Abstract
The plant-specific GRAS/SCL transcription factors play diverse roles in plant development and stress responses. In this study, a poplar SCL gene, PeSCL7, was functionally characterized in Arabidopsis thaliana, especially with regard to its role in abiotic stress resistance. Expression analysis in poplar revealed that PeSCL7 was induced by drought and high salt stresses, but was repressed by gibberellic acid (GA) treatment in leaves. Transient expression of GFP-PeSCL7 in onion epidermal cells revealed that the PeSCL7 protein was localized in the nucleus. Transgenic Arabidopsis plants overexpressing PeSCL7 showed enhanced tolerance to drought and salt treatments. The activity of two stress-responsive enzymes was increased in transgenic seedlings. Taken together, these results suggest that PeSCL7 encodes a member of the stress-responsive GRAS/SCL transcription factors that is potentially useful for engineering drought- and salt-tolerant trees.
Keywords: Abiotic stress, GRAS/SCL, overexpression, Populus euphratica Oliv
Introduction
Environmental abiotic stresses, such as cold, drought, and high salinity negatively impact plant growth and development. To reduce the adverse effects of these conditions, plants have evolved multifaceted strategies, including morphological, physiological, and biochemical adaptations. During the response and adaptation to diverse abiotic stresses, many stress-related genes are induced, and a variety of stress resistance-related functional proteins are accumulated. Numerous genes have been reported to be up-regulated under stress conditions in vegetative tissues (Seki et al., 2002; Zhu, 2002). Among these genes, transcription factors play important regulatory roles in stress responses by regulating their target genes via binding to the cognate cis-acting elements (Casaretto and Ho, 2003; Fujita et al., 2005; Yamaguchi-Shinozaki and Shinozaki, 2006). Members of the APETELA2 (AP2), bZIP, NAC, and MYB families have been shown to have regulatory roles in stress responses. Transgenic plants overexpressing these genes could enhance their tolerance to various stresses (Abe et al., 1997; Ito et al., 2006; Nakashima et al., 2007; Li et al., 2008).
The SCARECROW (SCR) gene was identified in Arabidopsis and is expressed specifically in root progenitor tissues of plant embryos and in certain root and stem tissues. The SCR gene encodes a novel putative transcription factor and is required for asymmetric cell division in an Arabidopsis root (DiLaurenzio et al., 1996). The modulation of SCR expression levels can be used advantageously to modify root and aerial structures of transgenic plants and to enhance the agronomic properties of such plants (Lim et al., 2005). A mutation of the SCR gene results in a radial pattern defect and the loss of a ground tissue layer in the root (Scheres et al., 1995; Sabatini et al., 2003; Heidstra et al., 2004). Pysh et al. (1999) identified a number of Arabidopsis expressed sequence tags (ESTs) that show similarity to the Arabidopsis SCR amino acid sequence and designated them the SCARECROW-LIKE genes (SCL). The SCL genes comprise a novel gene family, referred to as the GRAS gene family, based on the locus designations of three genes: the gibberellin-acid insensitive (GAI) locus, the repressor of GA1 (RGA) locus, and the SCARECROW (SCR) locus. The GRAS/SCL gene products have been reported to be plant-specific proteins that participate in various developmental processes. Members of the GRAS/SCL family have a variable N-terminus and a highly conserved C-terminus that contains five recognizable motifs: the leucine heptad repeat I (LHR I), the VHIID motif, the leucine heptad repeat II (LHR II), the PFYRE motif, and the SAW motif. The GRAS/SCL proteins function as transcription factors, but are not restricted to their role in asymmetric cell division. For example, the PAT1 protein was shown to be involved in phytochrome A signal transduction events of Arabidopsis thaliana (Bolle et al., 2000), and the tomato gene Lateral suppressor (Ls) functions in the formation of lateral branches (Schumacher et al., 1999). Two members of the GRAS family, the GAI and the RGA genes, play important roles in the gibberellic acid (GA) signal transduction pathway. Arabidopsis plants with a mutation at the GAI locus do not respond to exogenously applied GA and have a reduced stature (Koorneef et al., 1985); Peng et al. (1997, 1999) found that the mutation in the GAI locus is dominant negative leading to a GA-independent permanent accumulation of this protein that represses the GA pathway. The SLR1 gene of rice has been identified as a GAI orthologue and was demonstrated to be involved in the GA-signalling pathway in corn, rice, barley, grape, and wheat (Hynes et al., 2003). Overexpression of the Arabidopsis GAI in tobacco and rice produced a dwarf phenotype, compared with the wild-type plant (Hynes et al., 2003).
