Abstract
Innate immunity is vital for protection from microbes and is mediated by both humoral effectors, such as cytokines, and cellular immune defenses, including phagocytic cells such as macrophages. After internalization by phagocytes, microbes are delivered into a phagosome, a complex intracellular organelle with a well-established and important role in microbial killing. However, the role of this organelle in cytokine responses and microbial sensing is less well defined. Here we assess the role of the phagosome in innate immune sensing and demonstrate the critical interdependence of phagocytosis and pattern recognition receptor signaling during response to the Gram-positive bacteria Staphylococcus aureus. We show that phagocytosis is essential to initiate optimal MyD88-dependent response to Staphylococcus aureus. Prior to TLR-dependent cytokine production bacteria must not only be engulfed but also delivered into acidic phagosomes. Here acid-activated host enzymes digest the internalized bacteria to liberate otherwise cryptic bacterial-derived ligands that initiate responses from the vacuole. Importantly, in macrophages in which phagosome acidification is perturbed, the impaired response to Staphylococcus aureus can be rescued by addition of lysostaphin, a bacterial endopeptidase active at neutral pH that can substitute for the acid-activated host enzymes. Together these observations delineate the inter-dependence of phagocytosis with pattern recognition receptor signaling and suggest that therapeutics to augment functions and signaling from the vacuole may be useful strategies to increase host responses to Staphylococcus aureus.
INTRODUCTION
Phagocytosis is an evolutionarily ancient and conserved component of defense against pathogen invasion (1, 2). Material engulfed by phagocytosis is delivered into an intracellular organelle, the phagosome(3), that is constantly remodeled by fusion and limited fission events with endosomes and lysosomes (4). These changes ultimately deliver the internalized particle into a highly hydrolytic and bacteriocidal compartment known as the phagolysosome. Recent proteomic analyses have shown that over 600 proteins potentially associate with these organelles (5, 6). Some of the phagosome proteins reside in distinct flotillin-rich membrane domains that are likely to be dedicated regions for assembly of signaling complexes (7). Supporting this possibility, we have recently identified components of numerous signaling pathways associated with these organelles (6) and, our data suggest that the signals that emanate from the phagosome are likely to include defense pathways that are able to signal via both NFκB and MAP kinases.
Professional phagocytes such as macrophages and neutrophils not only destroy engulfed material but, after pathogen encounter, are also potent secretors of pro-inflammatory cytokines. The inflammatory response to pathogens is triggered by pattern recognition receptors, such as the Toll-like receptors (TLRs) that initiate inflammatory signaling cascades (8, 9). Although these receptors are highly expressed by phagocytic innate immune cells, it is clear that they are not bona fide phagocytic receptors that participate in the cytoskeletal changes required for particle internalization. Instead, TLRs function almost exclusively to sense microbes and regulate pro-inflammatory signaling cascades. TLRs are found both on the cell surface and in intracellular compartments such as endosomes (TLRs 3, 7 and 9) and phagosomes that form around internalized bacteria and other large particles (TLR2 and TLR4) (10–12). The observed recruitment of surface TLRs to phagosomes provides strong support for the proposition that these organelles might function not only to destroy internalized bacteria but also may contribute to pathogen sensing (10, 11, 13). Although subject to some debate, it has also been suggested that these phagosome-associated TLRs might regulate phagosome maturation in an organelle-autonomous manner (14, 15). However, despite nearly a decade passing since the original observation of the association of TLRs with phagosomes, the full contribution of these organelles and associated PRRs to innate immune signaling remains to be fully defined.
Here we have set out to formally assess the relationship between phagocytosis and innate immune signaling. We demonstrate the critical role of the phagosome in sensing and responding to Gram-positive S. aureus but not the Gram-negative bacteria E. coli. We show that phagocytosis is needed for digestion of the microbe and presentation of material derived from internalized S. aureus to trigger TLR-dependent responses. Importantly, TLR-dependent response to S. aureus occur only after phagosome maturation is complete as the vacuole must acidify to allow activation of pH-dependent host enzymes that liberate the bacterial-derived ligands required for the full immunogenicity of these microbes. Together these observations emphasize the interdependence of phagocytosis and PRR-signaling for optimal response to S. aureus and suggest that strategies to augment functions of the phagocytic vacuole might be potential adjunct therapies to increase host response to this important re-emerging pathogen.
MATERIALS AND METHODS
Mice and cell cultures
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Myd88−/− mice were from M. Freeman (Massachusetts General Hospital, Boston, MA), and Tlr2−/− mice and Tlr4−/− mice were from R. Medzhitov (Yale University School of Medicine, New Haven, CT). All mice were kept and handled under a protocol approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital). All experiments were performed on thioglycollate-elicited peritoneal macrophages unless otherwise stated. Macrophages were collected from mice by peritoneal lavage 4 days after intraperitoneal injection of 3% thioglycollate (Difco Laboratories, Detroit, MI), and maintained in Dulbecco’s Modified Eagle Medium (DMEM; Gibco BRL, Carlsbad, CA) containing 10% heat-inactivated FCS (Gibco BRL), and penicillin-streptomycin (50 IU/ml and 50 μg/ml; Cellgro, Herndon, VA). HEK293T and J774A.1 cell lines (ATCC, Manassas, VA) were maintained according to ATCC’s recommendations.
Bacterial strains
The strains of S. aureus used were Reynolds capsular serotype 5 (CP5) and capsule-negative mutant (provided by Dr. J. C. Lee, Brigham and Women’s Hospital, Boston, MA), and Newman (provided by Dr. Fred Ausubel, Massachusetts General Hospital, Boston, MA), and were grown at 37°C in Columbia media supplemented with 2% NaCl. Group B Streptococcus (strain GBS type III COH-1; provided by Dr. Michael Wessels, Childrens Hospital, Boston, MA), Escherichia coli (strain K12; ATCC) and Salmonella montevideo (strain SH5770; provided by Dr. Helena Mäkelä, National Public Health Institute, Helsinki, Finland) were grown as described previously (12, 16). The bacteria grown to the mid-exponential phase (OD600 = 0.6–0.8) were either heat-inactivated (65°C for 30 min) or used as live bacteria to stimulate cells as described below. In some experiments where capsule-negative mutant S. aureus were used, both Reynolds CP5 and capsule-negative mutant were grown to the stationary phase (OD600 > 1.0; 16 h) prior to heat-inactivation.
Bacteria digestion in vitro
Heat-inactivated S. aureus were incubated with 5 μg/ml of lysostaphin in the indicated pH. The digestion of the bacteria over the indicated time course was assessed by measuring OD600 of the bacterial suspensions. To assess LTA release, the supernatants from 2-h digestion with lysostaphin were collected, filtered with 2-μm filter membrane, and assayed for LTA concentrations by ELISA. In some experiments, the filtered digests from bacteria treated for 2 hr with 5 μg/ml lysostaphin in PBS (pH 7.5) or 5 μg/ml lysozyme in potassium phosphate buffer (pH 6.0) was used to stimulate macrophages.
Reagents and plasmids
Lipoteichoic acid (LTA; derived from S. aureus), Lipopolysaccharide (LPS; derived from E. coli 026:B6), lysozyme (from human neutrophils), and protease inhibitor panel were purchased from Sigma-Aldrich (St. Louis, MO). Peptidoglycan (PGN; derived from S. aureus) was from InvivoGen (San Diego, CA). Recombinant lysostaphin was from Abazyme (Needham, MA). Monoclonal anti-LTA antibody (clone 55) used to detect LTA release was from Cell Science (Canton, MA). YFP-tagged TLR2 (pcDNA3.1-TLR2-YFP) and CFP-tagged TLR6 (pcDNA3.1-TLR6-CFP) expression vectors were kindly provided by D. Golenbock (University of Massachusetts Medical school, Worcester, MA).
