Abstract
Somatic embryogenesis in Arabidopsis is achieved by culturing bending-cotyledon embryos on a 2,4-D-containing induction medium for 14 d followed by a transfer on to a hormone-free development medium. Several genes orthologous to Arabidopsis SHOOTMERISTEMLESS (STM), CLAVATA 1 (CLV1), and ZWILLE (ZLL) were isolated from Brassica oleracea (Bo), B. rapa (Br), and B. napus (Bn), and ectopically expressed in Arabidopsis to assess their effects on somatic embryogenesis. Ectopic expression of BoSTM, BrSTM, and BnSTM increased the number of somatic embryos, whereas a different effect was observed in lines overexpressing BnCLV1 in which somatic embryo formation was severely repressed. The introduction of BnZLL did not have any effects on Arabidopsis somatic embryogenesis. The increased embryo-forming capacity observed in lines overexpressing Brassica STM was associated with a lower requirement for the inductive signal 2,4-D, and a higher expression of WUSCHEL (WUS) which demarcates the formation of embryogenic cells. This was in contrast to the 35S::BnCLV1 lines which showed the highest requirement for exogenous 2,4-D and a reduced WUS expression. Microarray studies were conducted to monitor global changes in transcript levels during Arabidopsis somatic embryogenesis between the wild-type (WT) line and a BoSTM-overexpressing line, which showed the most pronounced enhancement of somatic embryo yield. The introduction of BoSTM affected the expression of many genes involved in hormone perception and signalling, as well as genes encoding DNA methyltransferases and enzymes of glutathione metabolism. Pharmacological experiments performed to confirm some of the microarray results showed that Arabidopsis somatic embryogenesis is encouraged by a global hypomethylation of the DNA during the induction phase and by a switch of the glutathione pool towards an oxidized state during the subsequent development phase. Both events occurred in the 35S::BoSTM line, but not in the WT line. Altered expression of Brassica STM also had profound effects on B. napus microspore-derived embryogenesis. The yield of microspore-derived embryos increased in lines overexpressing BnSTM and significantly decreased in antisense lines down-regulating BnSTM.
Keywords: Apical meristem, Arabidopsis, Brassica, embryogenesis, gene expression, glutathione
Introduction
Embryogenesis in higher plants is initiated by the fusion of the sperm cell with the egg, resulting in the formation of a diploid zygote (Willemsen and Scheres, 2004). An important event during embryogenesis is the establishment of the shoot apical meristem (SAM), which in Arabidopsis is regulated by several genes including SHOOTMERISTEMLESS (STM), ZWILLE (ZLL), and CLAVATA 1 (CLV1). STM encodes a knotted-like homeobox protein present throughout the vegetative and floral meristems (Barton and Poethig, 1993). Genetic studies (Barton and Poethig, 1993; Endrizzi et al., 1996) demonstrate the requirement for STM for the formation of embryonic meristems and for their post-embryonic maintenance by suppressing differentiation and maintaining an indeterminate cell fate within the SAM. As also observed for stm mutants, zll embryos produce terminally differentiated cells instead of stem cells at the site of the SAM (McConnell and Barton, 1995; Moussian et al., 1998; Lynn et al., 1999). However zll seedlings were able to form adventitious SAMs originating between the cotyledons and to generate shoots in vitro. These observations are consistent with the notion that ZLL, a member of the ARGONAUTE family, is also required for the formation of the primary embryonic SAM, but, unlike STM, it is not needed for post-embryonic meristem function (McConnell and Barton, 1995; Moussian et al., 1998; Lynn et al., 1999).
Another key player in SAM regulation is CLV1, a leucine-rich repeat receptor kinase protein (Clark et al., 1997), which promotes cell differentiation of the meristematic cells by limiting the expression of the WUSCHEL (WUS) domain through an elaborated signalling model involving other CLV members (reviewed by Dodsworth, 2009). A mutation in CLV1 disrupts the balance between cell division and differentiation within the SAM culminating in enlarged meristems as a result of an atypical expansion of the WUS expression domain (Schoof et al., 2000). Therefore, in contrast to STM which maintains stem cells in an undifferentiated state by preventing meristematic cells from adopting an organ-specific cell fate, the role of CLV1 is to limit the expansion of the undifferentiated stem cell population in the SAM and promote differentiation. Insights into the competitive role of STM and CLV1 in meristem activity have been documented by genetic studies (Clark et al., 1996).
Embryogenesis can also occur in vitro via somatic embryogenesis or androgenesis (Thorpe and Stasolla, 2001). In Arabidopsis, somatic embryogenesis can be induced from immature zygotic embryos (Mordhorst et al., 2002; Bassuner et al., 2007). This system is very effective in providing a large number of embryos through an intervening callus phase, and represents an attractive ‘proof of concept’ model to examine the molecular events associated with the transition of somatic cells into embryos (Mordhorst et al., 2002; Su et al., 2009).
An alternative propagation system is androgenesis which uses immature pollen grains, or microspores, to generate microspore-derived embryos (MDEs). In recent years Brassica napus androgenesis has received increasing attention as a model to investigate embryogenesis in plants especially because of the genetic similarities shared between Brassica and Arabidopsis, and for the absence of a callus phase which allows studies during the initial phases of embryo development. These characteristics have been exploited for the identification of transcripts delineating the initial transition of gametophytic cells into embryogenic cells (Malik et al., 2007), and the subsequent development of the MDEs (Stasolla et al., 2008). Despite the increasing interest in B. napus microspore-derived embryogenesis, no information is currently available on genes involved in the formation and maintenance of the SAM. This is unfortunate as this system has been proved to be extremely suitable for examining the physiological factors required for proper meristem development (Belmonte et al., 2006; Stasolla et al., 2008). Thus the identification of Brassica SAM molecular markers is critical for elucidating the mechanisms governing meristem development during microspore-derived embryogenesis.
Over the past few years attempts have been made to determine if genes regulating SAM activity in vivo are also involved in the formation of embryonic cells during in vitro embryogenesis. While the majority of studies have used loss-of-function mutants (Ogas et al., 1999; Mordhorst et al., 1998, 2002), very few deal with overexpression experiments (Zuo et al., 2002). The aim of the present work is to characterize Brassica genes orthologous to Arabidopsis STM, ZLL, and CLV1, and to investigate the effects of their ectopic expression on embryogenesis in vitro. The results indicate that (i) somatic embryogenesis in Arabidopsis is affected by the introduction of Brassica STM and CLV1, but not by Brassica ZLL; (ii) overexpression of BnSTM and BnCLV1 has antagonistic effects on Arabidopsis somatic embryo formation, with BnSTM having a promoting role and BnCLV1 a repressing role; (iii) the increased Arabidopsis somatic embryo production in tissue ectopically expressing Brassica STM entails profound changes in the expression of genes involved in different functions; and (iv) besides improving Arabidopsis somatic embryogenesis (an indirect embryogenic system requiring a callus phase), the introduction of Brassica STM also facilitates microspore-derived embryogenesis in B. napus (a direct embryogenic system without an intervening callus phase).
Materials and methods
Culture treatments
Production of B. napus cv. Topas DH4079 MDEs and Arabidopsis somatic embryos was achieved by following the method outlined in Belmonte et al. (2006) and Bassuner et al. (2007). Briefly, Brassica plants with flower buds were transferred to 12 °C day/7 °C night temperature cold treatment until bud collection. Flower buds (2–3 mm long) were collected and suspended in NLN medium (Lichter, 1982) with 13% sucrose (pH 5.8) where they developed into embryos after the imposition of a heat shock treatment at 32 °C for 72 h. Fully developed MDEs were observed after 25 d in culture (Belmonte et al., 2006).
Arabidopsis somatic embryogenesis was induced by culturing bending-cotyledon zygotic embryos on a 2,4-D containing induction medium for 14 d followed by transfer onto a hormone-free development medium in which somatic embryos developed. This procedure was identical to that described by Bassuner et al. (2007).
Isolation of BnSTM, BnCLV1, BnZLL-1, and BnZLL-2, and expression studies
Total RNA extracted from B. napus MDEs after 28 d in culture was utilized for cDNA synthesis using a SuperScript™ II Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA). PCR-based amplification of full-length BnSTM cDNA (GU480584) was obtained using primers (5′-ATGGAAAAGTGGTTCCAACA-3′ and 5′-ATCCGGGACAATGCTTTGA-3′) designed from the sequence of Brassica oleracea BoSTM (AF193813) identified by Teo and Swarup (1999). BnZLL-1 (EU329719) and BnZLL-2 (GU731230) were isolated using primers (5′-ATGCCGATTAGGCAAATGAA-3′ and 5′-GAGTCATGTTTTACTGCTAA-3′) based on the sequence of Arabidopsis ZLL (NM 123748). Brassica napus BnCLV1 (GU480585) was isolated by 5′ and 3′ RACE [rapid amplification of cDNA ends; First Choice RLM-RACE kit (Ambion, Austin, TX, USA)] using a partial CLV1 sequence (AY283519) from the same species.
Expression studies of AtSTM, AtCLV1, and AtZLL in different tissue of 28-d-old Arabidopsis plants and BnSTM, BnCLV1, BnZLL-1, and BnZLL-2 in 40-d-old Brassica plants were performed by semi-quantitative reverse transcriptase-PCR (RT-PCR). Briefly, total RNA was extracted using a QIAGEN RNeasy® Plant mini Kit (catalogue no. 74904) and used to generate cDNA with Invitrogen SuperScript® II Reverse Transcriptase (catalogue no. 19064-002). For RT-PCR studies in Brassica the following primers were used: BnSTM (5′-TGATGGTCCGATGTGTCCTA-3′ and 5′-GCACCAGAGGAAGGAGAACA-3′), BnCLV1 (5′-CGGCGGGGACTATTCACTA-3′ and 5′-AAGGAAGTGTCGTGAAGACTAGG-3′), BnZLL-1 (5′-CGACGACGACGCCGCCAGCTCAGA-3′ and 5′-CAGACTCTTTGTATAGTCTCACTA-3′), and BnZLL-2 (5′-GCCAAGCTTCTTCACCTTCTCCTC-3′ and 5′-CAGACTCTTTGTATAGTCTCACTA-3′). As an internal control actin (AF111812) was amplified using primers 5′-TAAAGTATCCGATTGAGCATGGTAT-3′ and 5′-GACATTAAAGAGAAGCTTGCCTACG-3′.
