Abstract
PRKAR1A encodes the regulatory subunit type 1-alpha (RIα), of the main mediator of the cAMP effects in the eukaryotic cells – cAMP dependant Protein Kinase A (PKA). Inactivating PRKAR1A mutations are known to be responsible for the multiple neoplasia and lentiginosis syndrome Carney complex (CNC). To date, at least 117 pathogenic variants in PRKAR1A have been identified. The majority of them are subject to non-sense mediated mRNA decay (NMD), leading to PRKAR1A haploinsufficiency and, as a result, activated cAMP signaling in the affected tissues. In recent years it became apparent that CNC may be caused not only by RIα haploinsufficiency, but also by the expression of altered PRKAR1A protein, as proven by analysis of the relatively small proportion of expressed mutations in the gene, consisting of aminoacid substitutions and in-frame genetic alterations. In addition, a new subgroup of mutations that potentially escape NMD and result in CNC through altered rather than missing protein has been analyzed – frame-shifts in the 3′end of the coding sequence that shift the stop codon downstream of the regular one and result in the expression of longer than the wild type protein. The PRKAR1A mutation detection rate in CNC patients is recently estimated at above 60%; PRKAR1A mutation negative CNC patients are characterized by significant phenotypic heterogeneity. In this paper, we present a comprehensive analysis of all 117 known to date pathogenic PRKAR1A sequence variations, and discuss their molecular context and clinical relevance.
Keywords: PRKAR1A, PKA, mutations, polymorphisms, Carney complex
Introduction
Cyclic AMP (cAMP) dependent Protein Kinase A (PKA, EC 2.7.1.37) is the major mediator of the cAMP effects in the eukaryotic cells. In its inactive state, the PKA holoenzyme is a tetramer comprised of dimer of two regulatory subunits bound to two catalytic subunits (Kinderman, et al., 2006; Tasken, et al., 1997). Upon elevated cellular cAMP concentrations, two molecules of cAMP bind each of the regulatory subunits, which leads to the dissociation of the tetramer into regulatory dimers and two free catalytic subunits (Tasken, et al., 1997). The free catalytic subunits represent active serine threonine kinases which in turn phosphorylate series of cellular targets that regulate downstream effector enzymes, ion channels, and transcription of specific genes that mediate cell growth and differentiation (Shabb, 2001).
In the human, 4 different regulatory (PRKAR1A, PRKAR1B, PRKAR2A, and PRKAR2B) and 3 catalytic subunits (PRKACA; PRKACB, PRKACG) have been identified so far. It is now assumed that the multiple combinations of homo- or heterodimers of regulatory subunits with associated catalytic subunits may, at least in part, account for the specificity of the transduction of the signal generated by numerous different hormones, neurotransmitters, and other signaling through one sole second messenger, cAMP.
The only PKA subunit in which mutations have been found to lead to human disease is PRKAR1A: inactivating PRKAR1A mutations cause the multiple neoplasia and lentiginosis syndrome Carney complex (CNC) (Casey, et al., 1998; Kirschner, et al., 2000a). CNC (MIM# 160980) is an autosomal dominant disorder associated with skin lesions, cardiac and other myxomas, and different types of endocrine tumors including primary pigmented nodular adrenocortical disease (PPNAD), growth hormone–secreting pituitary tumors, gonadal and thyroid neoplasias; rarely, malignant psammomatous melanotic schwannomas (PMS) may also be present (Carney, 1995; Carney, et al., 1986; Stratakis, et al., 1998). The vast majority of the reported PRKAR1A mutations lead to premature stop codons and subsequent degradation of the mutant mRNAs species by NMD (Kirschner, et al., 2000b). Inactivating PRKAR1A mutations lead to excess PKA signaling in affected tissues (Robinson-White, et al., 2006a).
PRKAR1A is located on chromosome 17q23-q24 and extends to a total genomic length of approximately 21 kB. The gene is composed of 11 exons, 10 of which coding, with a coding region of 1143 bp starting from exon 2. The RIα protein consists of 384 AA organized in a dimerization/docking domain at the amino-terminal, followed by a PKA inhibitor site, two tandem binding domains for cAMP at the carboxyl terminus (cAMP:A and cAMP:B), and a linker region that contains the main docking site for the C subunit (Zawadzki and Taylor, 2004). Until recently, it was widely accepted that, in contrast to RIIα, which typically is docked at discrete cellular locations through A Kinase Anchoring Proteins (AKAPs), RIα tends to diffuse in the cytoplasm (Kinderman, et al., 2006; Skalhegg, et al., 1994; Wong and Scott, 2004). In the past several years, however, evidence has accumulated that RIα can be dynamically recruited as well localized at certain compartments of the cell such as the plasma membrane, mitochondria, cytoskeleton, and centrosomes, and AKAPs capable of binding it have been described (Huang, et al., 1997a; Huang, et al., 1997b).
PRKAR1A orthologues have been characterized in many species, and high level of preservation can be tracked back to invertebrates (Aedes aegipty, 73% homology) and protozoa (Ciona intestinalis, 63% homology). The high degree of amino acid sequence conservation underlines the significance of PRKAR1A, the functional “receptor” for cAMP. Indeed, both cloning and gene knockout studies on the individual regulatory subunits have confirmed the essential role of PRKAR1A in maintaining cAMP control of PKA activity through compensatory mechanisms assuring balanced regulation of the free catalytic subunits (Amieux and McKnight, 2002).
Since the identification of PRKAR1A as the gene causing CNC, more than a hundred different pathogenic mutations have been reported, and their molecular and functional analysis has been investigated in several major studies (Bertherat, et al., 2009; Greene, et al., 2008; Groussin, et al., 2002; Kirschner, et al., 2000a; Veugelers, et al., 2004). Herein we review all described to date PRKAR1A sequence variations, including 17 that have not been published before, in an attempt for better understanding of their molecular context and clinical relevance.
Mutation spectrum
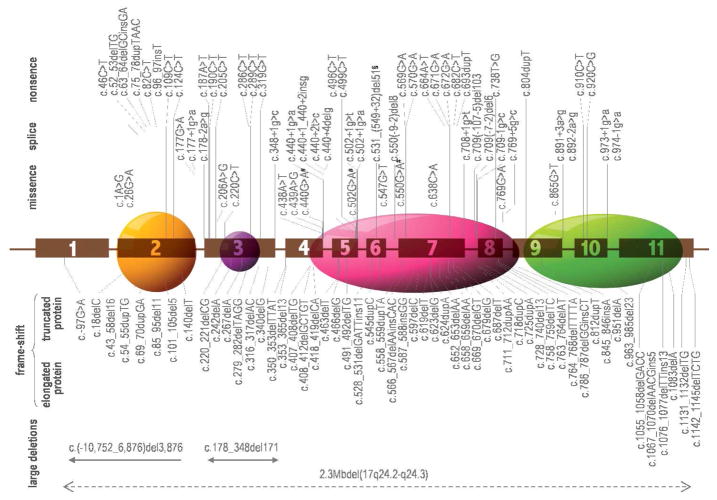
To date, a total of 117 different PRKAR1A mutations have been identified in 387 unrelated families of various ethnic origin; they are summarized in Table 1. The molecular changes involve single base substitutions and small (up to 15bp) deletions, insertions or combined rearrangements that are spread along the whole open reading frame of the gene; in addition, several relatively large deletions are reported (Blyth, et al., 2008; Horvath, et al., 2008a). A schematic representation of the PRKAR1A mutations’ type and location is shown on Figure 1.
Table 1.