Cell stimulations and treatments
Peritoneal macrophages in DMEM medium with 1% FCS were stimulated with heat-inactivated or live bacteria at the indicated MOIs, or bacterial ligands (i.e. LPS, LTA or PGN) at the indicated concentrations, at 37°C in 5% CO2 for 2 to 4 h. To assess the role of phagocytosis in the induction of cytokine responses, prior to the stimulation with bacteria, macrophages were pretreated with 6 μM cytochalasin D (Sigma-Aldrich) for 60 min to inhibit phagocytosis. Inhibition of phagosomal acidification was done by preloading the cells with 50 nM Bafilomycin A (Calbiochem, San Diego, CA) 60 min before the stimulation. Cells pretreated with same volume of vehicle, DMSO, were used as a negative control for both inhibitors. In experiments where live bacteria were used to infect macrophages, viability at 2 hours was assessed by trypan exclusion and showed no significant cell death (<5%) in control or treated macrophages. Additionally, survival of different S. aureus mutants was also determined by gentamicin protection assay. No significant differences in bacterial numbers and survival were noted over the first 2 hours. Where indicated, the cells were preloaded with 15 μg/ml lysostaphin, 15 μg/ml lysozyme, or protease inhibitors (Protease Inhibitor Panel; Sigma) at the indicated concentrations, 15 min before the stimulation. NF-κB activity was measured using a dual luciferase reporter assays for NFκB activation in HEK293T cells transfected with TLR2/6 as previously described (17).
ELISA
Cytokine secretion in cell culture supernatants were assayed for mouse IL-1β, IL-6 and TNF-α (DuoSet ELISA Development System; R&D System, Minneapolis, MN) in accordance with manufacturer’s protocol.
Quantitative real-time PCR
Total RNA was extracted from stimulated macrophages using TRIzol Reagents (Invitrogen). cDNA was synthesized from total RNA by reverse transcription (RT) using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and gene expression of tnf-α and il-6 were measured by quantitative real-time PCR (qPCR) using SYBR Green PCR core reagents (Applied Biosystems). The level of expression of each gene was determined by normalizing its mRNA quantity to the quantity of the glyceraldehyde-3-phosphate dehydrogenase (gapdh) mRNA at the same samples. The primer sequences used for qPCR were as follows: tnf-α-F: 5′-GCACAGAAAGCATGATCCG; tnf-α-R: 5′-GCCCCCCATCTTTTGGG; il-6–F: 5′-TGTTCTCTGGGAAATCGTGGA; il-6–R: 5′-AAGTGCATCATCGTTGTTCATACA; gapdh-F: 5′-TGTTCCTACCCCCAATGTGT; gapdh-R: 5′-TGTGAGGGAGATGCTCAGTG.
Phagocytosis and intracellular TNF-α
Phagocytosis and simultaneous cytokine response was measured as described previously (12, 16). Briefly, peritoneal macrophages in DMEM medium with 1% FCS were incubated with heat-inactivated bacteria, labeled with TAMRA (Molecular Probes, Eugene, OR), at MOI of 25 for 30 min on ice allowing the synchronization of bacteria binding onto the cell. In all cases, before the incubation with macrophages, bacterial clusters were disrupted by passing them through a 30-gauge needle. After 30 min on ice, the cells were further incubated for the indicated times at 37°C in the presence of GolgiStop (BD Bioscience, San Diego, CA) to accumulate intracellular TNF-α. The cells were washed twice with ice-cold PBS containing 5 mM EDTA (PBS/EDTA), detached with scrapers, and fixed in 3% paraformaldehyde. The cells were permeabilized and stained with allophycocyanin (APC)-conjugated anti-mouse TNF-α antibody (BD Bioscience) diluted in PBS with 0.2% saponin. After washing, the cells were analyzed by flow cytometry performed on FACS Calibur (Becton Dickinson); analysis was performed with CellQuest Pro software (Becton Dickinson) to determine phagocytosis and intracellular TNF-α production at the single-cell level. To estimate the number of bacteria engulfed by a single cell, TAMRA-labeled bacteria used in the same experiment were also analyzed by the flow cytometry to quantify the mean fluorescence intensity (MFI) of a single bacterial particle. The amount of TAMRA fluorescence internalized by macrophages was then quantified by FACS. As the TAMRA dye was stable and, knowing the MFI of an individual bacteria, estimates of the number of bacteria internalized could be extrapolated from the total internalized fluorescence. This allowed us to then determine, using the mean intracellular TNF-α production in cells with same bacterial loads (see Figure SF2), cytokine production normalized for bacterial load. In some cases, TNF-α induction at the transcriptional level was also determined in cells with different bacterial loads. To do that, phagocytosis of TAMRA-labeled bacteria was measured simultaneously by flow cytometry, and cells that had engulfed different amount of bacteria were sorted for total RNA extraction and subsequent measurement of TNF-α gene expression by RT-qPCR as described above.
Phagosomal pH
Peritoneal macrophages in DMEM medium with 1% FCS were incubated with S. aureus labeled with FITC (pH-sensitive) and Alexa Fluor 647 (pH-insensitive) fluorescent dyes (see Figure 4), at low MOI (≤ 10) for 30 min on ice allowing the synchronization of bacteria binding onto the cell. In all cases, before the incubation with macrophages, bacterial clusters were disrupted by passing them through a 30-gauge needle. After 30 min on ice, the cells were further incubated for the indicated times at 37°C. The cells were washed twice with ice-cold PBS/EDTA, detached and immediately analyzed by flow cytometry to determine the MFI emission between FITC and Alexa Fluor 647. To calculate pH using the ratiometric assay, values were compared with a standard curve obtained by resuspending and permeabilizing the cells that had phagocytosed bacteria for 2 h in buffers at a fixed pH (ranging from pH 3.5 to 8) and containing 0.05% Triton X 100. The cells were immediately analyzed by flow cytometry to determine the emission ratio of the two fluorescent dyes at each pH (see Figure 4C). In some cases, before the incubation with the bacteria, macrophages were pretreated with Bafilomycin A (Calbiochem). Cytokine secretion and gene expression in the cells arrested at different phagosomal pH were also determined by ELISA in the culture supernatants and RT-qPCR of the total RNA extracted from the same samples as described above.
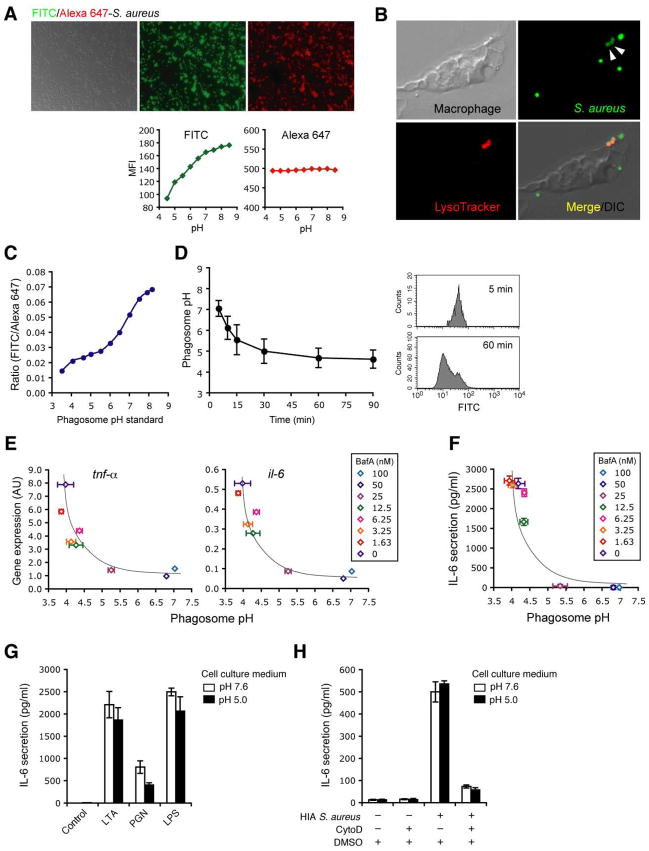
Figure 4. Phagosome acidification is required, but is not sufficient, for response to S. aureus.