For Arabidopsis the following primers were used: AtSTM (5′-TGTCAGAAGGTTGGAGCACCA-3′ and 5′-TTTGTTGCTCCGAAGGGTAA), AtCLV1 (5′-CTTAAATACCTCTCTTTCGGTGGA-3′ and 5′-ATCTCTATGCAAGATCAATGGTGA), and AtZLL (5′-CTGGTAAACGGGCAGATTGT-3′ and 5′-TTTTTCTTTCCCGTTCTCGTGATAC-3′). As an internal control ubiquitin (NM_178968) was amplified using the primers 5′-GATCTTTGCCGGAAAACAATTGGAGGATGGT-3′ and 5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGC-3′. The PCR products (during the exponential phase of amplification) were separated on 1% (w/v) agarose gels, and the intensities of ethidium bromide-stained bands were detected using ImageJ software (1.36b National Institutes of Health).
Light microscopy and localization studies
Histological studies and localization of BnSTM, BnCLV1, BnZLL-1, and BnZLL-2 were performed by RNA in situ hybridization using the procedure outlined in Belmonte et al. (2006). For histological examination, the tissue was stained with periodic acid–Schiff (PAS) reagent for total carbohydrates and counterstained with toluidine blue (TBO) for general histological organization.
Transformation studies and in vitro regeneration
Arabidopsis transformation experiments were carried out using Gateway technology (Invitrogen, Carlsbad, CA, USA) as reported by Karimi et al. (2002). Seeds of the Arabidopsis ecotype Columbia were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH, USA).
Full-length BoSTM, BrSTM, BnSTM, BnCLV1, BnZLL-1, and Bn-ZLL-2 cDNAs were inserted in the Gateway entry clone vector pDONR™221 (Invitrogen) according to the instruction manual and then transferred into the pK2GW7 (for sense transformation) or pK2WG7 (for antisense transformation) vectors carrying the 35S promoter and the 35S terminator (http://www.psb.ugent.be/gateway/index.php). The constructs were introduced into the Agrobacterium tumefaciens strain GV3101 which was utilized to spray transform wild-type (WT) Arabidopsis plants via standard Agrobacterium-mediated transfection techniques (Weigel and Glazebrook, 2002). Many transformed plants were generated and the level of the transgene was measured by quantitative RT-PCR (Supplementary Fig. S1 available at JXB online).
Brassica transformation with BnSTM in sense or antisense orientation was performed as described by Bhalla and Singh (2008). Briefly, surface-sterilized seeds were germinated for 5 d on 1/2 MS-B5 medium supplemented with 1% sucrose. Hypocotyls (2–3 mm) were inoculated on co-cultivation medium [MS-B5 medium supplemented with 3% sucrose and 2 mg l−1 benzyladenine (BA)] for 4 d, and then incubated with the same Agrobacterium strain used for Arabidopsis transformation, harbouring the pK2GW7 (for sense transformation) or pK2WG7 (for antisense transformation) vector for 2 min. Following a 4 d co-cultivation period, the explants were plated on shoot induction medium (MS-B5 supplemented with 5 mg l−1 AgNO3, 2 mg l−1 BA, 3% sucrose, 20 mg l−1 timentin, and 50 mg l−1 kanamycin) for 8 weeks. The emerging shoots were first transferred on elongation medium (MS-B5 supplemented with 3% sucrose, 0.1 mg l−1 GA3, and 1 mg l−1 BA) for 4 weeks and then placed on rooting medium [MS-B5 supplemented with 1% sucrose, 2 mg l−1 1-naphthaleneacetic acid (NAA)] for 4 weeks. Rooted shoots were transferred onto soil to generate fully mature plants (F0). Seeds from F0 plants were germinated on 1/2 MS basal medium with 1% sucrose and 50 mg l−1 kanamicin, and the resulting F1 generation was screened for the presence of the transgene by RT-PCR using a forward 35S promoter primer (5′-TGGACCCCCACCCACGAG-3′) and a reverse gene-specific primer (5′-GCACCAGAGGAAGGAGAACA-3′). Microspores harvested from the positive F1 plants were cultured to produce haploid MDEs which were germinated on 1/2 MS medium supplemented with 1% sucrose and 70 mg l−1 kanamycin for 4 weeks. The haploid seedling were then treated with 0.2% colchicine for 6 h in order to generate homozygous (double haploid) plants. After checking the ploidy level by chromosome counting and confirming the presence of the transgene by RT-PCR, selected plants were used as a source of seeds to generate the next (F2) generation. The expression level of BnSTM in these plants was measured by quantitative RT-PCR (Supplementary Fig. S2 at JXB online).
Microarray development: experimental design
Differences in global changes in transcript levels during somatic embryogenesis of WT Arabidopsis plants and plants ectopically expressing BoSTM (35S::BoSTM) were measured using the full genome Arabidopsis oligo microarrays [Quiagen-Operon Arabidopsis Ready Oligo Set (AROS) version 3.0] containing 29 000 elements and available through the University of Arizona. The experimental design was identical to that utilized in previous studies (Stasolla et al., 2008). Tissue during Arabidopsis somatic embryogenesis was collected at the beginning of the induction period (stage 0), at the end of the induction period (stage 1), and after 3 d and 9 d in development medium (stages 2 and 3, respectively). Three biological replicates were used for each of the four WT samples and four 35S::BoSTM samples. Each replicate sample was hybridized on the same slide with a common reference sample comprising equal amounts of amplified RNA derived from one biological replicate of each of the eight samples. This experimental design provided information on gene expression during the progression of somatic embryo development (stage 0, 1, 2, and 3) in the WT and 35S::BoSTM lines, i.e. time course experiment, as well as between stages of development (WT versus 35S::BoSTM at stage 0, 1, 2, and 3), i.e. direct comparison analysis. A swapped-dye experiment (stage 1 versus common reference) was also conducted to verify the effect of the dye on the hybridization outcome (Stasolla et al., 2008).
Microarray hybridization and scanning
Total RNA extracted from developing somatic embryos produced by the WT and 35S::BoSTM lines was amplified, labelled with fluorescent Cy3 or Cy5 dyes, and fragmented, using an RNA ampULSe: Amplification and Labelling kit (Kreatech Biotechnology, Amsterdam, The Netherlands) following the manufacturer's instructions. The integrity of the amplified RNA and the degree of labelling were determined by gel electrophoresis and by measuring Cy5 and Cy3 using a Nano-Drop ND-1000 spectrophotometer (Nano Drop Technologies, Thermo Scientific).
Microarray hybridization was performed as described at http://ag.arizona.edu/microarray/methods.html. Briefly, after a pre-hybridization treatment in a buffer containing 5× SSC, 0.1% SDS, and 0.1 mg ml−1 bovine serum albumin (BSA) at 37 °C for 1 h, the slides were washed three times in 0.1× SSC at 22 °C for 5 min and once in water at 22 °C for 30 s. The slides were dried by centrifugation at 1000 g for 2 min and stored until hybridization. The slides were then hybridized using 200 ng of Cy5-labelled target and 350 ng of Cy3-labelled target added to warm (42 °C) hybridization solution containing 2% formamide, 5× SSC, 0.1% SDS, and 0.1 mg ml−1 salmon sperm DNA. Hybridization was carried out for 16 h at 42 °C in a hybridization oven. Post-hybridization washes were performed by immersing the slides in a solution composed of 2× SSC, 0.1% SDS for 15 min at 37 °C, followed by two washes in 1× SSC for 2 min at 22 °C, and three washes in 0.1× SSC for 2 min at 22 °C. The slides were dried by centrifugation, and fluorescence levels of the Cy dyes were determined using an Axon GenePix 4000B laser scanner (MDS Analytical Technologies) with GenePix® Pro. Software Version 6.
Data analysis
Differences in gene expression were determined using a mixed model approach developed by Wolfinger et al. (2001) and used in previous studies (Stasolla et al., 2004). Prior to mixed model analysis of normalized log ratio expression data, flagged and error-tagged observations were removed. To improve normality of the data, outliers were removed based on studentized residuals (Lund, 1975). Although the experimental design was a two-way factorial treatment structure with main factors stage (0–3) and lines (WT versus 35S::BoSTM), the data were analysed as a one-way treatment structure. This reduced the impact of missing data points on the key comparisons that were required. Data were analysed within each gene considering treatment as the fixed effect and array as the random effect. Within each analysis, a priori expression differences were examined between 10 pairs of treatments. Differences were evaluated with four direct comparisons of WT versus transformed tissue at each time point (WT versus 35S::BoSTM at stages 0, 1, 2, and 3), and changes in gene expression over time were evaluated with three comparisons among successive time points (stage 0 versus 1, 1 versus 2, and 2 versus 3) within WT and 35S::BoSTM lines. To limit the experiment-wise type 1 error rate, experiment-wise (i.e. all genes combined) P-values were adjusted using the FDR (false discovery rate) adjustment option in the MULTITEST procedure in SAS (e.g. Saama et al., 2006). The FDR option adjusts P-values based on the Benjamini and Hochberg step-up procedure (Benjamini and Hochberg, 1995). The FDR adjusted P-values were used to verify whether a difference observed between stages or lines was statistically significant. A first selection was performed by comparing the expression ratios of signal intensities at different stages or between WT and 35S::BoSTM lines, and selecting only those probes having an adjusted P-value ≤0.01 and an absolute difference in expression ratios >4. Furthermore, in order to exclude possible dye effects, all probes exhibiting an opposite behaviour in the swapped-dye experiment were eliminated from the final analysis as per Stasolla et al. (2008).
The normalized expression ratios of the differentially expressed probes were imported in Tree View (http://sourceforge.net/projects/jtreeview/) and used to perform hierarchical cluster analysis, cluster analysis by self-organizing maps (SOMs), and principal component analysis (PCA).
Validation of microarray data
Microarray data were validated by quantitative real-time RT-PCR according to Stasolla et al. (2003) for several selected genes (Supplementary Table S1 at JXB online) that showed differential expression patterns during embryo development. The relative level of gene expression was analysed with the 2–ΔΔCT method described by Livak and Schmittgen (2001) using actin (AY139999) as a reference.
