Abstract
Embryonic stem cells (ESCs) are capable of undergoing self-renewal, and their developmental ability is known as the stemness. Recently, microRNAs (miRNAs) as regulators have been isolated from ESCs. Although Dicer and DiGeorge syndrome critical region gene 8 (DGCR8) are essential factors for the biogeneration of miRNA, Dicer-knockout (KO) ESCs have showed to fail to express differentiation markers and DGCR8-KO ESCs have showed to be arrest in the G1 phase. Furthermore, Dicer-KO ESCs lost the ability to epigenetically silence retroelemtns (REs). REs are expressed and transposed in ESCs, whose transcripts control expression of miRNAs, and their transposable retroelement (TE) expression is, therefore related to ESC proliferation and differentiation, suggesting that the interplay between miRNAs and REs may have a deep responsibility for the stemness including a short G1/S transition and for RE regulation in ESCs.
Keywords: ES cell, HIV-1, microRNA, retrotransposon, RNA wave.
INTRODUCTION
Today we know that the human genome encodes non-coding RNA (ncRNA) from its non-coding DNA (ncDNA) sections. It is known that ncDNA is predominantly composed of multiple segments of transposable retroelements (TEs) such as, long interspersed elements (LINEs) and, short interspersed elements (SINEs, human Alu), retrotransposons, DNA transposons and satellites (Fig. 1A) [1]. The ncRNAs contain ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), short interfering RNA (siRNA), microRNA (miRNA), other classes of small RNAs and longer RNA intermediates including complex patterns of interacting and overlapping sense and antisense transcripts from both coding and non-coding regions of genomic DNA [2, 3]. The small RNAs are functional and their sizes fall within a range of 19 to 31 nucleotides (nts). The most common small RNAs are miRNAs, which consist of genomic and resident miRNA genes, which are subsequently produced by digestions on two kinds of the nuclease III, Drosha and Dicer, and then directly target messenger RNAs (mRNAs) or possibly epigenetic machinery in human cells [4].
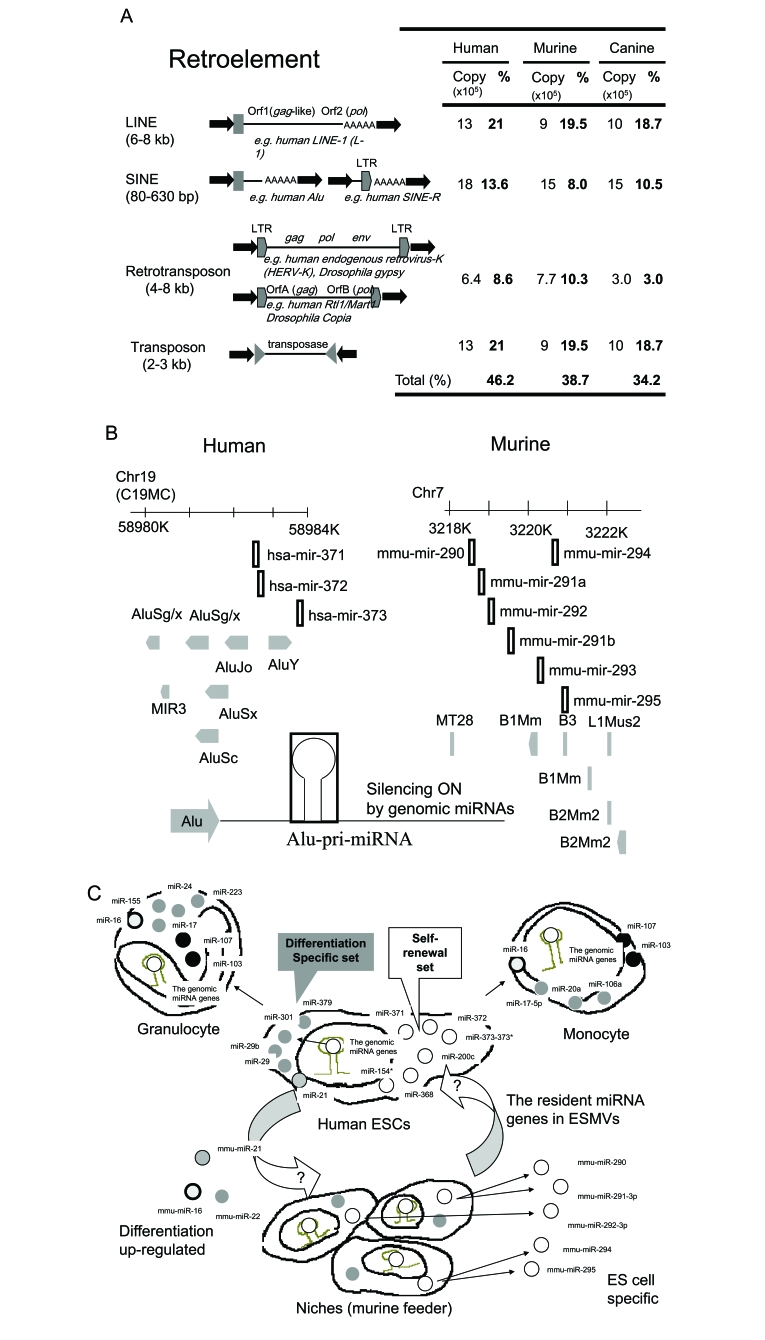
Fig. (1).
A model of TE-controlled miRNA expression mechanisms. (A) Transposable elements (TEs) in the genome. The architecture of the genome for TEs was represented. (B) Alu-genomic miRNA switching. Clusters of miRNAs in human chromosome 19 (Chr19) and murine chromosome 7 are represented. These genomic miRNA genes are closely or overlapping localized in TEs, such as Alu and B1. The direction of TEs may not matter. TE could promote the expression of pri-miRNA (Alu-pri-miRNA) and genomic miRNAs from the pri-miRNA can be silence on translation of target mRNAs in ESCs. (C) ESC specific resident miRNAs for self-renewal and differentiation. Human ESCs express self-renewal specific set of miRNAs. The cluster of miR-371, 372 and 373 is related to the self-renewal of ESCs. The cluster of miR-290 in murine is also specific for ESC self-renewal and the murine miR-290 cluster genes are transferred from ESCs to neighboring cells by ESC microvesicles (ESMVs). For differentiation of ESCs, other resident miRNAs are presented and then the profile of miRNAs in differentiated cells, such as granulocytes and monocytes is greatly different from that of ESCs.
