Summary
Myxococcus xanthus is a gram-negative bacterium capable of complex developmental processes involving vegetative swarming and fruiting body formation. Social (S-) gliding motility, one of the two motility systems employed by M. xanthus, requires at least two cell surface structures: type IV pili (TFP) and extracellular polysaccharides (EPS). Extended TFP which are composed of thousands of copies of PilA retract upon binding to EPS and thereby pull the cell forward. TFP also act as external sensor to regulate EPS production. In this study, we generated a random PilA mutant library and identified one derivative, SW1066, which completely failed to undergo developmental processes. Detailed characterization revealed that SW1066 produced very little EPS but wild-type amounts of PilA. These mutated PilA subunits, however, are unable to assemble into functional TFP despite their ability to localize to the membrane. By preventing the mutated PilA of SW1066 to translocate from the cytoplasm to the membrane, fruiting body formation and EPS production was restored to the levels observed in mutant strains lacking PilA. This apparent connection between PilA membrane accumulation and reduction in surface EPS implies that specific cellular PilA localization are required to maintain the EPS level necessary to sustain normal S-motilityin M. xanthus.
Keywords: Myxococcus xanthus, type four pili, PilA, extracellular polysaccharide
Introduction
Myxococcus xanthus is a gram-negative soil bacterium that swarms on solid surfaces without the help of flagella and coordinates thousands of cells to aggregate into multi-cellular fruiting bodies during starvation. Gliding motility is important for both functions. M. xanthus cells utilize two genetically distinct systems for their gliding motility: adventurous (A)-motility and social (S)-motility (Hodgkin & Kaiser, 1979). Single cell movements via A-motility is the preferred type of locomotion on dry surfaces while coordinated movement in groups via S-motility is mainly utilized on moist surfaces, enabling the bacterium to adapt to a variety of physiological and ecological environments (Shi & Zusman, 1993). S-motility is important for cell aggregation, cohesion and fruiting body morphogenesis (Hodgkin & Kaiser, 1979, MacNeil et al., 1994). The components that are known to be essential for S-motility are: type IV pili (TFP) (Kaiser, 1979, Wu & Kaiser, 1995), the extracellular polysaccharides (EPS) portion of the fibril material (Shimkets, 1986, Arnold & Shimkets, 1988a, Lu et al., 2005) and lipopolysaccharide (LPS) O-antigen (Bowden & Kaplan, 1998).
The TFP of M. xanthus are filaments of about 3–5 μm in length and less than 10 nm in diameter that extend from the bacterial cell poles and have been recognized as the motor for S-motility (Kaiser, 1979, Wu & Kaiser, 1995). TFP-dependent S-motility is achieved by extending the pili at one cell pole, attaching to surfaces or to another cell and then retracting to pull the cell forward (Merz et al., 2000, Sun et al., 2000, Skerker & Berg, 2001). The cell surface EPS was found to be the anchor for TFP and trigger retraction (Li et al., 2003). Mutants lacking TFP showed severely delayed development, reduced EPS production (Black et al., 2006) and the resulting fruiting bodies exhibit unstructured appearance compared to wild type (Wu et al., 1998).
TFP are composed of thousands of copies of the type IV pilin protein PilA encoded by the pilA gene (Wu & Kaiser, 1995). Since the organization of the pil operon in M. xanthus is highly homologous to the ones present in Pseudomonas aeruginosa and Neisseria gonorrhoeae, it is likely that the biogenesis of M. xanthus TFP employs similar mechanism as described for these organisms (Wolfgang et al., 2000, Nudleman & Kaiser, 2004). PilA is synthesized as prepilin in the cytoplasm and directly secreted across the cytoplasmic membrane via the Sec system (Pelicic, 2008). Due to its highly hydrophobic N-terminus the secreted PilA inserts into the inner membrane for processing by the PilD peptidase to form the mature pilin which likely remains membrane bound to form a readily available pool for TFP extension. The assembly and retraction of TFP involves a complex machinery composed of several Pil proteins including PilB, PilT and PilC (Mauriello & Zusman, 2007). Hydrolysis of ATP by PilB facilitates the assembly of TFP (Jakovljevic et al., 2008) and the assembled pili are extruded out of the cell through an outer membrane channel formed by the secretin protein PilQ (Wall et al., 1999). TFP retraction involves the disassembly mediated by the ATPase activity of PilT (Jakovljevic et al., 2008). Although not demonstrated in M. xanthus, it has been suggested in P. aeruginosa and N. gonorrhoeae that the mature pilin subunits are translocated from the base of the TFP to the cytoplasmic membrane during TFP retraction, thus dissolving the TFP into a membrane-bound PilA monomer pool from which they can be recycled (Skerker & Berg, 2001, Morand et al., 2004, Merz et al., 2000).
The anchoring substrate to enable movement upon TFP retraction in M. xanthus has been identified as the EPS portion of the extracellular fibrils (Li et al., 2003) which were originally described as peritrichous, filamentous cell surface appendages (Arnold & Shimkets, 1988a) comprised of equal amounts of protein and polysaccharides (Behmlander & Dworkin, 1994). The production of EPS is developmentally regulated and essential for fruiting body formation. Mutants lacking EPS are defective in S-motility and fruiting body formation (Shimkets, 1986, Weimer et al., 1998, Yang et al., 2000, Lu et al., 2005). However, the phenotype can be at least partially rescued by addition of wild-type cells or isolated wild-type EPS(Shimkets, 1986, Chang & Dworkin, 1994). Genetic analyses indicated that the biogenesis of EPS involve the DnaK homolog SglK (Weimer et al., 1998, Yang et al., 1998); the chemotaxis-like dif operon (previously known as dsp) (Arnold & Shimkets, 1988b, Yang et al., 2000, Lancero et al., 2002); the eps operon, which contains genes associated with carbohydrate transport and biosynthesis (Lu et al., 2005); and the transcriptional regulator Nla24 (Lancero et al., 2004). Interestingly, all mutants lacking surface TFP were found to have reduced EPS production. Assembled TFP, in addition to their role as the motor engine in S-motility, appear to function as an extracellular sensor to modulate EPS production, probably through the Dif pathway in a retraction independent manner (Black et al., 2006). Underscoring the importance of PilA in cellular processes of M. xanthus, transcription of pilA is strictly regulated during vegetative and developmental stages (Wu & Kaiser, 1997) and a threshold concentration of PilA must be reached to allow assembly into TFP (Jelsbak & Kaiser, 2005). Extracellularly, specific amounts of surface TFP are required for normal S-motility (Jelsbak & Kaiser, 2005) and modulation of EPS biogenesis (Black et al., 2006). However, at this point, little is known regarding the detailed mechanisms of these important cellular functions.
In this study, we have generated a PilA mutant library and identified the mutant derivative SW1066 that accumulates PilA in the membrane but is unable to extrude TFP. By studying this mutant, we demonstrated that membrane accumulation of PilA affects EPS production and thus fruiting body formation, suggesting that proper localization and concentration of PilA is essential for the coordination of EPS production and other S-motility related functions.
Results
Construction of a PilA mutant library
PilA, the subunit for TFP, plays a significant role in S-motility and fruiting body formation during the M. xanthus life cycle (Wu & Kaiser, 1995) and was more recently found to be involved in regulation of EPS production (Black et al., 2006). In order to genetically dissect PilA function, we constructed a PilA mutant library. Two separate in vitro mutagenesis approaches were used for introducing random mutations in the pilA gene to at least partially offset the intrinsic bias of each method.
