Abstract
Under various conditions mammals have the ability to maintain serum glucose concentration within a narrow range. SIRT1 plays an important role in regulating gluconeogenesis and fat metabolism; however, the underlying mechanisms remain elusive. Here we show that SIRT1 forms a complex with FOXO3a and NRF1 on the SIRT6 promoter and positively regulates expression of SIRT6, which in turn negatively regulates glycolysis, triglyceride synthesis and fat metabolism by deacetylating histone H3 lysine 9 in the promoter of many genes involved in these processes. Liver specific deletion of SIRT6 in mice causes profound alterations in gene expression, leading increased glycolysis, triglyceride synthesis, reduced β-oxidation, and fatty liver formation. Human fatty liver samples exhibited significantly lower levels of SIRT6 than normal controls. Thus, SIRT6 plays a critical role in fat metabolism, and may serve as a novel therapeutic target for treating fatty liver disease, the most common cause of liver dysfunction in humans.
Keywords: SIRT6, SIRT1, NRF1, triglyceride, fatty liver
Introduction
In budding yeast and Drosophila, the Sir2 histone deacetylase acts as a chromatin silencer to regulate recombination, genomic stability, and aging (Guarente and Kenyon, 2000). In mammals, seven sirtuin proteins (SIRT1-7) have been found to share homology with Sir2 and are suspected to have some similar functions to Sir2 (Finkel et al., 2009; Mantel and Broxmeyer, 2008; Saunders and Verdin, 2007; Vaquero et al., 2007). These proteins are primarily localized in different subcellular compartments with SIRT1 and SIRT2 being both in the nucleus and the cytoplasm, SIRT3, SIRT4 and SIRT5 in the mitochondrion, and SIRT6 and SIRT7 in the nucleus (Blander and Guarente, 2004; Haigis and Guarente, 2006; Saunders and Verdin, 2007). Sirtuins have emerged as broad regulators of many important processes, including cell fate determination, DNA damage repair, neuronal protection, adaptation to calorie restriction, organ metabolism and function, age-related diseases, and tumorigenesis, although much of the information has come from studies of SIRT1 (Ahn et al., 2008; Deng, 2009; Finkel et al., 2009; Jacobs et al., 2008; Kim et al., 2010; Saunders and Verdin, 2007; Vaquero et al., 2007; Wang et al., 2008a; Wang et al., 2008b).
In response to fasting and calorie restriction (CR), SIRT1 is induced, which then interacts with and deacetylates peroxisome proliferator-activated receptor gamma-coactivator-1α (PGC-1α) and controls gluconeogenic and glycolytic pathways in the liver (Rodgers et al., 2005; Rodgers and Puigserver, 2007). For example, it was demonstrated that SIRT1 and the histone acetyltransferase p300 form a fasting-inducible switch that maintains energy balance and regulates hepatic glucose production in mice through sequential induction of CREB regulated transcription coactivator 2 (CRTC2) and PGC-1α/FOXO1 (Liu et al., 2008). SIRT1 deacetylates CRTC2 which promotes its ubiquitin-dependent degradation, thereby attenuating the CRTC2-stimulated hepatic glucose production. In addition, SIRT1 deacetylates PGC-1α and FOXO1 subsequently augmenting their transcriptional activity to modulate the downstream gluconeogenic program. This switch strongly correlates with a transition from a high level of gluconeogenesis in the early phase of fasting to a lower level of glucose production at a later time. SIRT6 may also play a role in glucose metabolism as SIRT6-null mice suffer hypoglycemia before they die at 3–4 weeks of age (Mostoslavsky et al., 2006; Zhong et al. 2010). SIRT6 protein expression can be induced by nutritional stress (Kanfi et al., 2008); however, the physiological role of SIRT6 in response to nutritional status and its relationship with SIRT1 remains unclear.
To elucidate the function of SIRT6 and its relationship with SIRT1, we performed an in vitro study to understand the regulation of SIRT6 by SIRT1, generated liver specific SIRT6 knockout mice and performed a comprehensive phenotypic analysis in gene expression and acetylation associated with SIRT6 deficiency. Our data revealed that SIRT1 regulates SIRT6 by forming a complex with FOXO3a and NRF1 on the promoter of SIRT6. In turn, SIRT6 deacetylates lysine 9 of histone H3 (H3K9) on the promoters of many genes, which have an essential role in glycolysis and lipid metabolism.
Results
SIRT1 positively regulates SIRT6
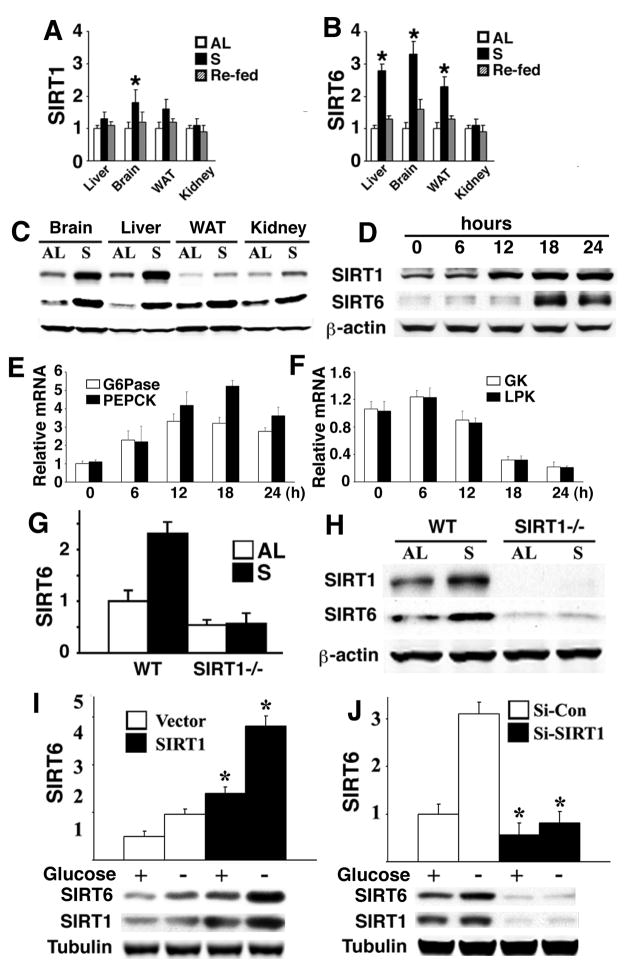
We first investigated the relationship between SIRT1 and SIRT6 in mice under fed, fasted, and re-fed conditions. Analysis of multiple organs revealed increased SIRT1 protein in the brain, liver, white adipose tissue (WAT) and kidney of fasted mice to a varying degree, although SIRT1 mRNA was only increased in the brain (Fig. 1A,C). In contrast, SIRT6 mRNA and protein were coordinately increased in the brain, WAT, and liver in fasted mice (Fig. 1B,C). Next we performed a time course study in the liver after fasting. We detected a positive correlation of SIRT1 induction and SIRT6 induction, and in addition, we found that the increase in SIRT1 protein occurred earlier than that of SIRT6 (Fig. 1D). For example, an obvious increase in SIRT1 occurred at 12 hours and peaked at 18 hours post fasting while a significant increase in SIRT6 was detected at 18 hours. Of note, fasting also induced expression of the gluconeogenic genes G6Pase and phosphoenolpyruvate carboxykinase 1 (Pepck) (Fig. 1E), and inhibited expression of glycolytic genes glucokinase (Gk) and liver pyruvate kinase (Lpk) (Fig. 1F). A similar expression pattern of SIRT1 and SIRT6 induction was also detected in cultured primary hepatocytes and hepatoma cell lines in the absence of glucose (Supplementary Fig. 1A-D).
Fig. 1. Regulation of SIRT1 and SIRT6 expression by nutrient deprivation.
