Abstract
Anaerobic ammonium-oxidizing (anammox) bacteria are divided into three compartments by bilayer membranes (from out- to inside): paryphoplasm, riboplasm and anammoxosome. It is proposed that the anammox reaction is performed by proteins located in the anammoxosome and on its membrane giving rise to a proton-motive-force and subsequent ATP synthesis by membrane-bound ATPases. To test this hypothesis, we investigated the location of membrane-bound ATPases in the anammox bacterium ‘Candidatus Kuenenia stuttgartiensis’. Four ATPase gene clusters were identified in the K. stuttgartiensis genome: one typical F-ATPase, two atypical F-ATPases and a prokaryotic V-ATPase. K. stuttgartiensis transcriptomic and proteomic analysis and immunoblotting using antisera directed at catalytic subunits of the ATPase gene clusters indicated that only the typical F-ATPase gene cluster most likely encoded a functional ATPase under these cultivation conditions. Immunogold localization showed that the typical F-ATPase was predominantly located on both the outermost and anammoxosome membrane and to a lesser extent on the middle membrane. This is consistent with the anammox physiology model, and confirms the status of the outermost cell membrane as cytoplasmic membrane. The occurrence of ATPase in the anammoxosome membrane suggests that anammox bacteria have evolved a prokaryotic organelle; a membrane-bounded compartment with a specific cellular function: energy metabolism.
Introduction
Anammox bacteria perform anaerobic ammonium oxidation (anammox) to dinitrogen gas and are applied for removal of ammonium from wastewater. They are also important in nature where they contribute significantly to oceanic nitrogen loss (Devol, 2003; Kuypers et al., 2003; 2005; Arrigo, 2005; Brandes et al., 2007). Anammox bacteria belong to the phylum Planctomycetes. One of the unique properties of species within this phylum is that they do not conform to the prokaryotic cell plan: their cells are compartmentalized and are proposed to lack a typical bacterial cell wall (Fuerst, 1995; Lindsay et al., 2001). The planctomycete compartmentalization is in some cases complex but always involves a single intracytoplasmic membrane defining a major cell compartment. Electron microscopy observations, chemical analysis, genome sequencing and resistance to beta-lactam and other cell wall-targeting antibiotics have revealed that planctomycetes lack the otherwise universal bacterial cell wall polymer peptidoglycan (König et al., 1984; Liesack et al., 1986; Stackebrandt et al., 1986; Fuerst, 1995) and an outer membrane typical of Gram-negative bacteria. The outermost membrane in planctomycetes has been defined as the cytoplasmic membrane based on the detection of RNA directly on its inner side by immunogold labelling and the position of the membrane and membrane surfaces relative to the cell wall visualized in thin sections of cryofixed and freeze-substituted cells and in freeze-fracture replicas (Lindsay et al., 1997; 2001;). The other, innermost, membrane has been defined as an intracytoplasmic membrane as it is within the cytoplasm and topologically inside the space bounded by the cytoplasmic membrane. The two compartments that are thus formed have been named ‘paryphoplasm’– the outermost compartment between cytoplasmic and intracytoplasmic membrane – and ‘riboplasm’– the innermost compartment inside the space bounded by the intracytoplasmic membrane. The riboplasm is the standard cytoplasmic compartment containing ribosomes and in most cases the nucleoid.
Anammox planctomycetes have an additional intracytoplasmic membrane compared with the standard planctomycete cell plan described above. The anammox cell is thus divided into three compartments by three individual bilayer membranes, from out- to inside; the paryphoplasm, riboplasm and anammoxosome compartment (Fig. 1). Anammox bacteria also contain unusual membrane lipids with ladderane moieties (linearly concatenated cyclobutane rings). These ladderane lipids are major membrane lipids of anammox membranes and are hypothesized to make these membranes highly impermeable (Sinninghe Damstéet al., 2002; 2005;) and to provide structural integrity to the cell.
Fig. 1.
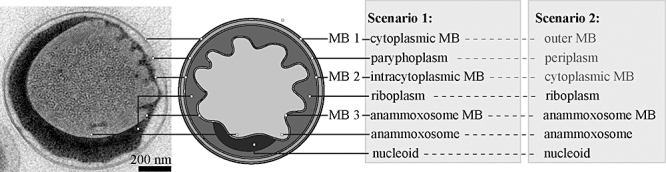
Electron micrograph (left) of an anammox bacterium and schematic overview (right) of the different scenarios regarding its cell plan. MB, membrane. Modified from van Niftrik et al. (2008b).