Glutathione metabolism
Measurements of glutathione content, glutathione reductase (GR) activity, and glutathione applications were performed according to Belmonte et al. (2006).
Methylation assays
DNA methylation was estimated using coupled restriction enzyme digestion and random amplification (CRED-RA), as reported by Leljak-Levanic et al. (2004). Briefly, the DNA extracted from WT and 35S::BoSTM lines at stage 1 of somatic embryogenesis was digested with HpaII and MspI endonucleases (which cut the sequence 5′-C/CGG-3′ with different sensitivity to cytosine methylation; MspI cuts if the inner C is methylated, whereas HpaII cannot cleave in the presence of methyl groups), and then used as a template in a PCR employing six random 10-base primers: 5-GACTGCACAC-3 (primer 1), 5-AGGCGGGAAC-3 (primer 2), 5-ACGATGAGCC-3 (primer 3), 5-ACCGCCTGCT-3 (primer 4), 5-AGGCCGGTCA-3 (primer 5), and 5-CAACCGGTCT-3 (primer 6). The PCRs were performed in a total volume of 25 μl using: 1 μl (50 ng) of digested DNA, 1 μl of 10 nM primer, 0.5 μl of 10 nM MgCl2, 11.25 μl of nuclease-free H2O, and 11.25 μl of 2× BioMix master mix (GE Healthcare UK Limited, Amersham, UK). The PCR conditions and measurements of the methylated bands were carried out exactly as described by Leljak-Levanic et al. (2004).
Results
Isolation and characterization of Brassica napus BnSTM, BnCLV1, BnZLL-1, and BnZLL-2
Using the available nucleotide sequence of STM from Arabidopsis and of CLV1 and ZLL from other Brassica species, full-length B. napus BnSTM, BnCLV1, BnZLL-1 and BnZLL-2 were isolated through a PCR-RACE approach. The four genes encode proteins with a high degree of amino acid sequence identity to their respective Arabidopsis orthologues (91% for BnSTM, 88% for BnCLV1, and 96% for BnZLL-1 and BnZLL-2) and with all the conserved domains. These include the KNOX1, KNOX2, ELK, and a homeodomain for BnSTM, a serine/threonine kinase domain for BnCLV1, and the PIWI and PAZ domains for BnZLL-1 and BnZLL-2 (Supplementary Fig. S3 at JXB online). Phylogenetic analyses further confirm the similarities of the Brassica genes to their Arabidopsis orthologues (Supplementary Fig. S4).
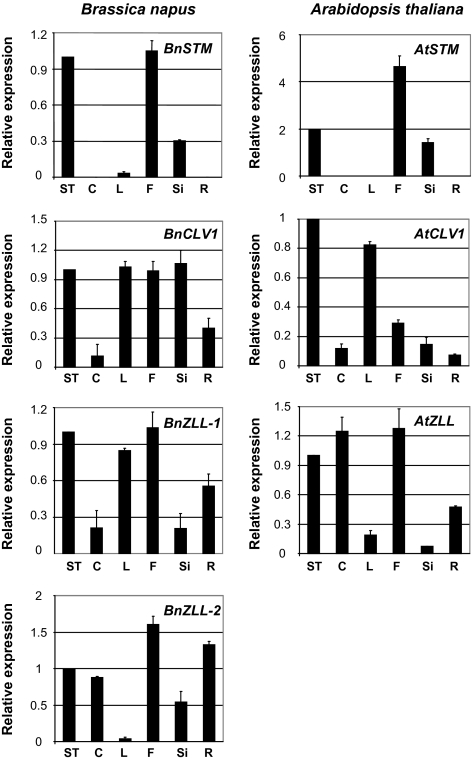
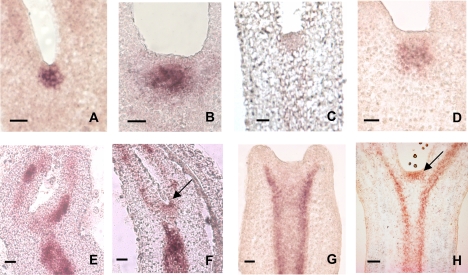
The relative expression of the B. napus genes in B. napus plants was measured in different tissues and compared with the expression pattern of the Arabidopsis orthologues measured in Arabidopsis plants (Fig. 1). Like AtSTM in Arabidopsis plants, the highest levels of BnSTM expression in B. napus plants were observed in shoot tips, flowers, and siliques. Transcripts of BnCLV1, BnZLL-1, and BnZLL-2 were detected in all those tissues where their respective Arabidopsis orthologues were also expressed (Fig. 1). Both BnSTM and BnCLV1 were expressed in the SAM of B. napus zygotic embryos and MDEs (Fig. 2). The combined signal of BnZLL-1 and BnZLL-2 (the high degree of similarity in the sequences of the two genes did not allow for the design of specific probes) was first localized in the vascular tissue of immature zygotic embryos and then extended to the SAM of fully developed cotyledonary embryos. A similar localization pattern to that observed for zygotic embryos was also observed in MDEs (Fig. 2). The expression domains of the Brassica genes in B. napus zygotic embryos and MDEs coincided perfectly with those described for the Arabidopsis orthologues, which are also expressed in the SAM of Arabidopsis embryos (reviewed by Dodsworth, 2009).
Fig. 1.
(A) Expression studies by semi-quantitative RT-PCR of BnSTM, BnCLV1, BnZLL-1, and BnZLL-2 in Brassica napus tissues, and AtSTM, AtCLV1, and AtZLL in Arabidopsis tissues. Shoot tips (ST), cotyledons (C), leaves (L), flowers (F), siliques (Si), and roots (R) harvested from B. napus (40-d-old plants, with the exception of C harvested from 10-d-old plants) and Arabidopsis (28-d-old plants with the exception of C harvested from 10-d-old plants). Values ±SE (n=3) were normalized to the expression level in ST set at 1.
Fig. 2.
RNA in situ hybridization of BnSTM, BnCLV1, BnZLL-1 and BnZLL-2 in Brassica napus zygotic embryos at the late cotyledonary stage (A, C, and F) or at the middle cotyledonary stage (E) of development, and in microspore-derived embryos (MDEs) cultured for 15 d (G) or 25 d (B, D, and H). The localization patterns of both BnSTM (A, B) and BnCLV1 (C, D) are restricted to the apical pole of both zygotic embryos and MDEs. The combined expression of BnZLL-1 and BnZLL-2 (the high degree of similarity in the sequences of the two genes did not allow for the design of specific probes) is first localized in the vascular tissue of both middle cotyledonary zygotic embryos (E) and 15-d-old MDEs (G), and then extended to the apical regions (arrows) of late cotyledonary zygotic embryos (F) and 25-d-old MDEs (H). All scale bars=40 μm
Effects of ectopic expression of Brassica genes on Arabidopsis somatic embryogenesis
Given the very low efficiency of B. napus Topas (DH4079 embryogenic line) transformation, the effects of ectopic expression of the four Brassica genes during in vitro embryogenesis were first analysed in Arabidopsis. Gene(s) promoting somatic embryogenesis were then introduced in sense and antisense orientation in B. napus to verify if their effects were also retained during microspore-derived embryogenesis.
Independently transformed Arabidopsis lines ectopically expressing BnSTM, BnCLV1, BnZLL-1, or BnZLL-2 were generated (Supplementary Fig. S1 at JXB online) and utilized to produce somatic embryos by first culturing bending-cotyledon zygotic embryos on a 2,4-D-enriched induction medium for 14 d and then transferring them onto a hormone-free development medium (Fig. 3A). Differences in the initial explants, i.e. bending-cotyledon zygotic embryos, were observed among the transformed lines (Fig. 3B). Compared with WT embryos, the introduction of BnCLV1 decreased the size of the SAM and induced the accumulation of storage products in the subapical cells. This was in contrast to 35S::BnSTM zygotic embryos which showed well-defined, dome-shaped SAMs (Fig. 3B).
Fig. 3.
(A) Schematic representation of the somatic embryogenic process in Arabidopsis and stages, with relative time points, used for the microarray experiment. Bending-cotyledon zygotic embryos were cultured on induction medium for 14 d and then transferred on development medium. Embryogenic cells formed from the adaxial sides of the cotyledons during the induction phase and meristemoids (arrowhead) appeared on the surface of the callus. Growth of the meristemoids resulted in the production of fully developed somatic embryos (stage 3). (B) Structure of the apical pole in Arabidopsis bending-cotyledon zygotic embryos used as explants to induce somatic embryogenesis. Compared with WT embryos (B1), the SAM of 35S::BnCLV1 embryos (B2) was smaller in size and was characterized by the accumulation of storage products (arrow) within the subapical cells. Dome-shaped SAMs were observed in 35S::BnSTM embryos (B3). Sections were stained with periodic acid–Schiff (PAS) reagent which stains carbohydrates red and subsequently counterstained with toluidine blue (TBO) for general histological organization. All scale bars=15 μm. (C) Effects of the ectopic expression of BnSTM, BoSTM, BrSTM, BnCLV1, BnZLL-1, and BnZLL-2 on the number of Arabidopsis explants with somatic embryos and on the number of somatic embryos originating from each explant. Values ±SE are averages of at least three independent experiments.
The number of explants (bending-cotyledon zygotic embryos) producing somatic embryos doubled in three 35S::BnSTM lines (2–4) analysed (Fig. 3C). This prompted the ascertainment of whether these results could be replicated, and possibly further improved, by overexpressing the STM orthologues from B. oleracea (AF193813) and B. rapa (GU480585) (denoted as BoSTM and BrSTM, respectively), which are the progenitor species of B. napus. The two cDNAs were isolated using a PCR-based approach followed by RACE. Protein sequence alignment revealed a high degree of identity between BnSTM and the two Brassica orthologues (97% for BrSTM and 99% for BoSTM). Ectopic expression of both BoSTM and BrSTM (Supplementary Fig. S1 at JXB online) increased the response of the explants to the culture conditions (Fig. 3C). The number of embryo-forming explants in the 35S::BnZLL-1 and 35S::BnZLL-2 lines was similar to control levels, although a small increment was observed for one 35S::BnZLL-2 line. All the 35S::BnCLV1 lines analysed showed a decrease in the number of explants able to generate embryos (Fig. 3C).