In one approach, plasmid pSWU359 (Table 1), a derivative of pSWU19 carrying the pilA gene (Wu & Kaiser, 1996), was mutagenized with hydroxylamine. This plasmid was chosen since the pSWU19 vector backbone contains the attP region which enables site specific recombination into the attB site on the M. xanthus chromosome. In a second approach, we performed error prone PCR of the pilA gene and cloned the PCR product into pSWU19 (Wu & Kaiser, 1995). Experimental conditions were chosen to favor 1–2 mutations in the pilA gene per plasmid (See experimental procedures). The resulting plasmid library was electroporated into the pilA in-frame-deletion strain DK10410 and selected for KanR colonies.
Table 1.
Strains and plasmids used in this study
| Strain/plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strain | ||
| DK1622 | wildtype | Kaiser (1979) |
| DK10410 | ΔpilA | Wu and Kaiser (1996) |
| DK10416 | ΔpilB | Wu et. al. (1997) |
| DK10409 | ΔpilT | Wu and Kaiser (1997) |
| SW504 | ΔdifA | Yang et. al. (1998b) |
| SW1066 | ΔpilA/pilA(A32V,D104N) | This study |
| SW1066A | ΔpilA/pilA(A32V) | This study |
| SW1066B | ΔpilA/pilA(D104N) | This study |
| SW1247 | ΔpilA/pilA (A136D,C215T) | This study |
| SW1260 | ΔpilA/pilA (A23T,A58T,S189T) | This study |
| SW1424 | ΔpilA/pilA (E175G) | This study |
| SW1426 | ΔpilA/pilA (V99M) | This study |
| SW2001 | ΔpilT ΔpilA | This study |
| SW2002 | ΔpilT ΔpilA/pilA (A32V,D104N) | This study |
| SW2003 | ΔpilT ΔpilA/pilA (A32V) | This study |
| SW2004 | ΔpilT ΔpilA/pilA (D104N) | This study |
| SW2009 | ΔpilA/pilA (sps) sps: PilA signal peptide shortened, the last glycine in the signal peptide was deleted |
This study |
| SW2010 | ΔpilA/pilA (spd) spd: PilA signal peptide deleted, all 12 amino acids in signal peptide was deleted |
This study |
| SW2011 | ΔpilA/pilA (sps,A32V,D104N) | This study |
| SW2012 | ΔpilB ΔpilA | This study |
| Plasmids | ||
| pSWU19 | Cloning vector with Mx8 attP locus for integration into Myxococcus xanthus genome, KmR | Wu and Kaiser (1995) |
| pSWU359 | pSWU19 with 872bp pilA gene and its promoter inserted, KmR | Wu and Kaiser (1996) |
| pBJ113 | Cloning vector, KmR, galK | Julien et al. (2000) |
| pMX01 | pilA in-frame-deletion PCR product cloned into pBJ113 using EcoRI and HindIII sites | This study |
| pMX02 | pilA (A32V,D104N) cloned into pSWU19 at EcoRI-BamHI sites | This study |
| pMX03 | pilA (A32V) cloned into pSWU19 at EcoRI-BamHI sites | This study |
| pMX04 | pilA (D104N) cloned into pSWU19 at EcoRI-BamHI sites | This study |
| pMX05 | pilA (sps) cloned into pSWU19 at EcoRI-BamHI sites | This study |
| pMX06 | pilA (spd) cloned into pSWU19 at EcoRI-BamHI sites | This study |
| pMX07 | pilA (sps,A32V,D104N) cloned into pSWU19 at EcoRI-BamHI sites | This study |
More than 1000 colonies carrying a mutagenized copy of pilA were isolated and screened for S-motility, fruiting body formation and whole cell PilA production. According to their various phenotypes the mutants were categorized into five distinct types (Fig. 1). Type I mutants closely resembled DK10410 (ΔpilA). They did not produce PilA, were defective in S-motility and formed immature fruiting bodies. Type II mutants were phenotypically similar to DK1622 (wild type). They produced PilA at wild-type level and exhibited normal S-motility and fruiting body formation. Type III mutants had reduced PilA production as well as swarming, and formed immature fruiting bodies. More interestingly, Type IV and type V mutants both produced wild-type level of PilA but were unable to swarm. However, type IV mutants still formed immature fruiting body while type V mutants were defective in fruiting body formation. Among all mutants in the library, about 70% were type I, 30% were type II while less than 1% of the mutants belonged to the other three categories.
Fig. 1.

Summary of PilA mutant library phenotypes. All M. xanthus mutants in the library were characterized and categorized into five different types (Type I – Type V) and representative phenotypes are shown: (A) swarming ability on 0.3% CYE agar, (B) fruiting body formation on CF agar and (C) whole-cell PilA production as measured by western blotting. Columns 1 and 2 are DK1622 (wild type) and DK10410 (ΔpilA). Columns 3–7 are representatives of Type I to Type V mutants: SW1424 (ΔpilA/pilAE175G), SW1426 (ΔpilA/pilAV99M), SW1247 (ΔpilA/pilAA136D, C215T), SW1260 (ΔpilA/pilAA23T, A58T, S189T) and SW1066 (ΔpilA/pilAA32V, D104N).
Two mutations are required for the non-fruiting phenotype of mutant SW1066
Among the PilA mutants, one type V mutant, SW1066 (ΔpilA/pilAA32V, D104N) resulting from the hydroxylamine mutagenesis approach was of interest due to its complete lack of fruiting body formation. This is unusual since even mutants lacking pilA are able to form fruiting bodies albeit delayed and less structured (Wu et al., 1998). To ensure that the observed phenotype was not caused by an unrelated secondary mutation, the mutant SW1066 was reconstructed by electroporating the genomic DNA of SW1066 into the pilA deletion mutant DK10410. All resulting KanR colonies exhibited the same phenotype as SW1066, indicating that the mutation in pilA is likely the only genetic locus responsible for the observed behavior. Sequencing of the pilA gene in SW1066 revealed two mutated sites: an A to V mutation at position 32 in the proposed N-terminal alpha helix domain and a D to N mutation at position 104 in the center of the protein (Fig. 2A). In order to test if one of these two mutations was dominant and sufficient to cause the non-fruiting phenotype, we separated these two mutations by generating the following strains: SW1066A (ΔpilA/pilAA32V) and SW1066B (ΔpilA/pilAD104N). Surprisingly, both SW1066A and SW1066B were able to form fruiting bodies albeit irregular shaped, indicating that neither one of the two mutations was sufficient and that the combination of the two mutations was required to completely abolish development in SW1066. In addition, the two derivatives carrying the respective individual mutations exhibited very different swarming phenotypes. Mutant SW1066B swarmed like wild-type while SW1066A was defective in S-motility (Fig. 2B).
Fig. 2.

The combination of two mutations is required for the non-fruiting phenotype of SW1066. (A) Complete PilA amino acid sequence of wild-type M. xanthus. Grey letters indicate the predicted 12 amino acid signal peptide of PilA in the prepilin. Underlined sequences correspond to the N-terminal alpha helix in the mature pilin predicted using SCRATCH protein predictor (http://scratch.proteomics.ics.uci.edu/). Dots under the amino acids indicate the two mutated sites in the PilA of SW1066. SW1066A only contains mutation A32V and SW1066B only contains mutation D104N. (B) Swarming and fruiting body formation phenotypes of mutants SW1066 (ΔpilA/pilAA32V, D104N), SW1066A (ΔpilA/pilAA32V) and SW1066B (ΔpilA/pilAD104N). The first row shows the swarming ability on 0.3% CYE agar. The second and third rows depict fruiting body formation on CF agar at a total magnification of 20 × and 200 ×, respectively.