The anammoxosome compartment is hypothesized to be the site where the anammox catabolism takes place. Based on the genome of the anammox bacterium ‘Candidatus Kuenenia stuttgartiensis’, a biochemical model (Fig. 2) has been proposed where the anammox reaction is catalysed by several cytochrome c proteins (Strous et al., 2006). In this model, nitrite is first reduced to nitric oxide, then nitric oxide is combined with ammonium to form hydrazine and finally hydrazine is oxidized to dinitrogen gas. The four electrons derived from the oxidation of hydrazine to dinitrogen gas are fed into a respiratory chain at the level of ubiquinone via soluble cytochrome c electron carriers and a hypothetical cytochrome c: ubiquinone oxidoreductase (Cirpus et al., 2005; Huston et al., 2007). The anammox reaction thus establishes a proton gradient by the translocation of protons from the riboplasm to the anammoxosome, giving rise to a proton-motive-force. This proton-motive-force could then be used by membrane-bound ATPases to produce ATP. Experimental evidence that supports the proposed model is the immunogold localization of the enzyme hydrazine/hydroxylamine oxidoreductase (HAO) to the anammoxosome (Lindsay et al., 2001) and the demonstration that all, or almost all, cytochrome c proteins are located in the anammoxosome by cytochrome peroxidase staining (van Niftrik et al., 2008a). Furthermore, preliminary results from 31P nuclear magnetic resonance (NMR) spectroscopy showed the presence of two phosphate peaks in K. stuttgartiensis cells indicating an intracytoplasmic pH gradient (van der Star et al., 2010).
Fig. 2.
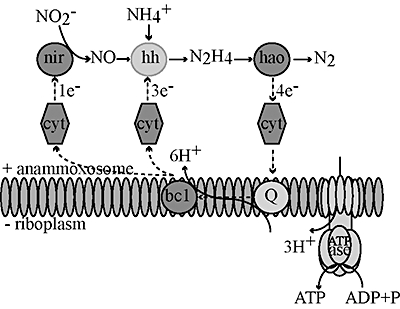
Postulated model for catabolic anammox reactions coupled over the anammoxosome membrane in anammox bacteria resulting in a proton-motive-force and subsequent ATP synthesis. bc1, cytochrome bc1 complex; cyt, cytochrome; hao, hydrazine/hydroxylamine oxidoreductase; hh, hydrazine hydrolase; nir, nitrite reductase; Q, co-enzyme Q. Modified from Strous et al. (2006).
Some questions concerning the anammox cell plan remain. Although it has been assumed that anammox bacteria share the planctomycete cell plan, there are at least two possibilities with respect to the nature of the paryphoplasm (or region equivalent to the paryphoplasm) in anammox bacteria. As in other planctomycetes, the anammox paryphoplasm may represent a space between a true cytoplasmic membrane and an intracytoplasmic membrane (Fig. 1; scenario 1). On the other hand, the paryphoplasm might represent a space similar to the periplasm of Gram-negative bacteria if the outermost membrane of the cell is more like an outer membrane of a Gram-negative cell wall and the intracytoplasmic membrane is actually the cytoplasmic membrane (Fig. 1; scenario 2).
To explore such potential similarities to a Gram-negative cell plan, the genome of K. stuttgartiensis (Strous et al., 2006) was examined by comparative genomic analysis, which indicated that K. stuttgartiensis may be genetically capable of the biogenesis of a periplasm and outer membrane. First, a number of open reading frames (ORFs) were homologous to outer membrane porins. These porin homologues were absent in the genome of the planctomycete Rhodopirellula baltica. Second, the K. stuttgartiensis genome encoded the complete TonB system, a protein complex that relays energy from the cytoplasmic membrane to the outer membrane to drive a number of outer membrane receptors, five of which were also encoded in the genome. Third, K. stuttgartiensis encoded a number of typical three-component Gram-negative multidrug exporters, which consist of a cytoplasmic membrane, a periplasmic and an outer membrane subunit (‘gated porins’). Fourth, a partial peptidoglycan biogenesis pathway was encoded, including a number of penicillin-binding proteins. The only step not present in the peptidoglycan pathway of this bacterium was the ability to cross-link the glycan. With respect to all these four points, R. baltica, another planctomycete with a publicly available genome, contains hardly any genetic potential for Gram-negative cell wall structure or peptidoglycan synthesis. This might be consistent with the view that the paryphoplasm (Fig. 1; scenario 1) in K. stuttgartiensis may actually be more similar to a ‘regular’ periplasm (Fig. 1; scenario 2). However, the presence of these genes could also be a result of lateral gene transfer or be remainders of the evolutionary ancestor of anammox bacteria, which would then be a Gram-negative bacterium.