Embryonic stem cells (ESCs) including somatic and germline stem cells (GSCs), employ their capacity for unlimited self-renewal and differentiation for tissue development and homeostasis. In mammalian embryo, the first stage of this differentiation involves differentiation into either the cells which form the inner cell mass and those which form the trophoblast [5-7]. The cells of the inner cell mass, which are known as ESCs, are pluripotent, meaning that they can give rise to all the cells of the animal. This is quality known as stemness and this stemness retained by the ESCs themselves. Thus, the stem cells can produce differentiated daughter cells, as well as replicate themselves. The key transcriptional regulators Oct4, Sox2 and Nanog, etc. have been identified as the self-renewal and differentiation-related factors in ESCs [8-12]. Artificial pluripotent stem (iPS) cells demonstrate the roles played by these transcriptional regulator genes [13], but these cells developed into tumors (due to the operation of myc oncogene and oncoviral vectors). Recent cancer research indicates that alteration of miRNA profiles, such as those involved in oncogenic miRNA (oncomirs) upregulation or tumour suppressor miRNA downregulation could induce tumors even without an oncovirus or oncogenes [14]. This suggests that tumourigenesis of iPS cells might be due to dysregulation of miRNAs induced by transfection of transcriptional factors rather than transduction of the oncogene myc or oncovirus vectors themselves [15-17] as evidenced by tumourigenesis in iPS cells without oncovirus vectors and myc oncogenes [18]. Since miRNA genes are mobile and functional genetic elements [19], the miRNA profile of ESCs is likely to be different in depending on the niche cells in which they are located when it is self-renewal or differentiation conditions. The character of ESCs may be very different from that of cancer such as embryonic carcinoma and carcinoma-like cells including iPS cells [20]. Based on research with skin cell iPS cells and ESCs, it has become clear that miRNAs play an important role in the precise mechanisms regulating self-renewal and differentiation in these cells [21].
It appears that miRNAs may be the master regulators of stemness. Further, it also appears that tumourigenesis may be rooted in something other than the transcriptional and cellular signaling factors of oncoproteins. The ESC transcripts, including those of miRNAs, contain many retrovirus-like retrotransposon-derived repeats (about 50%) [22, 23]. De novo LINE-1 (L1) retrotransposition events occurred in the human brain [24]. Further, L1 retrotransposition events have been shown to occur at early stages in human embryogenesis [25] and L1 transcripts are related to the mobilization of human SINE Alu [26]. The miRNA genes are located within Alu and L1, and about 20% of known human miRNAs are derived from these repetitive-element sequences [27]. It has been reported that over 1,000 genes of the human genome may be controlled by an Alu-associated promoter and that a dual relationship exists between the miRNA gene clusters and the miRNA targets in Alu regions that may have evolved together [28, 29]. Furthermore, epigenetic chromatin remodeling of Alu sequences plays an important role in the regulation of miRNA expression [30]. Taken together, these findings suggest that miRNAs may be the master regulator of ESC self-renewal and differentiation and that the miRNA genes may control both abilities through expression of TEs. Therefore, it appears that the activity of Alu and L1 is more complex than previously documented in artificially-induced transcriptional factors because ESCs specific transcriptional factors do not control the epigenetic state except for that of polycomb-group proteins [31].
The seed sequences of 2- to 7 or 8-nts in the 5’-end of miRNAs recognize not only the 3’ untranslated regions (3’UTRs) of its mRNAs but also the coding regions of Oct4, Sox2 and Nanog [32] through an imperfect match to repress target mRNA and probable stability of the mRNA target. Computer analysis has shown that miRNAs target the 3’UTR as well as the 5’UTR and protein coding regions (CDS) [33]. Further, experimental evidence indicates that the miRNA genes could target the CDS and/or 3’UTR of OCT4, Sox2 and Nanog [34] and could regulate all multi-gene expression. The miRNA-induced silencing complex (miRISC) appears to regulate this process. The centre of miRISC is an Argonaute protein (Ago) and the seed region of miRNAs in the miRISC targets the sequences of mRNA with incomplete paring. However, a miRNA gene superposes to multiple-targets and a mRNA is targeted by multiple-miRNAs. Therefore, elucidation about how the targets of miRNA selected and how the miRNA genes switched on and off requires additional insights from theoretical ciphering of RNA codes even if suppose a quantum computing model independent of the nucleic acid pairing [19].
In this paper, we explore the significant expression of ESC miRNAs induced by TEs and we propose a model involving TEs forming ‘RNA wave’ in ESCs. To further understand an importance role of fine-tuning by the miRNA genes in ESC epigenetic silencing, we also explore an extended model of quantum theory for RNA coding operating without the operon involvement.
ALU TEs AS A SWITCH FOR STEMNESS
TEs in the human genome have been shown to occupy ~46% of its genomic sequences (Fig. 1A) [1]. Since TEs transcribe non-coding RNA and this RNA is reverse-transcribed and re-integrated into its own genome, if miRNA is encoded in TEs, the miRNA genes in TEs could also be duplicated according to the same process as TEs. It has been reported that human miRNAs may be derived from retroelements (REs) [29, 35, 36]. It has been suggested that around half of all human genomic information is derived from RNA. Further, human MADE1, miniature inverted-repeat TE (MITE) involves total 550 paralogs upon miR-548a-1 (Chromosome 6: Chr6), miR-548a-2 (Chr6), miR-548a-3 (Chr8), miR-548b (Chr6), miR-548c (Chr12), miR-548d-1 (Chr8) and miR-548d-2 (Chr17) as numbers of 24, 81, 82, 23, 124, 71 and 145 paralogs in individual genome of humans, respectively. MADE1 emerged along the primate evolutionary lineage and the miR-548 gene is an orthologs in the human, chimpanzee and rhesus macaque genomes [35]. Further, miR-548 targets MITEs of the human genome. In turn, MITEs including MADE1 are widely distributed among eukaryotes and miRNA and endogenous siRNA genes may exist due to the palindrome-like RNA structures form a hairpin, which can be catalyzed by Dicer. Thus, a self-regulation system may be involved in MADE1 and the miRNA genes. This system may be necessary for the maintenance of the small RNA-derived human genome. Subsequently, transposition of TEs seems to be deleterious to the genome. However, the genome itself may be constructed of and evolved by small RNAs in TEs. Previously, we proposed “RNA wave model,” which holds that retroviruses control their own amplification rate in order to minimize their deleterious effects on their hosts, as exemplified by the latent state of human immunodeficiency virus type 1 (HIV-1) [37]. According to RNA wave, RNA tags can transport miRNA genes, which are a mobile genetic element that allows cross-talk among cells, organs and individuals from vertical to horizontal. Further, it has been reported that human TE Alu evolved symbiotically with miRNAs [29, 38].
The relationship between Alu retroelements, one of the SINE TEs, and a miRNA cluster on chromosome 19 (C19MC) in humans is compatible with RNA wave [28, 39]. As mentioned above, members of the C19MC-gene family, the miR-515-1, miR-517a and miR-519a-1 genes, are transcribed by Pol III from the upstream section of Alu TE as miRNA promoters, suggesting that some interplay between miRNAs and TEs may act to the circuit on and off. The human miRNA gene cluster, including SINEs, miR-371, miR-372 and miR-373 in the down stream of C19MC, and the murine miR-290, miR-291a, miR-291b, miR-292, miR-293, miR-294 and miR-295 cluster are similar to ESCs in their self-renewal capability (Fig. 1B, C). Furthermore, murine miR-290 cluster genes are transferred from ESCs to neighbouring cells by microvesicles (Fig. 1C) [40, 41]. Recently, synthetic miRNAs, viral miRNAs and endogenous miRNAs have been reported to be transferred between cells and to be able to spread their functionally silencing signals across cell boundaries [42, 43]. In addition, the vault particles implicated in intracellular transport contain small RNAs and these small RNAs may be related with the multidrug resistance mechanisms [44]. Thus, the direction of self-renewal or differentiation in ESCs and their neighbors may be commonly mediated through the information stored in the retroposable miRNA genes. Additionally, TEs is not a tag in this case, TE itself may be a RNA gene.