Arabidopsis somatic embryo composition was further examined in all transformed plants, and the results of representative lines are shown in Fig. 3C. The number of embryos produced by each responsive explant increased in all lines overexpressing the Brassica (Bo, Bn, and Br) STM gene especially in line 5 (35S::BoSTM) in which almost 90% of the responsive explants generated ≥5 embryos. No appreciable differences in embryo composition were observed between the 35S::BnZLL-1 and BnZLL-2 lines and the WT counterpart, whereas the number of explants producing only one somatic embryo almost doubled in all 35S::BnCLV1 lines.
Effects of BnSTM overexpression or down-regulation on B. napus MDE
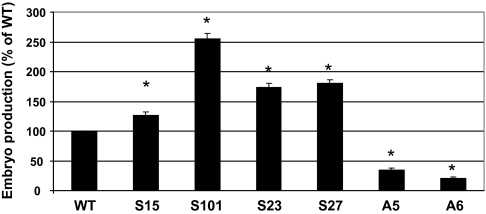
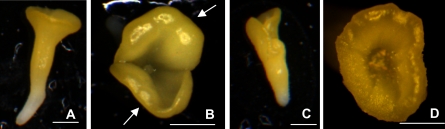
To evaluate if the beneficial effects of Brassica STM overexpression were only restricted to embryogenic systems with an intervening callus phase, such as Arabidopsis somatic embryogenesis, microspore-derived embryogenesis was induced in B. napus lines overexpressing or down-regulating BnSTM (Supplementary Fig. S2 at JXB online). In this system microspores develop directly into embryos (MDEs) without the formation of callus. Differentiation of microspores into MDEs increased significantly in sense lines overexpressing BnSTM, whereas an inhibition of embryo-forming capacity was observed in antisense lines with reduced BnSTM levels (Fig. 4). The down-regulation of BnSTM resulted in profound morphological abnormalities (Fig. 5). While MDEs produced by the WT line were characterized by two distinct cotyledons and an elongated embryonic axis, MDEs with reduced BnSTM expression were shorter and often exhibited fused cotyledons. No morphological alterations were observed in MDEs produced by lines overexpressing BnSTM (Fig. 5).
Fig. 4.
Production of microspore-derived embryos (MDEs) from Brassica napus plants with altered BnSTM expression. Antisense lines (A, line 5 and 6) with lower BnSTM expression or sense lines (S, line 15, 101, 23, and 27) with increased BnSTM expression were utilized to generate MDEs. For each experiment 40 000 microspores were plated and embryo number was counted after 25 d in culture. Values ±SE (n=3) are expressed as a percentage of the WT. Asterisks above each bar indicate values which are significantly different from the WT value (P ≤0.05).
Fig. 5.
Morphology of microspore-derived embryos (MDEs) produced by B. napus lines with altered BnSTM expression. MDEs were collected after 25 d in culture. MDEs obtained from the WT line (A) were characterized by two distinct cotyledons (arrows) (B). MDEs produced by antisense lines and with lower BnSTM expression were smaller (C) and often produced fused cotyledons (D). No morphological differences were observed in MDEs produced by lines overexpressing BoSTM. All scale bars=0.3 mm. (This figure is available in colour at JXB online.)
Transcriptome analysis during Arabidopsis somatic embryogenesis
Microarray analysis was conducted during stages of Arabidopsis somatic embryos produced by the WT line and the Brassica STM-overexpressing line showing the most pronounced enhancement of somatic embryogenesis, i.e. 35S::BoSTM (line 5). This comparison would allow identification of genes responsive to the introduction of the Brassica STM gene and important to the formation of somatic embryos.
In the WT line, 885 probes were differentially expressed (fold change >4 and P ≤0.01) between stage 0 and 1, and this number declined during the successive comparisons (315 between stage 1 and 2, and 56 between stage 2 and 3). A similar decline in expression profile was also observed in the 35S::BoSTM line, despite a larger number of probes being expressed during the first two stage comparisons (1334 between stage 0 and 1, and 1032 between stage 1and 2). To facilitate the analysis, the differentially expressed elements were grouped into categories based on their putative function, and a list of representative probes is shown in Table 1. Many probes implicated in hormone synthesis and signalling showed different behaviour, whereas probes involved in meristem function (STM, WUS, PINHEAD/ZWILLE, AGO1, and FIDDLEHEAD) were induced only in the 35S::BoSTM line between stage 0 and 1. Differences between WT and BoSTM-overexpressing lines were also observed for GR and glutathione synthase (GSH2), both repressed in the transformed line during the first days of development (stage 1 versus 2; Table 1). Two probes encoding DNA (cytosine-5)-methyltransferases were also repressed in the 35S::BoSTM line during the induction phase (stage 0 versus 1; Table 1).
Table 1.
List of selected genes showing differential expression (fold change >4 and P ≤0.01) during progressive stages (1–3) of Arabidopsis somatic embryogenesis in the wild-type (WT) line and the 35S::BoSTM line
| WT |
35S::BoSTM |
|||||
| Stage 0–1 | Stage 1–2 | Stage 2–3 | Stage 0–1 | Stage 1–2 | Stage 2–3 | |
| Cytokinin | ||||||
| At3g23630 Adenylate isopentenyltransferase 7/IPST7 | – | – | – | +6.5 | – | – |
| At3g48100 Response regulator 5 (ARR5) | – | – | – | +40 | – | – |
| Ethylene | ||||||
| At5g03280 Ethylene-insensitive 2 (EIN2) | – | – | – | –7.6 | – | – |
| At5g25190 Ethylene-responsive element | –10.9 | – | – | –33.5 | +5.6 | – |
| At4g29100 Ethylene-responsive element | – | – | – | – | –12.9 | – |
| Gibberellins | ||||||
| At5g07200 Gibberellin 20-oxidase | +8.6 | – | – | +16.9 | – | – |
| At3g02885 Gibberellin-regulated protein 5 (GASA5) | – | – | – | –16.7 | – | – |
| Abscisic acid | ||||||
| At3g19290 ABA-responsive element-binding protein 2 (AREB2) | – | – | – | – | –7.4 | – |
| Auxin | ||||||
| At1g70940 Auxin transport protein (PIN3) | –6.0 | – | – | – | – | – |
| At1g19850 MONOPTEROS transcription factor | –9.8 | – | – | –50.1 | +8.7 | – |
| At2g33860 Auxin-responsive factor (ARF3) | – | –6.2 | – | –7.4 | – | – |
| At4g14560 IAA-induced protein 1 (IAA1) | –13.3 | +4.3 | – | +45.2 | +6.7 | – |
| At2g22670 IAA-induced protein 8 (IAA8) | – | – | – | +13.7 | –9.4 | – |
| At5g65670 IAA-induced protein 9 (IAA9) | – | – | – | +7.4 | –22.8 | – |
| At3g15540 IAA-induced protein 19 (IAA19) | – | +24.8 | – | – | +41.2 | – |
| At5g25890 IAA-induced protein 28 (IAA28) | – | – | – | +4.3 | –16.4 | – |
| Brassinosteroids | ||||||
| At4g39400 Brassinosteroid-insensitive 1 (BR1) | – | – | – | –6.7 | – | – |
| At3g50750 Brassinosteroid signalling-positive regulator | – | – | – | –7.1 | – | – |
| Signal transduction | ||||||
| At5g19280 KAPP | – | – | – | +4.9 | – | – |
| Transcription | ||||||
| At1g62630 SHOOTMERISTEMLESS (STM) | – | – | – | +5.2 | – | – |
| At2g17950 WUSCHEL (WUS) | – | – | – | +4.6 | – | – |
| At5g17430 Ovule development protein (homologue to BABY BOOM) | – | – | – | +7.5 | – | – |
| Post-transcription | ||||||
| At5g43810 PINHEAD/ZWILLE | – | – | – | +6.8 | – | – |
| At1g48410 ARGONAUTE 1 (AGO1) | – | – | – | +7.5 | – | – |
| Disease and defence | ||||||
| At4g35000 L-Ascorbate peroxidase 3 (APX3) | – | – | – | –6.3 | – | – |
| At5g21105 L-Ascorbate oxidase | –8.3 | – | – | –25.2 | – | – |
| At3g27820 Monodehydroascorbate reductase | – | – | – | –9.7 | – | – |
| At3g10920 Superoxide dismutase [Mn]-mitochondrial | – | – | – | – | –6.1 | – |
| At3g56350 Superoxide dismutase [Mn]-putative | –4.5 | +8.0 | – | – | +9.6 | – |
| At5g27380 Glutathione synthetase (GSH2) | – | – | – | – | –9.3 | – |
| At3g24170 Glutathione reductase (GR) | – | – | – | – | –6.7 | – |
| Metabolism | ||||||
| At2g26250 β-KetoacylCoA synthase (FIDDLEHEAD) | – | – | – | +13.7 | – | – |
| At3g17820 Glutamine synthetase (GS1) | – | – | – | – | –9.0 | – |
| At5g35630 Glutamine synthetase (GS2) | – | – | – | –8.0 | – | – |
| At5g49160 DNA (cytosine-5)-methyltransferase | – | – | – | –4.5 | – | – |
| At5g25480 DNA (cytosine-5)-methyltransferase | – | – | – | –9.2 | – | – |
Positive values indicate an up-regulation of gene expression between two adjacent stages.