In contrast to the genomic evidence that could support the paryphoplasm being a periplasmic-like space, there is experimental evidence that supports the paryphoplasm being a cytoplasmic compartment with the cytoplasmic membrane on its outer side and the absence of a typical bacterial cell wall. First, neither peptidoglycan nor a typical outer membrane can be observed in transmission electron micrographs of all known species of anammox bacteria when examined after cryofixation and freeze-substitution or via classical chemical fixation (van Niftrik et al., 2008a,b;). Second, anammox bacteria and other non-anammox planctomycetes are prone to osmotic collapse under both hypotonic and hypertonic conditions (see Lindsay et al., 1995; 2001; van de Graaf et al., 1996), an indication that their structural integrity, normally derived from the presence of a cell wall, is not optimal. Third, the cell division ring of anammox bacteria is situated in the paryphoplasm compartment (van Niftrik et al., 2009). In general, the bacterial cell division ring is on the inside of, and closely opposed to, the cytoplasmic membrane indicating that the membrane on the outside of the paryphoplasm (Fig. 1; membrane 1) is the cytoplasmic membrane. Fourth, the apparent absence of cytochrome c proteins in the paryphoplasm as indicated by cytochrome peroxidase staining (van Niftrik et al., 2008a) supports the notion that this cannot be a typical periplasmic space analogous to that of Gram-negative bacteria.
A third scenario may reconcile the experimental and genomic evidence. In this scenario there is a periplasmic-like space outside the outermost membrane (Fig. 1; membrane 1), as proposed for some Archaea with S-layer walls (Küper et al., 2010) and for some Gram-positive bacteria (Zuber et al., 2008). This periplasmic-like space could be lost during sample preparation for electron microscopy.
The localization of anammox membrane-bound ATPases would resolve both the function of the anammoxosome and the location of the cytoplasmic membrane. If an ATPase was present on the innermost (anammoxosome) membrane (Fig. 1; membrane 3) this would strongly suggest that this compartment is indeed used for the generation of energy analogous to the function of mitochondria in Eukaryotes. The localization of an anammox ATPase to the outermost membrane of the anammox cell (Fig. 1; membrane 1) would indicate that this is indeed an energized, cytoplasmic membrane, and not an outer membrane typical of a Gram-negative cell wall, and that the middle anammox membrane (Fig. 1; membrane 2) is indeed an intracytoplasmic membrane as was initially proposed on structural grounds (Lindsay et al., 2001).
Three different types of membrane-bound ATPases have been described: F-ATPase, A-ATPase and V-ATPase (Grüber et al., 2001; Grüber and Marshansky, 2008). V-ATPase (vacuolar-type) functions as an ATP-dependent proton-pump in eukaryotes (Drory and Nelson, 2006). F-ATPase and prokaryotic V-ATPase (the latter found in Bacteria and Archaea) can function both as ion-pump or as ATP synthase (Mitchell, 1961; Müller et al., 1999). The prokaryotic V-ATPase is also known as A-ATPase (archaeal-type) or V/A-ATPase (Grüber et al., 2001; Yokoyama and Imamura, 2005). Subunit composition can differ, but in general the prokaryotic V-ATPase consists of nine subunits and the bacterial F-ATPase consists of eight subunits (Stock et al., 2000; Lolkema et al., 2003) (Fig. 3). Membrane-bound ATPases contain two components: a membrane-bound, hydrophobic part (F0/A0/V0) containing the ion channel that is connected by a central stalk to the cytoplasmic, hydrophilic part (F1/A1/V1) containing the catalytic sites. The way F-ATPase functions is described by the binding change (Boyer, 1993; 1997;) and rotary mechanism (Stock et al., 2000). F1-ATPase contains three catalytic β subunits that perform ATP synthesis (or in some cases hydrolysis). F0-ATPase translocates protons (or sodium ions) across the membrane down the electrochemical gradient. In coupling proton transport to ATP synthesis, the F0 and F1 domains function as a pair of rotary motors linked by a common central rotor (γεc) and a peripheral stator (bδ). The multiple c-subunits (proteolipids) form a ring and rotation of this c-ring allows protons to be carried between two partial subunit-a channels that lead to opposite sites of the membrane. Each c-subunit contains two transmembrane helices that are connected by a cytoplasmic loop and one protonizable, carboxylate group (glutamate or aspartate; active carboxylate of Escherichia coli subunit c is Asp-61). The active carboxylate undergoes protonation/deprotonation cycles during proton transport and is located in helix 2 (Rastogi and Girvin, 1999). Either three or four protons need to be transported in sequence for a group of 12 c-subunits to move 120 degrees and promote the release of one ATP. Among the different membrane-bound ATPase types, the number of proteolipid transmembrane helices, and the number of proteolipid subunits per enzyme, differs.