Alu transposition is required for LINE TEs but that is not dependent on LINE-encoded protein [26]. Two miRNAs, miR-95 and miR-151 in LINE2 target its LINE2 sequences [45]. The transcripts including miRNAs in ESCs contains many TE-derived repeats (~50%) [22, 23]. LINE1 retrotransposition events occur at early stages in human embryogenesis and human ESCs in vitro [24, 25]. Thus, the miRNA genes in TEs are completely disregarded in ESC investigations for human embryogenesis; this is a mistake at all and TEs may be a switch of self-renewal and differentiation of ESCs via miRNAs. Therefore, nobody has asked what is the code of RNA intermediates, such as miRNA, to elucidate the switch operation for ESC self-renewal and differentiation much shorter nanoRNA and TEs, furthermore intron and protein coding RNAs.
TRANSFORMATION AND REPROGRAMMING IN STEMNESS
Over 10 years ago, embryonic clone cells were produced from the nuclei of cultured cells [46] and then the first mammalian clone was produced [47]. DOLLY, the cloned ovine, proved that genome modification has been considered as irreversible event, but the genome can be reversible to able to reprogram by factors in the oocytes to make them pluripotent once again. The question is what is the factor(s). Recent research indicates that it may be miRNA genes [48, 49]. The initial profile of cloned bovine embryos was changed after somatic cell nuclear transfer as the reprogramming [49]. Further, the nuclear transfer with minimal contamination by the donor cytoplasm was accomplished by metaphase II G1-arrested G0 quiescent state cells [47]. This technological finding as reprogramming has forced us to accept that two factors may be in play here: resident miRNA genes and genomic miRNA genes.
‘Reprogramming’ was also used to create artificially induced adult somatic iPS cells. Recent papers by Werning et al. [50], Hanna et al. [51] or Takahashi et al. [52] gave the results that iPS cells from skin fibroblasts were isolated upon transformation with a combination of the transcription factor genes, Nanog, Sox2, Oct4 and/or Myc. These iPS cells were problematic in that Myc or vector retroviruses induced tumour formation, and iPS cells can essentially allow for isografting only, while ESCs can be considered for use of variable allografting [51-54]. Together with above nuclear transfer, experiments with iPS have caused biologist to completely reassess their thinking regarding the role of transcriptional factors, miRNA genes and other small nucleic acids (miRNA assessment).
It was originally thought that tumourigenesis might be avoided in iPS cells because it was believed that tumourigenesis was mediated by the oncogene Myc and other oncovirus vectors. However, it is now clear that it is not oncoviruses but oncomirs that can induce tumours and that oncogenes are controlled by the oncomirs or tumour suppressor miRNAs [17]. Further, iPS and ESCs up-regulate the expression of the miR-302 and 17-92 cluster, which is the oncomir in leukemia and lung cancer, and iPS cells show lower miR-371-cluster expression than ESCs, in which cluster expression is involved in self-renewal [55]. MiR-222 promotes continuous proliferation of human carcinoma cells [56-58]. In contrast, miR-222 is involved in helping human endometrial stromal cells to exit the cell cycle and enter the differentiation [59]. Therefore, at least miR-222 and likely other miRNA sets play an important role in reprogramming cells. Although the miRNA profiles represent clear difference from iPS cells to ESCs, the ability to reprogram cells throws the existing understanding of stemness into disarray.
Since transcriptional and post-transcriptional regulatory network could be determined in silico [60, 61], miR-145 controls Nanog, Sox2 and Oct4 in ESCs as well as in tumourigenesis [62, 63]. Murine miR-134, miR-296 and miR-470 target the protein-coding sequence of Nanog, Oct4 and Sox2 in various combinations, leading to transcriptional and morphological changes in differentiating murine ESCs [32, 64]. Although it has been shown that Nanog, Sox2 and Oct4 transcriptional regulators collaborate to form regulatory circuitry consisting of autoregulatory and feed-forward loops, which contribute to pluripotency and self-renewal of human ESCs [65], the miRNA findings discussed above suggest that reprogramming of differentiated adult cells by transduction may be a “phantom of the operon.” Later to further distinguish between transformation and reprogramming, the DGCR6 and Dicer gene were knocked out in ESCs.
DGCR8 OR DICER-DEFICIENT ESCs
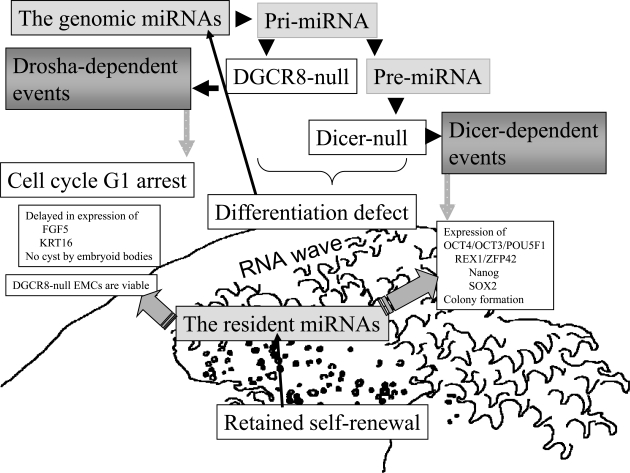
DCGR8 cooperates with the RNase III enzyme Drosha in the biogenesis of pre-miRNA from pri-miRNA transcribed from non-coding regions of the human genome. As shown in Fig. (2), ESCs in which the DCGR8 gene had been knockout (KO) are arrested in the G1 phase of the cell cycle, but these DGCR8-null ESCs remain alive [66]. It has been suggested that resident miRNAs are present independently in the cytoplasm, and these miRNAs may be able to self-proliferate the division of ESCs. Expression of markers of primitive ectodermal-FGF5, and epithelial-KRT18 was delayed in embryoid bodies (EBs), and DGCR8-KO EBs did not form a cyst. Therefore, DGCR8-null ESCs have a defect in their differentiation ability. The pluripotent markers of Oct4, Rex1, Sox2 and Nanog were not expressed [66]. It has been suggested that the DGCR8 gene is essential for the self-renewal silencing that normally occurs with the induction of differentiation. Surprisingly, it has been reported that by aberration of another RNase III enzyme Dicer ESCs are viable and retain their colony-forming ability and the ability to express Bmp4, HINF4A, GATA1, Brachyury and HPRT ESC-specific genes [67]. The Dicer-null ESCs show a prolonged G1 phase of the cell cycle and differentiation defects [68]. These results strongly support two elements of the RNA wave model: 1) the presence of resident miRNAs that might play an important role the self-renewal function of ESCs, and 2) the existence of genomic miRNAs, which are necessary for ESC differentiation (Fig. 2).
Fig. (2).
DGCR8- or Dicer-null ESCs. DGCR8-null ESCs lack differentiation ability, and, in turn DGCR8 is essential for the silencing of self-renewal. Dicer-null ESCs also lack differentiation ability. But both null cells remain alive and pluripotent markers still continue to be expressed. In Fig. (1B), there is self-renewal set of human miRNAs (miR-371 cluster in C19MC), suggesting that the resident miRNAs are present independently and the resident miRNAs might play important role in the ESCs’ self-renewal program. These data are based on the criteria of the RNA wave.