(A-C) Levels of mRNA (A,B) and protein (C) of SIRT1 and SIRT6 in multiple organs of mice under fed ad libitum (AL), starved (S), or re-fed for 24 hours. (D-F) Levels of SIRT1 and SIRT6 protein (D), G6Pase and PEPCK mRNA (E), and GK and LPK mRNA (F) in the liver of wild type mice during a time course of fasting. (G,H) Levels of SIRT6 mRNA revealed by Real-Time RT-PCR (G) and protein (H) in the liver from wild type and Sirt1−/− mice. (I,J) Over-expression of SIRT1 increased endogenous SIRT6 in the presence and absence of glucose (I), while RNAi-mediated knockdown of SIRT1 blocked SIRT6 induction in the absence of glucose (J) revealed by Real-Time PCR. * represents p < 0.05 by Student T-test in all figures.
To further elucidate the relationship between SIRT1 and SIRT6, we examined SIRT6 expression in the liver of Sirt1−/− mice (Wang et al., 2008a). Our data revealed that both SIRT6 mRNA and protein were reduced by 50% and failed to be induced by fasting (Fig. 1G,H). Furthermore, SIRT1 ectopic expression in Hepa1-6 cells induced SIRT6 mRNA in glucose containing medium, which was further increased in the absence of glucose (Fig. 1I). Conversely, siRNA-mediated SIRT1 knockdown blocked SIRT6 induction in the absence or presence of glucose (Fig. 1J) indicating that SIRT1 is involved in maintaining SIRT6 expression under the physiological conditions and is required for SIRT6 induction during fasting.
Regulation of SIRT6 by SIRT1 through NRF1 binding sites in the SIRT6 promoter
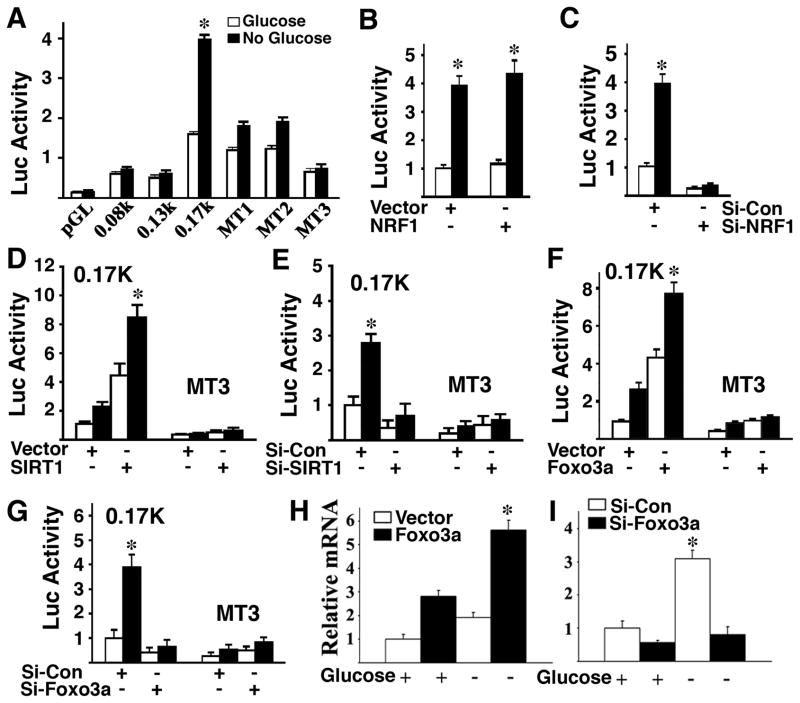
To understand the mechanism of SIRT6 induction by SIRT1, we examined the activity of the SIRT6 promoter. Our data using serial deletions of the SIRT6 promoter revealed that a region between 0.17 kb and 0.13 kb was essential for the stimulatory effect by glucose starvation (Supplementary Fig. 2A). Using a software program for candidate transcription factors, we identified two predicted nuclear respiratory factor 1 (NRF1) binding sites in this region (Supplementary Fig. 2B). Mutation of either site significantly impaired SIRT6 induction in the absence of glucose, while mutation of both sites completely blocked the induction (Fig. 2A). These data indicate that NRF1 binding sites are involved in the regulation of the SIRT6 promoter by glucose starvation.
Fig. 2. Regulation of SIRT6 by SIRT1, FOXO3a, and NRF1.
(A) Absence of glucose in Hepa1-6 cells induced luciferase activity of a SIRT6 promoter reporter, while mutation of NRF1 binding sites abolished the induction. (B,C) Effect of NRF1 ectopic expression (B) and RNAi-mediated knockdown (C) on a SIRT6 promoter reporter. (D,E) Ectopic expression of SIRT1 increased SIRT6 promoter activity in the presence or absence of glucose (D), while mutation of NRF1 binding sites (D), or RNAi-mediated knockdown of SIRT1 (E) abolished the induction. (F,G) Ectopic expression of FOXO3a increased SIRT6 promoter activity in the presence or absence of glucose (F), while RNAi-mediated knockdown of FOXO3a inhibits it (G). (H,I) Absence of glucose induces expression of SIRT6 mRNA, which is further increased by overexpression of FOXO3a (H), while RNAi-mediated knockdown of FOXO3a abolished the induction (I).
NRF1 is a transcription factor and its expression is regulated by caloric restriction (Mahishi and Usdin, 2006; Nisoli et al., 2005). To understand the role of NRF1 in the regulation of SIRT6 expression, we performed both NRF1 overexpression and knockdown experiments. Our data indicate that ectopic expression of NRF1 did not have a significant effect on a SIRT6 promoter reporter (Fig. 2B), while RNAi-mediated knockdown of NRF1 blocked the induction of the SIRT6 promoter by glucose starvation (Fig. 2C). A similar effect was observed true for endogenous SIRT6 mRNA expression (Supplementary Fig. 2C,D). The amount of endogenous NRF1 is sufficient to maintain SIRT6 expression either with or without glucose, and thus, an increase in NRF1 by ectopic expression does not have additional effects.
Next, we showed that ectopic expression of SIRT1 increased SIRT6 promoter activity by approximately three fold either with or without glucose, and this stimulatory effect was lost when the NRF1 binding sites in the SIRT6 promoter were mutated (Fig. 2D). Furthermore, SIRT6 promoter activity induced by the absence of glucose could be blocked by RNAi-mediated knockdown of SIRT1 (Fig. 2E). These data suggest that SIRT1 plays an important role in regulating SIRT6 through NRF1 binding sites in the SIRT6 promoter.
Regulation of SIRT6 by SIRT1, FOXO3a, and NRF1 protein complex upon nutritional stress
FOXO3a plays an important role in SIRT1 induction by glucose and serum starvation (Nemoto et al., 2004). Therefore, we determined whether FOXO3a is also involved in the regulation of SIRT6 expression. We found that SIRT6 promoter activity was increased four fold after transfection of a FOXO3a expression vector as compared with a GFP expression vector in the presence of glucose, and the induction was increased further in the absence of glucose (Fig. 2F). Conversely, knockdown of FOXO3a by siRNA significantly reduced SIRT6 promoter activity (Fig. 2G). Our analysis of endogenous SIRT6 mRNA (Fig. 2H) expression in Hepa1-6 cells confirmed these changes. Thus, both SIRT1 and FOXO3a were capable of regulating the expression of SIRT6 mRNA through the NRF1 binding sites in both fed and fasting conditions.
