Abstract
Although epitope tagging has been widely used for analyzing protein function in many organisms, there are few genetic tools for epitope tagging in Tetrahymena. In this study, we describe several C-terminal epitope tagging modules that can be used to express tagged proteins in Tetrahymena cells by both plasmid- and PCR-based strategies.
Keywords: Tetrahymena, Epitope tagging
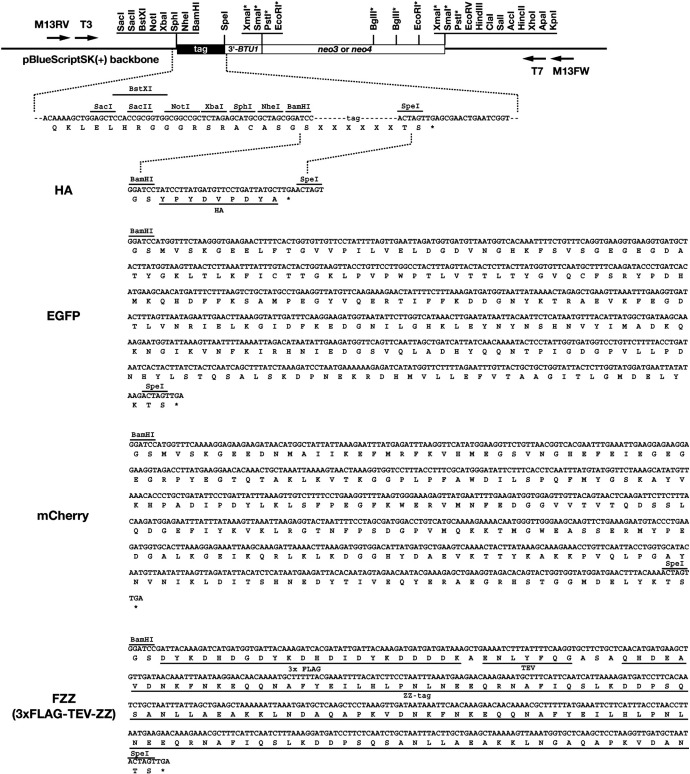
In the ciliated protozoan Tetrahymena thermophila, chromosomal integration of transgenes predominantly occurs by homologous recombination (Cassiby-Hanley et al., 1997). Therefore, epitope tags can be efficiently introduced into endogenous chromosomal loci in this organism. However, only a few tools for epitope tagging in Tetrahymena have been reported (Wloga et al., 2006, Yao et al., 2007) and these were designed and produced individually for each gene (for example, see the following: Bowman & Turkewitz, 2001, Dou et al., 2002, Aronica et al., 2008, Couvillion et al., 2009, Pearson et al. 2009). In this study, we generated several C-terminal epitope tagging modules for Tetrahymena. The general design of the modules is presented in Fig. 1. The epitope tags are followed by the TGA stop codon, the 3′-flanking sequence of beta-tubulin 1 (BTU1), which contains a 3′-UTR and a transcriptional terminator and a neomycin-resistance marker (neo) cassette. HA (a Hemagglutinin epitope), EGFP (Enhanced Green Fluorescent Protein), mCherry (a red fluorescent protein) and FZZ (3× FLAG followed by TEV protease cleavage site and ZZ domain of protein A) were selected as epitope tags for the modules. HA, EGFP and mCherry can be used for protein localization and for protein purification; FZZ can be used for tandem affinity protein purification (Lee and Collins, 2007). All of these tags were synthesized for optimal codon usage in Tetrahymena. New modules can be created by replacing tags in the existing module with any other epitope tags via the BamHI and SpeI sites. Either neo3 (Shang et al., 2002) or neo4 (Mochizuki, 2008) were used as drug resistance markers for the modules. Both markers confer paromomycin resistance to cells in the presence of cadmium ions. All modules are cloned into pBlueScript SK(+) vector (Stratagene). The nucleotide sequences of pHA-neo3, pHA-neo4, pEGFP-neo4, pmCherry-neo4, pFZZ-neo3, and pFZZ-neo4 can be found in DDBJ/EMBL/GenBank AB570107–AB570112AB570107AB570108AB570109AB570110AB570111AB570112.
Fig. 1.