By the high through-put analysis of the miRNA, ESC-specific miRNAs have been cloned and isolated. When human ESCs-specific miRNAs grouped by ‘AAAGUGCU’ seed sequence were shown, miR-17-5p, miR-18a, miR-18b, miR-20a, miR-20b, miR-93, miR-100a, miR-106b, miR-302a, miR-302b, miR-302c, miR-302d, miR-373, miR-519b, miR-519c, miR-519d, miR-520b and miR-520e including the same seed region were upregulated. These prominent upregulated miRNAs were clustered in human Chr 4, 13, 19 and X [69]. Comparison between human and mouse ESCs revealed that human miR-302b, 302c and miR-302d on Chr 4 are related to miR-302 on murine Chr 3. Another cluster of miRNAs, miR-371, miR-372, miR-373*, and miR-373 on human Chr 19 is the murine homologues of miR-290, miR-291a, miR-291b, miR-292, miR-293, miR-295 on Chr 7 [70, 71]. These miRNAs belong to a miRNA gene family common to human and murine (miRBase 13.0; see Fig. 1A). The miR-302 and miR-371 genes are regulated by KLF4, OCT4 and SOX2 transcriptional factors and miR-145 suppresses these three transcriptional factors in human ESCs [63, 70]. In the case of murine, miR-470 and miR-134 target the coding region of Oct4 and Sox2 mRNAs, respectively. Further, miR-470, miR-296 and miR-134 target to 3’UTR of Nanog mRNA [64]. According to miRBase, Nanog and SOX2 are targeted by 12 and 26 kinds of multi-miRNAs in human ESCs, and 17 and 26 kinds of multi-miRNAs in mouse, respectively (miRBase 13.0). In these miRNAs, miR-21 of both human and murine has binding sites in the 3’UTR of the mRNA that codes for SOX2, and probably OCT4 [72]. Like polycomb [73], the RE1-silencing transcriptional factor (REST) interacts with cis-elements in promoter of the miR-21 gene, REST represses expression of miR-21. Not only REST, but also the ESC-related transcriptional factors OCT4/SOX2/Nanog/TCF3 bind to the promoter of ESC-specific miRNAs [74]. For example, expression of pri-let-7g is dependent on OCT4 and let-7g is abundant in ESCs [75]. Although Lin28 inhibits expression of let-7g, ectopical expression of Lin28 promotes the reprogramming of human somatic cells into iPS cells [76] and is associated with human multiple cancer types [77]. Further, miR-302 reprogrammed human skin cancer cells into iPS cells [78]. Although quite recently it has been reported that down-regulation of the p53 guardian by siRNAs of p53 can increase the emergence rate of iPS cells produced by skin cells [79-83], we well know that the dominantly inherited Li-Fraumeni multi-carcinogenesis syndrome (MIM151623) as a case of mutations of p53 in human [84, 85]. Since the p53 was regulated by miR-34a and miR-125b [86, 87], these data suggest that production and proliferation of iPS cells may closely be controlled by miRNAs, and that ESCs are maintained not just by ESC-specific transcriptional factors but also by ESC-specific miRNAs as multiple factors. However, it is still not clear how the miRNA genes switch stemness on or off for a change and tuning of cell phenotypes.
One possible answer is an epigenetic regulation mechanism such as that involving miR-290. MiR-290-295 cluster in murine and are indirectly involved in epigenetic gene regulation. This cluster repressed DNA methyltransferase (DNMT), retinoblastoma like-2 (RBL2) and CpG methylation of the promoter by suppression of the DNMT regulated expression of OCT4 [88, 89]. Further, Lin28 regulates the production of the histone, H2a [63], suggesting that miRNAs may be indirectly involved in epigenetic gene regulation in ESCs. But there is a further question: why was only Lin28 finally suppressed via regulation of histone?
THE KEY FOR STEMNESS MAY BE FOUND IN THE LATENCY OF HIV-1
DNA and histone methylation is one of the epigenetic modifications of chromatin, which may be related to ESC programming [88, 89]. In ESCs, programming of cells results in loss of competency for the abbreviated G1 phase, such as independence of the release of RB protein from E2F transcriptional factor [90]. Since complete silencing of the typical cell cycle gene expression is necessary for the programming of ESCs, it is of great interest that miRNAs could be related to the epigenome. The heterochromatin domains are highly methylated and are also show histone modifications characteristic of silenced chromatin, such as trimethylated histone H3 lysine 9 (H3K9me+) [91]. Further, Dicer is essential for the formation of heterochromatin [92], suggesting that miRNAs control the methylation mechanisms of chromatin [93, 94]. In order to change from self-renewal to differentiation, self-renewal set of miRNAs derived from Alu-miRNAs must be switched off and differentiation set of miRNAs from Alu-miRNAs should be switched on. Recently, human miRNA-offset RNAs (moRNA) have been discovered [95]. Although chordate moRNAs are a relative small fraction of the miRNA family, moRNAs are produced from pri-miRNAs probably in Alu or L1 and could target to miRNAs and mRNAs, suggesting that in the case of human, TEs may play an important role in the epigenetic silencing of miRNA and/or mRNA expression. As in the above-described MADE1, human siRNA candidates from MADE1 have been revealed by computer analysis. Therefore, these data suggest that human moRNAs or siRNAs in TEs sought to be related to epigenetic silencing. Thus, the resident miRNAs, moRNAs, siRNAs and unusually small RNAs (usRNAs), which is much smaller than canonical miRNA [96] and the so-called nanoRNAs (nnRNAs) source from TEs are speculated as precursors of transcripts from retrovirus in ncDNA regions (Fig. 3A), suggesting that the function of HIV-1 proviruses may be similar to that of TEs in epigenetic regulation.
The typical model of complete and epigenetic silencing in TEs is like to be the latency of HIV-1 infection [97]. The integrated provirus corresponding to MADE1 could produce miRNAs from the 3’UTR (Fig. 3B). The targeting the 5’UTR by miRNAs can lead to histone 3 protein H3K9 methylation (H3K9me+) (Fig. 3A) and/or DNA methylation. Subsequently, epigenetic LTR-miRNA (Alu-miRNA) switch-off is occurred following histone deacetylation by histone deacetylase (HDAC) and the profile of the resident miRNAs changes in the cells. This is how the latent state is induced and the latent state is in control of the TEs. In the case of ESCs, programming should be initiated from the TE silencing like the latency.
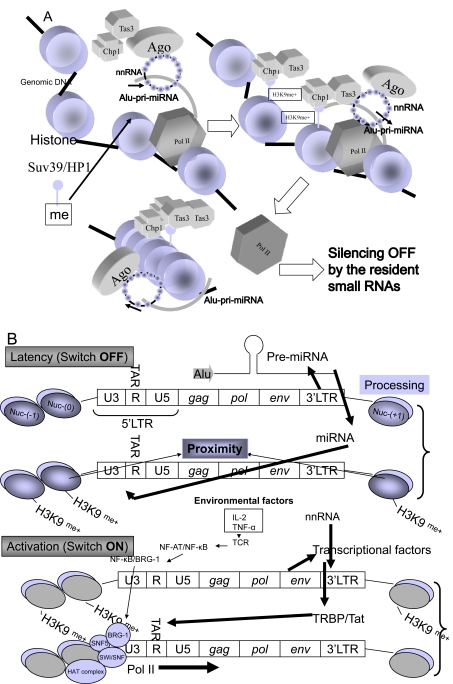
Fig. (3).