Populus euphratica Oliv., the most important species for large-scale afforestation projects on saline desert sites in China, can tolerate salt concentration up to 450 mM NaCl (Brosché et al., 2005). By analysing the cDNA-AFLP data (GH611858) in public databases at NCBI, an SCL gene was found, designated PeSCL7 according to Ma et al. (unpublished data). The characterization of SCL genes from P. euphratica would help us to understand its unexpected tolerance to environmental stresses. In this work, a SCL homologue, PeSCL7, from salt-treated P. euphratica is reported. Its expression pattern and subcellular localization were investigated. Transgenic Arabidopsis plants overexpressing PeSCL7 showed enhanced drought and salt tolerance.
Materials and methods
Plant materials and growth conditions
Two-year-old Populus euphratica plants were collected from Xinjiang in China. For expression analysis, uniformly developed seedlings of P. euphratica (50–60 cm high, with 40–50 leaves) were subjected to various stress treatments. Salt stress was conducted by watering young plants with 350 mM NaCl. The ABA and GA treatments were performed by spraying the leaves of the young plants with a 200 μM ABA and 200 μM GA solution. Dehydration was induced by removing plants from the plots and exposing them on filter paper to air with 70% RH at 25 °C under dim light for 24 h. For all treatments, leaves were collected at different time points as indicated in Fig. 1. Three leaves from a different plant were collected per harvest and were immediately frozen in liquid nitrogen for later use. Arabidopsis thaliana [ecotype Columbia-0 (Col-0)] plants were grown in soil under a short photoperiod (9 h of light, 8500 lux, 22 °C, 70% RH) for 3–4 weeks, followed by a long photoperiod (16 h of light, 8500 lux, 22 °C, 70% RH).
Fig. 1.
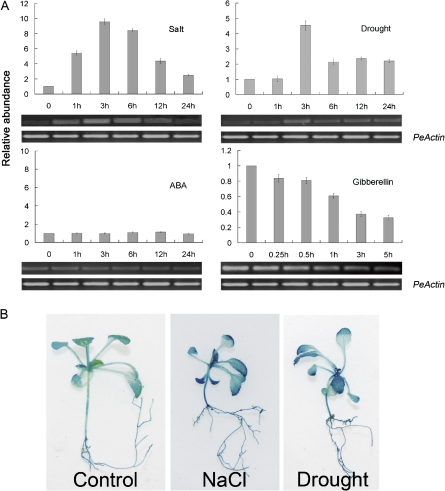
Expression patterns of PeSCL7. (A) qRT-PCR and RT-PCR assay of the accumulation of PeSCL7 gene transcripts in response to 350 mM NaCl, dehydration (removing plants from the plots and placing them under dim light for 24 h), 200 μM ABA, and 200 μM gibberellin (GA) treatment. The expression levels were normalized to that of PeActin, and the level of the PeSCL7 transcript in the controls was set at 1.0. Values are the mean ±SE (n=3 experiments). (B) PeSCL7 promoter–GUS expression pattern in transgenic Arabidopsis plants. Tissue localization of enhanced GUS expression in PeSCL7 promoter–GUS transgenic seedlings treated with 350 mM NaCl for 3 h and drought for 3 h. (This figure is available in colour at JXB online.)
PeSCL7 isolation
Total RNA was extracted by the CTAB method from leaves of P. euphratica treated with 350 mM NaCl for 3 h. cDNA synthesis were performed using M-MLV Reverse Transcriptase and an Oligo(dT) primer (Promega, U.S.A) according to the manufacturer's instructions. The cDNA sequence was amplified by PCR using the primers PeRT Fw and PeRT Rev. The primer sequences used are shown in Supplementary Table S1 at JXB online. PCRs were also performed with pairs of primers designed from the 5′ and 3′ ends of PeSCL7 and using poplar genomic DNA as the template.
RT- and qRT-PCR analysis
Total RNA from each sample was prepared from leaves indicated in Fig. 1 by the CTAB method, and 1 μg of RNA was used for the reverse transcription reaction. Subsequently, 1 μl of the reverse transcription products was used as the template for PCR amplification. The PCR products were examined on a 0.8% agarose gel stained with ethidium bromide. The same RNA samples and primers were used for real-time PCR analysis. The procedure described by Chen et al. (2009) was followed for real-time PCR and the statistical analysis. SYBR green was used to monitor the kinetics of PCR product formation in real-time RT-PCR. As an internal control, the PeActin transcript was used to quantify the relative transcript level of PeSCL7 in each sample, and Tubulin transcript levels served as an equal loading standard for the AMY1 and Cu/Zn SOD mRNA levels. The primer sequences used are shown in Supplementary Table S1 at JXB online. Each PCR assay was carried out for three biological replicates, and each replicate corresponded to three technological repeats of separate experiments.