Fig. 3.
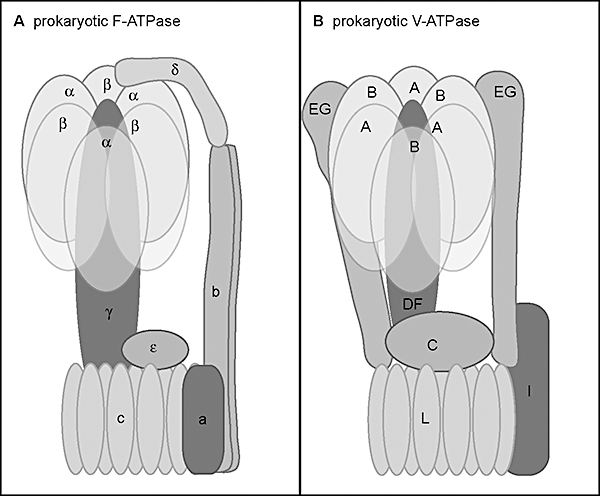
Schematic model of (A) a prokaryotic F-ATPase and (B) a prokaryotic V-ATPase. Assembled from Grüber et al. (2001), Lolkema et al. (2003), Yokoyama and Imamura (2005) and Lau and Rubinstein (2010).
Here, we identified and analysed four ATPase gene clusters in the K. stuttgartiensis genome. We show by transcriptomic, proteomic and immunoblot analyses that only one of these four ATPase gene clusters is likely to be expressed under the conditions investigated. Antiserum targeting this typical F-ATPase was used to locate this anammox membrane-bound ATPase in the anammox cell using immunogold localization. The typical F-ATPase was detected on all three anammox cell membranes but was predominantly present on both the innermost (anammoxosome) membrane and outermost membrane of the anammox cell. This indicates that the anammoxosome is used for the generation of ATP, probably vectorially from a proton-motive-force across the anammoxosome membrane itself, and is consistent with the identification of the outermost membrane of the anammox cell as the cytoplasmic membrane.
Results
ATPase gene clusters in the K. stuttgartiensis genome
Four putative gene clusters encoding membrane-bound ATPase complexes were identified in the genome assembly of K. stuttgartiensis (Strous et al., 2006) (Fig. 4). The syntheny (similarity of gene order) of gene cluster 1 was similar to the F-ATPase operon of E. coli with 23–69% sequence identities of individual orthologous genes to closest homologous genes with experimentally validated function. Note that in the databases where the K. stuttgartiensis genome has been deposited, the ORFs encoding for subunit b and subunit delta are translated in the wrong frame (3′5′−2 frame) as one ORF (kuste3792, unknown protein). Subunit b is actually encoded in the 5′3′+1 frame (169 aa, nucleotide position 1 430 441–1 430 947) and subunit delta is encoded in the 5′3′+2 frame (182 aa, nucleotide position 1 430 940–1 431 485). Putative gene clusters 2 and 3 were similar to each other and strongly resembled the atypical putative F-ATPase of Methanosarcina barkeri (Sumi et al., 1997) with 22–53% sequence identities of individual orthologous genes to closest homologous genes with experimentally validated function. The structure of gene cluster 4 resembled that of the Borrelia burgdorferi prokaryotic V-ATPase (Lolkema et al., 2003) with 30–72% sequence identities of individual orthologous genes to closest homologous genes (in this case all without experimentally validated function).