A direct comparison analysis was also performed to identify differentially expressed probes between WT and 35S::BoSTM lines at each stage of somatic embryo development. The most significant differences in expression between lines occurred at stage 1, when many ribosomal proteins and enzymes participating in the glycolytic and tricarboxylic acid (TCA) cycle were repressed in the WT line (Supplementary Fig. S5A at JXB online). A similar tendency was also observed for several transcription factors, including the lateral organ boundary (LOB) genes 12, 16, and 41 (LOB 1, 2, and 3) and two homeobox (HB) genes WUS and REVOLUTA (HB 6 and 7) (Supplementary Fig. S5B). Different profiles were detected for probes clustered in the categories of disease and defence (D&D), metabolism (M), and hormonal control. A probe (M 11) coding for S-adenosylmethionine synthase was significantly repressed in the WT line (Supplementary Fig. S5C)
WUS expression and localization during Arabidopsis somatic embryogenesis
Expression of WUS was measured by semi-quantitative RT-PCR at stage 1 of Arabidopsis somatic embryogenesis (Fig. 3A) in the WT line as well as in lines overexpressing BoSTM, BnSTM, BrSTM, and BnCLV1 (Fig. 6). Compared with the WT line, WUS expression increased in all lines overexpressing Brassica STM (confirming the microarray data, Table 1 and Supplementary Fig. S5B), whereas it decreased in the two BnCLV1 lines (Fig. 6). In order to assess if these alterations in expression levels were associated with changes in localization patterns, RNA in situ hybridization was also performed. In the WT line (stage 1) WUS signal was restricted to small clusters of cells originating from the callus on the adaxial side of the cotyledons (Fig. 6B1) and to the meristemoids (Fig. 6B2). WUS was expressed in the meristematic cells which stained blue with TBO (Fig. 6B3, 4). In the 35S::BoSTM line (line 5) the localization of WUS was extended to all apical and subapical cells of the callus (Fig. 6B5). Compared with the subtending parenchyma cells, the WUS-expressing cells were smaller and highly cytoplasmic (Fig. 6B6). The extent of WUS expression and the formation of meristematic cells at stage 1 correlated to the number of somatic embryos produced by the explants at the end of the culture period, i.e. stage 3 (Fig. 6B7, 8). The expression of WUS in the 35S::BnCLV1 line (line 3) at stage 1 of somatic embryogenesis was often limited to very small clusters of cells (Fig. 6B9), although in many instances no expression level was detected (Fig. 6B10).
Fig. 6.
WUSCHEL (WUS) expression (measured by semi-quantitative RT-PCR) and localization (by digoxigenin-labelled RNA in situ hybridization) at the end of the induction period (stage 1) of Arabidopsis somatic embryogenesis in the WT line and in lines overexpressing BoSTM (line 5), BnSTM (line 2), BrSTM (line 15), and BnCLV1 (line 3 and 6). Values ±SE are means of three independent experiments and are normalized to the value of the WT line set at 1. Asterisks indicate values which are significantly different from the WT value (P ≤0.05) (A). In the WT Arabidopsis line at stage 1 of somatic embryogenesis WUS expression (red stain) was initially localized in distinct clusters of cells on the adaxial side of the cotyledons of the explant (B1) and throughout the developing globular embryos (B2). WUSCHEL expression demarcated the formation of embryogenic cells which stained blue with toluidine blue (TBO) (B3, 4). In the line overexpressing the BoSTM, the WUS domain was extended to the surface of the cotyledons (B5) corresponding to apical and subapical layers occupied by embryogenic cells (B6). The extent of the WUS domain at stage 1 of somatic embryogenesis correlated with the number of somatic embryos produced by the WT line (B7) and 35S::BoSTM line (B8) at the end of the culture period (stage 3). In lines overexpressing the Brassica CLV1, the WUS signal at stage 1 of somatic embryogenesis was either localized in a very small cluster of cells (B9) or completely absent (B10). Scale bars=40 μm (1–6 and 9, 10) and 1 mm (7, 8).
Auxin requirement for the induction of Arabidopsis somatic embryogenesis
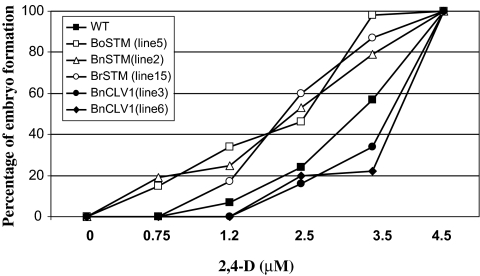
The requirement for auxin during the acquisition of embryogenic competence in the induction phase (stage 0–1, Fig. 3A) of Arabidopsis somatic embryogenesis was evaluated in the WT line and lines ectopically expressing BoSTM (line 5), BnSTM (line 2), BrSTM (line 15), and BnCLV1 (lines 3 and 6). Somatic embryo production in the WT line more than halved when the level of 2,4-D was reduced from the optimal concentration of 4.5 μM to 2.5 μM and declined further at lower auxin levels (Fig. 7). The introduction of STM attenuated the decline of somatic embryo formation in a low auxin environment. In some lines (35S::BoSTM line 5 and 35S::BnSTM line 2) embryo formation was still observed when 2,4-D levels were lowered to 0.75 μM. The highest requirement for auxin for the induction of somatic embryogenesis was observed in the lines ectopically expressing BnCLV1. The introduction of this gene caused a substantial decrease in embryo formation when the level of 2,4-D was lowered from 4.5 μM to 3.5 μM (Fig. 7).
Fig. 7.
Auxin requirement for the induction of Arabidopsis somatic embryogenesis. The WT line and lines ectopically expressing BoSTM (line 5), BnSTM (line 2), BrSTM (line 15), and BnCLV1 (lines 3 and 6) were cultured on the induction medium with different levels of 2,4-D. Values (n=3) are presented as a percentage of the embryo production occurring at the optimal 2,4-D level (4.5 μM).
DNA methylation and analysis of glutathione metabolism during Arabidopsis somatic embryogenesis
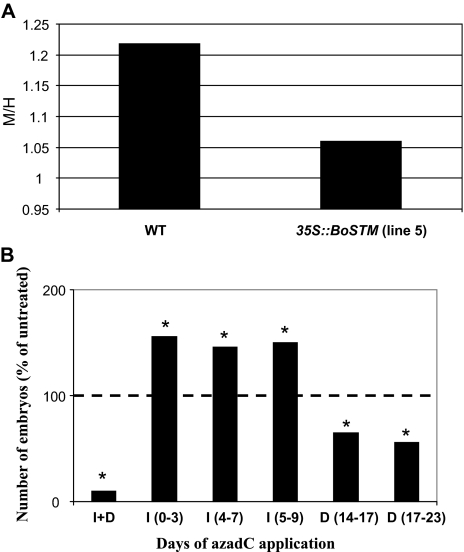
To ascertain whether the down-regulation of two DNA (cytosine-5)-methyltransferases observed in the 35S::BoSTM line during the induction phase (stage 0–1, Table 1) of Arabidopsis somatic embryogenesis affected the pattern of DNA methylation, a CRED-RA was performed in the WT line and the 35S::BoSTM line 5 at stage 1 of somatic embryogenesis. As shown in Fig. 8, the degree of methylation of cytidine residues was lower in the transformed line. The role of DNA methylation during somatic embryogenesis was further tested to assess if 5-azadeoxycytidine (azadC), a hypomethylating agent which inhibits DNA methyltransferase activity (Sano et al., 1989), would increase the somatic embryo-forming capacity of the WT line. Inclusions of this compound at different days during the induction phase favoured the production of WT somatic embryos. An opposite trend was observed if azadC was added in the development medium (Fig. 8B).
Fig. 8.
(A) DNA methylation analysis in the WT line and in the 35S::BoSTM line 5 at stage 1 of Arabidopsis somatic embryogenesis. Ratios of consistently amplified bands following digestion with MspI (M) and HpaII (H). DNA was extracted at stage 1, digested with HpaII and MspI endonucleases (which cut the sequence 5'-C/CGG-3' with different sensitivity to cytosine methylation; MspI cuts if the inner C is methylated, whereas HpaII cannot cleave in the presence of methyl groups), and then used as a template in a PCR employing six random 10-base primers (Leljak-Levanic et al., 2004). (B) Effects of the DNA-hypomethylating agent 5-azadeoxycytidine (azadC) on the somatic embryo-forming capacity of the WT line. Data are expressed as percentage changes in somatic embryo production following applications of azadC on different days in the induction (I) or development (D) medium. AzadC was also added throughout the culture period (I+D). Values (n=3) are expressed as percentages of the value of untreated tissue (set at 100%). Asterisks indicate values which are significantly different from the untreated value (P ≤0.05).
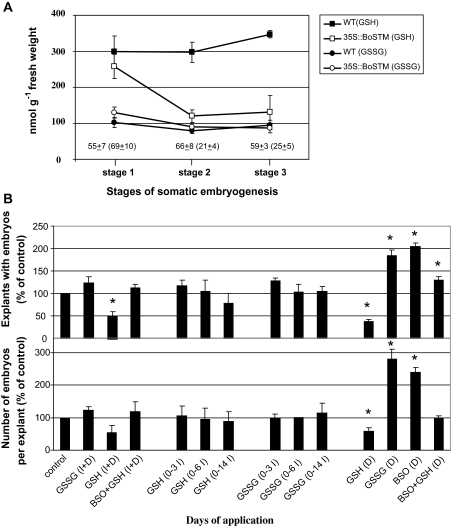
Transcriptional studies showed that during the initial days in the development medium (stage 1–2) GR and GSH2 are repressed in the 35S::BoSTM line (Table 1). This observation prompted the investigation of the role of glutathione metabolism during Arabidopsis somatic embryogenesis. Compared with the WT line, the endogenous level of reduced glutathione (GSH) in the 35S::BoSTM line 5 decreased sharply within the first 3 d in the development medium (from stage 1 to stage 2) and remained low during the following days in culture (Fig. 9A). This profile correlated with the reduced activity of GR in the 35S::BoSTM line after stage 1. No differences in the endogenous content of oxidized glutathione (GSSG) were observed between lines during somatic embryogenic (Fig. 9A).
Fig. 9.
Glutathione metabolism during Arabidopsis somatic embryogenesis. (A) Endogenous levels of reduced (GSH) and oxidized (GSSG) glutathione during different stages (0–3) of somatic embryogenesis in the WT line and the 35S::BoSTM line 5. Numbers above the abscissa (values of the 35S::BoSTM line in parentheses) indicate the activity of glutathione reductase (nmol g−1 fresh weight). Values ±SE are averages of at least three independent experiments. (B) Effects of exogenous applications of GSSG (1 mM), GSH (1 mM), and BSO (0.1 mM) on the number of explants with embryos and on the number of embryos per explant during somatic embryogenesis of the WT line. Compounds were added throughout the culture period (I+D) or on different days during induction (I) or development (D). Values ±SE (n=3) are expressed as percentages of the value in the untreated (control) tissue set at 100%. Asterisks above each bar indicate values which are significantly different from the control value (P ≤0.05).