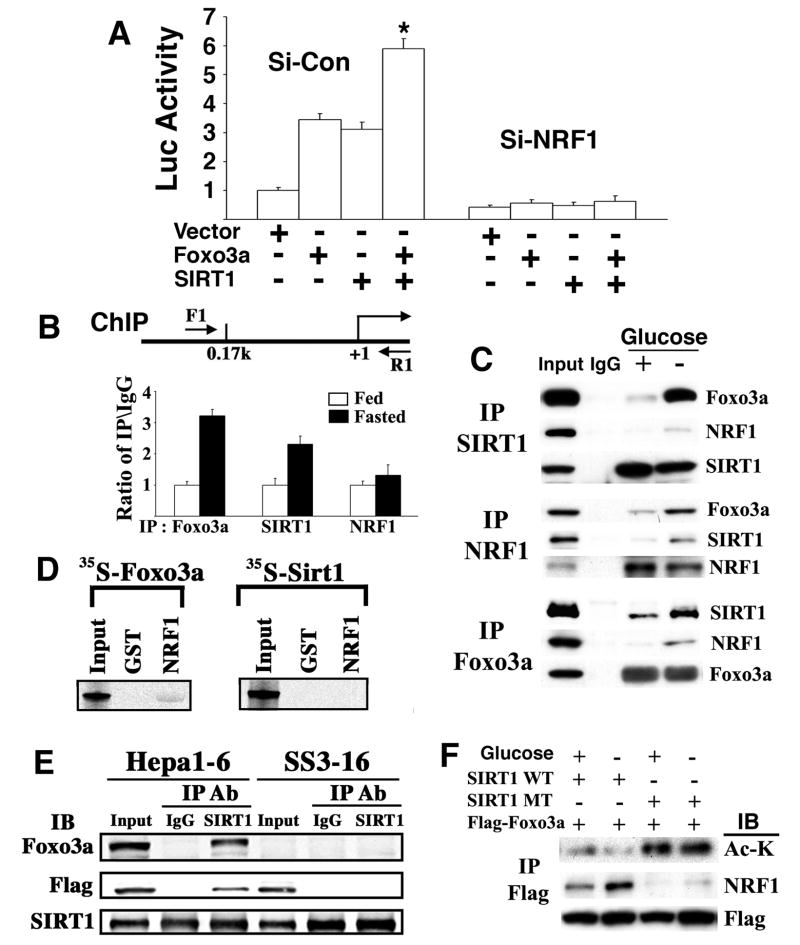
Next, we studied potential interactions among SIRT1, FOXO3a, and NRF1 in regulating SIRT6 expression. We showed that co-transfection of SIRT1 and FOXO3a enhanced SIRT6 expression compared with transfection of these genes alone, as revealed by a SIRT6 promoter reporter assay (Supplementary Fig. 3A,C) and endogenous SIRT6 gene expression (Supplementary Fig. 3B,C) either with or without glucose. We found that although ectopic over-expression of NRF1 did not have an apparent effect on SIRT6 induction by SIRT1 and/or FOXO3a (data not shown), siRNA-mediated NRF1 knockdown completely blocked such induction (Fig. 3A). This suggests that the amount of endogenous NRF1 is sufficient for SIRT6 induction by SIRT1 and/or FOXO3a. In addition, FOXO3a knockdown blocked induction of SIRT6 by SIRT1 (Supplementary Fig. 3D) and absence of SIRT1 blocked induction of SIRT6 by FOXO3a (Supplementary Fig. 3E). Collectively, these data suggest that SIRT1, FOXO3a and NRF1 might form a complex on the SIRT6 promoter with a defined configuration.
Fig. 3. SIRT1, FOXO3a and NRF1 form a protein complex on the NRF1 binding sites of the SIRT6 promoter.
(A) Expression of SIRT1 and FOXO3a synergistically activated a SIRT6 promoter reporter, which is blocked by siRNA specific to NRF1. (B) Binding of NRF1, SIRT1, and FOXO3a to the SIRT6 promoter in the presence or absence of glucose as revealed by ChIP assay. (C) Interaction of FOXO3a, SIRT1, and NRF1 in cultured cells in either the presence or absence of glucose as revealed by immunoprecipitation (IP). (D) Pull-down of 35S-FOXO3a, but not 35S-SIRT1, by GST tagged-NRF1. (E) SIRT1 pulls down both FOXO3a and NRF1 in Hepa1-6 cells. However, SIRT1 could not pull down NRF1 in Hepa1-6 cells carrying a stably transfected shRNA for FOXO3a (SS3-16). (F) Wild-type SIRT1 deacetylates FOXO3a, which is enhanced in the absence of glucose in Hepa1-6 cells, and the deacetylated form of FOXO3a interacts more abundantly with NRF1.
In light of these findings, we examined whether these factors could bind to the SIRT6 promoter using chromatin immunoprecipitation (ChIP) assay. We showed that both SIRT1 and FOXO3a bind to a fragment containing NRF1 sites on the SIRT6 promoter. This binding was enhanced by approximately three-fold in the absence of glucose, whereas the binding of NRF1 was not affected (Fig. 3B). Using an oligonucleotide pull-down assay, it was determined that the NRF1 binding sites were critical for complex formation (Supplementary Fig. 3F). We further showed that siRNA-mediated knockdown of either FOXO3a or SIRT1 (Supplementary Fig. 3G,H) blocked interaction of these factors at the NRF1 sites, while knockdown of either of them did not affect the binding of NRF1 to the SIRT6 promoter (bottom lane of Supplementary Fig. 3G,H). These data suggest that both SIRT1 and FOXO3a bind cooperatively to the SIRT6 promoter, while binding of NRF1 to the SIRT6 promoter does not require SIRT1 or FOXO3a.
Next, we performed experiments to examine protein-protein interaction. We showed that all three proteins (SIRT1, FOXO3a and NRF1) interacted with each other in cells in the presence of glucose, while the interaction was markedly enhanced in the absence of glucose (Fig. 3C). Thus, glucose deprivation enhanced the formation of a SIRT1-FOXO3a-NRF1 (SFN) protein complex on the SIRT6 promoter that is responsible for the induction of SIRT6 expression. Notably, our study revealed that GST tagged-NRF1 only pulled down 35S-FOXO3a but not 35S-SIRT1 (Fig. 3D). Furthermore, shRNA-mediated knockdown of FOXO3a in Hepa1-6 cells blocked the interaction between SIRT1 and NRF1 (Fig. 3E). Altogether, these data indicate that NRF1 binds directly to the SIRT6 promoter irrespective of glucose concentration, while SIRT1 binds to NRF1 through FOXO3a.
Previous studies have shown that SIRT1 binds and deacetylates FOXO3a (Brunet et al., 2004; Motta et al., 2004). Since we found that expression of a dominant-negative mutant form of SIRT1, SIRT1 (HY) that lacks deacetylase activity, blocked the induction of SIRT6 by SIRT1 and/or FOXO3a (Supplementary Fig. 3A-C), we investigated whether the acetylation status of FOXO3a could affect its interaction with NRF1. Our data showed that the presence of wild-type SIRT1 repressed FOXO3a acetylation as compared with expression of the mutant SIRT1 (HY) (Fig. 3F). The absence of glucose further decreased FOXO3a acetylation (compare the first 2 lanes in Fig. 3F) while no reduction of FOXO3a acetylation was observed in the SIRT1 (HY) transfected cells (compare the last 2 lanes in Fig. 3F). The data also revealed increased interaction between FOXO3a with NRF1 in the cells with expression of wild-type SIRT1 when compared to cells with mutant SIRT1 (Fig. 3F). A stronger band was also detected in the wild-type SIRT1 expressed cells in the absence of glucose (Fig. 3F). Altogether, these data indicate that the absence of glucose enhances the interaction between SIRT1 and FOXO3a, as well as the deacetylation of FOXO3a. Deacetylated FOXO3a has higher binding affinity for NRF1, which results in the increased quantity of SFN complex on the SIRT6 promoter that accounts for the increased expression of SIRT6 upon nutritional stress.