List of known mutations in the PRKAR1A (sorted by their position along the gene).
| # | Position | DNA Change | Protein/RNA Change | Type | Effect on the protein | Phenotype | Reference |
|---|---|---|---|---|---|---|---|
| 1 | exon 1B | c.-97G>A | Pseudo ATG | frame-shift | NMD, no protein | CNC | (Groussin, et al., 2002) |
| 2 | exon 2 | c.1A>G | p.M1V | missense | abolished initiation | CNC | (Kirschner, et al., 2000b) |
| 3 | exon 2 | c.18delC | p.A7PfsX122 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 4 | exon 2 | c.26G>A | p.S9N | missense | altered protein | CNC | (Greene, et al., 2008) |
| 5 | exon 2 | c.43_58del16 | p.L15SfsX109 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 6 | exon 2 | c.46C>T | p.R16X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 7 | exon 2 | c.52_53delTG | p.C18X | nonsense | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 8 | exon 2 | c.54_55dupTG | p.E19VfsX111 | frame-shift | NMD, no protein | CNC | This study |
| 9 | exon 2 | c.63_64delCGinsGA | p.Y21X | nonsense | NMD, no protein | CNC | (Almeida, et al., 2008) |
| 10 | exon 2 | c.69_70duplGA | p.K24RfsX106 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 11 | exon 2 | c.75_78dupTAAC | p.l27X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 12 | exon 2 | c.82C>T | p.Q28X | nonsense | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 13 | exon 2 | c.85_95del11 | p.A29RfsX12 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 14 | exon 2 | c.96_97insT | p.D33X | nonsense | NMD, no protein | CNC | This study |
| 15 | exon 2 | c.101_105del5 | p.S34CfsX9 | frame-shift | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 16 | exon 2 | c.109C>T | p.Q37X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 17 | exon 2 | c.124C>T | p.R42X | nonsense | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 18 | exon 2 | c.140delT | p.M46RfsX82 | frame-shift | NMD, no protein | CNC | (Imai, et al., 2005) |
| 19 | exon 2 | c.177G>A | retains part of intr 2 | splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 20 | intron 2 | c.177+1 g>a | retains part of intr 2 | splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 21 | intron 2 | c.178–2 a>g | skips ex 3 | splice | shorter prot. (324AA) | CNC | (Kirschner, et al., 2000b) |
| 22 | intron 2 | c.178_348del171 | p.E60_K116del | In-frame del | shorter prot. (328AA) | CNC | (Horvath, et al., 2008) |
| 23 | exon 3 | c.187A>T | p.K63X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 24 | exon 3 | c.190C>T | p.Q64X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 25 | exon 3 | c.205C>T | p.Q69X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 26 | exon 3 | c.206A>G | p.Q69R | missense | altered protein | CNC | This study |
| 27 | exon 3 | c.220_221delCG | p.R74YfsX7 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 28 | exon 3 | c.220C>T | p.R74C | missense | altered protein | CNC | (Veugelers, et al., 2004) |
| 29 | exon 3 | c.242delA | p.E81GfsX48 | frame-shift | NMD, no protein | CNC | This study |
| 30 | exon 3 | c.267delA | p.V90WfsX39 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 31 | exon 3 | c.279_282delTAGG | p.R94GfsX34 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 32 | exon 3 | c.286C>T | p.R96X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 33 | exon 3 | c.289C>T | p.R97X | nonsense | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 34 | exon 3 | c.316_317delAC | p.T106GfsX9 | frame-shift | NMD, no protein | CNC | (Skamrov, et al., 2003) |
| 35 | exon 3 | c.319G>T | p.Q107X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 36 | exon 3 | c.340delG | p.V114LfsX15 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 37 | Inton 3 | c.348+1 g>c | retains intr 3 | splice | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 38 | exon 4 | c.350_353delTTAT | p.V117DfsX11 | frame-shift | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 39 | exon 4 | c.353_365del13 | p.I118TfsX6 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 40 | exon 4 | c.407_408delTG | p.V136AfsX6 | frame-shift | NMD, no protein | CNC | (Mabuchi, et al., 2005) |
| 41 | exon 4 | c.408_412delGCTGT | p.L137FfsX4 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 42 | exon 4 | c.418_419delCA | p.H140SfsX1 | frame-shift | NMD, no protein | CNC | This study |
| 43 | exon 4 | c.438A>T | p.R146S | missense | altered protein | CNC | (Greene, et al., 2008) |
| 44 | exon 4 | c.439A>G | p.S147G | missense | altered protein | CNC | (Bertherat, et al., 2009) |
| 45 | exon 4 | c.440G>A | p.S147N | missense, splice | altered protein | CNC | (Gu, et al., 2004) |
| 46 | intron 4 | c.440+1g>a | retains part of intr 4 | splice | NMD, no protein | CNC | This study |
| 47 | intron 4 | c.440+1_440+2insg | retains part of intr 4 | splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 48 | intron 4 | c.440+2t>c | retains part of intr 4 | splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 49 | intron 4 | c.440+4del g | retains part of intr 4 | splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 50 | exon 5 | c.463delT | p.S155RfsX10 | frame-shift | NMD, no protein | CNC | This study |
| 51 | exon 5 | c.466delG | p.V156SfsX9 | frame-shift | NMD, no protein | CNC | (Vargas-Alarcon, et al., 2008) |
| 52 | exon 5 | c.491_492delTG | p.V164DfsX5 | frame-shift | NMD, no protein | CNC | (Kirschner, et al., 2000a) |
| 53 | exon 5 | c.496C>T | p.Q166X | nonsense | NMD, no protein | CNC | This study |
| 54 | exon 5 | c.499C>T* | p.Q167X | nonsense | NMD, no protein | UDC | (Sandrini, et al., 2002) |
| 55 | exon 5 | c.502G>A | p.G168S | missense, splice | altered protein | CNC | (Bertherat, et al., 2009) |
| 56 | intron 5 | c.502+1g>t | retains part of intr 5 | splice | NMD, no protein | CNC | (Groussin, et al., 2002) |
| 57 | intron 5 | c.502+1g>a | retains part of intr 5 | splice | NMD, no protein | CNC | (Gennari, et al., 2008) |
| 58 | exon 6 | c.528_531delGATTins11 | p.I177MfsX13 | frame-shift | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 59 | exon 6 | c.531_(549+32)del51 | p.I177MfsX1 | frame-shift, splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 60 | exon 6 | c.545dupC | p.D182GfsX5 | frame-shift | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 61 | exon 6 | c.547G>T | p.D183Y | missense | altered protein | CNC | (Greene, et al., 2008) |
| 62 | intron 6 | c.550(-9-2)del8 | Skip ex 7 | splice | shorter prot. (328AA) | CNC | (Kirschner, et al., 2000b) |
| 63 | exon 7 | c.550G>A | p.V184I | missense, splice | altered protein | CNC | (Bertherat, et al., 2009) |
| 64 | exon 7 | c.558_559dupTA | p.N187IfsX20 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 65 | exon 7 | c.566_567delAAinsCAC | p.E189AfsX44 | frame-shift | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 66 | exon 7 | c.569G>A | p.W190X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 67 | exon 7 | c.570G>A | p.W190X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 68 | exon 7 | c.587_588insGG | p.G198EfsX10 | frame-shift | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 69 | exon 7 | c.597delC | p.F200LfsX6 | frame-shift | NMD, no protein | CNC | (Sasaki, et al., 2008) |
| 70 | exon 7 | c.619delT | p.Y207MfsX15 | frame-shift | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 71 | exon 7 | c.623delG | p.G208EfsX14 | frame-shift | NMD, no protein | CNC | (Casey, et al., 2000) |
| 72 | exon 7 | c.624dupA | p.T209NfsX24 | frame-shift | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 73 | exon 7 | c.638C>A* | p.A213D | missense | altered protein | ODM | (Perdigao, et al., 2005) |
| 74 | exon 7 | c.652_653delAA | p.K218DfsX14 | frame-shift | NMD, no protein | CNC | This study |
| 75 | exon 7 | c.658_659delAA | p.N220CfsX12 | frame-shift | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 76 | exon 7 | c.664A>T* | p.L222X | nonsense | NMD, no protein | ACA | (Bertherat, et al., 2003) |
| 77 | exon 7 | c.669_670delGT | p.W224GfsX8 | frame-shift | NMD, no protein | CNC | This study |
| 78 | exon 7 | c.671G>A | p.W224X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 79 | exon 7 | c.672G>A | p.W224X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 80 | exon 7 | c.679delG | p.D227TfsX14 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 81 | exon 7 | c.682C>T | p.R228X | nonsense | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 82 | exon 7 | c.687delT* | p.D229EfsX12 | frame-shift | NMD, no protein | ODM | (Perdigao, et al., 2005) |