Based on this observation, it was hypothesized that the somatic embryo-forming capacity of the WT line could be modulated by alterations of the glutathione pool, with an oxidized environment (low GSH/GSSG ratio) favouring somatic embryo development and a reduced environment (high GSH/GSSG) having the opposite effect. This hypothesis was tested by supplementing the induction and/or the development medium of the WT line with exogenous GSH or GSSG (which switch the cellular glutathione pool towards a reduced or oxidized glutathione environment; Belmonte et al., 2006) or BSO (DL-buthionine-[S,R]-sulphoximine), a specific inhibitor of GSH synthesis (Griffith and Meister, 1979). Applications of GSH in the development medium (after stage 1 of somatic embryogenesis) decreased both the number of explants able to form somatic embryos and the number of embryos produced by each explant (Fig. 9B). The inhibitory effect of GSH was reversed by the combined application of GSH+BSO. Somatic embryo formation in the WT line was encouraged by the application of BSO or GSSG in the development medium (Fig. 9B).
Discussion
Characterization of B. napus SAM genes
The putative orthologues of Arabidopsis STM, CLV1, and ZLL were isolated from B. napus tissue and denoted as BnSTM, BnCLV1, and BnZLL-1 and BnZLL-2. The Brassica genes displayed high levels of sequence identity with their respective Arabidopsis orthologues with which they share all the conserved domains (Supplementary Figs S3, S4 at JXB online). Unique features of BnSTM are the four conserved regions KNOX1, KNOX2, ELK, and the homeodomain, which also characterize AtSTM (Scofield and Murray, 2006). Besides occupying the central region of the SAM during both zygotic and microspore-derived embryogenesis (Fig. 2), as also observed for AtSTM (Long and Barton 1998), a down-regulation of BnSTM in B. napus plants induces SAMs with a determined fate (data not shown). These observations suggest that like AtSTM, BnSTM is involved in the establishment and maintenance of a functional SAM.
The B. napus BnCLV1 contains all the signatures of its respective Arabidopsis orthologue AtCLV1 receptor kinase, including the conserved C-terminal serine/threonine kinase domain (Supplementary Fig. S3 at JXB online). Similarities among the two genes (BnCLV1 and AtCLV1) were also observed in their localization patterns during embryogenesis (Fig. 2 and Clark et al., 1997) and expression during post-embryonic development (Fig. 1). The termination of the SAMs due to differentiation of the meristematic cells observed in Arabidopsis seedling overexpressing BnCLV1 (data not shown) suggests that BnCLV1 regulates SAM activity through mechanisms analogous to those ascribed to AtCLV1, which limits the accumulation of undifferentiated or uncommitted cells and promotes differentiation (reviewed by Dodsworth, 2009).
The two products of the BnZLL genes, denoted as BnZLL-1 and BnZLL-2, share a high degree of amino acid sequence identity (96%) with AtZLL and also contain two conserved regions, the PIWI and PAZ domains, which are unique features of the AGO members, including AtZLL (Cerruti et al., 2000). Additional similarities shared by BnZLL-1 and BnZLL-2 and the Arabidopsis AtZLL include the two distinct expression domains (vascular tissue and SAM) during embryogenesis. The localization of BnZLL-1 and BnZLL-2 in the vascular tissue of immature embryos and in the SAM of fully developed cotyledonary embryos both in vivo and in vitro (Fig. 2) is very similar to that described for AtZLL during Arabidopsis embryogenesis (Moussian et al., 1998). The participation of AtZLL in the formation of the primary embryonic SAM relies on the creation of positional cues for proper STM expression (Moussian et al., 1998) and the enhancement of WUS function during the acquisition of stem cell fate (Tucker et al., 2008). Involvement in SAM formation is a prerogative which is possibly retained by BnZLL-1 and BnZLL-2, as shown by the similarities shared between the Arabidopsis and Brassica genes
Based on sequence analyses, as well as on expression, localization, and phenotypic studies, the present results are consistent with the notion that BnSTM, BnCLV1, and BnZLL-1 and BnZLL-2 are the respective orthologues of AtSTM, AtCLV1, and AtZLL.
Ectopic expression of the Brassica genes affects embryo production in vitro
Somatic embryogenesis in Arabidopsis is a two-step process comprising an induction phase (in the presence of 2,4-D) in which the somatic cells of the explant acquire embryogenic competence and proliferate to produce a callus, and a development phase required for the growth of somatic embryos (Fig. 3A). The molecular mechanisms underlying the states of embryogenic competence acquisition and embryo development are poorly understood and it is still unclear what changes somatic cells must undergo in order to become embryogenic. Over the past few years attempts have been made to identify genes with altered expression patterns during somatic embryogenesis (Lin et al., 1996; Stasolla et al., 2004), and to assess their function using molecular approaches (reviewed by Namasivayam, 2007). Although the effects of several loss-of-function mutants on somatic embryogenesis have also been described (Chaudhury et al., 1993; Ogas et al., 1999; Mordhorst et al., 1998, 2002), there are only a few reports documenting changes in somatic embryogenic induction frequency as a result of ectopic gene expression (Hecht et al., 2001; Stone et al., 2001; Boutilier et al., 2002; Zuo et al., 2002). Furthermore among the genes tested in these studies only one, WUS, is related to SAM activity (Zuo et al., 2002).
The present work shows that somatic embryo formation in Arabidopsis is affected by the introduction of Brassica (Bn, Bo, and Br) STM and BnCLV1, but not by BnZLL-1 and BnZLL-2 (Fig. 3). In Arabidopsis, AtSTM and AtCLV1 have key roles in the maintenance of post-embryonic SAMs (Clark et al., 1997; Schoof et al., 2000), whereas AtZLL is only involved in the formation of a functional embryonic SAM without affecting its post-embryonic development (Moussian et al., 1998; Lynn et al., 1999). Although it is difficult to surmise function from gain-of-function phenotypes, the present overexpression experiments suggest that genetic factors regulating the maintenance of the SAM are also relevant for the generation of embryo-competent cells.
The STM-mediated redirection of cell fate is not a novel concept. De novo shoot meristems were produced in leaves of Arabidopsis plants overexpressing KNAT1, a member of the kn-1 like homeobox gene family which includes STM (Chuck et al., 1996), and tobacco plants expressing the maize STM homologue KNOTTED 1 (Sinha et al., 1993). These observations are consistent with the role of STM in the formation and maintenance of the SAM. However, what is novel is the ability of STM to induce somatic embryos and not shoots in the present system (the identity of the embryos was verified by the presence of the bipolar apical meristems through histological analyses). It is postulated that a function of this gene when ectopically expressed in new domains is to encourage cells to dedifferentiate and acquire ‘meristematic’ identity. Depending on the physiological environment, such cells can then embark on an organogenic or embryonic developmental pathway. This flexibility in cell fate acquisition is common during in vitro development where positional cues are not as well defined as in vivo. The inductive effect of ectopic STM expression on Arabidopsis somatic embryogenesis is consistent with previous studies documenting an increased number of somatic embryos in spruce lines overexpressing HBK3, a class I KNOX homeobox gene, isolated from Picea abies (Norway spruce) (Belmonte et al., 2007).
Another interesting point emerging from the present study is the different response in culture exhibited by the lines overexpressing Brassica (Bo, Bn, and Br) STM and BnCLV1. While the introduction of the Brassica STM encourages Arabidopsis somatic embryo formation, BnCLV1 represses somatic embryogenesis (Fig. 3C). This competitive role of the Brassica genes in vitro is analogous to that fulfilled by Arabidopsis AtSTM and AtCLV1 in vivo, where AtSTM acts as a positive regulator of SAM activity by increasing the domain of meristematic cells (Lenhard et al., 2002) and AtCLV1 as a negative regulator which represses meristem proliferation and promotes cellular differentaition (Clark et al., 1997). In line with this analogy it is therefore suggested that while Brassica STM encourages Arabidopsis somatic embryo formation by broadening the domain of embryogenic cells, BnCLV1 limits the production of embryogenic cells by inducing differentiation. This notion is also supported by the signs of cellular differentiation (as estimated by the accumulation of storage products) observed in the SAMs of explants (bending-cotyledon embryos) overexpressing BnCLV1 (Fig. 3B). The mechanisms underpinning the interaction between the Brassica STM and CLV1 in culture are elusive, although they might involve the participation of WUS (discussed in the next section), which is a signalling molecule component for the induction of somatic embryos (Chen et al., 2009).
The altered expression of the Brassica STM gene also had profound effects on B. napus microspore-derived embryogenesis. A high yield of MDEs was observed in lines overexpressing BnSTM, whereas an opposite trend was noticed in lines with reduced BnSTM levels (Fig. 4). The phenotypic deviations (reduced embryo size and fused cotyledons) observed in many MDEs generated from lines down-regulating BnSTM (Fig. 5) confirm the notion that this gene is implicated in embryo development and meristem formation.
Overall these results clearly show that the overexpression of Brassica STM encourages somatic embryo formation in Arabidopsis and MDE production in B. napus. It is therefore suggested that the beneficial effect of this gene on in vitro embryogenesis is independent of the system utilized (direct embryogenesis from single microspores in B. napus or indirect somatic embryogenesis through a callus step formation in Arabidopsis) and the type of induction conditions (hormone-free medium in B. napus and auxin in Arabidopsis).
Genes and regulatory pathways altered by the ectopic expression of BoSTM during Arabidopsis somatic embryogenesis
Global changes in transcript levels during somatic embryogenesis in the WT line and in the 35S::BoSTM line were analysed by microarray studies and validated by quantitative RT-PCR (Supplementary Fig. S6 at JXB online). Formation of embryos from Arabidopsis somatic cells requires a reprogramming in gene expression pattern, which is affected profoundly by the overexpression of BoSTM. In bending-cotyledon zygotic embryos (stage 0) the introduction of BoSTM resulted in the differential expression of 176 probes, some of which encoded cell structural components, hormone-responsive elements, stress-related enzymes, and transcription factors (data not shown). The most pronounced differences in expression levels occurred during the induction phase, demarcating the transition from stage 0 to stage 1. During the following development phase such differences were attenuated, despite a large variation in function of the expressed probes (Table 1).