A-D, S. aureus ratiometric assay to determine phagosome pH. HIA S. aureus was labeled with FITC (pH-sensitive) and Alexa 647 (pH-insensitive) (A, upper panels). Quantification of FITC (green) and Alexa 647 (red) fluorescence intensity of the dual labeled bacteria incubated for 1–2 min in a series of known pH buffers was assessed by FACS to confirm pH sensitivity of the dyes (A, lower panels). pH sensitivity of FITC was also confirmed within acidic compartment in macrophages. After 30 min, FITC-labeled S. aureus were internalized into acidic compartments by macrophages preloaded with acidophilic LysoTracker (red). The FITC signal from these compartments (B, arrow heads) was significantly reduced as compared with those non-internalized or surface bound FITC-labeled S. aureus. Phagosome pH standard curves (C) were obtained as described in Materials and Methods. Kinetics of phagosome pH (D, left panel) was measured in peritoneal macrophages using the dual label S. aureus ratiometric assay, and examples of phagosome FITC signals from the cells at 5 and 60 min was shown in the histograms (D, right panels). E and F, Cytokine response in macrophages with arrested phagosome pH. Peritoneal macrophages pretreated with BafA at the indicated concentrations were incubated with the dual labeled S. aureus at low MOI (≤10). After 30 min at 4°C to synchronize uptake, macrophages were incubated for further 90 min (E) or 6 h (F) at 37°C and phagosome pH were determined. Cytokine gene expression in the cells (E) and IL-6 secretion in the supernatants (F) were measured by quantitative PCR and ELISA respectively, and correlated with the phagosome pH. G and H, Low pH is insufficient to rescue the impaired cytokine response to S. aureus in the absence of phagocytosis. Peritoneal macrophages in a neutral or acidic extracellular pH were stimulated with bacterial ligand 2 μg/ml LTA, 10 μg/ml PGN or 10 ng/ml LPS (G). Peritoneal macrophages pretreated with DMSO (control) or CytoD as in Figure 1A were stimulated with HIA S. aureus at MOI 50 in a neutral or acidic extracellular pH (H). IL-6 secretion was measure by ELISA in culture supernatants at 4h. Data indicate mean ± SD of triplicates. Data are representative of three independent experiments.
S. aureus infection in vivo
C57BL/6 mice between 6 and 12 weeks old were used for all in vivo experiments. In vivo S. aureus infections were performed as described previously (18). Previous studies have reported in vivo blockade of the v-ATPase with single injections of 25 ng/g body weight or ~0.5 μg/mouse of Bafilomycin A (19). To determine the role of phagosome acidification in vivo, mice were injected intraperitoneally with 0.5 μg Bafilomycin A or with same volume of vehicle, DMSO, at −1 hr, 4 hr and 8hr to decrease vacuolar acidification. S. aureus (5×107 c.f.u.) was inoculated i.v. at 0 hr, and mice were sacrificed at 4 hours to measure cytokines and at 18 hours to measure bacterial load.
RESULTS
Response to S. aureus requires phagocytosis
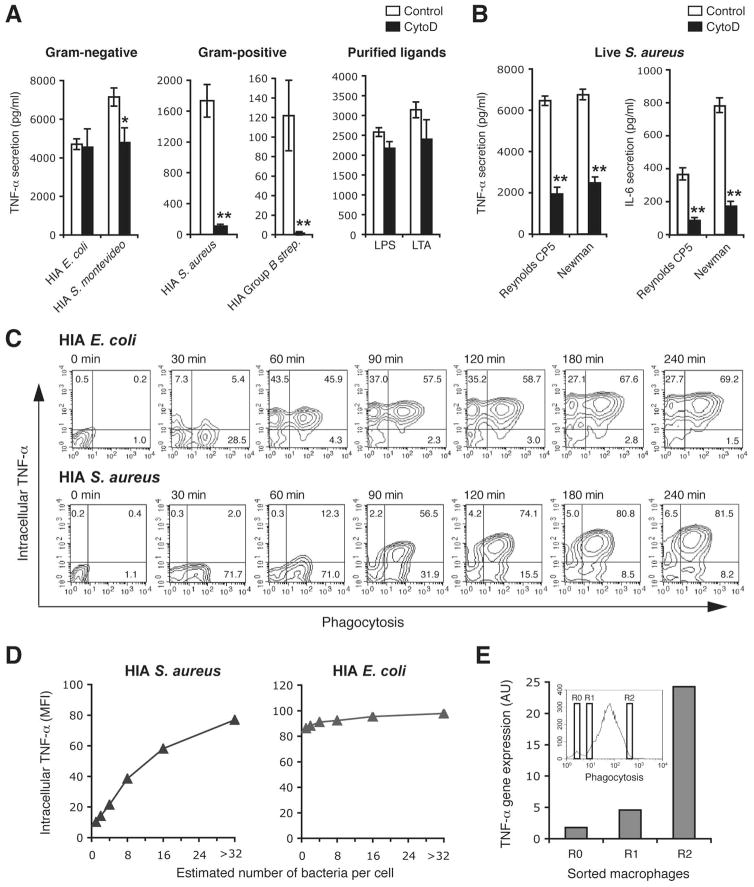
TLRs have been reported to associate with phagosomes containing a variety of particles, suggesting a relationship between engulfment and sensing of certain microbes. To determine the role of microbial uptake in initiating the innate immune response to different pathogens, we first tested whether it was required for cytokine production by blocking bacterial internalization with cytochalasin D. To obviate the problem of different rates of replication altering the effective MOIs during the course of the assay and hence confounding interpretation of the results, we first tested a number of heat-inactivated (HIA) bacteria. Inhibiting phagocytosis did not affect macrophage response to heat-killed Salmonella montevideo or Escherichia coli or purified TLR ligands, LPS and LTA. [Figure 1A]. In contrast, inhibition of phagocytosis completely blocked production of TNF-α [Figure 1A] and IL-6 [Supplementary Figure SF1] in response to heat-killed Gram-positive Group B Streptococcus and Staphylococcus aureus. These data suggested that response to Gram-positive bacteria was intimately associated with internalization and, to further explore this possibility, we chose to focus subsequent experiments on S. aureus. To ensure that the observed decreased stimulatory capacity in the absence of internalization was not a consequence of loss of immunogenicity during heat inactivation, we performed similar experiments and decreased internalization of live S. aureus using cytochalasin D. Similar to observations using heat inactivated bacteria, decreasing phagocytosis of live S. aureus decreased TNF-α and IL-6 [Figure 1B] production indicating that the majority of inflammatory signaling in response to S. aureus occurred after internalization of the bacteria.
Figure 1. Bacterial internalization is required for macrophages cytokine response to S. aureus and Group B streptococcus but not E. coli or S. montevideo.