Plasmid constructions
To produce the 35S-PeSCL7 and scl7-1/PeSCL7 plants, a 1779 bp BglII–SpeI fragment containing the PeSCL7 cDNA was cloned into the pCAMBIA-1304 binary vector (http://www.cambia.org/daisy/cambia/585.html), in which transgene expression is under the control of the CaMV 35S promoter. For the PeSCL7 promoter and GUS fusion construct, a 5′ flanking sequence (a 1 kb promoter region just upstream of the ATG start codon of PeSCL7) was amplified from genomic DNA by PCR and verified by sequencing. The PCR fragment was cloned into the PstI–SpeI site of the binary vector pCAMBIA-1304 to obtain a transcriptional fusion of the PeSCL7 promoter and the GUS coding sequence. Thus, the fragments were fused in-frame with the GUS–GFP fused gene.
Plant transformation and subcellular localization analysis
The constructs were introduced into the Agrobacterium tumefaciens strain LBA4404 and transformed into wild-type Arabidopsis (Columbia ecotype) (for gene overexpression, GFP and GUS staining assays) and scl7-1 mutants (for gene complementation) by the floral dip method (Zhang et al., 2006). T3 or T4 homozygous lines were used for the phenotypic analysis. T3 homozygous lines were used for detailed analysis. T2 seeds were germinated on MS plates containing 60 μg ml−1 hygromycin for pCAMBIA-1304 constructs, and the resistant plants were transferred to soil to obtain homozygous T3 seeds. GFP fusion proteins were expressed in onion epidermal cells by using particle-mediated DNA delivery (Von Arnim and Deng, 1994). After bombardment, the epidermal peels were incubated with liquid MS medium for 16 h in the dark, mounted on slides, and visualized using a laser-scanning confocal microscope (Leica Heerbrugg, Switzerland).
GUS activity assays
To test the induction of GUS expression by salt and drought, 10-d-old PeSCL7 promoter–GUS transgenic seedlings were transferred from agar plates to MS liquid medium containing NaCl for salt treatment or to a filter exposed to air with 70% RH for drought treatment. GUS staining was performed according to Nakashima and Yamaguchi-Shinozaki (2002).
Verification of the AtSCL7 T-DNA insertion mutant
T-DNA insertion lines were obtained from the ABRC (Alonso et al., 2003). The T-DNA insertion lines scl7-1 and scl7-2 (Salk_106909, Salk_106426) were both in the Col-0 ecotype background. The homozygous mutant was identified by PCR from genomic DNA using the AtSCL7 gene-specific forward primer P1, the T-DNA left border primers LBb1, and the AtSCL7 gene-specific reverse primer P2. The PCR products were analysed further by DNA sequencing to confirm the insertion of the T-DNA in the gene. For RT-PCR analysis of the AtSCL7 transcripts in wild-type and T-DNA insertion mutant seedlings, the primer pairs used were RT Fw and RT Rev. The primer sequences used are shown in Supplementary Table S1 at JXB online.
Water loss measurements
For water loss experiments, rosette leaves of wild-type, scl7 mutant and transgenic Arabidopsis plants that were grown under normal conditions for 25 d, were excised, weighed immediately (leaves weighing approximately 1 g were harvested and used immediately for experiments) and incubated on a bench at room temperature and at 60% humidity under dim light. Losses in fresh weight were monitored at the times indicated. Water loss is expressed as the percentage of initial fresh weight.
Measurement of α-amylase and SOD activity
The α-amylase activity was assayed by the starch azure method (Doehlert and Duke, 1983). Leaves were extracted in 15 mM calcium acetate at pH 7.2. The 500 μl reaction mixture contained 50 mM of MOPS at pH 6.6 and 10 mg ml−1 of starch azure. After incubation at 25 °C, the undigested starch azure was precipitated by adding 2.5% (w/v) of trichloroacetic acid and was removed by centrifugation. The absorbance of the supernatant at 595 nm was measured. The SOD activity was measured according to McCord and Fridovich (1969). Briefly, the inhibition of formazan formation at 560 nm in 50 mM potassium phosphate buffer at pH 7.5, supplemented with 0.5 mM NBT, was monitored. The SOD activity was determined by monitoring the inhibition of the reduction rate of Cytochrome c between the reaction mixture and the control without protein extract (up to 200 mg protein) at 500 nm.