Fig. 4.

Comparison of the four ATPase gene clusters identified in the K. stuttgartiensis genome to their closest homologue. A. Gene cluster 1 (kuste3787–3796): F-ATPase similar to E. coli F-ATPase with the exception of an unknown ORF between gene I and subunit a and two ORFs encoding subunit c. B. Gene cluster 2 (kuste4592–4600) and 3 (kustc0572–0579): F-ATPases similar to M. barkeri F-ATPase. Gene cluster 3 lacks the unknown ORF, urf2, between gene I and subunit a. C. Gene cluster 4 (kuste3864–3871): V-ATPase similar to B. burgdorferi prokaryotic V-ATPase with the exception of an unknown ORF between subunit B and subunit D. Genes marked with a star are annotated as (conserved) hypothetical proteins. These genes have a sequence identity of over 20% to a protein with an undefined function or to a protein with a defined function but without similar sequence length. ORF, open reading frame.
The protein sequences of the K. stuttgartiensis proteolipids were aligned using mega version 4 (Tamura et al., 2007) and were investigated for predicted transmembrane helices using tmhmm 2.0 (Krogh et al., 2001) and TMpred (Hofmann and Stoffel, 1993). The proteolipids of the F-ATPase gene clusters (subunit c, gene cluster 1–3) were aligned to the E. coli c-subunit (Fig. S1A). The K. stuttgartiensis c-subunits all consisted of two transmembrane helices and contained the subunit c signature motif. There was one discrepancy; subunit c from F-ATPase-2 contained glutamine (Q) instead of arginine (R) in the second position of the subunit c signature motif. The K. stuttgartiensis c-subunits all had glutamate (F-ATPase-1; both c-subunits Glu-58, F-ATPase-2 & -3; Glu-63) as the putative protonizable group. In F-ATPase-2 and -3 the protonizable group was not located in the predicted transmembrane helix 2 but instead was found one or two amino acids before. The proteolipid of the prokaryotic V-ATPase-4 gene cluster (subunit L) did not align very well to the E. coli proteolipid and was therefore aligned to its closest homolog; subunit L from B. burgdorferi (Fig. S1B). The prokaryotic V-ATPase-4 subunit L consisted of four transmembrane helices and helix 4 contained a putative skewed subunit c signature motif with Glu-137 as a putative protonizable group. In this motif, R-[NQ]-P was replaced by D-A-L. As in B. burgdorferi, no second protonizable group (D or E) was present in or near helix 2. Sequence analysis of the proteolipids indicated that the three F-ATPases are potentially capable of ATP synthesis. With respect to gene cluster organization, F-ATPase-1 contained the complete set of genes known to be part of the E. coli F-ATPase and therefore there is no reason why this gene cluster could not encode for a functional membrane-bound ATPase. F-ATPase-2 and -3 lacked a gene encoding the delta subunit that in F-ATPase connects the F1 and F0 static parts (Fig. 3), although it is unknown whether this would infer dysfunctionality. The prokaryotic V-ATPase-4 gene cluster contained both catalytic subunits (B and A), the proteolipid (L), ion channel (I), part of the rotor (D) and part of the stator (E), indicating that it could encode for a functional membrane-bound ATPase.
K. stuttgartiensis transcriptomic and proteomic analysis
To investigate the expression of the four ATPase gene clusters identified in the genome, the transcriptome and proteome of K. stuttgartiensis were analysed for the presence of mRNA or detected peptides respectively (Table 1). The analysis showed that F-ATPase-1, the typical ATP synthase, was detected at high levels in both the transcriptome and proteome. This indicates that this ATPase gene cluster most likely encodes a functional membrane-bound ATPase in K. stuttgartiensis. Both atypical F-ATPases-2 and -3 were not detected in the proteome and showed very low transcription levels (relative expression < 1.0), making it unlikely that these two ATPase gene clusters function in energy generation in K. stuttgartiensis cells at least under the cultivation conditions of this study. For the prokaryotic V-ATPase-4 the catalytic subunits (A and B) were detected in the transcriptome (relative expression > 1.0) and one peptide (subunit A) was detected in the proteome. More data will be needed to make a functional assignment possible for the prokaryotic V-ATPase.