A and B, TNF-α production by peritoneal macrophages pretreated with DMSO (control) or 6 μM cytochalasin D (CytoD) for 30 min to block internalization, and incubated with heat-inactivated (HIA) E. coli, S. montevideo, S. aureus (Reynolds CP5 strain) or Group B streptococcus at MOI 50, or bacterial ligand 10 ng/ml LPS or 2 μg/ml LTA (A), or exposed to live S. aureus strain Reynolds CP5 or Newman at MOI 10 (B). Induction of cytokine responses at 2 (TNF-α) or 4 (IL-6) h was measured by ELISA in culture supernatants. Data represents mean ± SD of triplicates. C, Single-cell analysis by FACS determining bacterial engulfment and intracellular TNF-α production. Peritoneal macrophages were pre-incubated with TAMRA-labeled HIA E. coli or HIA S. aureus at MOI 25 for 30 min at 4°C to synchronize phagocytosis and the further indicated time course at 37°C. Phagocytosis and intracellular TNF-α production was measured simultaneously by FACS. Contour plots show the percentages of TNF-α producing (top right quadrant) or -nonproducing (bottom right quadrant) cells that had phagocytosed bacteria, or TNF-α producing cells without bacterial internalization (top left quadrant). D, Correlation of intracellular TNF-α with the number of internalized bacteria in which the FACS analysis of the 120-min time point from (C) was used to estimate the number of internalized E. coli or S. aureus and to determine the correlation with intracellular TNF-α production (see Supplementary Figure S2). E, TNF-α gene expression in macrophages with low- or high-numbers of the phagosome containing S. aureus. Macrophages exposed to HIA S. aureus for 90 min were sorted into cells with no internalized bacteria (R0) and cells with either low (R1) or high numbers of internalized bacterial (R2) (as shown in the insert histogram). TNF-α gene expression in sorted cells was determined by quantitative PCR. Data represent TNF-α gene expression levels normalized to GAPDH. Data are representative of three (A and B) or two (C and D) independent experiments. *p ≤ 0.05; **p < 0.01.
To determine the relationship between phagosome formation and innate immune activation, a FACS based assay in which phagosome number and cytokine production could be simultaneously measured at a single cell level was used [Figure 1C] (12, 16). To allow accurate measurement of internalized bacteria we used heat inactivated bacteria labelled with TAMRA, a fluorescent dye stable within the phagosome for >4 hours (data not shown), to permanently mark macrophages that had internalized bacteria. Single-cell analysis indicated that following incubation with E. coli, TNF-α was detected in both the phagocytosing macrophages (top right quadrant of each plot) and macrophages that had been cultured with, but not internalized, bacteria (top left quadrant of each plot). In contrast, cytokines were produced only by macrophages that had internalized S. aureus (top right quadrant in each plot). As an example, 43.5% of macrophages that had been in contact with E. coli for 60 minutes, but had not internalized them, produced TNF-α, whereas only 0.3% of macrophages that had contacted but not internalized S. aureus had detectable TNF-α expression at this time point (Figure 1C, top left quadrant of the 60 minute plots). To further analyze the contribution of phagocytosis to the innate immune response, this assay was used to estimate the number of phagosomes (see methods and Supplementary figure SF2) and to determine the correlation with cytokine production. Unlike E. coli, cytokine response was proportional to the estimated number of S. aureus containing phagosomes, reaching maximal when approximately 16–32 phagosomes per cell had formed [Figure 1D]. Furthermore, when macrophages were sorted into those without S. aureus-containing phagosomes or those containing low or high numbers of phagosomes, cytokine expression (as determined by semi-quantitative RT-PCR) correlated with phagosome number [Figure 1E]. These data indicated that E. coli was able to trigger cytokine production from the cell surface whereas engulfment was required for response to S. aureus.
Both the TLR2-dependent and TLR2-independent component of the response to S. aureus occurs from the phagosome
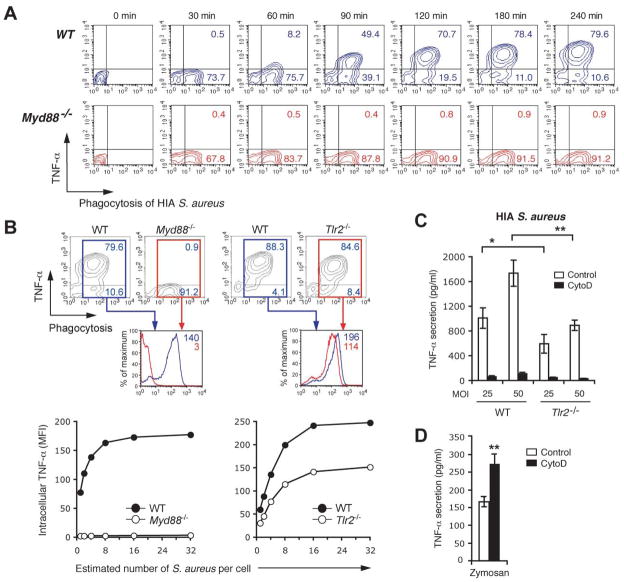
To determine the relative contributions of the TLR2/MyD88-signals to phagocytosis-dependent cytokine response to S. aureus, macrophages unable to signal through these pathways were analyzed using our FACS based assay. As it has been suggested that TLR signaling is involved in the rate of phagocytosis and phagosome maturation (14), we first tested the kinetics of S. aureus uptake in Myd88−/− macrophages. No defect in bacterial internalization was detected in the absence of this adaptor [Figure 2A], indicating that these Myd88-dependent signals did not participate in the initial uptake process and hence would not confound our interpretation of subsequent results. Consistent with a critical role of Myd88-dependent signals for response to S. aureus, phagosome-associated cytokine production was abolished in macrophages lacking MyD88 [Figure 2A and 2B]. In contrast, Tlr2−/− cells demonstrated approximately 50% reduction in the production of TNF-α [Figure 2B and 2C], confirming the role of this receptor in S. aureus response but also suggesting that other receptors that use the MyD88 adaptor are involved in sensing these bacteria, potentially by responding to nucleic acids liberated after bacterial destruction in the vacuole.
Figure 2. The relative contributions of TLR2 and MyD88 to phagosome-dependent response to S. aureus.
A and B, Phagocytosis and cytokine response in Myd88- or Tlr2-deficient macrophages. Peritoneal macrophages from wild-type (WT), Myd88−/− or Tlr2−/− mice in C57BL6 background were pre-incubated with TAMRA-labeled HIA S. aureus at MOI 25 for 30 min at 4°C, and incubated for the further indicated time course (A) or 4 h (B) at 37°C. Phagocytosis and intracellular TNF-α was measured simultaneously by FACS. Contour plots show the percentages of TNF-α-producing (top right quadrant) or TNF-α-non-producing cells (bottom right quadrant) after internalization of the bacteria. Lower histograms in (B) indicate the TNF-α production in macrophages of different genotypes that had engulfed bacteria in the different genotypes (WT in blue; Myd88−/− or Tlr2−/− in red). The number of internalized S. aureus in the macrophages was also estimated and correlated with intracellular TNF-α production as in Figure 1D (B, lower panels). C and D, The requirement for internalization on TLR2-dependent and TLR2-independent responses to S. aureus. Peritoneal macrophages from WT (C and D) or Tlr2−/− mice (C) were pretreated with DMSO (control) or 6 μM CytoD, and incubated with HIA S. aureus at MOI 50 (C) or zymosan (100 μg/ml) (D). TNF-α production at 2 h was measured by ELISA in culture supernatants. Data represent mean ± SD of triplicates. Data are representative of two (A and B) or three (C and D) independent experiments. *p ≤ 0.05; **p < 0.01.