A model of epigetic on/off switching mechanism of transposable miRNAs. (A) Epigenetic silencing off by the resident small RNAs is shown. The function of nnRNA is similar to that of siRNA. Alu-pri-miRNAs as the TE could be targeted by nnRNA during transcription epigenetically. This process induces complete silencing off, that starts translation of mRNAs for differentiation of ESCs. The Ago/nnRNA complex circulized as a Buddhist rosary (‘Jyuzu’ in Japanease) and the ring of nnRNA may epigentically lock Alu-pri-miRNA transcription by methylation of histone by Clr4-like human methyltransferase (HMT) enzyme, Suv39 associated with Swi6/HP1 and chromo domain-containing protein 1 (Chp1) plus RNA-induced transcriptional silencing complex protein 3 (Tas3) binding to H3K9me+ histone following CpG methylation by DNA methyltransferase (DNMT) [111]. (B) The HIV-1 latency model. The Weinberg and Morris model [97] is shown with minor modifications. The HIV-1 3’LTR U3 region contains at least three genomic miRNA genes: MIR#4, MIRN367 and MIRH1. The 5’LTR/gag/pol/env region may be corresponding to Alu TE. These genomic miRNAs yeild silencing of HIV-1 proviral expression (Switch OFF). Environmental factors, such as the resident miRNA in ESMV, IL-2, X-ray, dietary food etc. affect the profile of miRNAs in cells, nnRNA from digestion of the resident miRNAs, retroposable miRNAs or mobile miRNAs completely stopped expression of Alu-pri-miRNA described in (A). This is the mechanism of latency. Dominant transcriptional factor induces HIV-1 mRNA transcription (Switch ON). In this state, iPS cells may be produced; however, since there is no step of complete silencing on, iPS cells may occasionally change into tumour cells. Therefore, the differentiation set of the resident miRNAs should be completely switched off to reprogram the somatic cell.
On the other hand, started up reprogramming in iPS cells, it is resembles the reactivation state of HIV-1. Activation of LTR-like Alu promoter and/or environmental stimulation such as that with IL-2 and TNF-α induce transcriptional factor HIV-1 Tat corresponding to OCT4, SOX2 and Nanog in iPS cells (Fig. 3B). High concentration of transcriptional factor in the cells inhibits the activity of normal expression of genomic miRNA genes via suppression of Dicer activities, like Tat as an activator of RAS [37]. The H3K9me+ is reversed [98] and H3K4 plus H3K36me+ as well as CpG DNA methylation induce silencing of the LTR-miRNA (Alu-miRNA) (Fig. 3A). Therefore, transduction of OCT4, SOX2 and Nanog might induce an aberrant TE-expression state in differentiated skin cells and iPS cells may be occasionally turned into the tumourigenic cells by an aberrant miRNA gene-expression in TEs because Dicer loss promotes tumourigenesis [99]. To maintain the self-renewal state, the Alu-miR371 family is needed to switch on of self-renewal and to control TEs. In transcriptional factors dominant state, the Alu-miR371 cannot work at all. Consequently, it is clear that reprogramming could never be achieved only by transformation such as that in iPS cells because even ESCs decrease their pluripotency by epigenetic alteration of the expression of TEs in murine cells [100]. The human genome in ESCs always represents latent states between miRNAs and TEs. In turn, ESC genome robustness may be based on a circuit of fragile miRNAs in TEs, in turn based upon the capacious memory of retrotransposons, which may be built on the RNA wave including the niches’ mobile miRNA information.
MOBILE miRNA CODE
Observations of the expression of tens of thousands of miRNAs-TEs suggest that the classical promoter-transcription-protein view is contradicted because the miRNAs in TEs identified have been studied in relative depth and the functions have gradually been investigated in genomic in vivo and in silico studies. In light of the low number of validated miRNAs, bioinformatics may play a key role albeit in small ncRNAs [101], not obeying simple Watson-Crick DNA base pairing. A subset of the Watson-Crick model in the seed sequences is currently used to explain the incomplete target pairing of miRNA. A rather broader set of additional rules may be useful to explain how some small RNAs to develop bioinformatics approaches. The seed sequences of miRNAs target the 3’UTR as well as the 5’UTR and protein coding regions (CDS) based on computer analysis [98]. The miRNA genes could target CDS and/or 3’UTR of OCT4, Sox2 and Nanog [34]. Further, a miRNA, such as miR-24 regulates expression of E2F2 by recognizing out of the seed rule [102]. The resident miR-24 gene with miR-155 and miR-17 is specifically involved in the differentiation of granulocytic, monocytic megakaryocytic, and pro-B lymphoid as somatic stem cells [103]. In zebrafish, miR-214 without canonical seed pairing can effectively target a mRNA for silencing [104]. These data suggest that tuning by miRNAs is probably a high-speed event. Therefore, except for complete and partial seed base-pairing, we should add a neo-mechanism for the regulation by miRNA.
Recently, Fabian et al. [105] have reported that miRNA-mediated translational repression precedes target mRNA deadenylation. However, their model still depends on the seed sequences of miRNA-the target ones of mRNA. Further, there is no specificity of miRISC to bind to a specific miRNA. This is different from aminoacyl-tRNA synthetase, which recognizes a specific amino acid and the tRNAs corresponding to that amino acid. This yields the hypothesis that circular shaped mRNA produced by the cap-binding factor eIF4E plus eIF4G and poly(A)-binding protein PABP could make miRISC into a linear strand of mRNA. Since it is well known that episomal HIV-1 DNA forms the ring, if miRNA is also circular and miRISC including helicase activities links a miRNA ring, as shown in Fig. (4A), the miRNA rings may enter into the mRNA strand, resulting in probable inhibition of mRNA circularization [106].
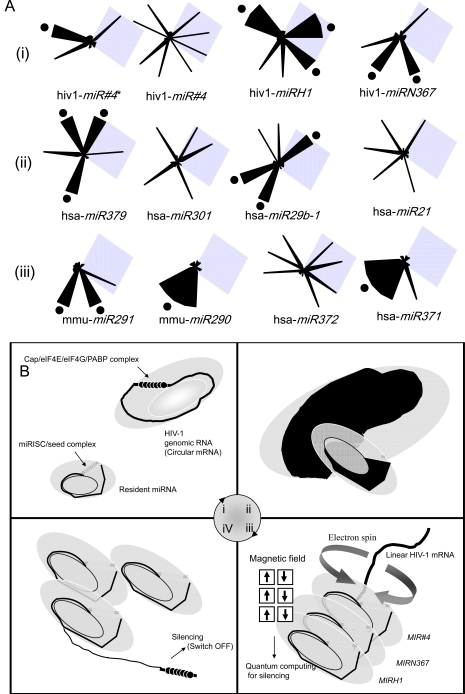
Fig. (4).