Liver specific knockout of SIRT6 results in fatty liver
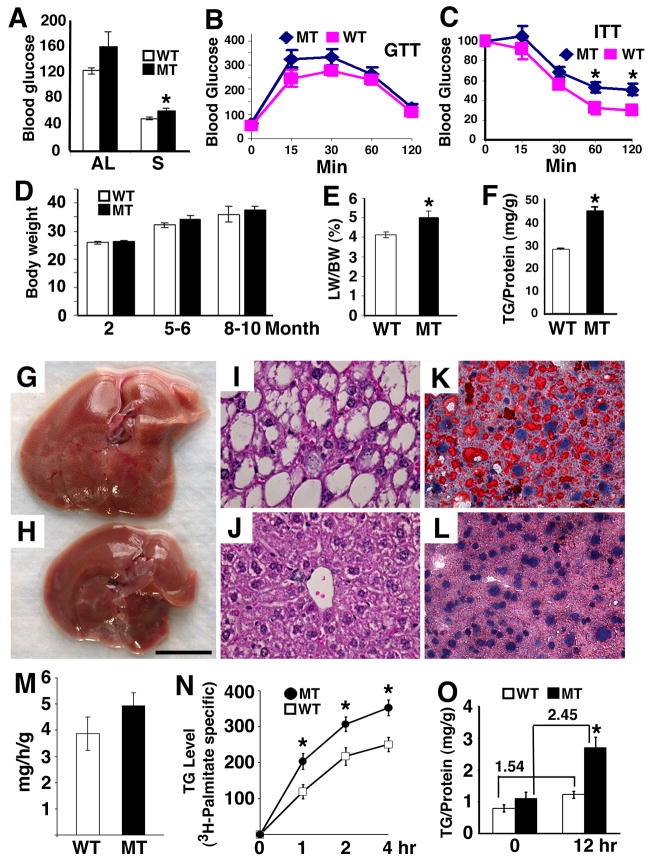
The observation that up-regulation of SIRT6 through the SFN complex under fasting conditions suggests that SIRT6 plays a role in glucose metabolism in the liver. Since Sirt6−/− mice die shortly after weaning (Mostoslavsky et al., 2006), we generated liver specific SIRT6 knockout mouse using albumin-Cre (Yakar et al., 1999) to overcome this early postnatal lethality (Supplementary Fig. 4). Sirt6Co/Co;Alb-Cre mice appeared morphologically normal and displayed comparable levels of blood glucose at one month of age (data not shown), suggesting that the hypoglycemia and lethal phenotype observed in Sirt6−/− mice (Mostoslavsky et al., 2006) are not caused by the lack of SIRT6 in the liver. Consistent with this observation, our study using older mice ranging in age from 1 to 8 months, also failed to reveal a significant difference in blood glucose between SIRT6 mutant and control mice after feeding (data not shown). Of note, our analysis of older Sirt6Co/Co;Alb-Cre mice (≥8 months of age) revealed slightly increased serum glucose (Fig. 4A). The mutant mice also exhibited slightly higher levels of glucose in the glucose tolerance test (GTT) (Fig. 4B) and insulin tolerance test (ITT) (Fig. 4C), although it did not reach a significant level at most time points. The elevation in glucose might be caused by an increase in hepatic glucose production. However, additional studies on hepatic gluconeogenesis, including the pyruvate tolerance test and clamp analysis, failed to detect increased hepatic gluconeogenesis in these mutant mice (data not shown), suggesting this phenotype might not be a direct consequence of SIRT6 deficiency in the liver.
Fig. 4. Phenotypic analysis of mice carrying a liver specific knockout of SIRT6.
(A) Blood glucose level (mg/dL) of 8–9 months old SIRT6 MT and WT mice under fed or 24 hours fasting condition. (B) Glucose tolerance test. Mutant mice had a slightly higher, but not significantly different glucose levels at 15 and 30 minutes than wild type mice. (C) Insulin tolerance test expressed as percentage of basal glucose level. We have also measured glucose value in the Area Under the Curve (AUC) for both GTT and ITT, and no difference is found between wild type and mutant mice. (D) Body weight (gram) of SIRT6 mice at 2 months: WT 17, MT 15; 5–6 months: WT 18, MT 15; and 8–10 months: WT 15, MT 22. (E,F) Percent of liver weight/body weight (E) and TG levels (F) of 8–9 months old SIRT6 MT and WT mice. (G-L) Morphology (G,H), H&E sections (I,J) and Oil Red O staining (K,L) of livers from MT (G,I,K) and WT (H,J,L) mice. Bar in (G,H) is 1 centimeter. At least 6 pairs of mice were analyzed in each experiment. (M) TG secretion rate to plasma (mg/hour/gram of liver). (N,O) TG content in primary hepatocytes measured by 3H-palmitate incorporation (N) and TG level (O) at different times. In panel (O), TG production increases 1.54 fold in wild type cells and 2.45 fold in mutant cells from 0 hour to 12 hours, respectively. This increase is statistically significant with p<0.04.
Sirt6Co/Co;Alb-Cre mice gradually increased in body weight beginning at 5 month of age although such an increase did not reach a statistically significant level (Fig. 4D). There was no significant difference in body fat, plasma concentration of lipids and insulin, as well as insulin singling between SIRT6 mutant and wild type mice (data not shown). However, our examination revealed that 43% (3/7) of Sirt6Co/Co;Alb-Cre mice started to develop fatty liver when they were at 5–6 months of age, and reached 90% (9/10) from 7.5–13 months of age, as revealed by increased liver weight (Fig. 4E), hepatic triglyceride (TG) (Fig. 4F), gross morphology (Fig. 4G,H), histopathology (Fig. 4I,J), and Oil Red O staining (Fig. 4K,L). In contrast, only 12% (2/17) of control animals exhibited mild fatty liver from 5–13 months of age.
Theoretically, an increase in hepatic TG could be caused by reduced VLDL export, increased TG synthesis, and/or increased TG uptake. Our experiments indicated that SIRT6 mutant mice had a slightly higher rate of TG secretion from the liver to the blood compared with wild type mice (Fig. 4M). To measure TG synthesis, we cultured primary hepatocytes in the presence of oleic acid, which stimulates TG synthesis, with or without 3H-Palmitate. The data revealed significantly higher levels of TG in the mutant cells than controls in both conditions (Fig. 4N,O). These data suggest that the increased hepatic TG in SIRT6 mutant mice may be caused by increased TG synthesis, rather than reduced TG secretion.