Results
Identification of the PeSCL7 gene
A gene encoding a member of the GRAS protein family was cloned; the latter show the highest identity to SCL7 in Arabidopsis, and the gene was named PeSCL7. The P. euphratica SCL7 (PeSCL7) protein is predicted to be 64.7 kDa in size and possesses a typical GRAS domain with two conserved LHR and VHIID motifs. A database search revealed that PeSCL7 is 49% and 46% identical to the Arabidopsis thaliana and Oryza sativa SCARECROW-LIKE 7 (SCL7) (Bolle, 2004; Tian et al., 2004) proteins, respectively. The genomic PCR products amplified by primers designed from the 5' and 3' untranslated region indicated that PeSCL7 had no intron interrupting its coding region. Our cDNA-AFLP data indicated that its transcript was induced to high levels after salt stress treatment, and this was later confirmed by RT- and qRT-PCR analysis (Fig. 1A). The cDNA-AFLP results suggested that PeSCL7 is involved in the stress response.
Expression profile of PeSCL7
To verify the cDNA-AFLP PeSCL7 expression result, the expression pattern of PeSCL7 under various stress treatments was investigated. PeSCL7 expression was up-regulated by salt and drought stress treatment and down-regulated by gibberellin (GA) but not ABA application in the leaves (Fig. 1A). Under the salt treatment, PeSCL7 expression was induced quickly, and the level of the product peaked at 3 h. Under the drought treatment, the PeSCL7 transcript was not induced immediately, but then reached its maximum at 3 h. For GA treatment, the expression of PeSCL7 became weaker within 5 h. Further analysis by qRT-PCR showed similar results to that of semi-quantitative RT-PCR. The expression pattern was also determined by analysing the expression of the PeSCL7 promoter–β-glucuronidase (GUS) fusion (Fig. 1B). Histochemical staining demonstrated that GUS activity increased throughout the plant in salt conditions and after drought treatment.
Targeting of PeSCL7 to the nucleus
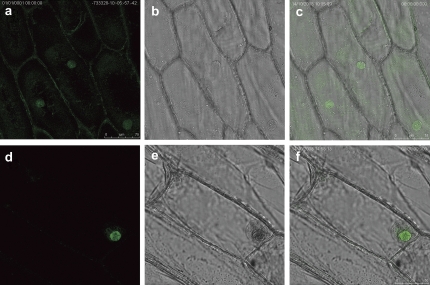
To determine the subcellular localization of PeSCL7, p35S:PeSCL7-GFP and p35S:GFP were transiently expressed in onion epidermal cells. As shown in Fig. 2, the onion cells transformed with the p35S:GFP vector displayed fluorescence throughout the whole cells (Fig. 2a–c). By contrast, fluorescence in the onion cell transformed with p35S:GFP-PeSCL7 was detected exclusively in the nucleus (Fig. 2d–f), indicating that PeSCL7 encoded a nuclear-localized protein.
Fig. 2.
Subcellular localization of PeSCL7 by GFP fusion expression in onion epidermal cells. p35S:GFP (as a control) and p35S:PeSCL7-GFP were transiently expressed in onion epidermal cells. The cells were analysed and photographed under a dark or light field by fluorescence microscopy for the p35:GFP control plasmid (a, b) and the p35S:PePeSCL7-GFP plasmid (d, e). (c, f) Merged images. (This figure is available in colour at JXB online.)
T-DNA insertion mutants of the AtSCL7 gene
To elucidate the in vivo functions of PeSCL7, reverse genetics, overexpression, and complementation approaches were applied. The loss-of-function mutant of the AtSCL7 gene and the two independent T-DNA insertion lines, scl7-1(Salk_106909) and scl7-2 (Salk_106426), were ordered from the ABRC seed stock centre. The T-DNA insertion positions are illustrated in Supplementary Fig. S1A at JXB online; the homozygous mutants were verified by diagnostic PCR using AtSCL7 gene-specific and T-DNA border primers (see Supplementary Fig. S1B at JXB online). Both the scl7-1 and scl7-2 null alleles were confirmed by RT-PCR (see Supplementary Fig. S1C at JXB online).