A model of tanslational tuning by virally mobile miRNAs. (A) G-G repeats in the miRNAs. The viral miRNAs of HIV-1 (i), self-renewal-related miRNAs (ii) and differentiation-related miRNAs (iii) contained GG repeats (black dots). The calculated FMO values were represented as a radar-circle, which could be represented as wings by Excel software (Microsoft corp.). Shaded diamonds are represented as the region of the seed. The FMO values of each atom of bases were calculated by winMOPAC software version 3 (SCIGRESS MO Compact Professional: Fujitsu Corp. Tokyo, Japan) and each panel was illustrated by EXCEL (Microsoft Corp. USA). (B) Quantum theory of the RNA wave in translational tuning. The model of Fabian et al. in the Sonenberg laboratory (2009) is modified and the modified model is represented. (i) Circularized viral HIV-1 RNA (mRNA) via eIF4G-PAGP interaction and viral miRNAs, such as MIR#4, MIRN367 and MIRH1 are circularized via Ago2 and GW182. (ii) miRNA targets mRNA. Black shaded regions represented appropriate mage of unbalanced electron distribution. (iii) Circularized mRNA by cap-binding factor eIF4E plus eIF4G and poly(A)-binding protein PABP could make miRISC into linear strand of mRNA. miRISC is the link of the miRNA ring and circularized miRNAs are superposed. Electrons on the virally circular miRNAs would move from one base to another neighboring base, whereas electrons may spin round. Depended on the direction of the rotating electron, the direction of the magnetic field would go up or down. The superposing of the magnetic direction of electrons according to quantum theory may be the code of RNA as in quantum computing (Fujii, 2008). (iv) The mobile miRNAs can tune silencing ON (Switch ON) and repeat to (i) ~ (iv) in very short time (see Movie S1).
Quite recently, it has been reported that HIV-1 replication and reactivation in patients with chronic HIV-1 infection are inhibited by shRNA (shN366) including HIV-1 miRNA MIRN367 [107]. This report suggests that TE-miRNA autoregulating system is involved in the latency model in humans. Therefore, for example of the miRNA gene ring, we represent here the miRNA rings in HIV-1 viral RNA [37], additionally in differentiation and in self-renewal specific sets into ESC (Fig. 4A). In Fig. (4A), according to electrostatic potential of each atom of the RNA bases with methyl residue instead of ribose 5-phosphate by the fragment molecular orbit methods (FMO) based on quantum theory [108], at first total electrostatic potential of A (approximate - 0.05 milli-Hartree), U (- 0.03), G (0.258) and C (- 0,002) base were calculated. Although in the original code, hypothesized long ago, the archaic code (G, C) would expand toward the complete variants (C, G, A, U) in the early evolution of mRNA [109, 110], only G had a positive potential. The calculated potencies were represented as a radar shape (Fig. 4A). We focused on the neighbouring bases and checked G multiplication in sequences of each miRNA (see black dots in Fig. 4A). In the seed sequences (see shaded diamonds in Fig. 4A), there was no specific relation between the G multiplication and miRNA specific sets in ESCs. But except for MIRH1 and MIR29b-1, black dots were not involved in the seed sequences of other miRNAs. Further, we found unbalanced and balanced electron potencies of the G base cluster in some miRNAs. In such miRNAs, electrons in the circular miRNAs move from one base to a neighboring base according to electrostatic potential of each atom of the RNA bases with FMO, whereas electrons may spin round. Depended on the direction of rotating electron, the direction of the magnetic field would go up or down. The superposing of the magnetic direction of electrons according to quantum theory may be the code of RNA (Fig. 4B), like quantum computing [19]. The pulse waves of the electrons in the organ hydrogen (H) atoms of water in magnetic resonance image (MRI), could, in part, be derived from a source of the magnetic waves produced by miRNA superposing for silencing gene expression, such as aberrant presence of miRNAs in tumor [17]. By measure miRNA electron spin, it may be available for investigation of the relationship between stemness and physicochemical characters of tuning by miRNAs and furthermore therapy of RNA diseases.
CONCLUSIONS
The RNA wave consists of four alternative concepts for crosstalk among cells: 1) mobile miRNAs induce transcriptional and post-transcriptional silencing processes; 2) the induced silencing information expands to intra-cellular, inter-cellular and intra-organ, inter-organ communication by free mobile miRNAs; 3) the mobile miRNAs can self-proliferate; and 4) cells contain both resident and genomic miRNAs. Thus, the non-selfish miRNAs are regarded as the basic unit of the evolution to move horizontally in genomic space. The miRNAs would horizontally transpose in TEs by feeding of miRNAs-containing foods, viruses, etc. The retroposable miRNAs might create phenotype by changing and tuning of the gene expression in the genomic space, in which its genotype can be vertically inherited under pressures of the environment corresponding to Darwininsm.
To clarify the capacity of ESCs to renew themselves and to differentiate into various cell lineages, miRNAs and TEs as RNA genes have an important role. The highlight of the importance of miRNAs in TEs has been shown in controlling ESC growth and differentiation by miRNAs. Although the significance of miRNA function has not yet been evaluated, retroposable miRNAs in TEs are expressed in ESCs, suggesting that TEs may switch on or off the alteration of ESCs from self-renewal, differentiation and reprogramming including the epigenetic regulation. This paper will aid in our understanding of the relationship between oncoviral miRNA genes and TEs. Understanding the seed theory for miRNA targets and further breaking the RNA code under the RNA wave will shed light on the quality of stemness in ESCs.
SUPPLEMENTARY MATERIAL
This article also contains supplementary material (video animation) and it can be viewed online along with the article at publisher’s website.
Table 1.
The General Abbreviations and Glossary for This Paper
| Abbreviation | Technical Terms | Explanation |
|---|---|---|
| ESC | Embryonic stem cell | Undifferentiated, pluripotent cells from a mammalian embryo. A key cell of retroelements and microRNAs interplay in this paper. |
| miRNA (miR) | MicroRNA | Mobile and small genetic RNA elements. Small RNA elements are encoded from non-coding DNA region including retroelements as genes. Exosomes and viruses contain microRNAs therefore that can be mobile and control transcription and translation in host. |
| RE or TE | Retroelement or transposable retroelement | Mobile genetic RNA element. RNA elements are reversetranscribed by reverse transcriptase that makes a cDNA copies. The DNA copies integrated into prototype genome by integrase that are composed of about half of human genome. The DNA elements in genome contain pol gene that encodes reverse transcriptase and integrase and can therefore be copied by its own. |
| (-) | Retrovirus | An RNA virus. The genetic composition and copying process of viruses are the same as that of retroelements. HIV-1 is a typical retrovirus. A particle of virus contains about 10 kilobases of two single-stranded (+) RNA. |
| Chr | Chromosome | The coiled coil DNA and histone proteins that is believed to be as the basic conformation. The number of chromosome in a human somatic cell is twice 22 plus X and X (female) or X and Y (male). The DNA of all chromosomes is composed of a genome. |
| ncRNA | Non-coding RNA | About 98% DNA of 3.2 gigabases of genome is protein non-coding region that therefore has been believed to be nonfunctional region as junk. The region encodes non-coding RNA containing retroelement and microRNA. |
| DNMT | DNA methyltransferase | The sequence 5'CG3' CpG dinucleotides within genomic DNA are target of a DNA methyltransferase. DNA methylation means conversion of cytosine into 5-methyl-cytosine. |
| CDS | Protein coding regions | 1-1.5% of DNA sequences in genome encode the amino acid sequences of a polypeptide via messenger RNA intermediates. The nucleotide triplet specifies an amino acid or translation stop signal or/and translation start signal. |
ACKNOWLEDGEMENT
I thank Dr. C. Rowthorn for his knowledgeable English editing.