| 83 | exon 7 | c.693dupT | p.R232X | nonsense | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 84 | intron 7 | c.708+1g>t | Skip ex 7 | splice | shorter prot. (328AA) | CNC | (Groussin, et al., 2002) |
| 85 | intron 7 | c.709(-5-107)del103 | skips ex 8 | splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 86 | intron 7 | c.709(-7-2)del6 | skips ex 8 | splice | NMD, no protein | CNC** | (Groussin, et al., 2002) |
| 87 | intron 7 | c.709-1g>c | skips ex 8 | splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 88 | exon 8 | c.711_712dupAA | p.S238KfsX4 | frame-shift | NMD, no protein | CNC | (Kirschner, et al., 2000b) |
| 89 | exon 8 | c.718dupC | p.L240PfsX8 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 90 | exon 8 | c.725dupA | p.R243AfsX5 | frame-shift | NMD, no protein | CNC | (Groussin, et al., 2002) |
| 91 | exon 8 | c.728_740del13 | p.K244NfsX9 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 92 | exon 8 | c.738T>G | p.Y246X | nonsense | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 93 | exon 8 | c.758_759delTC | p.S254YfsX15 | frame-shift | NMD, no protein | CNC | (Casey, et al., 2000) |
| 94 | exon 8 | c.763_764delAT | p.I255PfsX14 | frame-shift | NMD, no protein | CNC | (Groussin, et al., 2002) |
| 95 | exon 8 | c.764_768delTTTTA | p.I255RfsX12 | frame-shift | NMD, no protein | CNC | This study |
| 96 | exon 8 | c.769G>A | p.E257K | missense, splice | NMD, no protein | CNC | This study |
| 97 | intron 8 | c.769+5g>c | Retains part of intr 8 | splice | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 98 | exon 9 | c.786_787delGGinsCT | p.W262CfsX2 | frame-shift | NMD, no protein | CNC | (Kirschner, et al., 2000a) |
| 99 | exon 9 | c.804dupT | p.D269X | nonsense | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 100 | exon 9 | c.812dupT | p.L271FfsX7 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 101 | exon 9 | c.845_846insA | p.V283CfsX9 | frame-shift | NMD, no protein | CNC | (Bertherat, et al., 2009) |
| 102 | exon 9 | c.865G>T | p.G289W | missense | altered protein | CNC | (Greene, et al., 2008) |
| 103 | intron 9 | c.891+3a>g | Retains part of intr 9 | splice | skips ex 10 | CNC | (Kirschner, et al., 2000a) |
| 104 | intron 9 | c.892-2a>g | skips ex 10 | splice | skips ex 10 | CNC | This study |
| 105 | exon 10 | c.910C>T | p.Q304X | nonsense | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 106 | exon 10 | c.920C>G | p.S307X | nonsense | NMD, no protein | CNC | This study |
| 107 | exon 10 | c.951delA | p.R317SfsX14 | frame-shift | NMD, no protein | CNC | (Veugelers, et al., 2004) |
| 108 | exon 10 | c.963_985del23* | p.D322TfsX20 | frame-shift | NMD, no protein | ACA | (Bertherat, et al., 2003) |
| 109 | inton 10 | c.973+1g>a | Retains part of intr 10 | splice | NMD, no protein | CNC | This study |
| 110 | exon 11 | c.974-1g>a* | skips ex 11 | splice | NMD, no protein | ACA | (Bertherat, et al., 2003) |
| 111 | exon 11 | c.1055_1058delGACC | p.R352LfsX88 | frame-shift | elongated prot. (438AA) | CNC | (Bertherat, et al., 2009) |
| 112 | exon 11 | c.1067_1070delAACGins5 | p.E356GfsX71 | frame-shift | elongated prot. (425AA) | CNC | (Bertherat, et al., 2009) |
| 113 | exon 11 | c.1076_1077delTTins13 | p.L359QfsX86 | frame-shift | elongated prot. (443AA) | CNC | (Bertherat, et al., 2009) |
| 114 | exon 11 | c.1083delA | p.C362AfsX79 | frame-shift | elongated prot. (439AA) | CNC | This study |
| 115 | exon 11 | c.1131_1132delTG | p.S378TfsX48 | frame-shift | elongated prot. (425AA) | CNC | (Bertherat, et al., 2009) |
| 116 | exon 11 | c.1142_1145delTCTG | p.V381EfsX59 | frame-shift predicted | elongated prot. (439AA) | CNC | (Bertherat, et al., 2009) |
| 117 | promoter? | c.(-10,752_6,876)del3,876 | Before ex 1 | deletion of part of the promoter | decreased prot. expression | CNC | (Horvath, et al., 2008) |
somatic mutation (the presence of the mutation was ruled out in lymphocytes)
In the carriers of mutation c.709(-7-2)del6 and c.63_64delCGinsGA CNC phenotype was restricted to PPNAD, and, in some cases, skin manifestations
CNC+ phenotype is defined by the authors as „posterior laryngeal cleft and the presence of numerous freckles and lentigines, growth restriction, microcephaly and moderate mental retardation“.
CNC – Carney complex; ACA – adenocortical adenoma; UDC – undiferentiated thyroid carcinoma; ODM – odontogenic myxoma; NMD – Non-sense mediated m-RNA decay
Figure 1.
Schematic presentation of the type and the approximate location of the 117 identified to date mutations in PRKAR1A; a large deletion eliminating the whole PRKAR1A together with 13 more genes (2.3Mb del 17q24.2-q24.3) is also shown. The docking/dimerisation, the linker and the two cAMP functional domains (from left to right) of the protein are denoted with ovals.
The mutations in PRKAR1A are spread along the whole coding sequence, without significant preference for an exon or a domain. Most of them are unique – they are identified in single families only (Bertherat, et al., 2009). To date, only 3 mutations have been found in more than three unrelated pedigrees: c.82C>T, c.491_492delTG and c.709(-7-2)del6; these mutations have been seen in kindreds with different racial and ethnic background, suggesting that they are likely to result from more than one independent mutation events.
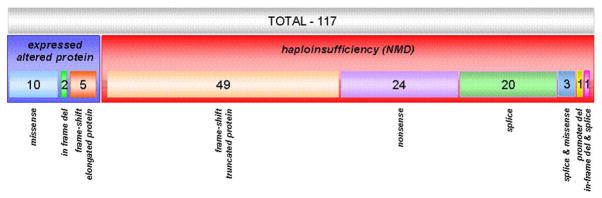
A graph representing the relationship between the molecular type of the PRKAR1A mutations’ and their functional effect is shown on Figure 2. Relatively small proportion (20/117, 17.1%) of the unique mutations result in the expression of an altered protein; these group convenes the 11 missense substitutions, 6 frameshift mutations affecting the last exon of the gene and thus escaping NMD, one in-frame deletion that eliminates exon 3 in frame, and one splice variant that leads to exon 7 skipping. The vast majority of mutations (97/117, 82.9%) result in premature stop codon (PSC) generation caused by nonsense and frameshift changes upstream of the last gene exon; the mutant mRNAs are degraded through NMD. In this second group fall also the majority of the splice variants – disrupted donor or acceptor sequences lead to exon skip and/or retaining of part of the intron, and, thus, direct or by frame-shift incorporation of PSC. The different mutations’ type functional effect and relation to the phenotype are discussed below.
Figure 2.
Quantitative schematic representation of the expressed vs NMD mutations in PRKAR1A.
Mutations generating premature stop codon
As mentioned, PSC are caused by nonsense, frame shift and splice variants located before the last exon of PRKAR1A. The expression of the shortened protein is abolished by the NMD, as shown by our experiments (Kirschner, et al., 2000b); thus, the overall effect of this type of mutation is a 50% reduction in cellular PRKAR1A levels. The resulting higher proportion of catalytic to regulatory subunits causes increased signaling and activation of the downstream cellular processes such as proliferation and differentiation, (Robinson-White, et al., 2006b). Hence, NMD mutations cause disease in the affected tissues through a mechanism of haploinsufficiency. This type of mutations is found in the majority of the CNC patients and the resulting phenotypes vary in a very wide range both as number of manifestation and severity of their expression. The factors to affect this variability, besides age, gender and environmental agents are likely to include other genetic determinants with modifying effects. The later is supported by family clustering of particular sets of manifestations as well as our recent studies showing influence of genetic variants in other cAMP-related molecules on the severity of the CNC phenotype (manuscript in preparation). Although the uniqueness of the most of the mutations limits genotype/phenotype analysis, it is now accepted that phenotype/genotype correlation is rare among carriers of NMD mutations in PRKAR1A - they all share haploinsufficiency at the protein level.