SW1066 PilA does not assemble into TFP
Since the two mutations in SW1066 were localized in the pilin protein encoding part of pilA (not the promoter or signal peptide encoding region) and the major known function of pilin in M. xanthus is to assemble into TFP and power S-motility, we tested whether PilA production and TFP assembly were affected in the SW1066 mutant strain. M. xanthus whole-cell lysates were probed using an anti-PilA antibody (Li et al., 2005) and mutants SW1066, SW1066A and SW1066B were all found to produce the same amount of whole-cell PilA as wild type (Fig. 3A). A more detailed analysis revealed that SW1066 had no detectable cell-surface PilA (Fig. 3B) despite producing wild-type levels of whole-cell PilA, indicating that the mutated PilA derivative produced by this strain likely cannot assemble into surface TFP. In contrast, SW1066B displayed similar amounts of cell-surface PilA as the wild type, indicating that SW1066B pilin can assemble into functional TFP consistent with its normal swarming phenotype. Interestingly, cell-surface PilA levels in SW1066A were increased. This strain, however, was unable to swarm (Fig. 2B), probably due to inability of the mutagenized TFP to retract (Yang et. al. unpublished data) (Fig. 3B). The above data demonstrated that the combination of the two mutations was necessary to alter the PilA structure in a way that prevented assembly into TFP, while neither of the two mutations in SW1066 alone affected TFP extension.
Fig. 3.

SW1066 PilA does not assemble into surface pili. (A) Immunoblot of M. xanthus whole-cell lysates obtained from 5 × 107 cells using anti-PilA antibody. Samples from left to right are: DK1622 (wild type), DK10410 (ΔpilA), SW1066 (ΔpilA/pilAA32V, D104N), SW1066A (ΔpilA/pilAA32V), SW1066B (ΔpilA/pilAD104N), DK10409 (ΔpilT), SW2002 (ΔpilT ΔpilA/pilAA32V, D104N), SW2003 (ΔpilT ΔpilA/pilAA32V) and SW2004 (ΔpilT ΔpilA/pilAD104N). (B) Cell-surface pili were sheared off from 1010 M. xanthus cells, precipitated with trichloroacetic acid (TCA) and probed using anti-PilA antibody. Lanes are identical to those in panel (A).
The absence of detectable cell-surface PilA in SW1066 could be caused by an inability of PilAA32V,D104N to assemble into TFP or an elevated retraction rate of the mutated TFP compared to wild type. In order to distinguish between these two possibilities, a ΔpilT mutation was introduced into SW1066, SW1066A and SW1066B to generate mutants SW2002 (ΔpilT ΔpilA/pilAA32V, D104N), SW2003 (ΔpilT ΔpilA/pilAA32V) and SW2004 (ΔpilT ΔpilA/pilAD104N). These three mutant strains each carried the respective mutagenized PilA of their parent strain but did not produce PilT, the ATPase that powers TFP retraction (Wu et al., 1997, Jakovljevic et al., 2008). Therefore all assembled and extruded TFP were not retracted into the cells resulting in an overpiliation phenotype. Whole-cell and cell-surface PilA were measured using DK10409 (ΔpilT) as the overpiliation control. Western blots showed that all strains produced PilA at wild-type level (Fig. 3A), however, SW2003 and SW2004 were overpiliated whereas SW2002 showed no detectable surface pili (Fig. 3B). These results confirmed that the mutant PilA in SW1066 indeed cannot assemble into TFP while SW1066A and SW1066B were still able to do so.
The non-fruiting phenotype of SW1066 is related to highly reduced EPS production
The finding that the mutagenized PilA in SW1066 did not assemble into TFP was consistent with the fact that SW1066 was defective in S-motility. However, it could not explain the non-fruiting phenotype because other mutants lacking surface TFP (ΔpilA, ΔpilG, ΔpilH, ΔpilI) still form fruiting bodies (Wu et al., 1998). In addition to their role in S-motility, TFP have been found previously to be involved in regulating the production of EPS (Black et al., 2006), a key component for fruiting body formation and S-motility. We therefore examined the cell surface EPS level of SW1066 during swarming on hard agar surface (the surface condition on which fruiting body morphogenesis is initiated) using a modification of the trypan blue binding assay (Black & Yang, 2004). Freshly grown cells from the agar surface instead of a vegetative shaking culture were harvested and the EPS levels of the different mutant strains were compared to wild type (Fig. 4). DK1622 (wild type) and SW504 (ΔdifA), a mutant deficient in EPS biogenesis, were used as positive and negative controls, respectively. SW504 EPS production was less than 20% of wild-type levels while lack of PilA in DK10410 (ΔpilA) reduced EPS production by about 50%. Surprisingly, the mutated PilA present in SW1066 reduced EPS production to only 20% of wild-type levels which was significantly less than DK10410 (ΔpilA) and rather comparable to SW504 (ΔdifA). In contrast, in SW1066A and SW1066B EPS production was only slightly reduced to about 90% of wild-type levels (Fig. 4). The amount of cell surface EPS was clearly correlated with the fruiting body formation phenotype observed in Fig. 2B. Reduction of EPS production below wild-type levels led to less compact and unstructured aggregates. A basal EPS threshold concentration appeared to be needed to enable fruiting body morphogenesis during development since mutant strains with very little EPS production failed to build up these structures.
Fig. 4.
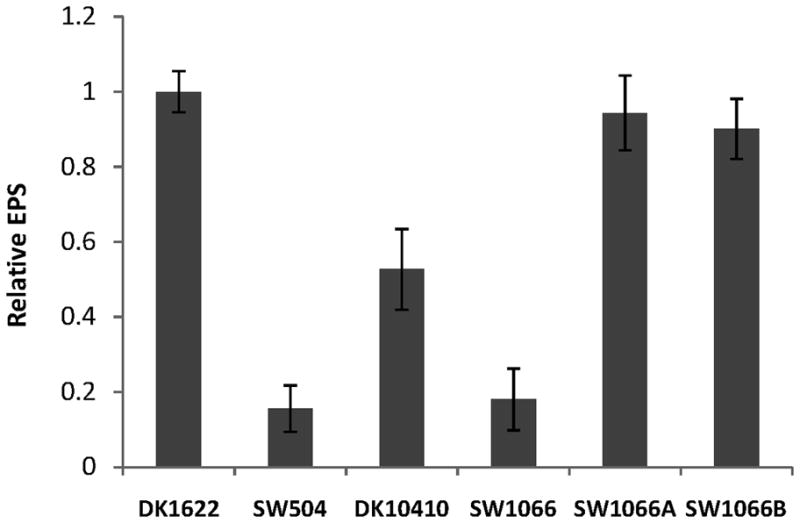
Examination of cell surface EPS production on agar. Freshly grown M. xanthus cells were harvested from 1.5% CYE agar plates and cell surface EPS was measured using a modified trypan blue binding assay (See experimental procedures). The data represent three independent experiments and the error bars show the standard deviation of three samples.