Salt and osmotic responses of 35S-PeSCL7 and scl7-1 plants
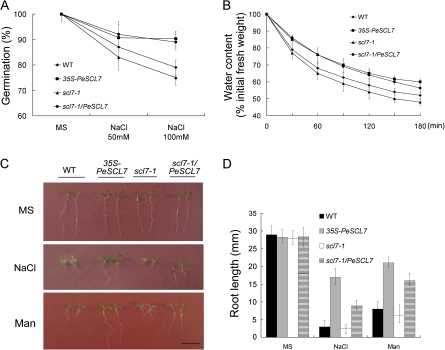
Salts inhibit germination and seedling growth in a concentration-dependent manner (Xiong et al., 2002). Since PeSCL7 is a salt-induced gene (Fig. 1), it is probable that PeSCL7 plays a role in plant responses to salt. Four different genotype seeds (WT, 35S-PeSCL7, scl7-1, scl7-1/PeSCL7) were germinated on MS medium containing 0, 50, and 100 mM NaCl, and differences were observed at both germination and post-germinative growth stages. Under our experimental conditions, the germination ratio of the wild-type plants was reduced about 12% by 50 mM NaCl and 20% by 100 mM NaCl, while the scl7-1 mutant was more affected. By contrast, the germination efficiency of the 35S-PeSCL7 and scl7-1/PeSCL7 plants was less affected (below 10% reduction) by 100 mM NaCl (Fig. 3A). To investigate PeSCL7 in plant salt or general osmotic effects further, the WT, 35S-PeSCL7, scl7-1, and scl7-1/PeSCL7 plants were germinated and grown on MS medium with or without 100 mM NaCl and 200 mM mannitol (an osmotic agent) (Fig. 3C). The growth of the WT and scl7-1 plants was strongly inhibited compared with the transgenic plants upon NaCl and mannitol treatment. The transgenic plants were much healthier and had more green cotyledons than the WT and scl7-1 plants (Fig. 3C). To quantify the difference between the four plant genotypes upon salt or general osmotic stress, the root length of the plants were measured (Fig. 3D). The result showed that the WT and scl7-1 mutant were dramatically affected by a decrease of about 75% of the control plants under the stress treatments, whereas the transgenic plants were less influenced. Thus, both the germination (Fig. 3A) and post-germination (Fig. 3C, D) growth of the 35S-PeSCL7 transgenic plants are more tolerant to high salinity and osmotic stress.
Fig. 3.
Salt and osmotic sensitivity of wild-type, 35S-PeSCL7, scl7-1, and scl7-1/PeSCL7 plants. (A) Germination ratio of the wild-type, mutant, and transgenic plants in the absence or presence of NaCl (50 or 100 mM). Values are mean ±SE (n=3 experiments). (B) Water loss in wild-type, mutant, and transgenic plants. Detached leaves from 25-d-old plants grown on soil were incubated on a bench and the fresh weight (FW) was measured at the time intervals indicated. Water loss is expressed as the percentage of initial fresh weight of detached leaves. Values are mean ±SE (n=3 experiments). (C) Salt and osmotic effects on newly germinated seedling growth. Seeds of four different genotypes were germinated for 5 d on MS medium and MS medium containing 100 mM NaCl or 200 mM mannitol (Man), transferred to the same type of medium, and grown in a vertical position for an additional 1 d. Representative seedlings are shown. (D) Root elongation of four different genotypes under normal condition or salt and osmotic treatment (described above). Values are mean ±SE (n=3 experiments). (This figure is available in colour at JXB online.)
Overexpression of PeSCL7 in transgenic Arabidopsis improved drought tolerance
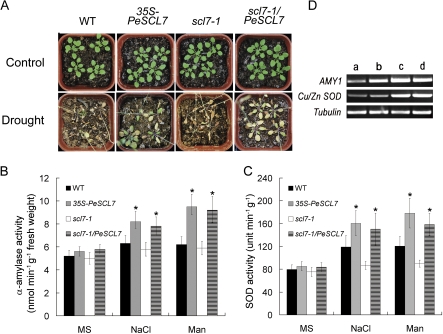
To characterize the in vivo function of PeSCL7, 18-d-old 35S-PeSCL7, scl7-1/PeSCL7, and scl7-1 mutant plants, as well as wild-type control plants, were not watered for 15 d to induce drought stress. As shown in Fig. 4A, most of the wild-type plants and all of the scl7-1 mutant plants were withered, but the 35S-PeSCL7 plants exhibited continued to survive and grow. Consistent with these results, the detached leaves of transgenic plants (35S-PeSCL7 and scl7-1/PeSCL7) lost water more slowly than did those of the wild-type and scl7-1 mutant plants (Fig. 3B). Thus, it is concluded that the 35S-PeSCL7 transgenic plants were highly tolerant to drought stress.
Fig. 4.