SIRT6 deficiency causes altered expression of genes involved in glycolysis and lipid metabolism
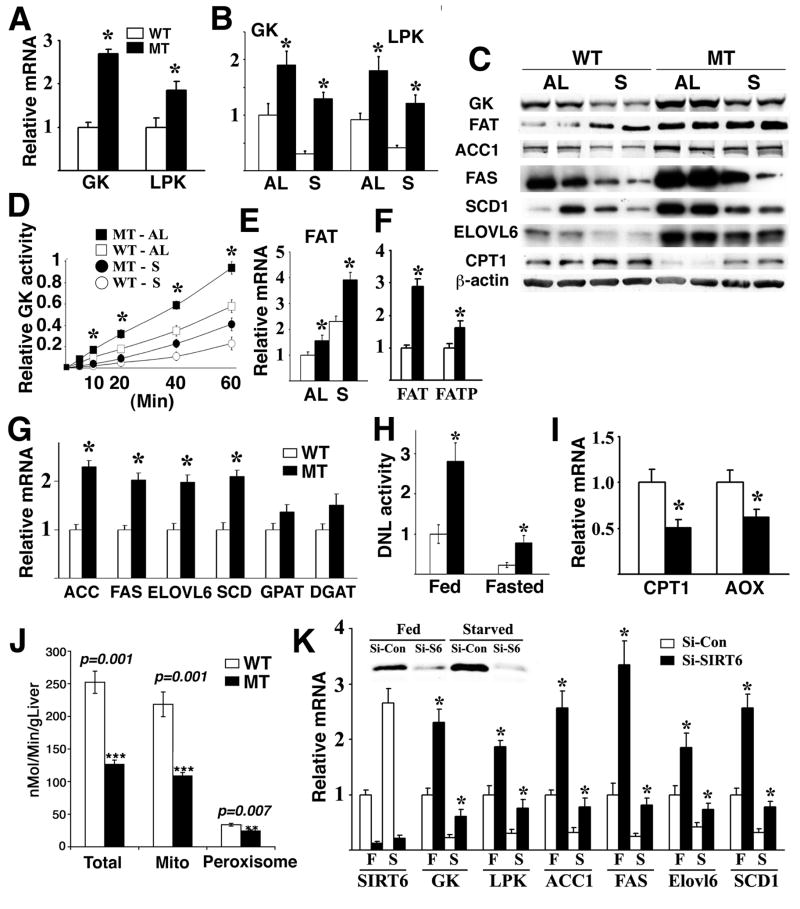
Fatty liver occurs at a high frequency in humans and may be caused by altered signaling in multiple biological pathways, including glycolysis, fatty acid uptake and synthesis, and TG synthesis (Postic and Girard, 2008). We observed an inverse correlation in gene expression between SIRT6 and glycolytic genes, GK and LPK (Fig. 1D,F) during the course of fasting, so we first investigated the impact of SIRT6 deficiency on glycolysis. Our analysis of the fatty liver of 8–9 month old mutant mice revealed significantly increased expression of both Gk and Lpk (Fig. 5A). To investigate whether this change in gene expression is directly related to a SIRT6 deficiency, we analyzed the liver of mutant mice at 2–3 months of age prior to fatty liver development. We found that SIRT6 deficiency resulted in approximately a two fold increase in Gk mRNA as compared with wild type mice (Fig. 5B). Upon fasting, Gk mRNA level was reduced to 20% of the fed level in wild type mice, however in SIRT6 mutant mice, this reduction was significantly attenuated, leading to about six fold higher Gk mRNA (Fig. 5B). Significantly higher levels of GK protein (Fig. 5C) and enzymatic activity (Fig. 5D) were observed in the SIRT6 mutant liver than control liver in both fed and fasting conditions. A similar increase in Lpk mRNA was also observed in the liver of Sirt6Co/Co;Alb-Cre mutants as compared with wild type mice (Fig. 5B). These data indicate that SIRT6 deficiency increases glycolysis under both fed and fasting conditions.
Fig. 5. SIRT6 regulates the expression of genes related to glycolysis and lipid metabolism.
(A,B) mRNA levels of GK and LPK in the liver of WT and MT mice under fed Ad Libitum (AL) or 24 hours starvation (S) detected by using Real-Time RT-PCR. (C) Protein expression of genes involved in glycolysis and lipid metabolism. (D) GK enzymatic activity in the liver of WT and MT mice. (E-G) mRNA levels of FAT and FATP (E,F) and several genes involved in TG synthesis (G). Mice used in panels (A,F,G) were 8–9 months old, and in panels (B-E) were 2–3 months old. At least 5 pairs of mice were used for each experiment. (H) De Novo lipogenesis activity in the primary hepatocytes under fed or fasted condition. (I,J) Absence of SIRT6 decreased the expression levels of genes involved in β-oxidation revealed by Real-Time RT-PCR (I) and fatty acid β-oxidation revealed by enzymatic activity (J). (K) Gene expression in primary hepatocytes upon acute knockdown of SIRT6 for 36 hours revealed by Real-Time PCR. Abbreviation of genes that is not mentioned in the text: ACC: acetyl-CoA carboxylase; Elovl6: long-chain elongase; FAS: Fatty acid synthase; GPAT: mitochondrial glycerol 3-phosphate acyltransferase; SCD1: stearoyl-CoA desaturase-1.
Next, we checked the expression level of lipid metabolism related genes. The liver of 2–3 month old Sirt6Co/Co;Alb-Cre mice exhibited moderate, yet statistically significant increased levels of fatty acid translocase (Fat), which uptakes long chain fatty acid from blood, than the control in both fed and fasting conditions (Fig. 5C,E). Fat transcripts increased about 3 fold in 8–9 month old mutant mice as compared with controls (Fig. 5F). Sirt6Co/Co;Alb-Cre mice also exhibited increased mRNA of fatty acid transport protein (Fatp) (Fig. 5F), and several genes involved in lipogenesis, fatty acid elongation, and desaturation of TG synthesis (Fig. 5C,G). Consistent with the increased expression of lipogenic genes, about a 3 fold increase in lipid production was detected in the primary hepatocytes of Sirt6Co/Co;Alb-Cre mice than that of WT mice under both fed and fasting conditions (Fig. 5H). SIRT6 mutant liver also exhibited significantly decreased levels of liver–carnitine palmitoyltransferase I (L-cpt1) and acyl-Coenzyme A oxidase 1, palmitoyl (Aox) (Fig. 5C,I), and lower levels of fatty acid β-oxidation (Fig. 5J), while expression of genes upstream of TG synthesis or involved in TG secretion were not affected (data not shown).
These data suggest that SIRT6 may regulate expression of many genes involved in various steps of glycolysis and lipid metabolism. To rule out a possibility that the expression change of these genes is secondary to a long-term loss of SIRT6, we performed acute knockdown of SIRT6 in primary hepatocytes. Our data showed that SIRT6 knockdown caused an increased expression of these genes (Fig. 5K). Conversely, infection of a SIRT6 expression vector using a lentiviral system into these cells resulted in a significant reduction in expression of these genes than the control vector under fed and fasting conditions (Supplementary Fig. 5A). These data provide convincing evidence that SIRT6 inhibits the expression of these genes.
SIRT6 binds and deacetylates H3K9 in the promoters of genes involved in glycolysis and lipid metabolism
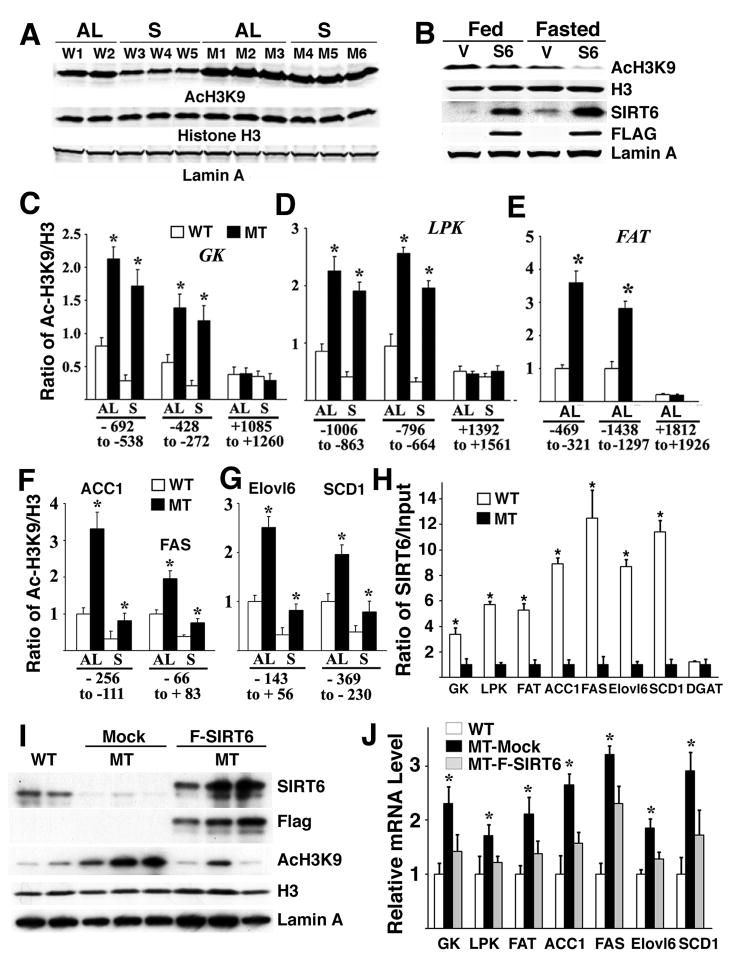
First, we investigated the mechanism underlying expression changes of these genes that are associated with SIRT6 deficiency. We found that levels of acetylated H3K9 (AcH3K9) were significantly higher in the liver of SIRT6 mutant mice than controls (Fig. 6A). Furthermore, fasting significantly reduced AcH3K9 in the liver of wild type mice but SIRT6 deficiency blocked such a reduction. A similar pattern of AcH3K9 levels was observed in SIRT6 mutant mouse embryonic fibroblast cells (MEFs) while modification of several other lysines in H3, and K16 of H4 was not affected (data not shown). Furthermore, primary hepatocytes that overexpressed FLAG-SIRT6 exhibited reduced levels of AcH3K9 (Fig. 6B). These data suggest that SIRT6 plays a specific role in deacetylation of AcH3K9 in both fed and fasting conditions.