Expressed mutations
The expressed missense substitutions in PRKAR1A are listed in Table 2. While the pathogenic effect of the NMD mutations is almost certain to destroy the PRKAR1A function, the expressed mutations that lead to altered protein clearly require estimation of their pathogenic potential. Eight of these mutations – six aminoacid substitutions (S9N, R74C, R146S, D183Y, A213D and G289W), and the two in-frame deletions of exons 3 and 7 - have been subject of detailed investigation of their effect in vitro (Greene, et al., 2008; Meoli, et al., 2008). The expressed mutations were spread over all functional PRKAR1A domains and every one of them led to variable extends of decrease of the cAMP binding and increase in the cAMP-specific PKA activation (Greene, et al., 2008; Meoli, et al., 2008). As expected, the highest impact on the protein function was measured for the in-frame deletion that eliminates exon 3, and, respectively, the primary binding site for the catalytic subunit (Greene, et al., 2008); this mutation leads to significant elevation in the PKA activity even in the absence of cAMP. Similar, although milder effect on the protein is caused by the R146S, which disrupts the secondary binding site for the catalytic subunit (residues 138-148 and 232-247 within the cAMP binding domain A (Greene, et al., 2008). Expressed mutations affecting the cAMP binding domains (A183Y, A213D and G289W), are shown, in line with their position, to significantly decrease the cAMP binding; the proposed mechanism of action is through conformational changes that alter the enzymatic function. Conformational changes are also proposed to take place for the last two of the studied expressed mutations: S9N, located in the dimerisation/docking domain, and R74C, which is positioned in the linker region, outside any known functional sequence.
Table 2.
In silico modeling and in vitro analysis of the effect of PRKAR1A missense substitutions on the protein function
| # | DNA Change | Protein Change | Domain | In silico modeling | In vitro analysis | Inter-species alignment | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| prediction | score* | cAMP binding** | cAMPspecific PKA activation** | Mus musculus | Gallus gallus | Anolis carolinensis | Xenopus laevis | Danio regio | Plasmodium falciparum | ||||
| 1 | c.1A>G | p.M1V | Initiation codon | Probably damaging | 2.055 | 0.31*** | 2.46*** | M | M | M | M | M | M |
| 2 | c.26G>A | p.S9N | Dimerization/docking | Benign | 0.243 | 0.28*** | 2.14** | S | S | S | S | S | - |
| 3 | c.206A>G | p.Q69R | Linker region | Probably damaging | 2.013 | na | na | Q | Q | Q | Q | Q | - |
| 4 | c.220C>T | p.R74C | Linker region | Possibly damaging | 1.686 | 0.69*** | 2.77 | R | R | R | R | R | - |
| 5 | c.438A>T | p.R146S | cAMP binding A | Benign | 0.733 | 0.49*** | 2.12*** | R | R | R | R | R | F |
| 6 | c.439A>G | p.S147G | cAMP binding A | Benign | 1.033 | 0.84*** | 2.16*** | S | S | S | S | S | E |
| 7 | c.440G>A | p.S147N | cAMP binding A | Benign | 0.145 | na | na | S | S | S | S | S | E |
| 8 | c.502G>A | p.G168S | cAMP binding A | Probably damaging | 2.250 | na | na | G | G | G | G | G | G |
| 9 | c.547G>T | p.D183Y | cAMP binding A | Probably damaging | 2.521 | 0.24*** | 1.63 | D | D | D | D | D | E |
| 10 | c.550G>A | p.V184I | cAMP binding A | Benign | 0.052 | na | na | V | V | V | V | V | I |
| 11 | c.638C>A | p.A213D | cAMP binding A | Probably damaging | 2.507 | 0.17*** | 2.06 | A | A | A | A | A | A |
| 12 | c.865G>T | p.G289W | cAMP binding B | Probably damaging | 2.592 | 0.43*** | 2.92*** | G | G | G | G | G | G |
greater score indicates higher probability to impair the protein function; the factors taken into account for the calculation of the score are
(1) difference in the thermo-physical properties of the wt and mutant protein, and
(2) evolutionary preservation of the residue in the corresponding position
both cAMP binding and PKA activity changes in response to cAMP are presented as ratio to the wt PRKAR1A
P<0.05
Notably, substantial correlation between the in vitro studies and the in silico modeling of the effect of the expressed missense substitutions on the protein function, along with the homology data, was observed (See Table 2). In silico prediction of the effect of the missense substitutions on the protein function (http://coot.embl.de/PolyPhen/) has estimated highest potential to impact the protein function generally for mutations residing in the cAMP binding domains (G168S, D183Y, A213D and G289D, Table 2).
Separate attention requires the missense substitution affecting the first coding aminoacid - M1V. In silico modeling predicted very severe effect on the protein function (See Table 2). Consistent with the above, the in vitro studies have shown significant effect of the mutation on both cAMP binding and cAMP specific PKA activation. However, although the mutant mRNA is found to be expressed in equal to the wt levels in carriers of the mutation, it is not clear if the mutant protein is expressed and stable in vivo (Kirschner, et al., 2000b). The possibility of alternate expression from a surrogate initiation site has been explored – an in-frame ATG in the context of a relatively good match for Kozak sequence is located 141 bp downstream from the original initiation codon; however, Western blot did not detect shorter protein forms.
Analysis of the phenotype characteristics in the carriers of expressed PRKAR1A mutations revealed severe CNC phenotype in the carrier of the in-frame deletion of ex3 and the c.708 +1G>T mutation, expressed both as number of manifestations and severity of their expression. Apart from this finding, no genotype-phenotype connection could be characterized within this relatively small group, which is expected taken the limited number of affected patients and the variable number CNC manifestations. Nevertheless, the finding of expressed PRKAR1A mutant variations that cause disease related changes in vitro, confirms that altered PRKAR1A function, not only haploinsufficiency, is enough to lead to the disease phenotype.
Frameshift mutations leading to elongated PRKAR1A protein
Six of the frameshift mutations (c.1055_1058delGACC, c.1067_1070delAACGins5, c.1076_1077delTTins13, c.1083delA, c.1131_1132delTG, and c.1142_1145delTCTG) are located in the last exon of PRKAR1A and are predicted to escape NMD. Each of the mutations causes a frame shift that abolishes the wild type termination codon and generates a new one further downstream. Compared to the wild type PRKAR1A, the predicted mutant proteins are longer and carry a different 3′ AA sequence, resulting from codon rearrangement. The functional significance of the elongated proteins versus the wild-type R1α is subject of another report under submission.
Large deletions
Several relatively large deletions in the region of PRKAR1A have been described so far (Blyth, et al., 2008; Horvath, et al., 2008a), including the above discussed expressed deletion that eliminates exon 3 in frame (c.178_348del171/p.E60_K116del). This mutation has been shown to result to the in vivo expression of shorter PRKAR1A protein lacking part of the linker region connecting the dimerisation/docking domain and the first cAMP binding domain; the deleted region includes the binding site for the catalytic subunit/inhibitor (Horvath, et al., 2008a). Only one patient was found to carry this mutation, and he presented with very severe phenotype, both as number of CNC manifestation and level of their expression (Horvath, et al., 2008a). In vitro studies with expression vectors harboring PRKAR1A ORF lacking exon 3 have shown extreme effect on the protein function (Greene, et al., 2008). The second identified deletion was located in the upstream regulatory region of PRKAR1A and did not affect the ORF of the gene; this mutation is expected to lead to decreased PRKAR1A mRNA levels but no other effects on the protein; the molecular phenotype is predicted to be PRKAR1A haploinsufficiency, consistent with the majority of PRKAR1A mutations causing CNC. The third noteworthy deletion encompasses significantly larger genomic region from 17q24.2-q24.3 and eliminates, along with PRKAR1A, 13 more genes (Blyth, et al., 2008). This mutation was identified as a de novo genomic rearrangement in a 12 years old patient presenting with posterior laryngeal cleft, moderate growth and mental retardation, microcephaly, and, as the only CNC related manifestation - multiple freckles and lentigines. However, other CNC manifestations can be underrepresented due to the severity of the main phenotype, or, more likely may occur later in the development. Although larger rearrangements involving PRKAR1A region are reported in the literature, in most of the cases they led to extremely severe phenotype and premature death that prevented comprehensive analysis of relatively mild CNC manifestation with later onset (Bridge, et al., 1985; Levin, et al., 1995; Olney, et al., 1999).
Splice mutations
Twenty-eight splice variants are identified to date in the close proximity of the exon-intron junctions of PRKAR1A (Table 3). As mentioned, the vast majority of them lead to a frame-shift and subsequent incorporation of PMS. Notably, for the most of the splice mutations (23, 82%), the in silico modeling predicted complete abrogation of the junction formation; in 2 of them accompanied by a shift of the predicted junction with 1 base (see Table 3). For the remaining five variants (c440+4delG, c.502G>A, c.550G>A, c.763delAT, and 891+3A>G) a significant decrease in the probability score to form junction compared to the wt was estimated (see Table 3). It is noteworthy that three of these last variants affected directly exonic sequences and their effect on the splice-site was complemented by a change in the approximate coding sequence, which additionally increased their pathogenic impact. Combined with the family data that show segregation of these variants with the CNC phenotype, all described 28 splice changes were classified as pathogenic.