Effect of PeSCL7 expression on drought tolerance in the Arabidopsis wild type, scl7-1 mutant, and transgenic plants. (A) Arabidopsis wild-type, scl7-1, and transgenic plants were grown for 18 d in a growth chamber under normal growth conditions (Control), as indicated. Thereafter, water was withheld for 15 d. A representative plant is shown. (B, C) Comparison of α-amylase and SOD activity among four different genotypes. 10-day-old seedlings were treated with 0 or 100 mM NaCl or 200 mM mannitol in liquid MS medium for 5 h, and then the enzyme activities were assayed. Values are mean ±SE (n=3 experiments, *P <0.05). (D) Estimation of AMY1 and Cu/Zn SOD transcript levels in the wild type (a) and 35S-PeSCL7 (b, c, d) by RT-PCR. Total RNA was isolated from leaves of 10-d-old seedlings treated with 0 (a, b), 100 mM NaCl (c), or 200 mM mannitol (d) in liquid MS medium for 5 h. The transcript levels were estimated by RT-PCR with specific primers for AMY1 and Cu/Zn SOD. Tubulin transcript levels served as an equal loading standard. The experiment was repeated three times, and the typical result of an ethidium bromide-stained agarose gel is presented. (This figure is available in colour at JXB online.)
To understand better the mechanisms of drought and salt tolerance conferred by overexpressing 35S-PeSCL7, the activity of several known enzymes related to abiotic stresses in transgenic Arabidopsis plants was investigated. As shown in Fig. 4B and C, the α-amylase and SOD activity assays showed that the transgenic plants exhibited the same level of activity as the WT and mutants under normal growth conditions. However, after stresses, the elevation of the enzymatic activity in transgenic plants was significantly larger than those of the WT and mutants (P <0.05). Since PeSCL7 is a transcription factor, the transcript levels of the AMY1 and Cu/Zn SOD enzymes were also investigated (Fig. 4D). The elevated transcript levels of AMY1 and Cu/Zn SOD were positively correlated with the α-amylase and SOD activities under stress conditions, as observed from the RT-PCR results. Under normal growth conditions, only the AMY1 transcript level was elevated in the transgenic plant, and no obvious difference was observed for the Cu/Zn SOD transcript level.
Discussion
Populus euphratica is a poplar species known for its high salt tolerance (Chen et al., 2003; Ottow et al., 2005). Thus, much interest has been focused on elucidating the molecular and cellular processes underlying its acclimation to abiotic stresses. A gene encoding a member of the GRAS protein family that is induced in cuttings of P. euphratica during the early stages of severe salt stress, has been characterized here. PeSCL7 is a homologue of SCL7, which belongs to the SCL4/7 subgroup in the GRAS family (Bolle, 2004). In this subgroup there are only two members (SCL4 and SCL7). By analysing the data of AtgenExpression (http://jsp.weigelworld.org/expviz/expviz.jsp) (Kilian et al., 2007), SCL4 and SCL7 have the same expression level under normal growth conditions, however, SCL7 was up-regulated, while SCL4 was down-regulated (data not shown) under stress conditions. In our experiment, expression analysis showed that the PeSCL7 mRNAs were concomitantly inducible in response to diverse environmental stresses in the P. euphratica seedlings (Fig. 1A). Promoter expression analysis of PeSCL7 provided further support for its role in stress tolerance (Fig. 1B); GUS gene expression was enhanced by drought and NaCl, and was observed throughout the entire plant after stress treatment.
GRAS proteins have been classified as transcriptional regulators and have been shown to be involved in gibberellic acid (GA) and phytochrome signalling, root and axillary shoot development, and the maintenance of the shoot apical meristem. GRAS proteins share signature VHIID motifs flanked by two leucine heptad repeats (LHRI and LHRII), and a PFYRE and SAW (designated after the profound amino acid residues in the conserved regions) motif with rather variable N-terminal regions (Pysh et al., 1999; Bolle, 2004). The VHIID domain of a GRAS protein from Brassica napus interacts with a histone deacetylase, supporting the notion that GRAS proteins function in regulating gene expression at the level of transcription (Gao et al., 2004). Using a transient expression assay, clear evidence is provided that the PeSCL7 protein localizes to the nucleus of the plant cell. Nuclear localization is consistent with the large body of evidence indicating that members of the GRAS family play key roles in transcriptional regulation (Bolle, 2004; Lee et al., 2008).