Fig. 6. SIRT6 is recruited to and deacetylates H3K9 in the promoter of many genes.
(A) Western blot analysis of nuclear extracts prepared from the liver of WT and MT mice under fed (AL) or 24 hours starvation (S). (B) Western blot analysis of nuclear extracts prepared from primary hepatoctyes infected with pCDH-FLAG-SIRT6 or vector control under fed or fasting condition. (C-G) ChIP analysis showing the acetylation pattern of H3K9 (C-G) in the promoter of several genes in samples extracted from the liver of WT and MT mice. Four fragments (two in promoter region, and two in exon/intron) for each gene were analyzed and in all cases the absence of SIRT6 increased acetylation in the promoter, but not in the exon/intron region (some data not shown). (H) ChIP analysis showing the binding of SIRT6 to the promoter of several genes in samples extracted from the liver of WT and MT mice. (I,J) Expression of Flag-tagged SIRT6 in SIRT6 mutant liver mediated by injection of lentivirus carrying Flag-tagged SIRT6 significantly reversed increased levels of AcH3K9 (I) and gene expression (J). At least three pairs of SIRT6 mutant and control mice were used for each experiment.
Next, we investigated whether altered AcH3K9 could be detected in the promoter of the genes that were studied earlier. ChIP assay of the proximal region revealed increased levels of AcH3K9 in the promoters of Gk, Lpk, Fat, ACC1, FAS, Elovl6 and SCD1 in the liver of SIRT6 mutant mice (Fig. 6C-G). There was no obvious change in the promoters of GPAT and DGAT, which is consistent with their expression (Supplementary Fig. 5B). To confirm whether SIRT6 can recruit and deacetylate H3K9 on these promoters, we performed ChIP assay after infection of a FLAG-SIRT6 vector into primary hepatocytes that were cultured under fed and fasted condition. SIRT6 infected cells had a lower level of AcH3K9 in the proximal promoter region of these genes than cells infected with vector control, but no difference was found in the coding or intron region of those genes (Supplementary Fig. 5C). We further investigated whether SIRT6 could bind to the promoter of these genes. ChIP analysis showed that SIRT6 was recruited to the proximal region but not to the coding/intron region of those ge nes (Supplementary Fig. 5D, and data not shown). Furthermore, ChIP analysis on endogenous genes also indicated that SIRT6 occupies the promoters of these genes (Fig. 6H).
Additionally we analyzed the changes in gene expression in WT and SIRT6 mutant livers at both 2 and 8 months of age by microarray analysis and observed that expression of many genes involved in lipid and carbohydrate metabolism were altered (Supplementary Table 1, and data not shown). The gene change involved in lipid metabolism was consistent with mRNA changes we detected by Real-Time RT PCR.
Finally, we reintroduced SIRT6 to the liver using a lentiviral mediated approach. The expression of SIRT6 in SIRT6 mutant liver significantly reversed increased levels of AcH3K9 (Fig. 6I) and increased gene expression (Fig. 6J) yielding strong evidence that the increased AcH3K9 and altered gene expression is a direct consequence of SIRT6 mutation. Of note, this short-term SIRT6 expression did not have an obvious effect on the fatty liver phenotype, which may reflect a view that the formation of fatty liver is a long process in the SIRT6 mutant liver. Altogether these data indicate that SIRT6 binds and deacetylates AcH3K9 in the promoter of many genes that are involved in glucose and lipid metabolism, and SIRT6 deficiency resulted in altered expression of these genes, which ultimately lead to fatty liver in the mutant mice.
Reduced expression of SIRT1 and SIRT6 in human fatty liver samples
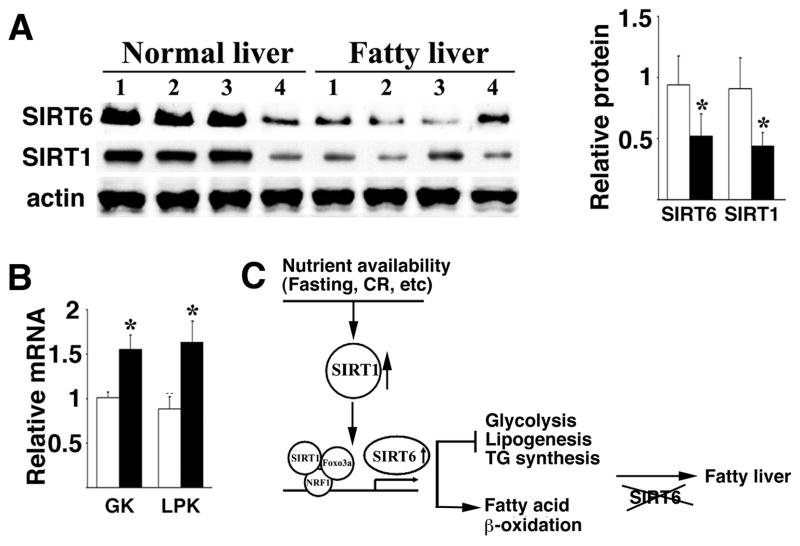
It has been shown that hepatocyte-specific deletion of SIRT1 resulted in fatty liver in mice (Purushotham et al., 2009), and that expression of SIRT1 was reduced in nonalcoholic fatty liver induced by high-fat diet in rats (Deng et al., 2007). These findings prompted us to examine expression of SIRT1 and SIRT6 in human fatty liver. Our data revealed a significant reduction of both SIRT1 and SIRT6 in human fatty liver samples as compared with normal controls (Fig. 7A). The fatty livers also exhibited significantly increased mRNA of GK and LPK (Fig. 7B). These data, combined with findings that SIRT1 regulates SIRT6 and liver specific deletion of SIRT6 in mice causes fatty liver, implicate an inhibitory role of SIRT6 in human fatty liver formation.
Fig. 7.
Reduced levels of SIRT1 and SIRT6, and increased levels of GK and LPK in human fatty liver samples. A, Western blot analysis of protein levels of SIRT1 and SIRT6 in fatty liver samples. Quantification of gel intensity of 8 normal livers and 8 nonalcoholic fatty livers was shown in right. B, mRNA levels of GK and LPK revealed by Real-Time RT-PCR in these samples. The fatty livers and controls were provided by the Liver Tissues Procurement and Distribution System (University of Minnesota). C, An integrated model for functions of SIRT6 in inhibiting fatty liver formation through regulation of glycolysis and lipid metabolism. SIRT6 deficiency, consequently, results in fatty liver fomation.