Table 3.
In silico modeling of the effect of PRKAR1A splice variants
| # | DNA Change | Exon affected | Spliceport WT Score | Spliceport mutation Score |
|---|---|---|---|---|
| 1 | c.177G>A | Incorporates intron2 | 0.679639 | 0.00 |
| 2 | c.177+1g>a | Incorporates intron2 | 0.679639 | 0.00 |
| 3 | c.178–2a>g | Skip exon 2 | 0.894923 | 0.00 |
| 5 | c.348 +1 g>c | Incorporates intron3 | 1.14417 | 0.00 |
| 6 | c.438A>T | Incorporates intron4 | 0.820566 | 0.00 |
| 7 | c.439A>G | Incorporates intron4 | 0.820566 | 0.00 |
| 8 | c.440G>A | Incorporates intron4 | 0.820566 | 0.00 |
| 9 | c.440+ 1 insG | Incorporates intron4 | 0.820566 | 0.268882 (+1 base) |
| 10 | c.440+ 4del g | Incorporates intron4 | 0.820566 | 0.142182 |
| 11 | c.440+2 t>c | Incorporates intron4 | 0.820566 | 0.00 |
| 13 | c.502 +1g>t | Incorporates intron5 | 1.69211 | 0.00 |
| 14 | c.502+1g>a | Incorporates intron5 | 1.69211 | 0.00 |
| 15 | c.502G>A | Incorporates intron5 | 1.69211 | 0.705629 |
| 17 | c.550(-8-2) del 7 | Skip exon 7 | 0.802186 | 0.00 |
| 18 | c.550G>A | Skip exon 7 (+ −) | 0.802186 | 0.478702 |
| 19 | c.708 +1g>t | Skip exon 7 | 1.74672 | 0.00 |
| 20 | c.709 (-7-2)del6(ttttta) | Skip exon 8 | 0.502791 | 0.0 |
| 21 | c.709 (-5-107)del103 | Skip exon 8 | 0.502791 | 0.0 |
| 22 | c.709-1G>C | Skip exon 8 | 0.502791 | 0.0 |
| 24 | c.763delAT | Incorporates intron8 | 1.02912 | 0.778435 |
| 25 | c.769+5g>c | Incorporates intron8 | 1.02912 | 0.00 |
| 26 | c.891+3A>G | Incorporates intron9 | 1.41361 | 0.158643 |
| 27 | c.892-2 a>g | Skip exon 10 | 1.64415 | 0.0 |
| 28 | c.974-1g>a | Skip exon 11 | 1.2552 | 0.112813 (+1 base) |
Interestingly, some splice-site alleles may express both mutant and wild type mRNA molecules in a proportion depending on the type of the mutation and its location relevant to the junction. Thus, the relative decrease of the cellular wt PRKAR1A for these mutations is expected to be lower, and, accordingly, to lead to a milder phenotype. This assumption is in line with a recent analysis of a “mild” splice variant (c.709(-7-2)del6), which is one of the very few mutations where incomplete penetrance of PRKAR1A mutation is seen, and, in the affected individuals, only PPNAD and lentiginosis – the most common disease manifestations - have been diagnosed (Groussin, et al., 2006).
Polymorphisms
Four nonpathogenic unique synonymous substitutions in the third base of the codon have been described so far in PRKAR1A: c.87G>A/p.A29A, c.204G>A/p.L68L, c.318G>C/p.T106T, and c.492G>A/p.V164V. From them, the first one has been seen in at least five unrelated kindreds, and the remaining three are detected in single families only. Multiple intronic variants in close proximity of the junctions have been described, from them the most frequent being c.349-5dup5 (detected in approximately 15% of the studied alleles), followed by c.769-24A>G and c.891-34G>T (in approximately 12% and 10% of the studied alleles, respectively). However, the extend to which different research groups analyze the exon flanking sequences widely vary and the frequency of these variants may therefore be significantly underreported; furthermore, both missense variants and splice changes may be as well underreported. All these polymorphic variants are seen in healthy control individuals and in both CNC families, and in the later, they do not show segregation with the disease status.
Mutation detection rate of PRKAR1A mutations in CNC
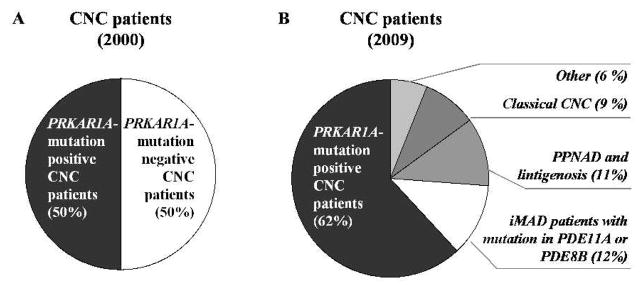
Since the first description of the disease, numerous CNC patients have been reported from all ethnic groups and presenting with various number, combination and severity of manifestations. The most recently re-evaluated diagnostic criteria for CNC are listed in Table 4; a definite diagnosis is given when two or more major manifestations are present. Soon after the identification of PRKAR1A and the initial patient screenings, the mutation detection rate was estimated to approximately 50% (Figure 3, A). After a decade of comprehensive phenotype analysis and re-evaluation of the diagnostic criteria, it is now assumed that mutations in PRKAR1A are responsible for the disease in more than 60% of the CNC patients (Figure 2B). The largest to date study of 185 families, detects PRKAR1A mutations in 114 (62%) (Bertherat, et al., 2009). Interestingly, this percent rises up to 80 among CNC patients presenting with Cushing syndrome due to PPNAD (Cazabat, et al., 2007). Among the mutation carriers, Cushing syndrome due to PPNAD is seen in 70% of the female patients before 45 years, but in only 45% of the male patients, likely reflecting the generally higher frequency of Cushing syndrome in females (Grumbach, et al., 2003). These numbers, however, may represent an underestimate of the true incidence of Cushing syndrome, since autopsy of CNC patients detects PPNAD in almost 100% of the cases (Stratakis, et al., 2001).
Table 4.
Diagnostic criteria for CNC
| Major Diagnostic criteria for CNC |
| 1. Spotty skin pigmentation with typical distribution (lips, conjunctiva and inner or outer canthi, vaginal and penile mucosal |
| 2. Myxoma* (cutaneous and mucosal) |
| 3. Cardiac myxoma* |
| 4. Breast myxomatosis* or fat-suppressed magnetic resonance imaging findings suggestive of this diagnosis |
| 5. PPNAD* or paradoxical positive response of urinary glucocorticosteroid excretion to dexamethasone administration during Liddle’s test |
| 6. Acromegaly due to GH-producing adenoma* |
| 7. LCCST* or characteristic calcification on testicular ultrasound |
| 8. Thyroid carcinoma* or multiple, hypoechoic nodules on thyroid ultrasound in a young patient |
| 9. Psammomatous melanotic schwannomas* |
| 10. Blue nevus, epithelioid blue nevus* |
| 11. Breast ductal adenoma* |
| 12. Osteochondromyxoma* |
| Supplementary criteria |
| 1. Affected first-degree relative |
| 2. Inactivating mutation of the PRKAR1A gene |
| Findings suggestive of or possibly associated with CNC, but not diagnostic for the disease |
| 1. Intense freckling (without darkly pigmented spots or typical distribution) |
| 2. Blue nevus, common type (if multiple) |
| 3. Café-au-lait spots or other “birthmarks” |
| 4. Elevated IGF-I levels, abnormal GTT, or paradoxical GH response to TRH testing in the absence of clinical acromegaly |
| 5. Cardiomyopathy |
| 6. Pilonidal sinus |
| 7. History of Cushing’s syndrome, acromegaly, or sudden death in extended family |
| 8. Multiple skin tags or other skin lesions; lipomas |
| 9. Colonic polyps (usually in association with acromegaly) |
| 10. Hyperprolactinemia (usually mild and almost always combined with clinical or subclinical acromegaly) |
| 11. Single, benign thyroid nodule in a young patient; multiple thyroid nodules in an older patient (detected on ultrasound) |
| 12. Family history of carcinoma, in particular of the thyroid, colon, pancreas, and ovary; other multiple benign or malignant tumors |
After histological confirmation
Figure 3.