Transgenic Arabidopsis overexpressing PeSCL7 showed significantly increased tolerance to abiotic stress at the seedling stage. Our data showed that transgenic plants had longer roots and a slower rate of water loss. The growing tips of the plant roots are critical tissues for establishing a root network. The root tip contains the root apical meristem, where directional cell divisions continuously produce new cells (Benfey and Scheres, 2000). Subsequently, the elongation and differentiation of these cells produce ordered root cell profiles. The developmental programme that regulates the plant root architecture has been studied intensively and involves layers of functions involving multiple transcription factors (Nakajima and Benfey, 2002; Petricka and Benfey, 2008). When plants are exposed to environmental stress, the root tips are usually the first to encounter the new environment. Therefore, the cell divisions and differentiations at the root tips are better able to respond to environmental signals. The primary roots of wild-type Arabidopsis under salt stress reduce the growth rate by reducing the number of dividing cells in the meristems, thus producing smaller mature cells without changing the duration of the cell cycle (West et al., 2004). By contrast, the salt-sensitive mutant stt3a undergoes cell cycle arrest under salt stress (Koiwa et al., 2003), indicating that sustaining cell cycle progression is an integral part of the stress tolerance mechanism in plant roots. Furthermore, the developmental programme that determines the identity of the cells determines the stress response outputs of individual root cells (Dinneny et al., 2008). Previous studies showed that transgenic plants overexpressing some stress-responsive genes, such as OsNAC6/SNAC2 (Nakashima et al., 2007; Hu et al., 2008), OsDREB1A, OsDREB1B, AtDREB1A, and AtDREB1B (Ito et al., 2006) led to growth retardation under normal conditions, and are probably responsible for the significant reduction in the potential yield. A similar phenotype in transgenic Arabidopsis overexpressing PeSCL7 was not observed. Next, an investigation will be made to assess whether transgenic poplar overexpressing PeSCL7 can enhance its stress tolerance without causing growth retardation.
It was observed that the activity of two stress-responsive enzymes, α-amylase and SOD, were elevated in transgenic plants under stress conditions (Fig. 4B, C). Amylase catalyses the conversion of starch. Doyle et al. (2007) reported that the AMY1 activity increases in Arabidopsis leaves under high temperature stress. In rice, the amylase gene AMY3D responds to osmotic stress (Hwang et al., 1998). The stress-inducible amylase is believed to play a role in the recovery of carbohydrates from damaged tissue to healthy tissues in plants. SOD belongs to a class of enzymes that catalyses the dismutation of superoxide into oxygen and hydrogen peroxide. As such, the enzymes are an important antioxidant defence in nearly all cells exposed to oxygen. Several previous studies have indicated that increasing endogenous SODs enhances stress tolerance (Samis et al., 2002; Yu et al., 2008). In this study, the α-amylase and SOD activities were also elevated by abiotic stresses (Fig. 4B, C); their transcript level (AMY1 and Cu/Zn SOD) also increased under stress conditions (Fig. 4D). However, under the normal growth conditions, the transcript level of AMY1 was higher in transgenic plants, while the transcript level of Cu/Zn SOD was similar to that of the wild type (Fig. 4D). The result suggests that AMY1 may be regulated by PeSCL7 directly, while Cu/Zn SOD is not. Further research is still needed to shed light on this complicated regulation network.
In conclusion, a stress-responsive GRAS gene, PeSCL7, was isolated. PeSCL7-expressing transgenic Arabidopsis plants showed improved tolerance to various environmental stresses, including high salinity, osmotic, and drought stresses. Further experiments are now required to define better the biochemical and physiological functions of PeSCL7 in response to adverse environmental factors in poplar.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Characterization of the homozygous Salk lines of AtSCL7 (At3g50650).
Supplementary Table S1. PCR primers used in this study.
Supplementary Material
Acknowledgments
This work was supported by the Hi-Tech Research and Development Program of China (2007AA10Z106), the National Natural Science Foundation of China (30730077, 30972339), the Program for New Century Excellent Talents in University of China (NCET-07-0083) and the ‘948’ Project of State Forestry Administration of China (2007-4-01).