PilA in SW1066 is accumulated at the membrane
The above results suggested that the lack of fruiting body formation in SW1066 was caused by its severe reduction in EPS production. Since the mutations in SW1066 are localized in pilA, the effect on EPS production was mediated by PilA. We have shown that SW1066 produced the same amount of whole-cell PilA compared to wild type but was unable to assemble the pilin subunits into cell-surface pili (Fig. 3). To obtain further evidence on PilA distribution in wild type and SW1066, cells were immunofluorescently labeled with anti-PilA antibody and visualized via fluorescence microscopy. Fig. 5 shows representative deconvoluted images of different mutant strains obtained from 20–30 images along the z-axis. Since the experimental conditions and the antibody used in this study did not stain PilA assembled into extracellular TFP filaments, all fluorescent signal observed represent unassembled intracellular or membrane-associated PilA. In DK1622 (wild type) PilA was predominantly localized at the membrane, consistent with the idea that mature pilins form a pool for TFP extension and recycling in the cytoplasmic membrane. In DK10410 (ΔpilA), a weak background fluorescence homogeneously distributed within the cell was observed which was probably due to non-specific binding by the polyclonal anti-PilA antibody. However, the intensity was very low compared to PilA producing strains. Similar to wild type, SW1066 showed membrane-associated localization of PilA (Fig. 5). Given the fact that wild type and SW1066 produced similar amounts of whole cell PilA (Fig. 1C and 3A) and wild type cells assemble a proportion of their PilA into TFP, it is very likely that more pilin subunits were accumulated in the membrane in SW1066. In addition, PilA in DK1622 and SW1066 were both found to be processed and only existed in the form of mature pilin (Fig. 7A). Therefore, both wild type and SW1066 cells formed a membrane-associated PilA pool that was composed of processed mature pilin and SW1066 possibly accumulated more PilA in this pool due to its inability to assemble TFP.
Fig. 5.
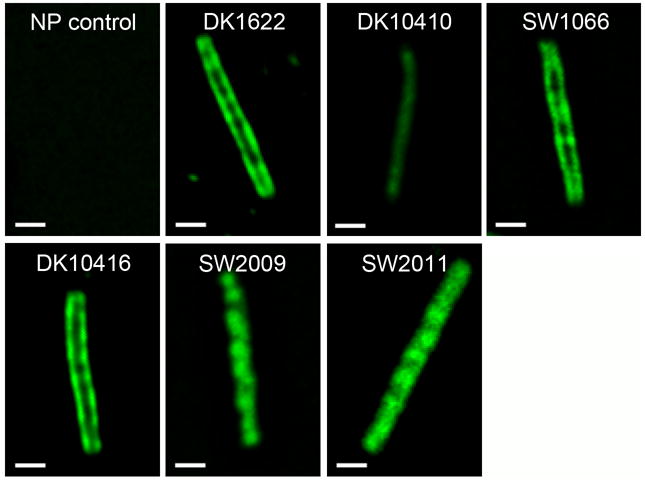
Localization of PilA in different M. xanthus strains. The localization of PilA in wild type and different M. xanthus mutant strains was determined using immunofluorescence labeling in combination with deconvolution microscopy. Fixed and permeabilized M. xanthus cells were stained using purified anti-PilA antibody (the secondary antibody is Alexa Fluor 488, green). Images were deconvolved from 20 to 30 cell sections along the Z axis. Samples from left to right, from top to bottom are: non-permeabilized DK1622 cell control, DK1622 (wild type), DK10410 (ΔpilA), SW1066 (ΔpilA/pilAA32V, D104N), DK10416 (ΔpilB), SW2009 (ΔpilA/pilAsps), and SW2011 (ΔpilA/pilAsps, A32V, D104N).The scale bar equals 1μm.
Fig. 7.

EPS and fruiting body formation defects were reduced by trapping PilA in the cytoplasm. (A) Whole-cell lysate of 5 × 107 M. xanthus cells were probed using anti-PilA antibody. Prepilin migrated more slowly (upper band in the gel) compared to mature pilin (lower band in the gel). (B) Fruiting body formation assay on CF agar. (C) Cell surface EPS levels were determined following the same protocol as in Fig. 4. The strains used have the following relevant genotypes: DK1622 (wild type), DK10410 (ΔpilA), SW2010 (ΔpilA/pilAspd), SW2009 (ΔpilA/pilAsps), SW1066 (ΔpilA/pilAA32V, D104N) and SW2011 (ΔpilA/pilAsps, A32V, D104N)
Membrane accumulation of wild-type PilA reduces EPS production
To examine if PilA accumulation in the cytoplasmic membrane per se can cause a more dramatic decrease in cell surface EPS than observed for the absence of PilA, or if this phenotype is specific to the mutated PilAA32V, D104N, we examined the effect of a pilB mutation on EPS production. Since the ATPase PilB powers PilA assembly and extension as TFP, lack of this protein should lead to accumulation of wild type PilA in the cytoplasmic membrane. Strain DK10416 (ΔpilB) was tested for swarming, fruiting body formation and EPS production (Fig. 6) and was found to exhibit a phenotype very similar to the one observed for SW1066 (Figs. 1, 2 and 4) including the expected membrane-associated PilA distribution (Fig. 5). In addition, we constructed and characterized a pilB pilA double mutant, SW2012. Lack of PilA in the pilB mutant background partially restored EPS production and fruiting body formation to the level of the pilA deletion mutant(Fig. 6), suggesting that the dramatic reduction of EPS levels in DK10416 (ΔpilB) is indeed due to accumulation of wild-type PilA in the membrane rather than missing the PilB protein. Taken together, these results demonstrated that the observed effects on EPS production were not exclusive to the mutated forms of PilA present in SW1066 but likely caused by accumulation of wild type PilA in the membrane.
Fig. 6.
Membrane accumulation of wild type PilA reduces EPS production. Swarming ability, EPS production and fruiting body formation were tested in DK1622 (wild type control), DK10416 (ΔpilB) and SW2012 (ΔpilB ΔpilA) mutants: (A) swarming ability on 0.3% CYE agar, (B) fruiting body formation on CF agar and (C) Cell surface EPS levels were determined following the same protocol as in Fig. 4.
EPS and fruiting body formation defects were reduced by trapping PilA in the cytoplasm
To further explore if the accumulation of PilA in the membrane per se was responsible for the significantly reduced EPS level and the non-fruiting phenotype in SW1066, we designed PilA mutant forms of wild type and SW1066 that were unable to translocate from the cytoplasm to the membrane and tested their EPS production and fruiting body phenotypes. PilA is synthesized as prepilin with a 12 amino acid hydrophilic signal peptide at the N-terminus (Fig. 2A) which is recognized and cleaved by the prepilin peptidase PilD for assembly into TFP after being secreted through the inner membrane by the Sec secretion system (Pelicic, 2008). In P. aeruginosa, the last glycine residue of the signal peptide was shown to be the only key amino acid for the efficient processing of PilA signal peptide (Strom & Lory, 1991). Since M. xanthus PilA shares a conserved amino terminal region with P. aeruginosa that includes the signal peptide (Nudleman & Kaiser, 2004), we hypothesized that the signal peptide especially the last glycine residue may also be important for M. xanthus prepilin recognition and processing. By deleting the last amino acid glycine in the signal peptide (sps – signal peptide shortened) or deleting the whole signal peptide (spd – signal peptide deleted) in wild-type M. xanthus PilA we generated two mutant strains: SW2009 (ΔpilA/pilAsps) and SW2010 (ΔpilA/pilAspd). Whole cell western blot of PilA showed that the PilA generated by SW2009 migrated more slowly in the SDS-PAGE compared to wild-type PilA, indicating that PilA was not processed but existed in its prepilin form. On the other hand, SW2010 was unable to produce PilA at all (Fig. 7A). Consistent with a failure to process and export PilAsps, immunofluorescence staining showed that PilA in SW2009 was not localized at the membrane but homogeneously distributed within the cell (Fig. 5). This indicated that the shortened sps signal peptide in SW2009 allowed “trapping” of PilA in the cytoplasm by preventing prepilin recognition, secretion and processing. Based on these findings, we replaced the signal peptide of PilAA32V, D104N in SW1066 with the sps signal peptide and generated mutant SW2011 in order to retain this mutated PilA in the cytoplasm. As expected, PilA of SW2011 appeared unprocessed in the western blot (Fig. 7A) and displayed cytoplasmic distribution upon immunofluorescence labeling (Fig. 5). EPS production of SW2011 was restored to 50% of wild type levels and fruiting body formation was similar to mutant strains lacking PilA (Fig. 7B and 7C). Therefore, preventing membrane accumulation of PilAA32V, D104N by retaining it in the cytoplasm partially rescued EPS and fruiting body formation defects observed for SW1066. The EPS production and fruiting body formation phenotypes of SW2009, the mutant strain in which wild-type PilA is retained in the cytoplasm were very similar to SW2011 (Fig. 7B and 7C). These results suggested that membrane-associated PilA accumulation triggered highly reduced EPS production and defects in fruiting body formation while cytoplasmic accumulation did not have such an effect.