C-terminal epitope tagging modules for Tetrahymena. HA, EGFP, mCherry, and FZZ (3× FLAG followed by TEV protease cleavage site and ZZ domain of protein A) coding-sequences, optimized for Tetrahymena codon usage, were synthesized and inserted into both the BamHI and SpeI sites of the common C-terminal tag module schematically drawn on top. Non-unique restriction enzyme sites are marked with asterisks. DNA sequences of individual modules can be found in DDBJ/EMBL/GenBank AB570107–AB570112AB570107AB570108AB570109AB570110AB570111AB570112.
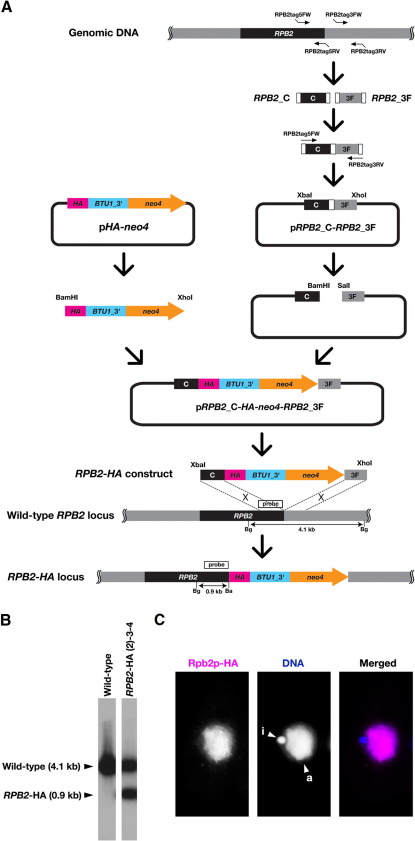
These modules were designed to be used for both plasmid- and PCR-based production of C-terminal tagging constructs. To confirm this, we first tested a plasmid-based method to produce C-terminal HA-tagging constructs for the RPB2 gene using pHA-neo4. The procedures we followed are presented in Fig. 2A; a list of the primers used in this study can be found in Supplemental information. We first amplified ~ 0.6 kb sequences from the C-terminus of the coding region of RPB2 (RPB2_C) with the primers RPB2tag5FW and RPB2tag5RV. We then amplified ~ 1.0 kb sequences from the 3′ flanking region of RPB2 (RPB2_3F) with the primers RPB2tag3FW and RPB2tag3RV. The 3′-terminus of RPB2tag5RV and the 5′-terminus of RPB2tag3FW had complementary sequences. Using these sequences, RPB2_C and RPB2_3F were combined by overlapping PCR with RPB2tag5FW and RPB2tag3RV. All of the PCR reactions described above were performed with PrimeStar HS DNA Polymerase (Takara). The PCR product was cloned into the XbaI and XhoI sites of pBlueScript SK(+). The overlapping PCR produced BamHI and SalI sites between RPB2_C and RPB2_3F. After digestion by both BamHI and XhoI, the HA-neo4 module was inserted into these sites. The resulting construct (RPB2_C-HA-neo4-RPB2_3F) was excised, using both XbaI and XhoI, and introduced into Tetrahymena cells by homologous recombination. Transfection was performed with a biolistic gun system (Cassiby-Hanley et al., 1997). Successful homologous recombination into the endogenous RPB2 locus was confirmed by Southern blot (Fig. 2B). Expression of Rpb2p-HA was analyzed by immunofluorescent staining (Loidl and Scherthan, 2004) using an anti-HA antibody (clone 16B12, Covance). RPB2 encodes the second largest subunit of RNA polymerase II, and therefore, its product is expected to localize to the transcriptionally-active macronucleus. We found that HA was localized to the macronucleus (Fig. 2C), indicating that the tagging of HA to Rpb2p was successful.
Fig. 2.
Plasmid-based C-terminal epitope tagging. (A) Production scheme for C-terminal HA tagging construct for the RPB2 gene. See text for details. (B) Southern blot analysis of an RPB2-HA strain of Tetrahymena. Genomic DNA from both the wild-type B2086 and RPB2-HA (2)-3-4 strains were digested with BamHI (Ba in panel A) and BglII (Bg panel A). The southern blot was prepared using the DNA from these strains and was probed with the DNA fragment shown in panel A. The bands corresponding to both the wild-type RPB2 and the RPB2-HA are marked by arrowheads. (C) Rpb2p-HA localization in a vegetative Tetrahymena cell. Rpb2p was localized using an anti-HA antibody (magenta). DNA was counter-stained with DAPI (blue). The macro- and micronuclei are marked with “a” and “i”, respectively.