Discussion
In this study, we showed that nutrient deprivation induces expression of both SIRT1 and SIRT6. While it has been shown that nutritional stress induces SIRT1 through p53 and FOXO3a (Nemoto et al., 2004), our data indicate that the induction of SIRT6 requires SIRT1, as the absence of SIRT1 blocked SIRT6 induction. Additionally, we showed that upon nutritional stress, SIRT1 deacetylates FOXO3a, which subsequently enhances the formation of a SIRT1/FOXO3a/NRF1 (SFN) protein complex on the promoter of SIRT6 that positively regulates SIRT6 expression. Thus, our study reveals, for the first time that SIRT1 regulates another sirtuin family member through interaction with other transcription factors.
After bypassing the early post-weaning lethality associated with SIRT6 deficiency (Mostoslavsky et al., 2006) using a Cre-loxP mediated liver specific knockout, we demonstrated that SIRT6 negatively regulates the glycolytic genes, Gk and Lpk, under both fed and fasting conditions. A previous study indicated that SIRT1 interacts with and deacetylates PGC-1α and controls gluconeogenic and glycolytic pathways in the liver (Rodgers et al., 2005). Our data indicate that the absence of SIRT6 does not affect expression of Pgc1α and its downstream gene Pepck. It also did not affect blood glucose level under fed condition, although a small, yet statistically significant increase in fasting glucose was observed in 8 months and older Sirt6Co/Co;Alb-Cre mice. Thus, SIRT6 mediates a part of SIRT1 function and plays an essential role in glycolysis. In the SIRT6 liver specific mutant mice, increased glycolysis eventually affected the health of mutant mice due to an imbalance in glucose metabolism.
An important finding is that the absence of SIRT6 results in accumulation of TG, which is associated with fatty liver (den Boer et al., 2004; Postic and Girard, 2008). Our data demonstrated that SIRT6 deficiency resulted in increased expression of genes responsible for hepatic long chain fatty acid uptake and reduced expression of genes for β-oxidation. SIRT6 deficiency also increased expression of several genes involved in multiple steps of TG synthesis, while expression of genes upstream of TG synthesis and TG secretion were not affected. Increased TG synthesis was also observed in SIRT6 mutant primary hepatocytes as compared with WT hepatocytes when cultured in vitro. These data suggest that SIRT6 serves as a negative regulator of TG synthesis. Thus, the combined effect of SIRT6 deficiency in the liver causes increased glycolysis, elevated uptake of long chain fatty acid, reduced fat acid β-oxidation, and increased TG synthesis and lipogenesis, eventually leading to fatty liver formation (Fig. 7C).
It was reported that SIRT6 modulates telomeric chromatin and expression of downstream genes in the NF-κB signaling pathway through its H3K9 deacetylase activity (Kawahara et al., 2009; Michishita et al., 2008). Two recent studies also found that SIRT6 deacetylates H3K56 at telomeres and at global DNA levels in response to cell cycle arrest, suggesting a possible mechanism by which SIRT6 promotes genomic stability (Michishita et al., 2009; Yang et al., 2009). Combined with the early post-weaning lethality associated with SIRT6 deficiency, these data underscore important functions of SIRT6 in many biological processes, including DNA damage repair, telomeric chromatin integrity, glucose uptake, and aging through its deacetylase activity and/or ADP-ribosyltransferase activity (Liszt et al., 2005; Lombard et al., 2008; Michishita et al., 2008; Mostoslavsky et al., 2006; Zhong et al.). In this study, although we tried to dissect the complexity of SIRT6 function by specifically deleting SIRT6 in the liver and studying its effects on deacetylation of H3K9 in the promoters of many genes that play a role in glycolysis and lipid metabolism, it is possible that the absence of SIRT6 could still have some effects on many other processes as revealed by earlier studies. For example, our analysis confirmed that SIRT6 deacetylates H3K56 in DNA isolated from the liver at the global level, but the biological relevance of this modification in metabolism is not clear and will be an interesting topic for future investigation. In addition, our microarray analysis of the liver revealed altered expression of many genes involved in multiple biological processes (Supplementary Table 1). Although many of the changes can be secondary, some others may be directly linked to SIRT6 mutation and their impact on histopathological onset and development should be carefully investigated in future studies.
The findings delineated herein have important clinical significance as fatty liver disease is the most common cause of liver dysfunction worldwide (Ahmed and Byrne, 2009; Rogers et al., 2008). We found that human fatty livers exhibited lower levels of SIRT6 compared to normal controls. In light of this finding, SIRT6 mutant and control mice could be used as a model in chemical screens for SIRT6 activators, and TG synthesis inhibitors, which may be beneficial for the prevention and/or therapeutic treatment of fatty liver in affected individuals in the near future.
Methods
Mating and Genotyping Mice
Chimeric mice were mated with NIH Black Swiss females (Taconic) to screen for germline transformation. Male mice bearing germline transmission were mated with female FVB EII-Cre mice(Lakso et al., 1996) to generate conditional SIRT6 mice according to the procedure described earlier (Xu et al., 2001). Mice carrying a SIRT6 conditional allele were genotyped by PCR using primers P1 (5′ GCTAATGGGAACGAGACCAA 3′) and P2 (5′ ACCCACCTCTCTCCCCTAAA 3′). This primer pair flanks the LoxP insertion site in intron 1, and amplifies a 390 bp fragment from the wild type Sirt6 gene and 444 bp from the conditional allele. The recombination allele of SIRT6 is amplified using primers P1 and P3 (5′-gcgtccacttctctttcctg-3′), which produces a fragment of 524bp. All experiments were approved by the Animal Care and Use Committee of the National Institute of Diabetes, Digestive and Kidney Diseases (ACUC, NIDDK).
Lentivirus Injection into Mouse Tail Vein
To prepare lentiviral particles for injection, 293T cells were transfected with PsPAX2, VSV-G, and either pCDH or pCDH-mSIRT6 Flag. After 2 days, 30 ml of virus-containing media was collected and concentrated by ultracentrifugation. Mice were then injected 2 times (once per week) by tail-vein injection with 100 μl of concentrated virus. Mouse liver was then harvested for analysis after 2 weeks.
Determination of Hepatic Triglyceride Secretion
We used a protocol modified from (Tietge et al., 1999). Briefly, mice were injected 100 μl of 10% Tyloxapol (Triton WR1339, which inhibits all lipoprotein lipases and therefore clearance of TG from the blood) per animal by I.V. injection, and collected blood to check TG at 0 min, 1 hr and 2 hrs. Plasma was separated and assayed for triglycerides. TG secretion rates were expressed as mg/kg per h after normalizing with their liver weight.
Triglyceride Assay in the Liver and Cultured Hepatocytes
The triglyceride level of liver was measured as described previously (Jeong et al., 2008; You et al., 2004). Liver extracts were prepared by homogenization in 0.25% sucrose with 1 mmol/L EDTA, and lipids were extracted using chloroform/methanol (2:1 v/v) and suspended with 5% fatty acid-free BSA. Triglyceride level was measured using triglyceride assay reagents (Sigma Chemical Co.). For measuring triglyceride synthesis in cultured primary hepatocytes, freshly isolated hepatocytes from 5 weeks old WT and MT mice were cultured for 24 hr with normal media (DMEM with 10% FBS) in the presence or absence of oleic acid (500 mM). Quantitative estimation of hepatic triglyceride accumulation was performed by extraction of hepatic lipids from cell homogenates using chloroform/methanol (2:1) and enzymatic assay of triglyceride mass using EnzyChrom™ Triglyceride assay kit (Bioassay Systems).
Glucokinase Assay
Glucokinase activity was determined by an enzyme-linked assay based on the NADP+/NADPH ratio (Gonzali et al., 2001). The proteins extracted from mouse liver were incubated in the buffer with 12 mM MgCl2, 6.5 mM ATP, 1 mM DTT, 0.9 mM NADP+, 1 IU/ml Glucose-6-Phosphate Dehdrogenase (Sigma), 45 mM Glucose, and 32 mM Sodium-Hepes buffer, pH 7.6, 22°C, and then the absorbance was measured at 340 nm for several time points.