The induction phase
The induction phase was characterized by alterations in expression levels of several genes involved in hormone synthesis/response. The BoSTM activation of adenylate isopentenyltransferase 7 (IPT7), a cytokinin biosynthesis gene, and ARR5, the primary cytokinin-responsive gene (Table 1), suggests that the role of this gene is to stimulate cytokinin synthesis and perception. A rapid elevation in the transcription of IPT7 and ARR5 was also observed in mature tissues of Arabidopsis plants ectopically expressing AtSTM and other KNOTTED1 genes (Yanai et al., 2006). Another role assigned to KNOTTED genes is to inhibit gibberellin synthesis through the repression of GA 20-oxidase (Sakamoto et al., 2001). In the present system this regulation does not seem to occur as the expression of this gene increased in both WT and 35S::BoSTM lines during the induction phase (Table 1).
Acquisition of embryogenic competence during Arabidopsis somatic embryogenesis relies on the ability of the tissue to respond to the exogenously supplied auxin (2,4-D). The altered expression of genes involved in auxin transport and signalling, together with the activation of several indole acetic acid (IAA)-induced elements (IAA1, 8, 9, and 28) during the induction phase (stage 0 versus 1) of the 35S::BoSTM line (Table 1), suggest that the introduction of BoSTM modifies the response of the tissue to 2,4-D, possibly by making it more sensitive to this growth regulator. This is demonstrated by the sustained production of somatic embryos in Arabidopsis lines overexpressing Brassica (Bo, Bn, or Br) STM when exposed to reduced levels of 2,4-D which preclude embryogenesis in the WT line (Fig. 7). The requirement for 2,4-D is higher in the 35S::BnCLV1 lines, thus suggesting opposite effects of Brassica STM and CLV1 on embryogenic competence acquisition and ultimately somatic embryo production. The mechanisms whereby auxin triggers embryonic competence are poorly understood (reviewed by Raghavan, 2004). Recent work (Su et al., 2009) emphasized the relevance of the PIN-induced auxin gradient in the activation of WUS which, besides defining the organizing centre in the SAM in vivo (Mayer et al., 1998), is associated with the production of totipotent embryogenic cells during in vitro morphogenesis (Chen et al., 2009; Su et al., 2009). A relationship between auxin requirement and WUS expression and localization is established in this study. The expression of WUS at the end of the induction period (stage 1) is higher in the Bo, Br, and BnSTM lines (Fig. 6A), which exhibit the lowest requirement for auxin, whereas it is repressed in the BnCLV1 lines, which have the highest requirement for auxin. This trend was also confirmed by localization studies revealing an extended domain of WUS (which demarcates the formation of embryogenic cells) in lines overexpressing the Brassica STM gene and a limited WUS signal in the 35S::BnCLV1 line (Fig. 6B). A plausible regulatory mechanism explaining the increased expression of WUS in the 35S::BoSTM line involves the kinase-associated protein phosphatase (KAPP), which is also activated during the induction phase of somatic embryogenesis (Table 1). Genetic studies have shown that during maintenance of the Arabidopsis SAM in vivo, KAPP activity attenuates the inhibitory effect of CLV signalling on the transcription of WUS by binding to the active CLV receptors (reviewed by Carles and Fletcher, 2003). This raises the intriguing possibility that mechanisms governing stem cell maintenance in vivo also regulate embryogenic cell formation in vitro. It must be said, however, that KAPP expression also increased in lines overexpressing Brassica CLV1 (data not shown), possibly as a feedback mechanism to limit the ectopic activation of CLV signalling.
The activation of WUS during the induction phase of the 35S::BoSTM line coincides with the increased expression of other elements playing a key role during the initiation of embryogenesis. These include transcription factors such as STM and BABY BOOM, the latter being responsible for triggering the conversion from vegetative to embryonic development (Boutilier et al., 2002), as well as two ARGONAUTE members PINHEAD/ZWILLE and AGO 1 (Table 1), both involved in meristem formation (Moussian et al., 1998).
Common embryogenesis-related programmes have been postulated to be under the control of genetic mechanisms, which often rely on the methylation state of the DNA regulated by auxin levels (Namasivayam 2007). Several studies on animal and plant systems showed that embryogenic cells or germline cells contain hypomethylated DNA (Monk et al., 1987), and experimental hypomethylation of DNA of somatic cells induces a state of differentiation similar to that observed during the early phases of zygotic embryogenesis (Okkels, 1988). The expression of two key DNA methyltransferases is repressed in the 35S::BoSTM line during the induction phase (stage 0–1, Table 1). One of these enzymes (At5g49160) has been identified as METHYLTRANSFERASE 1, which is responsible for maintaining CpG DNA methylation in Arabidopsis (Finnegan and Kovac, 2000). Investigations with isoschizomeric restriction endonucleases differentially sensitive to methylation of cytosine in CCGG sequences confirmed a lower DNA methylation state in the line overexpressing BoSTM at the end of the induction phase (Fig. 8A). This is in agreement with several independent studies suggesting an essential role for DNA hypomethylation in encouraging the acquisition of embryogenic competence of somatic cells (Lo Schiavo et al., 1989; Munksgaard et al., 1995). Pharmacological studies using azadC (Fig. 8B), a hypomethylating drug which inhibits METHYLTRANSFERASE 1 (Ghoshal et al., 2005), confirm the requirement for DNA hypomethylation during the induction of Arabidopsis somatic embryogenesis.
The development phase
Compared with the induction phase, the number of differentially expressed probes during somatic embryo development in WT and 35S::BoSTM lines decreased, despite a significant variation in function (data not shown). Major differences in expression levels during the transition from stage 1 to stage 2 were observed for probes related to disease and defence processes, which included superoxide dismutase, enzymes involved in ascorbate metabolism, and two enzymes involved in glutathione metabolism (GSH2 and GR) (Table 1).
A key signal encouraging somatic embryo development involves a switch of the cellular glutathione pool [composed of reduced (GSH) and oxidized (GSSG) forms] towards an oxidized state, i.e. a low GSH/GSSG ratio. This concept has been demonstrated in several culture systems of gymnosperm and angiosperm species (Belmonte et al., 2006, 2007). Somatic embryogenesis in spruce was favoured if the endogenous GSH/GSSG ratio was lowered through applications of either GSSG or BSO, a specific inhibitor of GSH de novo synthesis (Griffith and Meister, 1979). These conditions increased somatic embryo yield and quality (Belmonte et al., 2007). Similar improvements were also described in B. napus microspore-derived embryogenesis where a low cellular GSH/GSSG ratio encouraged zygotic-like embryonic development (Belmonte et al., 2006). Physiological and molecular studies have provided insights into the role of GSSG during in vitro embryogenesis (reviewed in Stasolla, 2010). The ectopic expression of BoSTM results in a drop in GSH levels during the first 3 d of somatic embryo development (stage 2) (Fig. 9A) possibly due to the transcriptional repression of GSH2, a biosynthetic enzyme catalysing the condensation of γ-glutamylcysteine and glycine to form GSH, and to a repression in transcription and activity of GR which recycles GSH from GSSG (Table 1, Fig. 9A). These changes, not observed in the WT line, would lower the GSH/GSSG ratio of the transformed line, thereby encouraging the development of somatic embryos. The beneficial effect of an oxidized glutathione environment was further demonstrated by the improvements in Arabidopsis somatic embryo production obtained in the WT line when GSSG or BSO, which lower the GSH/GSSG ratio (Belmonte et al., 2007), was included in the development medium. This was in contrast to the application of GSH, which increases the endogenous GSH/GSSG ratio (Belmonte et al., 2007) and represses the formation of explants able to form somatic embryos as well as the number of somatic embryos produced by each explant (Fig. 9B).
The relationship between BoSTM expression and glutathione redox state is difficult to define, although indirect evidence ties glutathione metabolism to the regulation of self-renewal and differentiation of stem cells in both plant and animal systems (Smith et al., 2000; Jiang et al., 2002).
Conclusion
This study demonstrates that in vitro embryogenesis, i.e. Arabidopsis somatic embryogenesis and B. napus microspore-derived embryogenesis, can be modulated by the overexpression of Brassica genes orthologous to the Arabidopsis AtSTM, AtCLV1, and AtZLL. The enhanced somatic embryogenesis observed in Arabidopsis lines overexpressing Brassica STM was associated with a lower requirement for the inductive signal 2,4-D, and a higher expression of WUS which demarcates the formation of embryogenic cells. This was in contrast to the 35S::BnCLV1 lines which, besides showing poor somatic embryo-forming capacity, had the highest requirement for exogenous 2,4-D and a reduced expression of WUS. The introduction of BoSTM (which showed the greatest improvement on Arabidopsis somatic embryogenesis among the other Brassica STM genes) was associated with the hypomethylation of the DNA during the induction phase of somatic embryogenesis, and with a switch of the glutathione pool towards an oxidized state during development. Such changes were critical for the successful production of somatic embryos. Altered expression of Brassica STM also had profound effects on B. napus microspore-derived embryogenesis. The number of MDEs increased in lines overexpressing BnSTM and significantly decreased in antisense lines down-regulating BnSTM. This suggests that the beneficial effect of the Brassica STM is independent of the embryogenic system utilized.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Identification of Arabidopsis lines ectopically expressing the Brassica genes.
Figure S2. Identification of Brassica napus lines transformed with BnSTM in the sense or antisense orientation.
Figure S3. Schematic representation of protein structure and amino acid sequence alignments of conserved domains of BnSTM, BnCLV1, and BnZLL-1.
Figure S4. Phylogenetic trees of several class-1 KNOX proteins (A), CLAVATA 1-related proteins (B), and ZLL-like proteins (C).
Figure S5 Expression of selected probes showing a different behaviour between the WT and 35S::BoSTM lines at stage 1 of Arabidopsis somatic embryogenesis.
Figure S6. Validation of microarray experiments by quantitative RT-PCR.
Table S1. Genes and primers used for validating the microarray results using quantitative RT-PCR.
Supplementary Material
Acknowledgments
This work was supported by a NSERC Discovery Grant to CS and by a scholarship from the Egyptian government to ME. The authors thank Professor E. C. Yeung for his critical suggestions.