Mutation detection rate among CNC patients A) soon after the identification of PRKAR1A as a gene responsible for the disease phenotype, and, B) after the most recent analyses. The mutation detection rate increased from ~50 (2000) to above 62% (2009), and, in addition, genetic defect in PDE11A and PDE8B were identified in a phenotypic subgroup of initially recognized as CNC patients.
When opposed to the PRKAR1A mutation carriers, CNC patients with no genetic defect in PRKAR1A presented with disease overall later in life and more frequently in a sporadic fashion suggesting implication of different genetic factors (Bertherat, et al., 2009). Further, several phenotype subgroups could be distinguished among the PRKAR1A mutation negative individuals: with typical CNC manifestations (9%); with PPNAD and lentigines only (11%); with isolated bilateral micronodular hyperplasia (12%); (see Figure 3, B). Recently, a genome wide based study on the last group identified inactivating mutations in two of the human genes encoding phosphodiesterases – PDE11A and PDE8B (Gunther, et al., 2004; Horvath, et al., 2006a; Horvath, et al., 2008b). Another CNC locus on region 2p16 has been suggested by linkage analysis, however, so far no other gene has been identified, and the genetic searches for additional molecular factors continues (Stratakis, et al., 1996; Stratakis, et al., 2001).
Penetrance of CNC mutations
The overall penetrance of CNC among PRKAR1A mutations’ carriers is above 95% by the age of 50; only few PRKAR1A carriers are reported who do not meet the diagnostic criteria. Only two mutations so far have been found to result in incomplete penetrance of CNC – the splice variant c.709(-7-2)del6 and the initiation alternating substitution p.M1Vp. Even more interesting is the observation that, when expressed, these two mutations lead to relatively mild CNC phenotype restricted mostly to PPNAD, in some cases accompanied by lentigines. These findings were confirmed in all 12 unrelated families affected by c.709(-7-2)del6 and in the two unrelated kindreds where c.1A>G/p.M1V was seen (Groussin, et al., 2006). The explanation of this mild, and, in some cases, missing phenotype is seek in the light molecular effect of these two mutations – as a splice variant, c.709(-7-2)del6 does not affect 100% of the molecules, and, respectively, leads to lower level of PRKAR1A insufficiency. The molecular effect of M1V substitution is not clear, but the same substitution in the first codon is suggested to be a mild mutation for other genetic disorders such as Smith-Lemli-Opitz syndrome (Witsch-Baumgartner, et al., 2005). Taking into account the particular sensitivity of the adrenal cells to even minor changes in the PKA activity and/or PRKAR1A levels, it is suggested that adrenal will be the first tissue to react on these PRKAR1A mutations.
PRKAR1A in unselected population
Sequencing of the entire coding region of PRKAR1A has not identified alterations in at least 200 unrelated chromosomes. In addition, genotyping of 745 unrelated healthy individuals enrolled in New York Cancer Prospective study (Mitchell, et al., 2004) for 51 unique PRKAR1A mutations (Supplementary Table 1) did not detect the presence of any of them. Thus, PRKAR1A mutations are concluded to be very rare in general population. It is an essential parallel to be made with the gene encoding phosphodiesterase type 11A (PDE11A) – despite the proven association between pathogenic mutations, including null alleles, with adrenal neoplasias, such mutations are seen in general population with lower frequency (Horvath, et al., 2006a; Horvath, et al., 2006b).
Genotype-phenotype correlations
Until recently, genotype-phenotype correlation studies were limited by the high variability of the CNC phenotype, both as number and combinations of manifestations, and the relatively limited series of patients. After more than 20 years of analysis and studying of CNC patients, as well as the fusion of the major databases from the National Institutes of Health, Bethesda, MD, and the Hospital Cochin, Paris, France, more than 380 patients are genotypically characterized and clinically evaluated. An overall phenotypic segregation could be seen when the PRKAR1A mutation carriers were opposed to the mutation negative CNC patients: the former presented more frequently and earlier in life with pigmented skin lesions, myxomas, thyroid and gonadal tumors (Bertherat, et al., 2009). Further, subgroups of patients presenting with particular genotype-phenotype correlation could be outlined. The first such a group was presented by patients with isolated PPNAD, in some cases accompanied with lentiginosis. Among these patients, the following tendencies were observed: (1) patients diagnosed before 8 years of age were rarely carriers of PRKAR1A mutation; (2) most of the patients with isolated PPNAD and presence of PRKAR1A mutation were carriers of either the c.709 (-7-2) del6(ttttta) mutation (p<0.0001) or the c.1A>G/p.M1V substitution affecting the initiation codon of the protein. These observations are in line with the discussed above mild, mainly adrenal or, in some cases absent phenotype associated with these two PRKAR1A mutations.
The second group of CNC patients with particular genotype-phenotype correlation was comprised by individuals with myxomas (affecting all locations - skin, heart, and breast), PMS, thyroid tumors, and LCCSCT. In these patients, PRKAR1A mutations were seen substantially more often. Related to this is the acknowledgment that certain tumors presented at significantly younger age in the PRKAR1A mutation carriers: cardiac myxomas (p=0.02), thyroid tumors (p=0.03) and LCCSCTs (p=0.04) (Bertherat et al., 2009). Within the group of PRKAR1A carriers the mutations located in exons associated more frequently with lentigines, PMS, acromegaly and cardiac myxomas, compared to the intronic ones (p = 0.04); this observation is in line with the above discussion of a milder phenotype associated with splice variants. And, finally, the “hot spot” c.491-492delTG mutation was most significantly associated with lentigines, cardiac myxoma, and thyroid tumors when opposed to all other PRKAR1A defects combined (p = 0.03).
Diagnostic relevance
Clinical and genetic studies of CNC patients have revealed considerable heterogeneity both in mutational spectrum and in phenotypic variation. Patients with two or more diagnostic criteria from Table 4 or with isolated PPNAD are referred to genetic analysis of PRKAR1A. Once a mutation is identified, genotyping for the particular variant is recommended for all at-risk relatives. The proportion of de novo mutations in PRKAR1A is estimated to be approximately 20% (our unpublished results). Germline mosaicism has not been seen so far and may not be frequent, and thus not of diagnostic concern currently.
Conclusions and future prospects
To date, most PRKAR1A studies are focused on its coding regions and on screening for point mutations and small rearrangements detectable by PCR-based techniques. The identification of large deletions in the gene, and the significant proportion of patients with characteristic CNC phenotype and not detectable PRKAR1A mutations have triggered the application of alternative technologies for detection of non-coding variations and larger re-arrangements that may account for the disease phenotype (our on-going project).
Apart from PRKAR1A, three other genes from the cAMP signaling pathway are known to date to cause related or overlapping to CNC phenotype. Germline mutations in PDE11A and PDE8B associate with iMAD (Horvath, et al., 2006a; Horvath, et al., 2008b). Somatic activation mutations in G-alpha stimulatory subunit of the G-protein coupled receptor (GNAS) are known to result in McCune Albright syndrome – a postzygotic somatic multiple neoplasia characterized by polyostotic fibrous dysplasia (POFD), cafe-au-lait skin pigmentation, and peripheral precocious puberty (Weinstein, et al., 1991). Identification of genetic defects in cAMP-signaling molecules in patients with similar and/or overlapping phenotype underlines the importance of this signaling in the normal functioning of the endocrine and other cells. Identification of genetic defects in cAMP-signaling molecules in patients with similar and/or overlapping with CNC features underscores the importance of this signaling pathway in endocrine and other tumorigenesis; on-going studies aim at characterizing this involvement better.