Discussion
PilA is the key subunit of TFP which are essential for S-motility during all stages of the M. xanthus life cycle (Wu & Kaiser, 1995) and have been implicated as sensors regulating EPS production (Black et al., 2006). In this study, we have generated a comprehensive PilA mutant library with more than 1000 mutants that resulted in five distinct types of mutants (Fig. 1). The type V mutant SW1066 was chosen for detailed characterization due to its singular phenotype. The mutated PilAA32V, D104N of SW1066 is processed into mature pilin (Fig. 7A), translocated into the membrane (Fig. 5,) but fails to assemble into TFP (Fig. 3). Interestingly, in this mutant membrane accumulation of PilA causes significant reduction in EPS production (Fig. 4) which causes the observed defects in fruiting body formation (Fig. 2B). Examination of pilB and pilB pilA double mutants demonstrated that membrane accumulation of PilA per se influences EPS levels, since wild type PilA has the same effect as the mutated PilA of SW1066 (Fig. 6). Furthermore, retention of PilA in the cytoplasm alleviated the observed defects to the level displayed by mutant strains lacking PilA (Fig. 7), thus confirming the regulatory effect of membrane accumulation of PilA on EPS production and consequently fruiting body formation. Additionally, we demonstrated that the signal sequence of prepilin is not only important for cellular trafficking of PilA but also provided protection from degradation (Fig. 7A) and allowed the cell to distinguish PilA destined for recycling/degradation from the subunits that will be translocated to the membrane.
Coordination of EPS production by PilA
EPS and TFP are two essential components for S-motility in M. xanthus. Previous genetics and phenotypic studies revealed that assembled TFP are an important factor in EPS production (Black et al., 2006), while EPS acts as extracellular anchoring substrate for TFP that triggers retraction (Li et al., 2003). Mutant cells without EPS are hyperpiliated while lack of TFP reduces EPS level, strongly suggesting a mutual regulation of these important cellular structures. It has been demonstrated previously that assembled TFP but not their ability to retract is an important factor in the regulation of EPS production (Black et al., 2006). The results of this study indicated the existence of an additional layer of PilA/TFP-mediated regulation of EPS biogenesis: the cellular localization of unassembled pilin subunits is equally important for the regulation of EPS production since in the absence of surface TFP, membrane accumulation of PilA resulted in a greater reduction in EPS production than observed for the complete lack of PilA (Fig. 4, 6C, 7C).
Above observations naturally lead to the question: How does accumulation of PilA at the membrane influence EPS production? EPS biogenesis in M. xanthus has been suggested to involve three steps: synthesis of monosaccharides in the cytoplasm, membrane-associated assembly of the monosaccharides into polysaccharides and transport of the polysaccharides to the cell surface (Lu et al., 2005). Several genomic loci including dif (Yang et al., 2000), stk (Dana & Shimkets, 1993), and sglK (Weimer et al., 1998) as well as the presence of cell surface TFP (Black et al., 2006) have been implicated in regulating EPS biogenesis but little is known about which of the steps is targeted and the mechanisms involved. The data presented here, leave open several possibilities for direct and indirect interference of PilA with certain steps in the EPS biogenesis pathway of M. xanthus (Fig. 8). Since TFP/PilA can directly bind to EPS and certain polysaccharides, this regulatory event could involve a direct interaction between PilA and EPS precursors (Fig. 8 - SW1066/DK10416 - 1). PilA has lectin-like properties (Hu et. al. unpublished results) and the PilA monomer pool in the membrane could titrate the EPS precursors prior to assembly. Another possibility would be that the amount of PilA in the membrane regulates the activity of enzymes involved in the assembly or transport of polysaccharides to the cell surface (Fig. 8 - SW1066/DK10416 - 2). Additionally, accumulation of PilA in the membrane may directly or indirectly (probably through the Dif pathway (Black et al., 2006)) trigger signal transduction events to regulate the genes and proteins involved in the synthesis and assembly of polysaccharides(Fig. 8-SW1066/DK10416 -3). Furthermore, the EPS reduction could result from a combination of above possible effects.
Fig. 8.
Coordination of EPS production by PilA in different M. xanthus strains. From left to right are: DK1622 (wild type), DK10410 (ΔpilA)/SW2012 (ΔpilBΔpilA), SW1066 (ΔpilA/pilAA32V,D104N)/DK10416 (ΔpilB) and SW2009 (ΔpilA/pilAsps)/SW2011 (ΔpilA/pilAsps, A32V, D104N). The table, from top to bottom shows approximate EPS production levels relative to wild type(100%), amount of PilA in the membrane pool, PilA assembly into TFP, and a model for coordination of EPS production, respectively. In the model, thin arrows indicate processing of PilA and thick arrows indicate processing of EPS. Dashed arrows represent proposed signal transduction and regulation. Numbers 1 – 3 specify three possible mechanisms of EPS regulation by membrane accumulation of PilA: 1) titration of the EPS precursors prior to assembly; 2)interference with the assembly or transport of polysaccharides to the cell surface; 3) triggering of signal transduction events. See text for details of the model.
The cellular localization of PilA of the different strains examined in this study and the effects on EPS production is summarized in Fig. 8. Based on the detailed analysis in this study, prepilin is completely processed to mature pilin in DK1622 (wild type), SW1066 (ΔpilA/pilAA32V, D104N) (Fig. 7A) and likely DK10416 (ΔpilB). According to studies in other organisms, the signal peptide directs the export of prepilin for cleavage by PilD to allow embedding of the N-terminal alpha helix into the hydrophobic inner membrane while exposing the C-terminal functional domain in the periplasmic space (Strom & Lory, 1987, Pelicic, 2008). The mature pilin forms a membrane-associated pool (Fig. 5) from which wild-type cells remove PilA to assemble TFP from PilA (Fig. 8 - DK1622). This reduces the size of the PilA monomer pool in the membrane which according to our model may interfere with the generation and transport of EPS precursor. At the same time the TFP sensors which have been proposed to stimulate EPS production through the Dif pathway (Black et al., 2006) are being extended. These combined actions allow wild type cells to produce sufficient EPS on the cell surface and in the environment to provide the anchoring substrate for S-motility and scaffold material for fruiting body formation. In DK10410, the mutant lacking pilA, no pilin membrane pool is present that could interfere with EPS biogenesis (Fig. 8 – DK10410). However, due to the lack of assembled TFP and thus the lack of extracellular signal for EPS production only a reduced level of EPS is detected on the cell surface, despite the absence of PilA in membrane which negatively regulates EPS levels.