Glossary
Abbreviations
- CLV
CLAVATA
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- MDE
microspore-derived embryo
- SAM
shoot apical meristem
- STM
SHOOTMERISTEMLESS
- WUS
WUSCHEL
- ZLL
ZWILLE
References
- Barton MK, Poethig RS. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development. 1993;119:823–831. [Google Scholar]
- Bassuner BMR, Lam R, Lukowitz W, Yeung EC. Auxin and root initiation in somatic embryos of Arabidopsis. Plant Cell Reports. 2007;26:1–11. doi: 10.1007/s00299-006-0207-5. [DOI] [PubMed] [Google Scholar]
- Belmonte M, Ambrose SJ, Ross ARS, Abrams SR, Stasolla C. Improved development of microspore derived embryo cultures of Brassica napus cv Topaz following changes in glutathione metabolism. Physiologia Plantarum. 2006;127:690–700. [Google Scholar]
- Belmonte MF, Tahir M, Schroeder D, Stasolla C. Over-expression of HBK3, a class I KNOX homeobox gene, improves the development of Norway spruce (Picea abies) somatic embryos. Journal of Experimental Botany. 2007;58:2851–2861. doi: 10.1093/jxb/erm099. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. Journal of the Royal Statistics Society, Series B. 1995;57:289–300. [Google Scholar]
- Bhalla PL, Singh MB. Agrobacterium-mediated transformation of Brassica napus and Brassica oleracea. Nature Protocols. 2008;3:181–189. doi: 10.1038/nprot.2007.527. [DOI] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, et al. Ectopic expression of BabyBoom triggers conversion from vegetative to embryonic growth. The Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles CC, Fletcher JC. Shoot apical meristem maintenance: the art of a dynamic balance. Trends in Plant Sciences. 2003;8:394–401. doi: 10.1016/S1360-1385(03)00164-X. [DOI] [PubMed] [Google Scholar]
- Cerrutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the PIWI domain. Trends in Biochemical Sciences. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham DS, Craig GS, Dennis ES. amp-1a mutant with high cytokinins levels and altered embryogenic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. The Plant Journal. 1993;4:907–916. [Google Scholar]
- Chen S-K, Kurdyukov S, Kereszt A, Wang X-D, Gresshoff PM, Rose RJ. The association of homeobox gene expression with stem cell formation and morphogenesis in cultured Medicago truncatula. Planta. 2009;230:827–840. doi: 10.1007/s00425-009-0988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when over-expressed in Arabidopsis. The Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOTMERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA 1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Dodsworth S. A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Developmental Biology. 2009;336:1–9. doi: 10.1016/j.ydbio.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOTMERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem gene WUSCHEL and ZWILLE. The Plant Journal. 1996;10:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA. Plant DNA methyltransferases. Plant Molecular Biology. 2000;43:189–201. doi: 10.1023/a:1006427226972. [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. Deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN Box, bromo-adjacent homology domain, and nuclear localization signal. Molecular and Cellular Biology. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Griffith OW, Meister A. Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (s-n-butyl homocysteine sulfoximine) Journal of Biological Chemistry. 1979;254:7558–7560. [PubMed] [Google Scholar]
- Hecht V, Viella-Calzada J-P, Hartog MV, Schmidt DL, Boutilier K, Grossniklaus U, de Vries SC. The Arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiology. 2001;127:803–816. [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Meng YL, Feldman LJ. Quiescent center formation in maize roots is associated with an auxin-regulated oxidizing environment. Development. 2002;130:1429–1438. doi: 10.1242/dev.00359. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Leljak-Levanic D, Bauer N, Mihaljevic S, Jelaska S. Changes in DNA methylation during somatic embryogenesis in Cucurbita pepo L. Plant Cell Reports. 2004;23:120–127. doi: 10.1007/s00299-004-0819-6. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Jürgens G, Laux T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfill complementary roles in Arabidopsis shoot apical meristem regulation. Development. 2002;129:3195–3206. doi: 10.1242/dev.129.13.3195. [DOI] [PubMed] [Google Scholar]
- Lin X, Hwang GJ, Zimmerman JL. Isolation and characterization of a diverse set of genes from carrot somatic embryos. Plant Physiology. 1996;112:1365–374. doi: 10.1104/pp.112.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter R. Induction of haploid plants from isolated pollen of Brassica napus. Zeitschreift für Pflanzenphysiologie. 1982;105:427–434. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long JA, Barton KM. The development of apical embryonic pattern in Arabidopsis. Development. 1998;125:3027–3035. doi: 10.1242/dev.125.16.3027. [DOI] [PubMed] [Google Scholar]
- Lo Schiavo F, Pitto L, Giuliano G, Torti G, Nuti-Ronchi V, Marazziti D, Vergara R, Orselli S, Terzi M. DNA methylation of embryogenic carrot cell cultures and its variations as caused by mutation, differentiation, hormones, and hypomethylating drugs. Theoretical and Applied Genetics. 1989;77:325–331. doi: 10.1007/BF00305823. [DOI] [PubMed] [Google Scholar]
- Lund RE. Tables for an approximate test for outliers in linear models. Techometrics. 1975;17:473–476. [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Malik MR, Wang F, Dirpaul JM, Zhou N, Polowick PI, Ferrie AMR, Krochko JE. Transcript profiling and identification of molecular markers for early embryogenesis in Brassica napus. Plant Physiology. 2007;144:134–154. doi: 10.1104/pp.106.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. Effect of mutations in the PINHEAD gene of Arabidopsis on the formation of the shoot apical meristem. Developmental Genetics. 1995;16:358–366. [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembronic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC. Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics. 1998;149:549–563. doi: 10.1093/genetics/149.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordhorst AP, Hartog MV, El Tamer MK, Laux T, de Vries S. Somatic embryogenesis from Arabidopsis shoot apical meristem mutants. Planta. 2002;214:829–836. doi: 10.1007/s00425-001-0700-6. [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jürgens G, Laux T. Role of ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO Journal. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munksgaard D, Mattsson O, Okkels FT. Somatic embryo development in carrot is associated with an increase in levels of S-adenosylmethionine, S-adenosylhomocysteine and DNA methylation. Physiologia Plantarum. 1995;93:5–10. [Google Scholar]
- Namasivayam P. Acquisition of embryogenic competence during somatic embryogenesis. Plant Cell, Tissue and Organ Culture. 2007;90:1–8. [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodelling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkels FT. A theory explaining formation of somatic embryogenic cells by auxin-induced inhibition of DNA methylation. Physiologia Plantarum. 1988;73:11A. [Google Scholar]
- Raghavan V. Role of 2,4-dichlorophenoxyacetic acid (2,4-D) in somatic embryogenesis on cultured zygotic embryos of Arabidopsis: cell expansion, cell cycling, and morphogenesis during continuous exposure of embryos to 2,4-D. Annals of Botany. 2004;91:1743–1756. doi: 10.3732/ajb.91.11.1743. [DOI] [PubMed] [Google Scholar]
- Saama PM, Patel OV, Bettegowda A, Ireland JJ, Smith GW. Novel algorithm for transcriptome analysis. Physiological Genomics. 2006;28:62–66. doi: 10.1152/physiolgenomics.00108.2006. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M. Expression of gibberelliin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiology. 2001;125:1508–1516. doi: 10.1104/pp.125.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Kamada I, Youssefian S, Wabiko H. Correlation between DNA under-methylation and dwarfism in maize. Biochimica and Biophysica Acta. 1989;1009:35–38. [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Meyer KF, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Scofield S, Murray JA. KNOX gene function in plant stem niches. Plant Molecular Biology. 2006;60:929–946. doi: 10.1007/s11103-005-4478-y. [DOI] [PubMed] [Google Scholar]
- Sinha NR, Williams RE, Hake S. Over-expression of the maize homeobox gene, KNOTTED1, causes a switch from determinate to indeterminate fate. Genes and Development. 1993;7:787–795. doi: 10.1101/gad.7.5.787. [DOI] [PubMed] [Google Scholar]
- Smith E, Ladi M, Mayer-Proschel M, Noble M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proceedings of the National Academy of Sciences, USA. 2000;97:10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C. Glutathione redox regulation of in vitro embryogenesis. Plant Physiology and Biochemistry. 2010;48:319–327. doi: 10.1016/j.plaphy.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Stasolla C, Belmonte M, Tahir M, Elhiti M, Khamiss K, Joosen R, Maliepaard C, Sharpe A, Gjetvaj B, Boutilier K. Buthionine sulfoximine (BSO)-mediated improvement in in vitro cultured embryo quality is associated with major changes in transcripts involved with ascorbate metabolism, meristem development and embryo maturation. Planta. 2008;228:255–272. doi: 10.1007/s00425-008-0735-z. [DOI] [PubMed] [Google Scholar]
- Stasolla C, Bozhkov PV, Chu T-Z, van Zyl L, Egertsdotter U, Suarez M, Craig D, Wolfinger RD, von Arnold S, Sederoff RR. A transcriptional pathway during somatic embryogenesis in gymnosperms. Tree Physiology. 2004;24:1073–1085. doi: 10.1093/treephys/24.10.1073. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepinec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON 2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences, USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Zhao XY, Liu YB, Zhang CL, O'Neill S, Zhang ZS. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. The Plant Journal. 2009;59:448–460. doi: 10.1111/j.1365-313X.2009.03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo W, Swarup S. Isolation and characterization of a cDNA clone encoding an STM homolog from rapid cycling Brassica oleracea. Plant Physiology. 1999;121:1383. [Google Scholar]
- Thorpe TA, Stasolla C. Somatic embryogenesis. In: Bhojwani SS, Soh WY, editors. Current trends in the embryology of angiosperms. Dordrecht: Kluwer Academic Publisher; 2001. pp. 279–336. [Google Scholar]
- Tucker MR, Hinze A, Tucker EJ, Takada S, Jürgens G, Laux T. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135:2839–2843. doi: 10.1242/dev.023648. [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Willemsen V, Scheres B. Mechanisms of pattern formation in plant embryogenesis. Annual Reviews of Genetics. 2004;38:587–614. doi: 10.1146/annurev.genet.38.072902.092231. [DOI] [PubMed] [Google Scholar]
- Wolfinger RD, Gobson G, Wolfinger ED, Bennet L, Hamadeh H, Bushel P, Afshari C, Pauls RS. Assessing gene significance from cDNA microarray expression data via mixed models. Journal of Computational Biology. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sanderberg G, Samach A, Ori N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Current Biology. 2006;15:1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Frugis G, Chua NH. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. The Plant Journal. 2002;30:349–359. doi: 10.1046/j.1365-313x.2002.01289.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.