Supplementary Material
References
- Almeida MQ, Brito LP, Domenice S, Costa MH, Pinto EM, Osorio CA, Latronico AC, Mendonca BB, Fragoso MC. Absence of PRKAR1A loss of heterozygosity in laser-captured microdissected pigmented nodular adrenocortical tissue from a patient with Carney complex caused by the novel nonsense mutation p.Y21X. Arq Bras Endocrinol Metabol. 2008;52(8):1257–63. doi: 10.1590/s0004-27302008000800009. [DOI] [PubMed] [Google Scholar]
- Amieux PS, McKnight GS. The essential role of RI alpha in the maintenance of regulated PKA activity. Ann N Y Acad Sci. 2002;968:75–95. doi: 10.1111/j.1749-6632.2002.tb04328.x. [DOI] [PubMed] [Google Scholar]
- Bertherat J, Groussin L, Sandrini F, Matyakhina L, Bei T, Stergiopoulos S, Papageorgiou T, Bourdeau I, Kirschner LS, Vincent-Dejean C, et al. Molecular and functional analysis of PRKAR1A and its locus (17q22-24) in sporadic adrenocortical tumors: 17q losses, somatic mutations, and protein kinase A expression and activity. Cancer Res. 2003;63(17):5308–19. [PubMed] [Google Scholar]
- Bertherat J, Horvath A, Groussin L, Grabar S, Boikos S, Cazabat L, Libe R, Rene-Corail F, Stergiopoulos S, Bourdeau I, et al. Mutations in regulatory subunit type 1A of cyclic AMP-dependent protein kinase (PRKAR1A): phenotype analysis in 353 patients and 80 different genotypes. J Clin Endocrinol Metab. 2009 doi: 10.1210/jc.2008-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyth M, Huang S, Maloney V, Crolla JA, Karen Temple I. A 2.3Mb deletion of 17q24.2-q24.3 associated with ‘Carney Complex plus’. Eur J Med Genet. 2008;51(6):672–8. doi: 10.1016/j.ejmg.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Bridge J, Sanger W, Mosher G, Buehler B, Hearty C, Olney A, Fordyce R. Partial duplication of distal 17q. Am J Med Genet. 1985;22(2):229–35. doi: 10.1002/ajmg.1320220203. [DOI] [PubMed] [Google Scholar]
- Carney JA. Carney complex: the complex of myxomas, spotty pigmentation, endocrine overactivity, and schwannomas. Semin Dermatol. 1995;14(2):90–8. doi: 10.1016/s1085-5629(05)80003-3. [DOI] [PubMed] [Google Scholar]
- Carney JA, Hruska LS, Beauchamp GD, Gordon H. Dominant inheritance of the complex of myxomas, spotty pigmentation, and endocrine overactivity. Mayo Clin Proc. 1986;61(3):165–72. doi: 10.1016/s0025-6196(12)61843-6. [DOI] [PubMed] [Google Scholar]
- Casey M, Mah C, Merliss AD, Kirschner LS, Taymans SE, Denio AE, Korf B, Irvine AD, Hughes A, Carney JA, et al. Identification of a novel genetic locus for familial cardiac myxomas and Carney complex. Circulation. 1998;98(23):2560–6. doi: 10.1161/01.cir.98.23.2560. [DOI] [PubMed] [Google Scholar]
- Casey M, Vaughan CJ, He J, Hatcher CJ, Winter JM, Weremowicz S, Montgomery K, Kucherlapati R, Morton CC, Basson CT. Mutations in the protein kinase A R1alpha regulatory subunit cause familial cardiac myxomas and Carney complex. J Clin Invest. 2000;106(5):R31–8. doi: 10.1172/JCI10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazabat L, Libe R, Perlemoine K, Rene-Corail F, Burnichon N, Gimenez-Roqueplo AP, Dupasquier-Fediaevsky L, Bertagna X, Clauser E, Chanson P, et al. Germline inactivating mutations of the aryl hydrocarbon receptor-interacting protein gene in a large cohort of sporadic acromegaly: mutations are found in a subset of young patients with macroadenomas. Eur J Endocrinol. 2007;157(1):1–8. doi: 10.1530/EJE-07-0181. [DOI] [PubMed] [Google Scholar]
- Gennari M, Stratakis CA, Hovarth A, Pirazzoli P, Cicognani A. A novel PRKAR1A mutation associated with hepatocellular carcinoma in a young patient and a variable Carney complex phenotype in affected subjects in older generations. Clin Endocrinol (Oxf) 2008;69(5):751–5. doi: 10.1111/j.1365-2265.2008.03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EL, Horvath AD, Nesterova M, Giatzakis C, Bossis I, Stratakis CA. In vitro functional studies of naturally occurring pathogenic PRKAR1A mutations that are not subject to nonsense mRNA decay. Hum Mutat. 2008;29(5):633–9. doi: 10.1002/humu.20688. [DOI] [PubMed] [Google Scholar]
- Groussin L, Horvath A, Jullian E, Boikos S, Rene-Corail F, Lefebvre H, Cephise-Velayoudom FL, Vantyghem MC, Chanson P, Conte-Devolx B, et al. A PRKAR1A mutation associated with primary pigmented nodular adrenocortical disease in 12 kindreds. J Clin Endocrinol Metab. 2006;91(5):1943–9. doi: 10.1210/jc.2005-2708. [DOI] [PubMed] [Google Scholar]
- Groussin L, Kirschner LS, Vincent-Dejean C, Perlemoine K, Jullian E, Delemer B, Zacharieva S, Pignatelli D, Carney JA, Luton JP, et al. Molecular analysis of the cyclic AMP-dependent protein kinase A (PKA) regulatory subunit 1A (PRKAR1A) gene in patients with Carney complex and primary pigmented nodular adrenocortical disease (PPNAD) reveals novel mutations and clues for pathophysiology: augmented PKA signaling is associated with adrenal tumorigenesis in PPNAD. Am J Hum Genet. 2002;71(6):1433–42. doi: 10.1086/344579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbach MM, Biller BM, Braunstein GD, Campbell KK, Carney JA, Godley PA, Harris EL, Lee JK, Oertel YC, Posner MC, et al. Management of the clinically inapparent adrenal mass (“incidentaloma”) Ann Intern Med. 2003;138(5):424–9. doi: 10.7326/0003-4819-138-5-200303040-00013. [DOI] [PubMed] [Google Scholar]
- Gu YY, Chen Y, Song HD, Li XY, Luo TH, Qiao JO, Zhang Y, Xiao JC, Zhu Y, Zhao YJ, et al. Clinical and molecular research in a case of familial Carney complex. Zhonghua Nei Ke Za Zhi. 2004;43(10):764–8. [PubMed] [Google Scholar]
- Gunther DF, Bourdeau I, Matyakhina L, Cassarino D, Kleiner DE, Griffin K, Courkoutsakis N, Abu-Asab M, Tsokos M, Keil M, et al. Cyclical Cushing syndrome presenting in infancy: an early form of primary pigmented nodular adrenocortical disease, or a new entity? J Clin Endocrinol Metab. 2004;89(7):3173–82. doi: 10.1210/jc.2003-032247. [DOI] [PubMed] [Google Scholar]
- Horvath A, Boikos S, Giatzakis C, Robinson-White A, Groussin L, Griffin KJ, Stein E, Levine E, Delimpasi G, Hsiao HP, et al. A genome-wide scan identifies mutations in the gene encoding phosphodiesterase 11A4 (PDE11A) in individuals with adrenocortical hyperplasia. Nat Genet. 2006a;38(7):794–800. doi: 10.1038/ng1809. [DOI] [PubMed] [Google Scholar]
- Horvath A, Bossis I, Giatzakis C, Levine E, Weinberg F, Meoli E, Robinson-White A, Siegel J, Soni P, Groussin L, et al. Large deletions of the PRKAR1A gene in Carney complex. Clin Cancer Res. 2008a;14(2):388–95. doi: 10.1158/1078-0432.CCR-07-1155. [DOI] [PubMed] [Google Scholar]
- Horvath A, Giatzakis C, Robinson-White A, Boikos S, Levine E, Griffin K, Stein E, Kamvissi V, Soni P, Bossis I, et al. Adrenal hyperplasia and adenomas are associated with inhibition of phosphodiesterase 11A in carriers of PDE11A sequence variants that are frequent in the population. Cancer Res. 2006b;66(24):11571–5. doi: 10.1158/0008-5472.CAN-06-2914. [DOI] [PubMed] [Google Scholar]
- Horvath A, Giatzakis C, Tsang K, Greene E, Osorio P, Boikos S, Libe R, Patronas Y, Robinson-White A, Remmers E, et al. A cAMP-specific phosphodiesterase (PDE8B) that is mutated in adrenal hyperplasia is expressed widely in human and mouse tissues: a novel PDE8B isoform in human adrenal cortex. Eur J Hum Genet. 2008b;16(10):1245–53. doi: 10.1038/ejhg.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc Natl Acad Sci U S A. 1997a;94(21):11184–9. doi: 10.1073/pnas.94.21.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J Biol Chem. 1997b;272(12):8057–64. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- Imai Y, Taketani T, Maemura K, Takeda N, Harada T, Nojiri T, Kawanami D, Monzen K, Hayashi D, Murakawa Y, et al. Genetic analysis in a patient with recurrent cardiac myxoma and endocrinopathy. Circ J. 2005;69(8):994–5. doi: 10.1253/circj.69.994. [DOI] [PubMed] [Google Scholar]