ABBREVIATIONS
- Ago
= Argonaute protein
- CDS
= Protein coding regions
- Chr
= Chromosome
- C19MC
= A miRNA cluster on chromosome 19
- DNMT
= DNA methyltransferase
- EB
= Embryonic body
- ESC
= Embryonic stem cell
- FMO
= Fragment molecular orbit method
- GSC
= Germline stem cell
- HDAC
= Histone deacetylase
- iPS
= Artificial pluripotent stem
- KO
= Knockout
- L1
= LINE-1
- LINE
= Long interspersed element
- miRISC
= miRNA-induced silencing complex
- miRNA (miR)
= MicroRNA
- mRNA
= Messenger RNA
- MITE
= Miniture inverted-repeat
- moRNA
= miRNA-offset RNA
- ncDNA
= Non-coding DNA
- ncRNA
= Non-coding RNA
- nnRNA
= nanoRNA
- nts
= nucleotides
- oncomir
= Oncogenic miRNA
- RBL2
= Retinoblastoma like-2
- RE
= Retroelement
- REST
= RE1-silencing transcriptional factor
- rRNA
= Ribosomal RNA
- SINE
= Short interspersed element
- siRNA
= Short interfering RNA
- snRNA
= Small nuclear RNA
- snoRNA
= Small nucleolar RNA
- TEs
= Transposable retroelements
- tRNA
= Transfer RNA
- usRNA
= Unusually small RNA
REFERENCES
- 1.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin LV. Non-coding RNA. Hum Mol Gene. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Valeri N, Vannini I, Fanini F, et al. Epigenetics, miRNAs, and human cancer: a new chapter in human gene regulation. Mamm Genome. 2009;20:573–80. doi: 10.1007/s00335-009-9206-5. [DOI] [PubMed] [Google Scholar]
- 5.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 6.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–8. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith A. Embryonic stem cells. In: Marshak DR, Gardner RL, Gottleib D, editors. Stem Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 205–230. [Google Scholar]
- 8.Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor OCT-4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 9.Pesce M, Gross MK, Scholer HR. In line with our ancestors: Oct-4 and the mammalian germ. BioEssays. 1998;20:722–32. doi: 10.1002/(SICI)1521-1878(199809)20:9<722::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiaton, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 11.Avilion AA, Nichols SK, Pevny LH, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Gene Dev. 2003;17:126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers I, Colby D, Robertson M, et al. Functional expression coning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 13.Holden C, Vogel G. A seismic shift for stem cell research. Science. 2008;319:560–3. doi: 10.1126/science.319.5863.560. [DOI] [PubMed] [Google Scholar]
- 14.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 15.Kota J, Chivukula RR, O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi JJ. New hope for a microRNA therapy for liver cancer. Cell. 2009;137:990–2. doi: 10.1016/j.cell.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 17.Fujii YR. Oncoviruses and pathogenic microRNAs in humans. Open Virol J. 2009;3:37–51. doi: 10.2174/1874357900903010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. PvtI-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujii YR. Formulation of new algrithmics for miRNAs. Open Virol J. 2008;2:37–43. doi: 10.2174/1874357900802010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurent LC, Chen J, Ulitsky I, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 21.Yi R, Pasolli HA, Landthaler M, et al. DGCR8-dependent microRNA biognenesis is essential for skin development. Proc Natl Acad Sci USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 23.Kikuchi K, Fukuda M, Ito T, et al. Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation. Nucleic Acids Res. 2009;15:4987–5000. doi: 10.1093/nar/gkp426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coufal NG, Garcia-Perez JL, Peng GE, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Perez JL, Marchetto MCN, Muotri AR, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–77. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 26.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–8. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 27.Borchert GM, Lanier W, Davidson B. RNA polymerase III transcribes human microRNAs. Nature. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 28.Oei S-L, Babich VS, Kazakov VI, et al. Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters. Genomics. 2004;83:873–82. doi: 10.1016/j.ygeno.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Lehnert S, Van Loo P, Thilakarathne PJ, et al. Evidence for co-evolution between human microRNAs and Alu-Rpeats. PloS ONE [serial on the Internet] 2009;4:e4456. doi: 10.1371/journal.pone.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito Y, Suzuki H, Tsugawa H, et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–44. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 31.Gieni RS, Hendzel MJ. Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochem Cell Biol. 2009;87:711–46. doi: 10.1139/O09-057. [DOI] [PubMed] [Google Scholar]
- 32.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–30. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Duan X, Qian J, Li F. Aboundant conserved microRNA target sites in the 5'-untranslated region and coding sequence. Genetica. 2009;137:159–64. doi: 10.1007/s10709-009-9378-7. [DOI] [PubMed] [Google Scholar]
- 34.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MixcroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 35.Piriyapongsa J, Marino-Ramirez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–37. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang R, Wang Y–Q, Su B. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol. 2008;25:1493–502. doi: 10.1093/molbev/msn094. [DOI] [PubMed] [Google Scholar]
- 37.Fujii YR, Saksena NK. Viral infection-related microRNAs in viral and host genomic evolution. In: Morris KV, editor. RNA and the regulation of gene expression. London: Horizon Scientific Press; 2008. pp. 91–107. [Google Scholar]
- 38.Cao H, Yang C–S, Rana TM. Evolutionary emergence of microRNAs in human embryonic stem cells. PloS ONE. 2008;3:e2820. doi: 10.1371/journal.pone.0002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;12:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 40.Luo S–S, Ishibashi O, Ishikawa G, et al. Human villous trophoblasts express and secerete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–29. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 41.Yuan A, Farber EL, Rapoport AL, et al. Transfer of microRNAs by embryonic stem cell microvesicles. PloS ONE. 2009;4:e4722. doi: 10.1371/journal.pone.0004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rechavi O, Erlich Y, Amram H, et al. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev. 2009;23:1971–9. doi: 10.1101/gad.1789609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwakiri D, Zhou L, Samanta M, et al. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from toll-like receptor 3. J Exp Med. 2009;206:2091–9. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persson H, Kvist A, Vallon-Christersson J, et al. The non-coding RNA of the multidrug resistance-linked vault particle encodes multidrug regulatory small RNA. Nat Cell Biol. 2009;11:1268–71. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 45.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–6. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Campbell KHS, McWhir J, Richie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–7. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 47.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Cmapbell KHS. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 48.Cui X–S, Zhang D–, Ko Y–G, Kim N–H. Aberrant epigenetic reprogramming of imprinted microRNA-127 and Rtl1 in cloned mouse embryos. Biochem Biophys Res Commun. 2009;379:390–4. doi: 10.1016/j.bbrc.2008.12.148. [DOI] [PubMed] [Google Scholar]
- 49.Castro FO, Sharbati S, Rodriqez-Alvarez LL, et al. MicroRNA expression profiling of elongated cloned and in vitro fertilized bovine embryos. Theriogen. 2009;73:71–85. doi: 10.1016/j.theriogenology.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Werning M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 51.Hanna J, Werning M, Markoulaki S, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–3. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 53.Lengner CJ, Camargo FD, Hochedinger K, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–15. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaenisch R, Young R. Stem cell, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–82. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson KD, Venkatasubrahmanyam S, Jia F, et al. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cell. 2009;18:749–58. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Sage C, Nagel R, Egan DA, et al. Regulation of the CDKN1B/p27 (Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Felicetti F, Errico MC, Bottero L, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–54. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 58.Medina R, Zaidi SK, Liu CG, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–80. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian K, Hu L, Chen H, et al. Hsa-miR-222 is involved in differentiation of endometrial stromal cells in vitro. Endocrinology. 2009;150:4734–43. doi: 10.1210/en.2008-1629. [DOI] [PubMed] [Google Scholar]