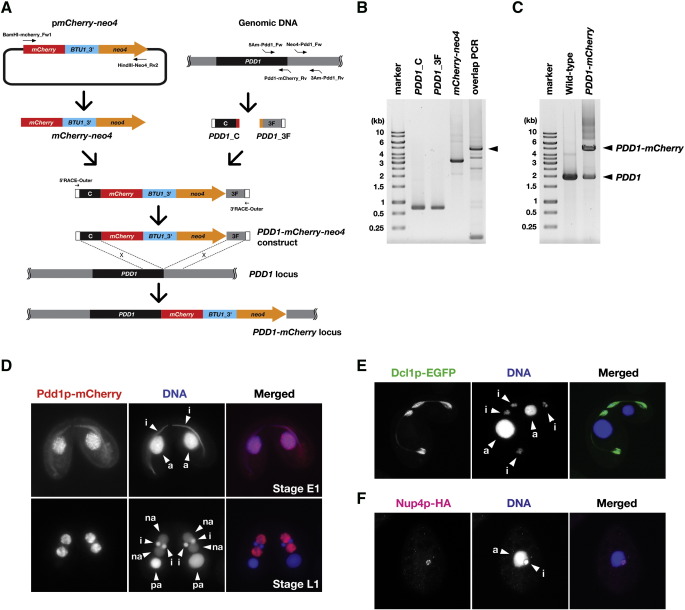
Next, we tested a PCR-based method to produce C-terminal tagging constructs for the PDD1 gene using pmCherry-neo4. The procedures are schematically presented in Fig. 3A and a detailed protocol can be found in Supplemental information. We first amplified ~ 0.8 kb sequences from both the coding (PDD1_C) and the 3′ flanking (PDD1_3F) regions of the PDD1 genomic locus (Fig. 3B). In parallel, the mCherry-neo4 module was amplified from plasmid DNA by PCR (Fig. 3B). We designed overlapping sequences between the 3′-terminus of PDD1_C and the 5′-terminus of mCherry-neo4, and between 3′-terminus of mCherry-neo4 and 5′-terminus of PDD1_3F. The three PCR products were combined by overlapping PCR (Fig. 3B). All of these PCR reactions were performed with PrimeStar HS DNA Polymerase (Takara). Typically, 50 μl of a PCR reaction provides ~ 0.5 μg of construct DNA, which is enough for several transformation experiments. The resulting construct (PDD1_C-mCherry-neo4-PDD1_3F) was introduced into Tetrahymena cells as described above. The successful integration of the construct into the endogenous PDD1 locus by homologous recombination was confirmed by PCR (Fig. 3C). Expression of Pdd1p-mCherry was analyzed by fluorescent microscopy (Fig. 3D). mCherry fluorescence was localized to both the micronucleus and the macronucleus at early conjugation (the sexual reproduction process of Tetrahymena) stages (Fig. 3D top) and to the new macronucleus at late stages (Fig. 3D bottom). These localization patterns are indistinguishable from those of endogenous Pdd1p (Coyne et al., 1999). Therefore, we concluded that the strategy presented here can successfully introduce mCherry tags to Pdd1p. Similar PCR-based methods were successful for tagging DCL1 with EGFP using the EGFP-neo4 module (Fig. 3E) and for tagging NUP4 (MicNUP98A) with HA using the HA-neo4 module (Fig. 3F). Dcl1p-EGFP was localized to the meiotic micronuclei in mating cells (Fig. 3E) and Nup4p-HA was localized to the micronucleus of vegetative cell (Fig. 3F). These patterns are indistinguishable to the previously reported localizations of Dcl1p-HA (Mochizuki and Gorovsky 2005) and GFP-Nup4p (Malone et al. 2008, Iwamoto 2009), respectively. These data suggest that a PCR-based method for the C-terminal tagging of genes with the modules established in this study can be used to quickly generate transformation constructs for epitope tagging experiments in Tetrahymena.
Fig. 3.