The mutations present in PilA of SW1066 still allow normal production, stability, processing and membrane translocation of the protein (Fig. 3A, 5, 7A) but prevent assembly of the individual pilin subunits into TFP(Fig. 3B). The concomitant build-up of PilA in the membrane pool possibly leads to interference by one or the combination of the different mechanisms discussed above (Fig. 8 - SW1066). Interestingly, this accumulation of PilA in the membrane pool triggers an even greater reduction in EPS production than observed for lack of the TFP sensor alone (Figs. 4 and 7C). This effect did not appear to be a peculiarity of the mutated PilA A32V,D104N of SW1066: retention and accumulation of wild type PilA in the membrane pool due to lack of PilB, the ATPase necessary for TFP extension in a mutant strain DK10416 resulted in the same phenotype observed for SW1066 (Fig. 6). Further evidence that the amount of PilA in the membrane pool indeed comprises an important negative regulator of EPS production was provided by additional PilA derivatives constructed in this study. When PilA subunits are prevented from entering the membrane pool by locking them in the prepilin state by altering the signal peptide in SW2009 (PilAsps) and SW2011 (PilAsps,A32V,D104N) (Fig. 8 - SW2009/SW1011), EPS production restored to levels found in DK10410 (ΔpilA) (Fig. 7C) which is equally unable to extend the TFP sensor (Fig. 8 - DK10410, SW2009 and SW2011). Precluding membrane translocation of PilA eliminates the regulatory effect on EPS production exerted by its membrane accumulation possibly by EPS precursor titration, blocking of transport and/or signal transduction effects. These findings suggest that in addition to the sensory function of TFP only the amount of PilA subunits accumulated in the membrane but not intracellular PilA levels is an important parameter in regulating EPS production. Furthermore, correct PilA localization and proper concentration are required to maintain the EPS level necessary to sustain normal S-motility.
PilA processing and localization
In addition to exploring the role of the PilA monomer pool in the membrane in regulating EPS production, the results of this study address several aspects of PilA processing and cellular localization. It was previously known that extracellular PilA exist in the form of polarly localized TFP (Wu & Kaiser, 1995, Wu & Kaiser, 1997, Li et al., 2005). In this study we provided information regarding the intracellular and membrane localization of PilA. SW2009 and SW2011 both have shortened signal peptides which prevent processing of the prepilin form. Western blot analysis showed that these unprocessed PilA migrated more slowly in SDS-PAGE compared to the processed PilA (Fig. 7A). In wild type, the faster migrating band (processed PilA) is the only band observed, indicating that in M. xanthus wild-type cells, most if not all PilA only exist in their mature pilin form. Moreover, PilA immunofluoresence showed that in wild type PilA are membrane associated, while in mutants SW2009 and SW2011, PilA are localized in the cytoplasm (Fig. 5A). The gel migration patterns together with the immunofluorescent images suggest that in wild type, most PilA are processed into mature pilin that are associated with the membrane while very little or no prepilin is maintained within the cell. This preferred membrane localization of the mature PilA pool could facilitate TFP assembly, TFP recycling during retraction and trafficking of PilA monomers between two cells poles during cellular reversals.
In M. xanthus, pilA expression is strictly regulated by PilR and PilS as well as an autoregulation mechansim (Wu & Kaiser, 1997). Proper PilA concentration is known to be essential for TFP assembly and swarming in S-motility (Jelsbak & Kaiser, 2005). By characterizing mutants in the PilA library and PilA mutants with altered or missing signal peptides, we found that the fidelity of PilA may be subject to a stringent surveillance system to determine its stability in M. xanthus. Most interestingly, the signal peptide of the prepilin appears to be essential for the stability of PilA in M. xanthus., The mutagenized PilA of SW2010that is missing the entire signal peptide was not detectable in western blots (Fig. 7A). Since the signal peptide is essential for the prepilin to be processed and secreted through the inner membrane, missing the signal peptide renders PilA unable to be secreted and form TFP. Therefore, in the cytoplasm, only the PilA with a signal peptide is stable while the ones that are already in the mature pilin form will not be maintained. However, PilA of SW2009, which is only missing the last amino acid in its signal peptide, is an exception and can be retained inside the cytoplasm. Therefore, the PilA surveillance system enables M. xanthus to only maintain potentially functional PilA in the cells.
In conclusion, the work in this study demonstrated that membrane accumulation of PilA affects EPS production and fruiting body formation. We proposed that proper PilA localization and concentration is essential for its normal function. In addition, we also provided information about the processing and localization of PilA in M. xanthus. We will continue to characterize other mutants in the PilA library and investigate additional roles of PilA and detailed molecular mechanism for TFP-dependent S-motility, fruiting body formation and regulation of EPS production in M. xanthus.
Experimental procedures
Bacterial strains, plasmids and culture conditions
All M. xanthus strains and plasmids used in this study are listed in Table 1. All primers used in this study are listed in Table 2. M. xanthus cells were grown in CYE medium (1% casitone, 0.5% yeast extract, 8 mM MgSO4 in 10 mM Mops buffer, pH 7.6) at 32°C on a rotary shaker at 300 rpm and were maintained using 1.5% CYE agar plates. The E. coli strain DH5α, used for cloning and plasmid construction was grown at 37°C using Luria-Bertani (Sambrook, 2001) medium. Medium and plates were supplemented with kanamycin at 100 μg ml−1 when needed.
Table 2.
Primers used in this study
| Primers | Sequence (5′–3′) |
|---|---|
| pilAgF_EcoRI | CCGGAATTCCGTCAGCGTCACTGCCGAAT |
| pilAgR_BamHI | CGCGGATCCGGACCCTCCACTGCTCATTC |
| pF4 | CACCCCAGGCTTTACACTTT |
| pR4 | GGCCTCTTCGCTATTACGC |
| pilAifdF | ATACCGGAATTCGTAAAGGCCGGACGCTTTCG |
| pilAifdR | ATACGCAAGCTTGCGGTGTTCGTAGAAGGCTC |
| spsF | GATGAGCGTGAAACGGTTGCGGGGGTTGAATC |
| spsR | CCCCGCAACCGTTTCACGCTCATCGAGCTCAT |
| spdF | GATGAGCGTGAACATGGGGGTCCTCAGAGAAG |
| spdR | AGGACCCCCATGTTCACGCTCATCGAGCTCAT |
Hydroxylamine mutagenesis
Hydroxylamine mutagenesis was modified from a previously described protocol (Sikorski & Boeke, 1991). Briefly, the hydroxylamine solution (1M hydroxylamine hydrochloride, 50 mM sodium pyrophosphate PH 7.0, 100 mM sodium chloride, 2 mM EDTA) was freshly prepared and filtered before mutagenesis. 100 μg target plasmid pSWU359 (containing the pilA gene, KanR and Mx8 attP locus) (Wu & Kaiser, 1996) were added to 1 ml hydroxylamine solution and mixed by pipetting. The reaction was started by incubating the mixture in a 75°C water bath. Aliquots of 100 μl were taken at different time points (20, 30, 40, 50, 60, 70, 80 and 90 min) and the reaction was stopped by placing the tube on ice. The excess hydroxylamine was removed from the plasmid DNA using Wizard SV Gel and PCR clean-up system (Promega). The degree of mutagenesis for the individual time points was determined by transforming 2 μl of plasmid from each time point into E. coli strain DH5α and sequencing of the pilA gene from 5 colonies for each time point. The mutagenized plasmid aliquot with the highest single mutation rate (30 min and 40 min in this study) was amplified in E. coli and the extracted plasmids were stored as pilA mutant plasmid library for electroporation into DK10410, a mutant derivative of M. xanthus lacking pilA.