To formally test whether phagocytosis was required for both the TLR2-dependent or TLR2-independent component of the response to S. aureus we used Tlr2−/− macrophages and determined the consequence of blocking internalization on their ability to respond. Consistent with our FACS assay, Tlr2−/− macrophages demonstrate a 50% reduction in TNF-α secretion confirming a TLR2-independent component of this response to S. aureus [Figure 2C]. Cytochalasin D completely blocked cytokine production by S. aureus, both in WT and Tlr2−/− macrophages, indicating that both TLR2-dependent and TLR2-independent components of the response to this Gram-positive bacteria were sensitive to inhibition of internalization [Figure 2C]. When similar experiments were performed using Tlr4−/− macrophages stimulated with E. coli we also observed a residual, TLR4-independent component of the response to this Gram-negative bacterium. However, in contrast to what was observed for S. aureus, but in keeping with our other observations, Cytochalasin D had no effect on either the TLR4-dependent or TLR4-independent response to E. coli [Supplementary Figure SF3].
Our observation that both the TLR2-dependent and TLR2-independent component of response was dependent on phagocytosis suggested that the nature of the particulate ligand, and not the receptor it engages for signaling, dictated the need for engulfment. To test this we used Zymosan, a yeast cell wall-derived particle that, similar to S. aureus, recruits TLR2 to its phagosomes and signals via this PRR (10). In contrast to what was observed for S. aureus, blocking internalization of zymosan increased cytokine production [Figure 2D]. These data indicate that signaling from the cell surface was more efficient than from the phagosome at triggering response to this particular TLR2 ligand. Thus, despite both stimulating TLR2, zymosan and S. aureus demonstrate opposite requirements for phagocytosis indicating that the cargo, not the PRR involved in response, determine the need for internalization.
Phagosome acidification is required for cytokine response to S. aureus
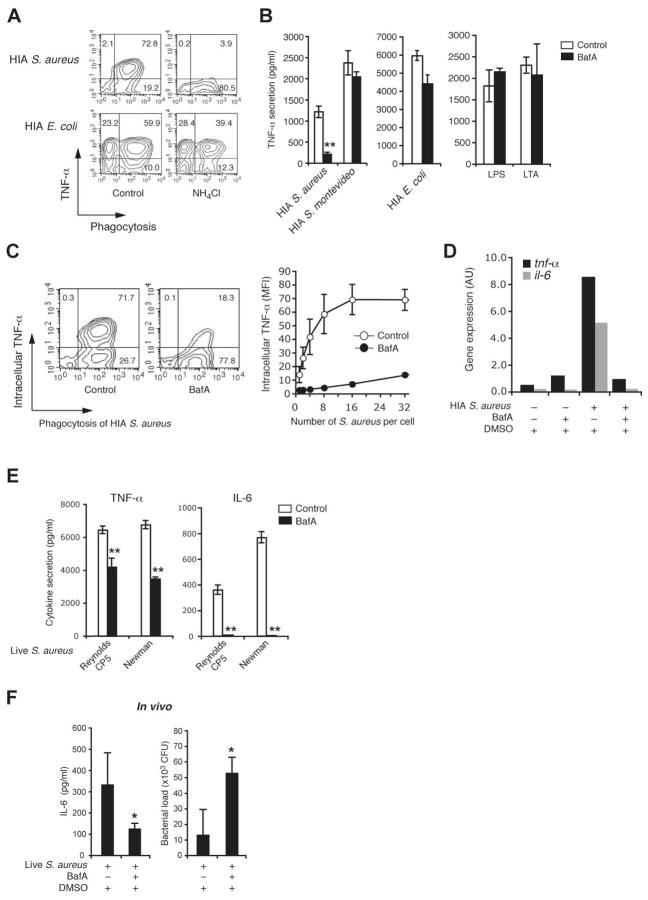
The nascent phagosome undergoes a process termed “maturation” by fusion and limited fission events with endosomes and lysosomes to generate an acidic and highly hydrolytic mature phagolysosome. To determine whether formation of a phagosome was sufficient for response to S. aureus, or whether phagosome acidification was also required, we neutralized phagosomes using a weak base, NH4Cl. NH4Cl blocked S. aureus but not E. coli induce TNF-α production [Figure 3A] suggesting that not only phagocytosis but also phagosome acidification was required for response to S. aureus. To further test this we perturbed phagosome acidification using Bafilomycin A, a specific inhibitor of the vacuolar-ATPase (V-ATPase). Similar to NH4Cl, Bafilomycin A had no effect on TNF-α produced by E. coli, S. montivideo or purified TLR ligands, LTA and LPS [Figure 3B]. In contrast, Bafilomycin A decreased cytokine response to S. aureus as measured by ELISA [Figure 3B], intracellular cytokine staining [Figure 3C] and QRT-PCR [Figure 3D]. Importantly, Bafilomycin A treatment decreased TNF-α and IL-6 secretion in response both HIA and live S. aureus [Figure 3E].
Figure 3. Phagosome acidification is required for response to S. aureus.
A, Effect of neutralizing the phagosome pH on cytokine response to E. coli and S. aureus. Peritoneal macrophages were pretreated without (control) or with NH4Cl (40 mM) for 30 min and incubated with TAMRA-labeled HIA E. coli or S. aureus at MOI 25 for 30 min at 4°C and the further 2 h at 37°C. Phagocytosis and intracellular TNF-α production was assessed simultaneously FACS as in Figure 1D. B–E, Impaired cytokine response to S. aureus in the absence of phagosome acidification. Peritoneal macrophages were pretreated with DMSO (control) or 50 nM bafilomycin A (BafA) for 60 min, and incubated with HIA S. aureus (Reynolds CP5 strain), E. coli or S. montevideo at MOI 50 (B and D), or TAMRA-labeled HIA S. aureus at MOI 25 (C), or live S. aureus strain Reynolds CP5 or Newman at MOI 10 (E). Alternatively cells were stimulated with purified bacterial ligands 10 ng/ml LPS or 2 μg/ml LTA (B). Induction of cytokine responses (B and E) at 2 (TNF-α) or 4 (IL-6) h was measured by ELISA in culture supernatants. Data represents mean ± SD of triplicates. Phagocytosis and intracellular TNF-α production (C, left panel) was assessed as in A, and the number of internalized S. aureus in the macrophages was also estimated and correlated with intracellular TNF-α production (C, right panel) as in Figure 1D. TNF-α and IL-6 gene expression at 3 h (D) was determined by quantitative PCR and normalized to GAPDH gene expression. F, Cytokine production (4 h) and bacterial load (24 h) were determined after in vivo S. aureus infection of mice in which phagosome acidification was blocked with i.p. BafA as described in Materials and Methods. Data are representative of three independent experiments. *p ≤ 0.05; **p < 0.01.
To determine if vacuolar acidification played a role in regulating cytokine responses in vivo, mice were injected at −1, 4 and 16 hours with Bafilomycin A i.p. to reduce the efficiency of the v-ATPase in vivo (20). These mice were then challenged with 5×107 S. aureus i.v. and cytokines measured at 4 hours. Although i.p. Bafilomycin A is unlikely to completely block acidification in vivo, Bafilomycin A treatment blunted the early IL-6 response [Figure 3F], consistent with our in vitro observations. This decreased early cytokine response was also associated with higher bacterial numbers at 24 hours [Figure 3F]. Together these data indicate that S. aureus must both be internalized and delivered into an acidic phagolysosome to trigger an inflammatory response.