- 60.Cui Q, Yu Z, Pan Y, Purisima EO, Wang E. MicroRNAs preferentially target the genes with high transcriptional regulation complexity. Biochem Biophys Res Commun. 2007;352:733–8. doi: 10.1016/j.bbrc.2006.11.080. [DOI] [PubMed] [Google Scholar]
- 61.Re A, Corá D, Taverna D, Caselle M. Genome-wide survey of microRNA-transcription factor feed-forward regulatory circuits in human. Mol BioSyst. 2009;5:854–67. doi: 10.1039/b900177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chivukula RR, Mendell JT. Abate and switch: miR-145 in stem cell differentiation. Cell. 2009;137:606–8. doi: 10.1016/j.cell.2009.04.059. [DOI] [PubMed] [Google Scholar]
- 63.Xu B, Huang Y. Histone H2a mRNA interact with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res. 2009;37:4256–63. doi: 10.1093/nar/gkp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tay TM–S, Tam W–L, Ang Y–S, et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- 65.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2006;122:1–10. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nature. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanellopoulou C, Muljo S, Kung AL, et al. Diceer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Gene Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic cells. Proc Natl Acad Sci USA. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laurent LC, Chen JC, Ulitsky I, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cell. 2008;26:1506–16. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 70.Suh M–R, Lee Y, Kim S–K, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 71.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNA. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 72.Singh SK, Kagalwala MN, Parker-Thomburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–7. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schuettengruber B, Ganapathi M, Leblanc B, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2008;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Viswanatham SR, Daley GQ, Gregory RI. Selective blockade of micreoRNA processing by Lin-28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 77.Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin S–L, Chang DC, Chang-Lin S, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:1–10. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hong H, Takahashi K, Ichisaka T, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–5. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Utikal J, Polo JM, Stadfeld M, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–8. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marión RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–53. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, Collado M, Villasante A, et al. The Link4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–9. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawamura T, Suzuki J, Wang YV, et al. Linking the p53 tumor suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–4. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olivier M, Goldgar DE, Sodha N, et al. Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res. 2003;63:6643–50. [PubMed] [Google Scholar]
- 85.Wu C–C, Shete S, Amos CI, Strong LC. Joint effects of germ-line p53 mutation and sex on cancer risk in Li-Fraumeni syndrom. Cancer Res. 2006;66:8287–92. doi: 10.1158/0008-5472.CAN-05-4247. [DOI] [PubMed] [Google Scholar]
- 86.Tarascov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing. Cell Cycle. 2007;13:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 87.Le MTN, The C, Shyh-Chang N, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–76. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benetti R, Gonzalo S, Jaco I, et al. A mammalian microRNA cluster controls DNA methylation and teromere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 89.Sinkkonen L, Hugenschmidt T, Beminger P, et al. MicroRNA control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;5:259–67. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 90.Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Human embryonic stem cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase upon lineage programming. J Cell Phys. 2009;222:103–10. doi: 10.1002/jcp.21925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 92.Fukagawa T, Nogami M, Yoshikawa M, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–91. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 93.Valeri N, Vannini I, Famini F, et al. Epigenetics, miRNAs, and human cancer: a new chapter in human gene regulation. Mamm Genome. 2009;20:573–80. doi: 10.1007/s00335-009-9206-5. [DOI] [PubMed] [Google Scholar]
- 94.Howell Jr PM, Liu S, Ren S, et al. Epigenetics in human melanoma. Cancer Cont. 2009;16:200–18. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- 95.Langenberger D, Bermudez-Santana C, Hertel J, et al. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics. 2009;25:2298–301. doi: 10.1093/bioinformatics/btp419. [DOI] [PubMed] [Google Scholar]
- 96.Li Z, Kim SW, Lin Y, et al. Characterization of viral and human RNAs smaller than canonical microRNAs. J Virol. 2009;83:12751–8. doi: 10.1128/JVI.01325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weinberg MS, Morris KV. Are viral-encoded microRNAs mediating latent HIV-1 infection? DNA Cell Biol. 2006;25:223–31. doi: 10.1089/dna.2006.25.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou M, Deng L, Lacoste V, et al. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol. 2004;78:13522–33. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lambertz I, Nittner D, Mestdagh P, et al. Monoallelic but not biallelic loss of Dicer promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–41. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramírez MA, Pericuesta E, Fernandez-Gonzalez R, et al. Transcriptional and post-transcriptional regulation of retrotransposons IAP and MuERV-L affect pluripotency of mice ES. Repr Biol End. 2006;4:55. doi: 10.1186/1477-7827-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alexiou P, Maragkakis M, Papadopopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–55. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 102.Lai A, Navarro F, Maher CA, et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3'UTR microRNA recognition elements. Mol Cell. 2009;35:610–25. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Georgantas RW 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–5. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li N, Flynt AS, Kim HR, Solnica-Krezel L, Patton JG. Dispatched Homolog 2 is targeted by miR-214 through a combination of three weak microRNA recognition sites. Nucleic Acids Res. 2008;36:4277–85. doi: 10.1093/nar/gkn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fabian MR, Mathonnet G, Sundermeier T, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Cell. 2009;135:868–80. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 107.Yamamoto T, Samri A, Marcelin A, et al. Effect of lentivirus encoding HIV-1 Nef-U3 shRNA on the function of HIV-specific memory CD4+ T cells in patients with chronic HIV-1 infection. AIDS. 2009;23:2265–75. doi: 10.1097/QAD.0b013e328332817c. [DOI] [PubMed] [Google Scholar]
- 108.When PM, Harrington PE, Eksterowicz JE. Facile synthesis of substituted 5-amino- and 3-amino-1,2,4-thiadiazoles from a common precursor. Org Lett. 2009;11:5666–9. doi: 10.1021/ol902371y. [DOI] [PubMed] [Google Scholar]
- 109.Hartman H. Speculations on the Evolution of the Genetic Code. Orig Life. 1975;6:423–7. doi: 10.1007/BF01130344. [DOI] [PubMed] [Google Scholar]
- 110.Rodin S, Ohno S. Four primordial modes of tRNA-synthetase recognition, determined by the (G, C) operational code. Proc Natl Acad Sci USA. 1997;94:5187–8. doi: 10.1073/pnas.94.10.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Debeauchamp JL, Moses A, Noffsinger VJP, et al. Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenace of centromeric heterochromatin. Mol Cell Biol. 2008;28:2154–66. doi: 10.1128/MCB.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This article also contains supplementary material (video animation) and it can be viewed online along with the article at publisher’s website.