- Kinderman FS, Kim C, von Daake S, Ma Y, Pham BQ, Spraggon G, Xuong NH, Jennings PA, Taylor SS. A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol Cell. 2006;24(3):397–408. doi: 10.1016/j.molcel.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner LS, Carney JA, Pack SD, Taymans SE, Giatzakis C, Cho YS, Cho-Chung YS, Stratakis CA. Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nat Genet. 2000a;26(1):89–92. doi: 10.1038/79238. [DOI] [PubMed] [Google Scholar]
- Kirschner LS, Sandrini F, Monbo J, Lin JP, Carney JA, Stratakis CA. Genetic heterogeneity and spectrum of mutations of the PRKAR1A gene in patients with the carney complex. Hum Mol Genet. 2000b;9(20):3037–46. doi: 10.1093/hmg/9.20.3037. [DOI] [PubMed] [Google Scholar]
- Levin ML, Shaffer LG, Lewis R, Gresik MV, Lupski JR. Unique de novo interstitial deletion of chromosome 17, del(17) (q23.2q24.3) in a female newborn with multiple congenital anomalies. Am J Med Genet. 1995;55(1):30–2. doi: 10.1002/ajmg.1320550110. [DOI] [PubMed] [Google Scholar]
- Mabuchi T, Shimizu M, Ino H, Yamguchi M, Terai H, Fujino N, Nagata M, Sakata K, Inoue M, Yoneda T, et al. PRKAR1A gene mutation in patients with cardiac myxoma. Int J Cardiol. 2005;102(2):273–7. doi: 10.1016/j.ijcard.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Meoli E, Bossis I, Cazabat L, Mavrakis M, Horvath A, Stergiopoulos S, Shiferaw ML, Fumey G, Perlemoine K, Muchow M, et al. Protein kinase A effects of an expressed PRKAR1A mutation associated with aggressive tumors. Cancer Res. 2008;68(9):3133–41. doi: 10.1158/0008-5472.CAN-08-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D. The New York Cancer Project: rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81(2):301–10. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olney PN, Kean LS, Graham D, Elsas LJ, May KM. Campomelic syndrome and deletion of SOX9. Am J Med Genet. 1999;84(1):20–4. [PubMed] [Google Scholar]
- Perdigao PF, Stergiopoulos SG, De Marco L, Matyakhina L, Boikos SA, Gomez RS, Pimenta FJ, Stratakis CA. Molecular and immunohistochemical investigation of protein kinase a regulatory subunit type 1A (PRKAR1A) in odontogenic myxomas. Genes Chromosomes Cancer. 2005;44(2):204–11. doi: 10.1002/gcc.20232. [DOI] [PubMed] [Google Scholar]
- Robinson-White A, Meoli E, Stergiopoulos S, Horvath A, Boikos S, Bossis I, Stratakis CA. PRKAR1A Mutations and protein kinase A interactions with other signaling pathways in the adrenal cortex. J Clin Endocrinol Metab. 2006a;91(6):2380–8. doi: 10.1210/jc.2006-0188. [DOI] [PubMed] [Google Scholar]
- Robinson-White AJ, Leitner WW, Aleem E, Kaldis P, Bossis I, Stratakis CA. PRKAR1A inactivation leads to increased proliferation and decreased apoptosis in human B lymphocytes. Cancer Res. 2006b;66(21):10603–12. doi: 10.1158/0008-5472.CAN-06-2200. [DOI] [PubMed] [Google Scholar]
- Sandrini F, Matyakhina L, Sarlis NJ, Kirschner LS, Farmakidis C, Gimm O, Stratakis CA. Regulatory subunit type I-alpha of protein kinase A (PRKAR1A): a tumor-suppressor gene for sporadic thyroid cancer. Genes Chromosomes Cancer. 2002;35(2):182–92. doi: 10.1002/gcc.10112. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Horikawa Y, Suwa T, Enya M, Kawachi S, Takeda J. Case report of familial Carney complex due to novel frameshift mutation c.597del C (p.Phe200LeufsX6) in PRKAR1A. Mol Genet Metab. 2008;95(3):182–7. doi: 10.1016/j.ymgme.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101(8):2381–411. doi: 10.1021/cr000236l. [DOI] [PubMed] [Google Scholar]
- Skalhegg BS, Tasken K, Hansson V, Huitfeldt HS, Jahnsen T, Lea T. Location of cAMP-dependent protein kinase type I with the TCR-CD3 complex. Science. 1994;263(5143):84–7. doi: 10.1126/science.8272870. [DOI] [PubMed] [Google Scholar]
- Skamrov AV, Feoktistova ES, Khaspekov GL, Kovalevskii DA, Goriunova LE, Bibilashvili R, Vinnitskii LI, Sheremet’eva GF, Nechaenko MA. PRKAR1A gene mutations in two patients with myxoma syndrome (Carney complex) Kardiologiia. 2003;43(7):77–82. [PubMed] [Google Scholar]
- Stratakis CA, Carney JA, Lin JP, Papanicolaou DA, Karl M, Kastner DL, Pras E, Chrousos GP. Carney complex, a familial multiple neoplasia and lentiginosis syndrome. Analysis of 11 kindreds and linkage to the short arm of chromosome 2. J Clin Invest. 1996;97(3):699–705. doi: 10.1172/JCI118467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratakis CA, Kirschner LS, Carney JA. Carney complex: diagnosis and management of the complex of spotty skin pigmentation, myxomas, endocrine overactivity, and schwannomas. Am J Med Genet. 1998;80(2):183–5. doi: 10.1002/(sici)1096-8628(19981102)80:2<183::aid-ajmg19>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Stratakis CA, Kirschner LS, Carney JA. Clinical and molecular features of the Carney complex: diagnostic criteria and recommendations for patient evaluation. J Clin Endocrinol Metab. 2001;86(9):4041–6. doi: 10.1210/jcem.86.9.7903. [DOI] [PubMed] [Google Scholar]
- Tasken K, Skalhegg BS, Tasken KA, Solberg R, Knutsen HK, Levy FO, Sandberg M, Orstavik S, Larsen T, Johansen AK, et al. Structure, function, and regulation of human cAMP-dependent protein kinases. Adv Second Messenger Phosphoprotein Res. 1997;31:191–204. doi: 10.1016/s1040-7952(97)80019-5. [DOI] [PubMed] [Google Scholar]
- Vargas-Alarcon G, Vargas-Barron J, Cruz-Robles D, Perez-Vielma N, Garcia-Trejo JJ, Aguilar-Gaytan R, Cortes-Hernandez P, Vazquez-Ortiz ZY, Romero-Cardenas A. A deletion in the PRKAR1A gene is associated with Carney complex. J Pediatr Endocrinol Metab. 2008;21(7):705–9. doi: 10.1515/jpem.2008.21.7.705. [DOI] [PubMed] [Google Scholar]
- Veugelers M, Wilkes D, Burton K, McDermott DA, Song Y, Goldstein MM, La Perle K, Vaughan CJ, O’Hagan A, Bennett KR, et al. Comparative PRKAR1A genotype-phenotype analyses in humans with Carney complex and prkar1a haploinsufficient mice. Proc Natl Acad Sci U S A. 2004;101(39):14222–7. doi: 10.1073/pnas.0405535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein LS, Shenker A, Gejman PV, Merino MJ, Friedman E, Spiegel AM. Activating mutations of the stimulatory G protein in the McCune-Albright syndrome. N Engl J Med. 1991;325(24):1688–95. doi: 10.1056/NEJM199112123252403. [DOI] [PubMed] [Google Scholar]
- Witsch-Baumgartner M, Clayton P, Clusellas N, Haas D, Kelley RI, Krajewska-Walasek M, Lechner S, Rossi M, Zschocke J, Utermann G. Identification of 14 novel mutations in DHCR7 causing the Smith-Lemli-Opitz syndrome and delineation of the DHCR7 mutational spectra in Spain and Italy. Hum Mutat. 2005;25(4):412. doi: 10.1002/humu.9328. [DOI] [PubMed] [Google Scholar]
- Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. 2004;5(12):959–70. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- Zawadzki KM, Taylor SS. cAMP-dependent protein kinase regulatory subunit type IIbeta: active site mutations define an isoform-specific network for allosteric signaling by cAMP. J Biol Chem. 2004;279(8):7029–36. doi: 10.1074/jbc.M310804200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.