Error Prone PCR
Error Prone PCR in vitro random mutagenesis was performed using GeneMorph II Random Mutagenesis Kit (Stratagene). First, the pilA gene as well as its promoter was amplified from DK1622 genomic DNA using primers pilAgF_EcoRI and pilAgR_BamHI (Table 2). The PCR product was cleaned with Wizard SV Gel and PCR clean-up system (Promega) and two different template concentrations of 0.5 and 1 μg were used for the mutagenesis reaction. Error prone PCR was carried out according to manufacturer’s instructions using primers pilAgF_EcoRI and pilAgR_BamHI. After the reaction, the PCR products were cleaned with Wizard SV Gel and PCR clean-up system (Promega), digested with EcoRI and BamHI (NEB), cloned into vector pSWU19 (Wu & Kaiser, 1995), and transformed into E. coli DH5α. The degree of mutagenesis in the resulting pilA mutant plasmid library was determined by sequencing the pilA gene in eight E. coli transformants for each template concentration used. The plasmid library with 1 μg starting DNA concentration resulted in a more desirable mutagenesis degree (most clones have single mutation) therefore was used to transform M. xanthus cells.
Construction of a pilA mutant library
The pilA random mutation plasmid libraries generated using hydroxylamine mutagenesis and error prone PCR were electroporated (Kashefi & Hartzell, 1995) into DK10410 (ΔpilA), selected for KanR on 1.5% CYE kanamycin agar plate. Single colonies were grown in CYE medium (plus 100 μg ml−1 kanamycin) and further screened for phenotypes of interest.
Construction of strains
In-frame-deletions of pilA were introduced into different strain backgrounds using a positive–negative selection cassette as previously reported (Ueki et al., 1996). A DNA fragment carrying a pilA in-frame deletion was amplified from DK10410 (ΔpilA) using primers pilAifdF and pilAifdR (Table 2), resulting in a PCR product that contains about 700bp upstream and 700bp downstream of pilA. This pilA in-frame-deletion DNA fragment was then digested with EcoRI and HindIII (NEB) and cloned into the corresponding sites in pBJ113 (Julien et al., 2000) to generate pMX01(ΔpilA). pMX01 was electroporated (Kashefi & Hartzell, 1995) into DK10409 (ΔpilT) and DK10416 (ΔpilB) and selected on kanamycin plate for integrants. Deletion mutants SW2001 (ΔpilA ΔpilT) and SW2012 (ΔpilB ΔpilA) were counter-selected by its resistance to galactose and sensitivity to kanamycin.
In order to transfer mutagenized pilA genes into different strain backgrounds, primers pilAgF_EcoRI and pilAgR_BamHI were used to amplify the pilA gene from genomic DNA of SW1066, SW1066A and SW1066B. PCR products were then cloned into the EcoRI and BamHI sites of pSWU19 to generate pMX02 (pilAA32V, D104N), pMX03 (pilAA32V) and pMX04 (pilAD104N). pMX02, pMX03 and pMX04 were electroporated (Kashefi & Hartzell, 1995) into SW2001 (ΔpilA ΔpilT) to generate SW2002, SW2003, SW2004, respectively.
Plasmids carrying pilA with mutations in the signal peptide (sps and spd) were generated using the two-step overlap PCR procedure (Sambrook, 2001). Overlap primers spsF and spsR were designed to delete the last amino acid (GGC in the DNA sequence) in the PilA signal peptide; spdF and spdR were designed to delete the whole 12 amino acids in the PilA signal peptide. For generating pMX05 (pilAsps), pSWU359 (pilA) was used as the template in the initial round of PCR. DNA regions upstream and downstream of the amino acid to be mutagenized were amplified using primers pF4 and spsF, pR4 and spsR, respectively. The two PCR products from the first round of PCR were used as the template for a subsequent round of overlap PCR using primers pF4 and pR4. The PCR products from the second round PCR were cleaned and cloned into the EcoRI and BamHI sites of pSWU19 to generate pMX05 (pilAsps). pMX06 (pilAspd) was generated in a similar way using pSWU359 (pilA) as the template, spdF and spdR as overlap primers in the first PCR reaction. pMX07 (pilAsps, A32V, D104N) was also generated in a similar way using pMX02 (pilAA32V, D104N) as template, spsF and spsR as overlap primers in the first PCR. pMX05, pMX06 and pMX07 were electroporated into DK10410 (ΔpilA) to generate SW2009, SW2010 and SW2011. All plasmids and mutants generated in this study were confirmed by PCR and sequencing using primers pF4 and pR4.
Swarming assay
M. xanthus cells were grown in CYE medium to exponential phase and concentrated to OD600=10 (5 × 109 cells ml−1) in CYE medium. An aliquot of 5μl concentrated culture was spotted onto 0.3% CYE agar plate (Shi & Zusman, 1993)and incubated for 3 days at 32°C. S-motility was analyzed by observing the colonies for expansion. Pictures were taken using a Nikon D50 camera.
Fruiting body formation assay
M. xanthus cells were grown in CYE to exponential phase and concentrated to OD600=5 (2.5 × 109 cells ml−1) in TPM buffer (10mM Tris-HCl pH 7.6, 1mM KH2PO4, 8mM MgSO4). 5 μl aliquot concentrated cells was spotted onto CF agar (Hagen et al., 1978) and incubated for 48 hours at 32°C. Fruiting body pictures were taken using Nikon eclipse TE200 inverted microscope and recorded using SPOT camera/software (Diagnostic Instruments).
Western blot analysis of whole-cell and cell-surface PilA
Western blots on whole-cell lysate as well as cell-surface PilA were performed following standard procedures (Harlow, 1988). For whole-cell lysate, 5 × 107 M. xanthus cells were lysed by boiling in SDS-PAGE loading buffer for 10min. Cell-surface pili were purified as previously described (Wu & Kaiser, 1997), with minor modifications. Briefly, 1010 mid-log phase M. xanthus cells were harvested and resuspended to OD600=10 (5 × 109 cells ml−1) in TPM buffer and passed through a 26G3/8 needle (Becton Dickinson & Co) for 30 times and vortexed for 3min to shear off surface pili. The suspension was sedimented at 16,000 × g for 1min. The supernatant were transferred to a clean tube, centrifuged at 16,000 × g for 5min and the supernatant was transferred to another clean tube. Pili were precipitated by adding trichloroacetic acid (TCA) to a final concentration of 10%, incubating on ice for 3h, and sedimenting at 16,000 × g for 30min at 4°C. Isolated pili were washed with acetone, resuspended in SDS-PAGE loading buffer and boiled for 10min before being subjected to western blot analysis. Primary anti-PilA antibody (Li et al., 2005) was used at a 1:10000 dilution, anti-rabbit horseradish peroxidase-conjugated secondary antibody (Pierce) was used at a 1:20000 dilution. Blots were developed using the Supersignal West Pico chemiluminescence reagent (Pierce). Images were taken with the ChemiDoc XRS system (Bio-Rad)
Examination of cell surface EPS on agar
Cell surface EPS was measured using trypan blue binding assay (Black & Yang, 2004) with modifications. In order to measure the extracellular EPS level on agar surface, 5 × 108 exponential phase cells (concentrated into 100μl CYE medium) were spotted onto 1.5% CYE agar plates and incubated at 32°C overnight to allow cells to swarm. Cells were then collected and resuspended in 0.5ml cohesion buffer (10mM MOPS, 1mM MgSO4, 1mM CaCl2). 300μg protein equivalent of cells (determined by Pierce BCA protein assay) were mixed with 1ml trypan blue working solution (25μg ml−1 trypan blue in cohesion buffer). The mixtures were incubated in the dark at room temperature for 30min. The cell suspensions were then pelleted at 16,000g in a bench-top centrifuge for 10 min and the absorbance of the supernatants was measured at 585 nm. The relative amounts of cell surface EPS were calculated by the quotient of dye bound to each sample compared to wild type.