PCR-based C-terminal epitope tagging. (A) Production scheme for C-terminal mCherry tagging construct for PDD1 gene. See text for details. (B) Production of C-terminal mCherry tagging construct for PDD1 gene. The PCR products of PDD1_C, PDD1_F, mCherry-neo4 and PDD1-mCherry-neo4 (overlapping PCR) were analyzed. The position of PDD1-mCherry-neo4 construct is marked with an arrowhead. (C) PCR analysis of a PDD1-mCherry strain. Genomic DNA from both the wild-type B2086 and the PDD1-mCherry strains were amplified by PCR using the primers 5Am-Pdd1_Fw and 3Am-Pdd1_Rv. (D) Pdd1p-mCherry localization in Tetrahymena. The PDD1-mCherry strain shown in (C) was mated to a wild-type strain. Localization of Pdd1p-mCherry at conjugation stages E1 (top) and L1 (bottom) was observed using fluorescent microscopy (red). For reference, see Aronica et al. (2008) for conjugation stages. DNA was counterstained by DAPI (blue). a: macronucleus, i: micronucleus, na: new macronucleus, pa: parental macronucleus. (E) Dcl1p-EGFP localization in Tetrahymena. A DCL1-EGFP strain was mated to a wild-type strain and localization of Dcl1p-EGFP at conjugation stage E2 was observed under fluorescent microscope (green). DNA was counterstained by DAPI (blue). a: macronucleus, i: micronucleus. (F) Nup4p-HA localization in a vegetative Tetrahymena cell. Nup4p-HA was localized using an anti-HA antibody (magenta). DNA was counter-stained by DAPI (blue). The macronuclei and micronuclei are marked by “a” and “i”, respectively.
Acknowledgments
This research was funded by the Naito Foundation to KK, the European Research Council (ERC) Starting Grant (204986) under the European Community's Seventh Framework Programme and by the Austrian Academy of Sciences to KM. UES was supported by Doktoratskolleg RNA Biology funded by the Austrian Science Fund (FWF).
Footnotes
Supplementary data associated with this article can be found, in the online version, at 10.1016/j.mimet.2010.07.009.
Appendix A. Supplementary data
References
- Aronica L., Bednenko J., Noto T., Desouza L.V., Siu K.W., Loidl J., Pearlman R.E., Gorovsky M.A., Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes & Development. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman G.R., Turkewitz A.P. Analysis of a mutant exhibiting conditional sorting to dense core secretory granules in Tetrahymena thermophila. Genetics. 2001;159:1605–1616. doi: 10.1093/genetics/159.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D., Bowen J., Lee J.H., Cole E., VerPlank L.A., Gaertig J., Gorovsky M.A., Bruns P.J. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion M.T., Lee S.R., Hogstad B., Malone C.D., Tonkin L.A., Sachidanandam R., Hannon G.J., Collins K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes & Development. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne R.S., Nikiforov M.A., Smothers J.F., Allis C.D., Yao M.C. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Molecular Cell. 1999;4:865–872. doi: 10.1016/s1097-2765(00)80396-2. [DOI] [PubMed] [Google Scholar]
- Dou Y., Bowen J., Liu Y., Gorovsky M.A. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. The Journal of Cell Biology. 2002;158:1161–1170. doi: 10.1083/jcb.200202131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M., Mori C., Kojidani T., Bunai F., Hori T., Fukagawa T., Hiraoka Y., Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate Tetrahymena. Curr Biol. 2009;19:843–847. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]
- Lee S.R., Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nature Structural & Molecular Biology. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- Loidl J., Scherthan H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. Journal of Cell Science. 2004;117:5791–5801. doi: 10.1242/jcs.01504. [DOI] [PubMed] [Google Scholar]
- Malone C.D., Falkowska K.A., Li A.Y., Galanti S.E., Kanuru R.C., Lamont E.G., Mazzarella K.C., Micev A.J., Osman M.M., Piotrowski N.K. Nucleus-specific importin alphas and nucleoporins regulate protein import and nuclear division in the bi-nucleate Tetrahymena thermophila. Eukaryotic Cell. 2008;7:1487–1499. doi: 10.1128/EC.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene. 2008;425:79–83. doi: 10.1016/j.gene.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes & Development. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Osborn D.P., Giddings T.H., Jr., Beales P.L., Winey M. Basal body stability and ciliogenesis requires the conserved component Poc1. The Journal of Cell Biology. 2009;187:905–920. doi: 10.1083/jcb.200908019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Song X., Bowen J., Corstanje R., Gao Y., Gaertig J., Gorovsky M.A. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D., Camba A., Rogowski K., Manning G., Jerka-Dziadosz M., Gaertig J. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Molecular Biology of the Cell. 2006;17:2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.C., Yao C.H., Halasz L.M., Fuller P., Rexer C.H., Wang S.H., Jain R., Coyne R.S., Chalker D.L. Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. Journal of Cell Science. 2007;120:1978–1989. doi: 10.1242/jcs.006502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.