Acidification is required but is not sufficient for response to S. aureus
To establish the exact pH required for induction of the inflammatory response we developed a FACS based ratiometric assay in which Alexa 647/FITC-labeled S. aureus act as a pH sensitive probe (21). Low pH quenches the fluorescence of FITC-S. aureus but not that of pH-stable dyes such as Alexa 647 [Figure 4A]. During phagocytosis this pH-dependent loss of fluorescence could be visualized by microscopy as reduced intensity of FITC-S. aureus when localized in lysotracker positive (and hence acidic) phagolysosomes [Figure 4B]. The ratio of FITC/Alexa-647 correlated with the pH of the phagosome [Figure 4C] and, using this to monitor vacuolar pH, we determined that phagosomes acidified to pH<5.0 by 1 hour [Figure 4D]. Notably, the kinetic of this acidification preceded the onset of cytokine production after S. aureus encounter as determined by intracellular staining [Figure 1C and 2A]. We next used different concentrations of Bafilomycin A to clamp phagosomes at different pHs (as measured using Alexa-647/FITC-labeled S. aureus) and simultaneously measured cytokine production. Arresting S. aureus phagosomes at a pH of >5 blocked cytokine production as determined either by Q-RT-PCR [Figure 4E] or by ELISA [Figure 4F].
Ligand binding to certain TLRs (such as TLR9) occurs preferentially in acidic environments (22, 23). We therefore determined if the sole role of the phagosome was to provide an acidic environment to allow S. aureus derived ligands to bind their cognate PRRs. However, cytokine response to the purified TLR ligands LPS, LTA and PGN was unaffected by an acidic environment indicating that, for these ligands, interactions with their cognate TLRs was pH-independent [Figure 4G]. We next determined whether low extracellular pH would obviate the need for internalization by permitting S. aureus to signal from the cell surface. Despite expression of TLR2 on the plasma membrane, acidification of the extracellular milieu failed to rescue the block in cytokine response to S. aureus that occurred after Cytochalasin D treatment [Figure 4H] indicating that an acidic environment was not sufficient to allow S. aureus to induce a cytokine response. Together these data indicate that the low pH (<5) in the mature phagolysosome is necessary, but not sufficient, for response to S. aureus.
Phagosome-associated pathogen processing liberates cryptic ligands
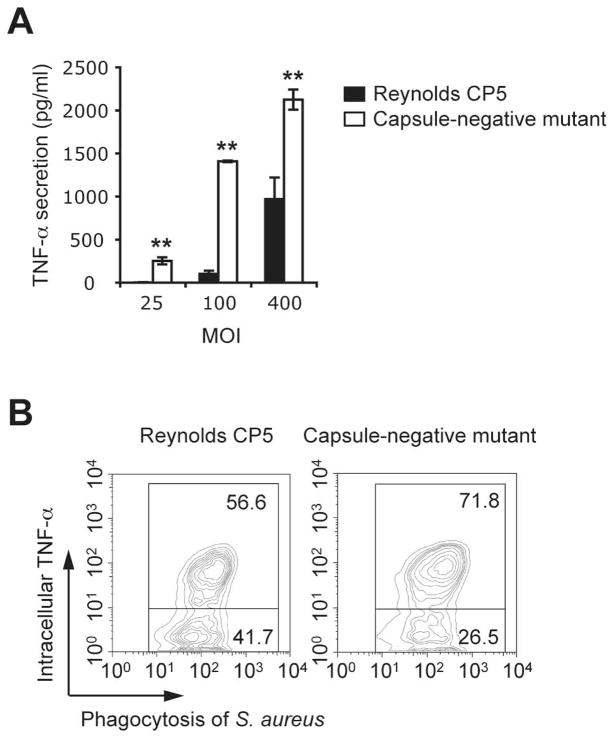
pH is likely to control the presence and activity of many enzymes within the phagosome (24) that might function to release of bacterial ligands within the vacuole. To test whether this was a possibility we first compared the macrophage response to capsulated and uncapsulated S. aureus strains. CP5- S. aureus induced more robust cytokine production than their capsulated counterparts [Figure 5A and 5B], suggesting that one function of the capsule might be to limit access of the host PRR to their agonistic ligands in the bacterial cell wall.
Figure 5. The S. aureus capsule limits access of PRRs to their agonistic ligands in the bacterial cell wall and cytokine response.
A, HIA encapsulated (Reynolds CP5) or unencapsulated (capsule-negative mutant) S. aureus were incubated with peritoneal macrophages at the indicated MOIs. TNF-α secretion at 2 h was measured by ELISA in culture supernatants. Data represent mean ± SD of triplicates. B, Phagocytosis and intracellular TNF-α were measured simultaneously in macrophages incubated with TAMRA-labelled HIA Reynold CP5 or capsule-negative mutant. Contour plots show the percentages of TNF-α-producing (top right quadrant) or TNF-α-non-producing cells (bottom right quadrant) after internalization of the bacteria Data are representative of three independent experiments. *p ≤ 0.05; **p < 0.01.
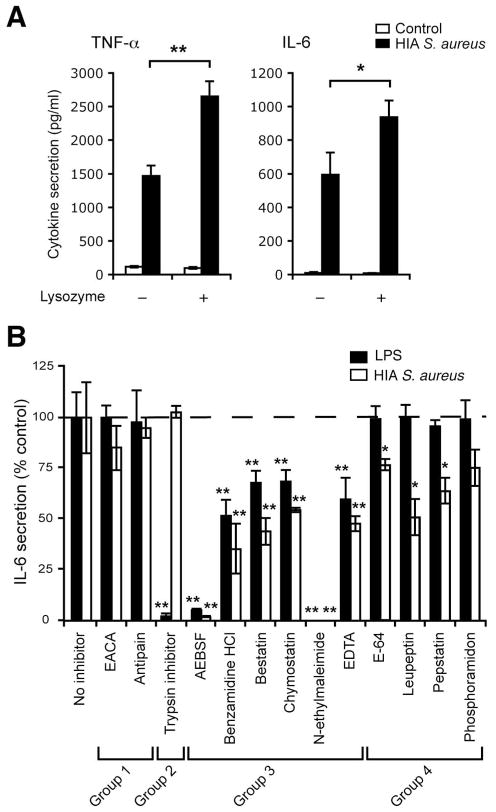
We next set out to identify host enzymes that might digest S. aureus and to test if they contributed to liberate bacterial ligands and increase bacterial sensing. Lysozyme, a prominent component of the mature phagolysosome of activated macrophages is a 1,4-N-acetylmuramidase, which catalyzes hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in peptidoglycan (25). It is likely that lysozyme digestion helps open the peptidoglycan back-bone and to release LTA and lipopeptides buried in this matrix. Consistent with this possibility, pre-loading macrophages with excess lysozyme increased the ability of S. aureus to induce proinflammatory cytokines, indicating that lysozyme-mediated digestion did facilitate sensing of S. aureus [Figure 6A]. However, lysozyme alone would not be predicted to be efficient for generation of a full repertoire of PRR ligands from S. aureus, especially as many strains are somewhat resistant to this enzyme (26). To identify other enzymes that might be involved in liberation of bacterial ligands we tested a collection of protease inhibitors for the ability to selectively block IL-6 response to S. aureus but not LPS. Response to S. aureus was inhibited by approximately 50% by leupeptin and pepstatin, and 25% by phosphoramidon and E-64 [Figure 6B], suggesting that serine, cysteine and acid proteases [Supplementary Table ST1] may also contribute in bacterial digestion and ligand release.
Figure 6. pH-regulated host enzymes digest bacteria in the phagosome and are required for maximal induction of cytokine response to S. aureus.