Immunofluorescence Deconvolution Microscopy
Immunofluorescence microscopy was performed as previously described (Mauriello et al., 2009). Briefly, 500 μl of 4 × 108 cells ml−1 vegetative culture were mixed with 100 μl 16% paraformaldehyde (w/v), 0.2 μl 25% glutaraldehyde (v/v) and 20 μl 1M NaPO4 pH7.4. Cells were fixed in the mixture for 20 min and then immobilized on freshly prepared poly-L-lysine treated slides. The cells were rendered permeable by a 4 min treatment with 1 μg ml−1 lysozyme, washed with PBS solution and probed in a 2% BSA PBS solution with anti-PilA antibody (Li et al., 2005)at a 1:1000 dilution. Alexa Fluor 488-coupled goat-anti-rabbit antibody (Molecular Probes) was used as secondary antibody at a 1:200 dilution. Slowfade gold antifade reagent (Molecular Probes) was added. Images were captured using an Olympus IX71 inverted microscope with a 100 X oil immersion objective lens. For each sample, 20 to 30 images were collected spaced at 0.1 μm through the specimen using a standard FITC filter set. The images were then deconvolved using SoftWoRx imaging workstation software (Applied Precision). Different samples were grown and treated under the same condition and immunofluorescence images were taken and deconvoluted using the same parameters. For the non-permeabilized cell control, DK1622 cells were treated using the same protocol without the lysozyme treatment step.
Acknowledgments
We would like to thank Mitch Singer for providing plasmids, James Gober for providing access to and help with deconvolution microscopy, Arturo Calderon Flores, Xuesong He and Lina Li for helpful discussion. This work was supported by National Institutes of Health (NIH) Grant GM54666to W. Shi.
References
- Arnold JW, Shimkets LJ. Cell surface properties correlated with cohesion in Myxococcus xanthus. J Bacteriol. 1988a;170:5771–5777. doi: 10.1128/jb.170.12.5771-5777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JW, Shimkets LJ. Inhibition of cell-cell interactions in Myxococcus xanthus by congo red. J Bacteriol. 1988b;170:5765–5770. doi: 10.1128/jb.170.12.5765-5770.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behmlander RM, Dworkin M. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J Bacteriol. 1994;176:6295–6303. doi: 10.1128/jb.176.20.6295-6303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WP, Xu Q, Yang Z. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol. 2006;61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- Black WP, Yang Z. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J Bacteriol. 2004;186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden MG, Kaplan HB. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol Microbiol. 1998;30:275–284. doi: 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- Chang BY, Dworkin M. Isolated fibrils rescue cohesion and development in the Dsp mutant of Myxococcus xanthus. J Bacteriol. 1994;176:7190–7196. doi: 10.1128/jb.176.23.7190-7196.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana JR, Shimkets LJ. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J Bacteriol. 1993;175:3636–3647. doi: 10.1128/jb.175.11.3636-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen DC, Bretscher AP, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Harlow ELD. Antibodies: a Laboratory Manual. Cold Spring Harbor. NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- Jakovljevic V, Leonardy S, Hoppert M, Sogaard-Andersen L. PilB and PilT are ATPases acting antagonistically in type IV pili function in Myxococcus xanthus. J Bacteriol. 2008 doi: 10.1128/JB.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsbak L, Kaiser D. Regulating pilin expression reveals a threshold for S motility in Myxococcus xanthus. J Bacteriol. 2005;187:2105–2112. doi: 10.1128/JB.187.6.2105-2112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien B, Kaiser AD, Garza A. Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci U S A. 2000;97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashefi K, Hartzell PL. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF-defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- Lancero H, Brofft JE, Downard J, Birren BW, Nusbaum C, Naylor J, Shi W, Shimkets LJ. Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J Bacteriol. 2002;184:1462–1465. doi: 10.1128/JB.184.5.1462-1465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancero H, Caberoy NB, Castaneda S, Li Y, Lu A, Dutton D, Duan XY, Kaplan HB, Shi W, Garza AG. Characterization of a Myxococcus xanthus mutant that is defective for adventurous motility and social motility. Microbiology. 2004;150:4085–4093. doi: 10.1099/mic.0.27381-0. [DOI] [PubMed] [Google Scholar]
- Li Y, Lux R, Pelling AE, Gimzewski JK, Shi W. Analysis of type IV pilus and its associated motility in Myxococcus xanthus using an antibody reactive with native pilin and pili. Microbiology. 2005;151:353–360. doi: 10.1099/mic.0.27614-0. [DOI] [PubMed] [Google Scholar]
- Li Y, Sun H, Ma X, Lu A, Lux R, Zusman D, Shi W. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc Natl Acad Sci U S A. 2003;100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Cho K, Black WP, Duan XY, Lux R, Yang Z, Kaplan HB, Zusman DR, Shi W. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol Microbiol. 2005;55:206–220. doi: 10.1111/j.1365-2958.2004.04369.x. [DOI] [PubMed] [Google Scholar]
- MacNeil SD, Mouzeyan A, Hartzell PL. Genes required for both gliding motility and development in Myxococcus xanthus. Mol Microbiol. 1994;14:785–795. doi: 10.1111/j.1365-2958.1994.tb01315.x. [DOI] [PubMed] [Google Scholar]
- Mauriello EM, Astling DP, Sliusarenko O, Zusman DR. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc Natl Acad Sci U S A. 2009;106:4852–4857. doi: 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EM, Zusman DR. Polarity of motility systems in Myxococcus xanthus. Curr Opin Microbiol. 2007;10:624–629. doi: 10.1016/j.mib.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- Morand PC, Bille E, Morelle S, Eugene E, Beretti JL, Wolfgang M, Meyer TF, Koomey M, Nassif X. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. Embo J. 2004;23:2009–2017. doi: 10.1038/sj.emboj.7600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudleman E, Kaiser D. Pulling together with type IV pili. J Mol Microbiol Biotechnol. 2004;7:52–62. doi: 10.1159/000077869. [DOI] [PubMed] [Google Scholar]
- Pelicic V. Type IV pili: e pluribus unum? Mol Microbiol. 2008;68:827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor. NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shi W, Zusman DR. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc Natl Acad Sci U S A. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets LJ. Role of cell cohesion in Myxococcus xanthus fruiting body formation. J Bacteriol. 1986;166:842–848. doi: 10.1128/jb.166.3.842-848.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom MS, Lory S. Mapping of export signals of Pseudomonas aeruginosa pilin with alkaline phosphatase fusions. J Bacteriol. 1987;169:3181–3188. doi: 10.1128/jb.169.7.3181-3188.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom MS, Lory S. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem. 1991;266:1656–1664. [PubMed] [Google Scholar]
- Sun H, Zusman DR, Shi W. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- Ueki T, Inouye S, Inouye M. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene. 1996;183:153–157. doi: 10.1016/s0378-1119(96)00546-x. [DOI] [PubMed] [Google Scholar]
- Wall D, Kolenbrander PE, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer RM, Creighton C, Stassinopoulos A, Youderian P, Hartzell PL. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J Bacteriol. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. Embo J. 2000;19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SS, Kaiser D. Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- Wu SS, Kaiser D. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J Bacteriol. 1996;178:5817–5821. doi: 10.1128/jb.178.19.5817-5821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SS, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SS, Wu J, Cheng YL, Kaiser D. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol Microbiol. 1998;29:1249–1261. doi: 10.1046/j.1365-2958.1998.01013.x. [DOI] [PubMed] [Google Scholar]
- Wu SS, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Geng Y, Shi W. A DnaK homolog in Myxococcus xanthus is involved in social motility and fruiting body formation. J Bacteriol. 1998;180:218–224. doi: 10.1128/jb.180.2.218-224.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ma X, Tong L, Kaplan HB, Shimkets LJ, Shi W. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J Bacteriol. 2000;182:5793–5798. doi: 10.1128/jb.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]




