-correlation analyses of the molecular interaction between an Aux/IAA protein, MSG2/IAA19 and protein–protein interaction domains of auxin response factors of Arabidopsis expressed in HeLa cells. Plant and Cell Physiology. 2006;47:1095–1101. doi: 10.1093/pcp/pcj080. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3: biochemical insight into an auxin mutant phenotype. The Plant Cell. 2001;13:829–841. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanelli MG, Morandi C. Importin alpha binds to an unusual bipartite nuclear localization signal in the heterogeneous ribonucleoprotein type I. European Journal of Biochemistry. 2002;269:2727–2734. doi: 10.1046/j.1432-1033.2002.02942.x. [DOI] [PubMed] [Google Scholar]
- Song Y, Wang L, Xiong L. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta. 2008;229:577–591. doi: 10.1007/s00425-008-0853-7. [DOI] [PubMed] [Google Scholar]
- Song Y, You J, Xiong L. Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant MolecularBiology. 2009;70:297–309. doi: 10.1007/s11103-009-9474-1. [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. The Plant Cell. 2004;16:379–393. doi: 10.1105/tpc.018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Jain M, Tyagi AK, Khurana JP. Exogenous auxin enhances the degradation of a light down-regulated and nuclear-localized OsiIAA1, an Aux/IAA protein from rice, via proteasome. Biochimica et Biophysica Acta. 2005;1730:196–205. doi: 10.1016/j.bbaexp.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. The Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. Aux/IAA proteins are active repressors and their stability and activity are modulated by auxin. The Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant and Cell Physiology. 2008;49:1025–1038. doi: 10.1093/pcp/pcn079. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds auxin response elements. Science. 1997a;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences, USA. 1999a;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. The Plant Journal. 1999b;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell. 1997b;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV. Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. The Plant Cell. 1992;4:1213–1227. doi: 10.1105/tpc.4.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y. Genome-wide analysis of the auxin response factor (ARF) gene family in rice (Oryza sativa) Gene. 2007;394:13–24. doi: 10.1016/j.gene.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Wang S, Tiwari SB, Hagen G, Guilfoyle TJ. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. The Plant Cell. 2005;17:1979–1993. doi: 10.1105/tpc.105.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO Journal. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jurgens G. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Developmental Cell. 2006;10:265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. The Plant Journal. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. Degradation of Aux/IAA protein is essential for normal auxin signaling. The Plant Journal. 2000;21:553–562. doi: 10.1046/j.1365-313x.2000.00703.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.