A, Lysozyme enhances the induction of cytokine response by S. aureus. Peritoneal macrophages preloaded with or without 15 μg/ml lysozyme for 15 min were incubated without (control) or with HIA S. aureus at MOI 50. Induction of cytokine responses at 2 h (TNF-α) or 4 h (IL-6) was measured by ELISA in culture supernatants. B, Effect of protease inhibitors on LPS or S. aureus induced IL-6 secretion. Peritoneal macrophages preloaded with the indicated inhibitor were stimulated with 50 ng/ml LPS or S. aureus at MOI of 50 for 4 h. IL-6 secretion was measured by ELISA of the culture supernatants. Data indicate mean ± SD of triplicates, and are shown as percentages of IL-6 secretion in (B) as compared with the response by control macrophages (no inhibitor). Group 1: no effect; Group 2: blocked LPS only; Group 3: blocked non-specifically (i.e. LPS and S. aureus); Group 4: blocked S. aureus only. Data are representative of three (A) or two (B) independent experiments. Statistical difference in the IL-6 response between no inhibitor and the indicated inhibitor treatments in (B) was indicated. *p ≤ 0.05; **p < 0.01.
Increasing digestion of S. aureus can rescue cytokine responses in the absence of phagosome acidification
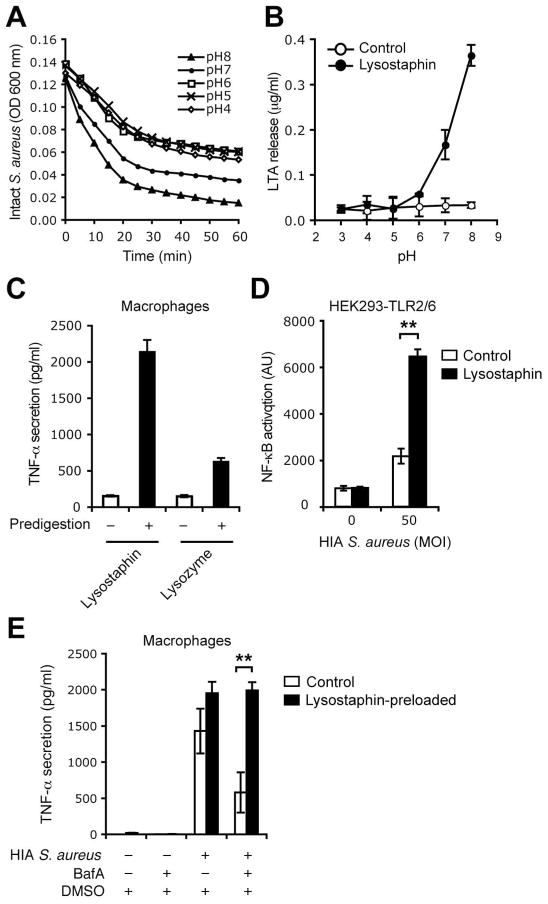
The above observations suggested that bacterial digestion might be a critical step required for response to S. aureus. We hypothesized that using an alternative pH-independent means of digesting the bacterial cell wall might increase the stimulatory capacity of S. aureus and also rescue the block seen after Bafilomycin A treatment. Lysostaphin, a bacteria-derived glycylglycine endopeptidase, which cleaves the pentaglycine cross bridges of peptidoglycan found in the bacterial cell wall (27), more efficiently digested S. aureus at neutral than at acidic pH, as measured by the kinetic loss of optical density (O.D.) [Figure 7A]. We therefore used lysostaphin to test our hypothesis that S. aureus digestion liberates cryptic TLR ligands. As determined by ELISA, addition of lysostaphin to bacteria caused pH-dependent liberation of the TLR2 ligand, LTA [Figure 7B]. Confirming that immunostimulatory ligands were released after bacterial digestion, filtered supernatants from lysostaphin pre-digestion of S. aureus were 5–10 fold more efficient at stimulating macrophage production of TNF-α than filtered supernatants from undigested S. aureus [Figure 7C]. When compared to lysozyme, lysostaphin digestion was 3–5 times more efficient at releasing ligands from S. aureus that stimulated macrophages. This was in part due to release of TLR2/6 ligands as S. aureus showed increased capacity to induce NFκB in HEK 293T cells transfected with TLR2/6 and an NFkB reporter construct in the presence of lysostaphin [Figure 7D]. As lysostaphin was optimally active at a neutral pH we hypothesized that it would efficiently digest bacteria and release PRR ligands even in Bafilomycin A treated cells, in which acid-activated endogenous phagolysosomal enzymes could not function. To test this, macrophages treated with or without Bafilomycin A were preloaded with Lysostaphin and stimulated with S. aureus. Consistent with our model Lysostaphin rescued the cytokine response in Bafilomycin A treated macrophages [Figure 7E]. These observations demonstrate that signaling can occur from a neutral phagosome once the pathogen has been digested and were consistent with our hypothesis that a critical role of phagosome acidification is to facilitate bacterial digestion and liberation of cryptic PRR ligands.
Figure 7. Lysostaphin-mediated digestion of S. aureus rescues cytokine responses in the absence of phagosome acidification.
A and B, Lysostaphin efficiently digests S. aureus at neutral pH. HIA S. aureus were incubated without (control) or with 5 μg/ml lysostaphin at the indicated pH. The digestion of the bacteria was assessed by measuring optical density (OD) of the bacterial suspensions at 600 nm over 60 min (A), and LTA release was measured by ELISA in the bacteria-free filtered supernatant at 2 h (B). C, Cryptic ligands released from lysostaphin- or lysozyme-treated S. aureus induce TNF-α response. HIA S. aureus were incubated in PBS (pH 7.5) without (−) or with (+) 5 μg/ml lysostaphin, or in potassium phosphate buffer (pH 6.0) without (−) or with (+) 5 μg/ml lysozyme, for 2 h. Soluble digests (filtered bacteria-free supernatants) were used to stimulate peritoneal macrophages and TNF-α secretion at 4 h was measured by ELISA of culture supernatant. D, Lysostaphin-mediated digestion of S. aureus increases immunostimulatory capacity of S. aureus by release of TLR2/6 ligands. HEK 293 cells stably expressing TLR2, co-transfected with NF-| B reporter system and TLR6, were incubated with heat-inactivated S. aureus in the absence (control) or presence of 15 μg/ml lysostaphin. Reporter gene activity at 4 h was measured by a luciferase assay. E, Preloading of lysostaphin in macrophages rescues cytokine response to S. aureus in the absence of phagosome acidification. Peritoneal macrophages pretreated with BafA were preloaded with 15 μg/ml lysostaphin for 15 min and incubated with HIA S. aureus. TNF-α secretion at 2 h was measured as in (C). Data are representative of three independent experiments.
DISCUSSION
Phagocytosis is an evolutionarily conserved and central component of host defense of many organisms (3). Critical to the process of bacterial killing is the maturation of phagosomes that fuse ultimately with lysosomal compartments that contain numerous lytic enzymes. Here we expand our understanding of the role of the phagolysosome by showing that it is required not only to destroy the internalized bacteria but also to liberate cryptic PRR ligands through digestion of the Gram-positive cell wall. Without phagocytosis and phagosome maturation the immuno-stimulatory ligands of S. aureus remain inaccessible and hence are unable to fully activate TLR-dependent responses. Consistent with recent work, our observations are also compatible with the proposal that phagosomes are a source of ligands for intracellular NLRs (28), which are also liberated during this “pathogen-processing”. Thus phagosome digestion of S. aureus regulates two important arms of the innate immune response: TLRs and NLRs.