De Novo Lipogenesis Assay
Primary hepatocytes were cultured with 10% DMEM with insulin (100 nM) and dexamethasone (1 μM) for overnight and then incubated with 74 KBq/ml (2-14C) Sodium Acetate (2.07 GBq/mmol; GE Healthcare Inc.) for 1hr. The cells were lysed with 1N NaOH and acidified (Harada et al., 2007). Then, the lipids were extracted with petroleum ether and radioactivity was measured by liquid scintillation counter (Beckman Inc.).
Fatty Acid β-oxidation Activity
Fatty acid β-oxidation activity was measured by the method with some modifications (Shindo et al., 1978). Briefly, 0.3 gram of fresh livers were homogenized in 1.2 ml of 0.25 M sucrose containing 1 mM EDTA in a Potter-Elvehjem homogenizer using a tight-fitting teflon pestle. Approximately 500 μg of homogenate (no centrifugation) was incubated with the assay medium in 0.2 ml of 150 mM potassium chloride, 10 mM HEPES, pH 7.2, 0.1 mM EDTA, 1 mM potassium phosphate buffer, pH 7.2, 5 mM Tris malonate, 10 mM magnesium chloride, 1 mM carnitine, 0.15% bovine serum albumin, 5 mM ATP, and 50 μM C14-palmitic acid (5.0 × 104 cpm of radioactive substrate). The reaction was run for 30 min at 25 °C and stopped by the addition of 0.2 ml of 0.6 N perchloric acid. The mixture was centrifuged at 2,000 × g for 10 min., and the unmetabolized fatty acids were removed by three extractions using 2 ml of n-hexane. Radioactive degradation products in the water phase were counted. In some experiments, 2 mM potassium cyanide was added to the incubation mixture to inhibit mitochondrial β-oxidation activity. Fatty acid β-oxidation activity was expressed as nmol/min/gram of liver.
Human liver clinical samples
All of these nonalcoholic fatty liver samples were collected from donor livers or recipient livers during liver transplantation from the Liver Tissue Procurement and Distribution System, University of Minnesota. These samples are #case 647: 50 years old, female white, AST 13 U/L; #case 753: 16 years old. Male, AST 43 U/L; #case 801: 44 years old, female white, AST 46; #case 935: 60 years old, female, AST 30; #case 970: 62 years old, female, AST 25; #case 1075: 56 years old, male, AST 77; #case 1098: 59 years old, female, AST 64; and #case 1095: 61 years old, female AST 59. Normal healthy liver samples were also provided by the Liver Tissue Procurement and Distribution System, and collected from the part of donor livers that was not used for transplantation. All of samples were collected by quickly freezing in liquid nitrogen, and diagnosed as nonalcoholic steatohepatitis by histology.
Statistics Analysis
Student t test is used for data analysis. Bars represent +-SD, and * represents p<0.05 in all corresponding figures.
High lights.
SIRT1 positively regulates expression of SIRT6 upon nutritional stress
SIRT6 regulates glycolysis, TG synthesis and fat metabolism by deacetylating H3K9
Expression of SIRT6 is reduced in human fatty liver samples
Liver specific disruption of SIRT6 in mice results in fatty liver formation
Supplementary Material
Acknowledgments
We thank Drs. E. Mueller, J. Wess, S. Tydlacka, and C. Chisholm for critical reading of the manuscript, and Drs. O. Gavrilova, W. Jou, D. Simon and C. Li for technical assistance. This work was supported by the intramural Research Program of National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed MH, Byrne CD. Current treatment of non-alcoholic fatty liver disease. Diabetes Obes Metab. 2009;11:188–195. doi: 10.1111/j.1463-1326.2008.00926.x. [DOI] [PubMed] [Google Scholar]
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- den Boer M, Voshol PJ, Kuipers F, Havekes LM, Romijn JA. Hepatic steatosis: a mediator of the metabolic syndrome. Lessons from animal models. Arterioscler Thromb Vasc Biol. 2004;24:644–649. doi: 10.1161/01.ATV.0000116217.57583.6e. [DOI] [PubMed] [Google Scholar]
- Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XQ, Chen LL, Li NX. The expression of SIRT1 in nonalcoholic fatty liver disease induced by high-fat diet in rats. Liver Int. 2007;27:708–715. doi: 10.1111/j.1478-3231.2007.01497.x. [DOI] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzali S, Pistelli L, De Bellis L, Alpi A. Characterization of two Arabidopsis thaliana fructokinases. Plant Sci. 2001;160:1107–1114. doi: 10.1016/s0168-9452(01)00350-8. [DOI] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Harada N, Oda Z, Hara Y, Fujinami K, Okawa M, Ohbuchi K, Yonemoto M, Ikeda Y, Ohwaki K, Aragane K, Tamai Y, Kusunoki J. Hepatic de novo lipogenesis is present in liver-specific ACC1-deficient mice. Mol Cell Biol. 2007;27:1881–1888. doi: 10.1128/MCB.01122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, Sun L, Nguyen P, Ahn BH, Leclerc J, Deng CX, Spitz DR, Gius D. SIRT3 interacts with the daf-16 homolog FOXO3a in the Mitochondria, as well as increases FOXO3a Dependent Gene expression. Int J Biol Sci. 2008;4:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WI, Osei-Hyiaman D, Park O, Liu J, Batkai S, Mukhopadhyay P, Horiguchi N, Harvey-White J, Marsicano G, Lutz B, Gao B, Kunos G. Paracrine activation of hepatic CB1 receptors by stellate cell-derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 2008;7:227–235. doi: 10.1016/j.cmet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, Cohen HY. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahishi L, Usdin K. NF-Y, AP2, Nrf1 and Sp1 regulate the fragile X-related gene 2 (FXR2) Biochem J. 2006;400:327–335. doi: 10.1042/BJ20060734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel C, Broxmeyer HE. Sirtuin 1, stem cells, aging, and stem cell aging. Curr Opin Hematol. 2008;15:326–331. doi: 10.1097/MOH.0b013e3283043819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Postic C, Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest. 2008;118:829–838. doi: 10.1172/JCI34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CQ, Ajmo JM, You M. Adiponectin and alcoholic fatty liver disease. IUBMB Life. 2008;60:790–797. doi: 10.1002/iub.124. [DOI] [PubMed] [Google Scholar]
- Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- Shindo Y, Osumi T, Hashimoto T. Effects of administration of di-(2-ethylhexyl)phthalate on rat liver mitochondria. Biochem Pharmacol. 1978;27:2683–2688. doi: 10.1016/0006-2952(78)90042-4. [DOI] [PubMed] [Google Scholar]
- Tietge UJ, Bakillah A, Maugeais C, Tsukamoto K, Hussain M, Rader DJ. Hepatic overexpression of microsomal triglyceride transfer protein (MTP) results in increased in vivo secretion of VLDL triglycerides and apolipoprotein B. J Lipid Res. 1999;40:2134–2139. [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008a;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, Lee MH, Xiao C, Vassilopoulos A, Chen W, Gardner K, Man YG, Hung MC, Finkel T, Deng CX. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008b;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Li C, Garrett-Beal L, Larson D, Wynshaw-Boris A, Deng CX. Direct removal in the mouse of a floxed neo gene from a three-loxP conditional knockout allele by two novel approaches. Genesis. 2001;30:1–6. doi: 10.1002/gene.1025. [DOI] [PubMed] [Google Scholar]
- Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci U S A. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Matsumoto M, Pacold CM, Cho WK, Crabb DW. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 2004;127:1798–1808. doi: 10.1053/j.gastro.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.