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. The Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Scheres B. Root development. Current Biology. 2000;10:R813–R815. doi: 10.1016/s0960-9822(00)00814-9. [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH. PAT1, a new member of the GRAS family, is involved in phytocrome A signal transduction. Genes and Development. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- Brosché M, Vinocur B, Alatalo ER, et al. Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biology. 2005;6:R101. doi: 10.1186/gb-2005-6-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaretto J, Ho TH. The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. The Plant Cell. 2003;15:271–284. doi: 10.1105/tpc.007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Xia XL, Yin WL. Expression profiling and functional characterization of a DREB2-type gene from Populus euphratica. Biochemical and Biophysical Research Communications. 2009;378:483–487. doi: 10.1016/j.bbrc.2008.11.071. [DOI] [PubMed] [Google Scholar]
- Chen SL, Li JK, Wang SS, Fritz E, Huttermann A, Altman A. Effects of NaCl on shoot growth, transpiration, ion compartmentation, and transport in regenerated plants of Populus euphratica and Populus tomentosa. Canadian Journal of Forest Research. 2003;33:967–975. [Google Scholar]
- DiLaurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- Doehlert DC, Duke SH. Specific determination of α-amylase activity in crude plant extracts containing β-amylase. Plant Physiology. 1983;71:229–234. doi: 10.1104/pp.71.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EA, Lane AM, Sides JM, Mudgett MB, Monroe JD. An α-amylase (At4g25000) in Arabidopsis leaves is secreted and induced by biotic and abiotic stress. Plant, Cell and Environment. 2007;30:388–398. doi: 10.1111/j.1365-3040.2006.01624.x. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. AREB1 is a transcription activator of novel ABRE-dependent ABA-signaling that enhances drought stress tolerance in Arabidopsis. The Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao MJ, Parkin I, Lydiate D, Hannoufa A. An auxin responsive SCARECROW-like transcriptional activator interacts with histone deacetylase. Plant Molecular Biology. 2004;55:417–431. doi: 10.1007/s11103-004-0892-9. [DOI] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes and Development. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Molecular Biology. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- Hwang YS, Karrer EE, Thomas BR, Chen L, Rodriguez RL. Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Molecular Biology. 1998;36:331–341. doi: 10.1023/a:1005956104636. [DOI] [PubMed] [Google Scholar]
- Hynes LW, Peng J, Richards DE, Harberd NP. Transgenic expression of the Arabidopsis DELLA proteins GAI and gai confers altered gibberellin response in tobacco. Transgenic Research. 2003;12:707–714. doi: 10.1023/b:trag.0000005145.68017.6e. [DOI] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant and Cell Physiology. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. The Plant Cell. 2003;15:2273–2284. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koorneef M, Elgersma A, Hanhart CJ, van Loenen-Martinet EP, Rijn L, Zeevaart JAD. A gibberellin insensitive mutant of Arabidopsis thaliana. Plant Physiology. 1985;65:33–39. [Google Scholar]
- Lee MH, Kim B, Song SK, et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Molecular Biology. 2008;67:659–670. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Jung JW, Lim CE, Lee MH, Kim BJ, Kim M, Bruce WB, Benfey PN. Conservation and diversification of SCARECROW in maize. Plant Molecular Biology. 2005;59:619–630. doi: 10.1007/s11103-005-0578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein) Journal of Biological Chemistry. 1969;244:6049–6055. [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K. Use of β-glucuronidase to show dehydration and high-salt gene expression. In: Jackson JF, Linskens HF, editors. Molecular methods of plant analysis, Vol. 22. Testing for genetic manipulation in plants. Berlin: Springer-Verlag; 2002. pp. 37–61. [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress responsive gene expression in rice. The Plant Journal. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Benfey PN. Signaling in and out: control of cell division and differentiation in the shoot and root. The Plant Cell. 2002;14:S265–S276. doi: 10.1105/tpc.010471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottow EA, Brinker M, Teichmann T, Fritz E, Kaiser W, Brosché M, Kangasjärvi J, Jiang X, Polle A. Populus euphratica displays apoplastic sodium accumulation, osmotic adjustment by decreases in calcium and soluble carbohydrates, and develops leaf succulence under salt stress. Plant Physiology. 2005;139:1762–1772. doi: 10.1104/pp.105.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes and Development. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Moritz T, Cano-Delgado A, Harberd NP. Extragenic suppressors of the Arabidopsis gai mutation alter the dose–response relationship of diverse gibberellin responses. Plant Physiology. 1999;119:1199–1207. doi: 10.1104/pp.119.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Benfey PN. Root layers: complex regulation of developmental patterning. Current Opinion in Genetics and Development. 2008;18:354–361. doi: 10.1016/j.gde.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROWLIKE genes. The Plant Journal. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes and Development. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samis K, Bowley S, McKersie B. Pyramiding Mn-superoxide dismutase transgenes to improve persistence and biomass production in alfalfa. Journal of Experimental Botany. 2002;53:1343–1350. [PubMed] [Google Scholar]
- Scheres B, DiLaurenzio L, Willemsen V, Hauser M-T, Janmaat K, Weisbeek P, Benfey PN. Mutations affecting the radial organization of the Arabidopsis root display specific effects throughout the embryonic axis. Development. 1995;121:53–62. [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. The Lateral supressor (Ls) gene of tomato encodes a member of the VHIID protein family. Proceedings of the National Academy of Sciences, USA. 1999;96:290–295. doi: 10.1073/pnas.96.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Tian C, Wan P, Sun S, Li J, Chen M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Molecular Biology. 2004;54:519–532. doi: 10.1023/B:PLAN.0000038256.89809.57. [DOI] [PubMed] [Google Scholar]
- Von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- West G, Inzé D, Beemster GTS. Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiology. 2004;135:1050–1058. doi: 10.1104/pp.104.040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. The Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. The Plant Cell. 2008;20:1134–1151. doi: 10.1105/tpc.108.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Proceedings of the National Academy of Sciences, USA. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.