The demonstrated relationship between the TLRs and the phagosome links these two arms of the innate immune system and confirms their critical interdependence for optimal host defense against certain pathogens. However, not all pathogens display a similar requirement for phagocytosis to trigger a proinflammatory response as we have demonstrated for S. aureus. Gram-negative microbes such as E. coli or S. montivideo do not require internalization presumably as LPS and the other immunostimulatory components of their bacterial cell wall are readily accessible and able to stimulate TLRs either at the cell surface, or within the vacuole without additional processing. Intriguingly, a different scenario exists for the yeast cell wall extract zymosan, in which inhibition of phagocytosis increases response (29, 30). Recent work has shown that Dectin-1, an important receptor that cooperates with TLR2 to mediate response to zymosan, rapidly dissociates from the phagosome suggesting that, in cases such as Dectin-1, internalization can also be a mechanism of receptor desensitization (31). This contrasts with the situation for S. aureus in which we have previously shown that two of the key TLR2 co-receptors, CD36 and MBL, need to be internalized along with the ligand to functionally cooperate with TLR2/6 in the phagosome (12, 17).
Our data indicate that phagosome acidification is an essential step for optimal MyD88-dependent responses to S. aureus and raise the question of how the rates of acidification of S. aureus-containing vacuoles are regulated. Although the subject of debate, TLR signals have been suggested to regulate phagosome maturation in an organelle autonomous manner (14). While we have not set out to test formally this possibility, our data do not support a model of TLR-dependent phagosome maturation for S. aureus. Notably, we find that phagocytosis and phagosome acidification must precede TLR signaling to S. aureus but not purified TLR ligands such as LTA or LPS. Instead our observations are more consistent with those of Yates and Russell, who failed to observe a role of TLR signaling in phagosome maturation (15). Nonetheless, our data indicating the critical role of phagosome acidification in regulating the host response to S. aureus emphasizes the importance of identifying mechanisms that regulate this process after microbial engulfment. Additionally, as perturbations in phagosome maturation underlie a number of clinical syndromes associated with increased susceptibility to S. aureus, such as chronic granulomatous disease (32) and cystic fibrosis (33), our observations suggest that augmenting functions of this organelle to increase the rate of acidification or microbial digestion may be important adjunct therapies in such conditions. Supporting this notion we show that Lysostaphin, a bacterial derived endopeptidase able to substitute for host enzymes and digest S. aureus at a neutral pH, increases host response to these bacteria and may therefore be a particularly useful adjunct antimicrobial in situations associated with impaired vacuolar function.
Supplementary Material
Acknowledgments
We would like to thank Alan Ezekowitz for his useful discussion.
References
- 1.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg S, Grinstein S. Phagocytosis and innate immunity. Curr Opin Immunol. 2002;14:1366–145. doi: 10.1016/s0952-7915(01)00309-0. [DOI] [PubMed] [Google Scholar]
- 3.Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity. 2005;22:539–550. doi: 10.1016/j.immuni.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. The phagosome proteome: insight into phagosome functions. J Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stuart LM, Boulais J, Charriere GM, Hennessy EJ, Brunet S, Jutras I, Goyette G, Rondeau C, Letarte S, Huang H, Ye P, Morales F, Kocks C, Bader JS, Desjardins M, Ezekowitz RA. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 7.Dermine JF, Duclos S, Garin J, St-Louis F, Rea S, Parton RG, Desjardins M. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J Biol Chem. 2001;276:18507–18512. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
- 8.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 9.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 10.Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 11.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ip WK, Takahashi K, Moore KJ, Stuart LM, Ezekowitz RA. Mannose-binding lectin enhances Toll-like receptors 2 and 6 signaling from the phagosome. J Exp Med. 20078;205:169–181. doi: 10.1084/jem.20071164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Underhill DM, Gantner B. Integration of Toll-like receptor and phagocytic signaling for tailored immunity. Microbes Infect. 2004;6:1368–1373. doi: 10.1016/j.micinf.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 15.Yates RM, Russell DG. Phagosome maturation proceeds independently of stimulation of toll-like receptors 2 and 4. Immunity. 2005;23:409–417. doi: 10.1016/j.immuni.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Stuart LM, Lucas M, Simpson C, Lamb J, Savill J, Lacy-Hulbert A. Inhibitory effects of apoptotic cell ingestion upon endotoxin-driven myeloid dendritic cell maturation. J Immunol. 2002;168:1627–1635. doi: 10.4049/jimmunol.168.4.1627. [DOI] [PubMed] [Google Scholar]
- 17.Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RA, Moore KJ. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol. 2005;170:477–485. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi L, Takahashi K, Dundee J, Shahroor-Karni S, Thiel S, Jensenius JC, Gad F, Hamblin MR, Sastry KN, Ezekowitz RA. Mannose-binding lectin-deficient mice are susceptible to infection with Staphylococcus aureus. J Exp Med. 2004;199:1379–1390. doi: 10.1084/jem.20032207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, Masselli A, Frisch SM, Hunton IC, Jiang Y, Wang JY. Blockade of tumor necrosis factor-induced Bid cleavage by caspase-resistant Rb. J Biol Chem. 2007;282:29401–29413. doi: 10.1074/jbc.M702261200. [DOI] [PubMed] [Google Scholar]
- 20.Myers MA, I, Mackay R, Rowley MJ, Zimmet PZ. Dietary microbial toxins and type 1 diabetes--a new meaning for seed and soil. Diabetologia. 2001;44:1199–1200. doi: 10.1007/s001250100617. [DOI] [PubMed] [Google Scholar]
- 21.Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, Lennon-Dumenil AM, Seabra MC, Raposo G, Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 22.Ahmad-Nejad P, Hacker H, Rutz M, Bauer S, Vabulas RM, Wagner H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur J Immunol. 2002;32:1958–1968. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Latz E, Visintin A, Espevik T, Golenbock DT. Mechanisms of TLR9 activation. J Endotoxin Res. 2004;10:406–412. doi: 10.1179/096805104225006525. [DOI] [PubMed] [Google Scholar]
- 24.Savina A, Amigorena S. Phagocytosis and antigen presentation in dendritic cells. Immunol Rev. 2007;219:143–156. doi: 10.1111/j.1600-065X.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 25.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 26.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 27.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinberg BE, Huynh KK, Grinstein S. Phagosomal acidification: measurement, manipulation and functional consequences. Biochem Soc Trans. 2007;35:1083–1087. doi: 10.1042/BST0351083. [DOI] [PubMed] [Google Scholar]
- 29.Gantner BN, Simmons RM, Underhill DM. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. Embo J. 2005;24:1277–1286. doi: 10.1038/sj.emboj.7600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, Reis e Sousa C. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Heinsbroek SE, Taylor PR, Martinez FO, Martinez-Pomares L, Brown GD, Gordon S. Stage-specific sampling by pattern recognition receptors during Candida albicans phagocytosis. PLoS Pathog. 2008;4:e1000218. doi: 10.1371/journal.ppat.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dri P, Presani G, Perticarari S, Alberi L, Prodan M, Decleva E. Measurement of phagosomal pH of normal and CGD-like human neutrophils by dual fluorescence flow cytometry. Cytometry. 2002;48:159–166. doi: 10.1002/cyto.10123. [DOI] [PubMed] [Google Scholar]
- 33.Di A, Brown ME, Deriy LV, Li C, Szeto FL, Chen Y, Huang P, Tong J, Naren AP, Bindokas CV, Palfrey HC, Nelson DJ. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat Cell Biol. 2006;8:933–944. doi: 10.1038/ncb1456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.