Table 1.
Presence of mRNA (transcriptome) and peptides (proteome) from the four ATPase gene clusters present in the genome of K. stuttgartiensis.
| Genome |
Transcriptome |
Proteome |
|||||
|---|---|---|---|---|---|---|---|
| ORF | Putative ATPase subunit | aa | nt | # unique reads detected | Relative expression | # unique peptides detected | Gene coverage (%) |
| F-ATPase-1 | |||||||
| kuste3787 | I | 84 | 255 | 50 | 0.9 | ND | NA |
| kuste3788 | Unknown | 124 | 375 | 136 | 1.6 | ND | NA |
| kuste3789 | a | 257 | 774 | 656 | 3.7 | 2 | 25 |
| kuste3790 | c | 108 | 327 | 204 | 2.8 | ND | NA |
| kuste3791 | c | 90 | 273 | 317 | 5.1 | 1 | 25 |
| kuste3792 | b | 169 | 510 | 882 | 7.6 | ND | NA |
| kuste3792 | Delta | 182 | 549 | 1013 | 8.1 | ND | NA |
| kuste3793 | Alpha | 504 | 1515 | 1881 | 5.5 | 23 | 66 |
| kuste3794 | Gamma | 290 | 873 | 415 | 2.1 | 8 | 29 |
| kuste3795 | Beta | 471 | 1416 | 1956 | 6.1 | 24 | 60 |
| kuste3796 | Epsilon | 134 | 405 | 413 | 4.5 | 3 | 27 |
| F-ATPase-2 | |||||||
| kuste4592 | Beta | 462 | 1389 | 74 | 0.2 | ND | NA |
| kuste4593 | Epsilon | 130 | 393 | 16 | 0.2 | ND | NA |
| kuste4594 | I | 111 | 336 | 31 | 0.4 | ND | NA |
| kuste4595 | Unknown | 101 | 306 | 18 | 0.3 | ND | NA |
| kuste4596 | a | 236 | 711 | 68 | 0.4 | ND | NA |
| kuste4597 | c | 93 | 282 | 15 | 0.2 | ND | NA |
| kuste4598 | b | 255 | 768 | 21 | 0.1 | ND | NA |
| kuste4599 | Alpha | 513 | 1542 | 101 | 0.3 | ND | NA |
| kuste4600 | Gamma | 299 | 900 | 67 | 0.3 | ND | NA |
| F-ATPase-3 | |||||||
| kustc0572 | Beta | 454 | 1365 | 177 | 0.6 | ND | NA |
| kustc0573 | Epsilon | 134 | 405 | 12 | 0.1 | ND | NA |
| kustc0574 | I | 92 | 279 | 32 | 0.5 | ND | NA |
| kustc0575 | a | 221 | 666 | 39 | 0.3 | ND | NA |
| kustc0576 | c | 88 | 267 | 34 | 0.6 | ND | NA |
| kustc0577 | b | 246 | 741 | 35 | 0.2 | ND | NA |
| kustc0578 | Alpha | 498 | 1497 | 78 | 0.2 | ND | NA |
| kustc0579 | Gamma | 279 | 840 | 16 | 0.1 | ND | NA |
| V-ATPase-4 | |||||||
| kuste3864 | E | 210 | 633 | 99 | 0.7 | ND | NA |
| kuste3865 | Unknown | 180 | 543 | 74 | 0.6 | ND | NA |
| kuste3866 | A | 589 | 1770 | 531 | 1.3 | 1 | 2 |
| kuste3867 | B | 437 | 1314 | 503 | 1.7 | ND | NA |
| kuste3868 | Unknown | 99 | 300 | 36 | 0.5 | ND | NA |
| kuste3869 | D | 202 | 609 | 21 | 0.2 | ND | NA |
| kuste3870 | I | 583 | 1752 | 218 | 0.6 | ND | NA |
| kuste3871 | L | 152 | 459 | 66 | 0.6 | ND | NA |
ORF, open reading frame; aa, amino acids; nt, nucleotides; ND, not detected; NA, not applicable. The relative expression (transcriptome) is calculated as: [# unique reads detected * length of the reads (75 nt)/gene length (nt)]/coverage of the transcriptome (= 17). The gene coverage (proteome) is calculated as: (# unique peptides detected/# unique peptides predicted) * 100%.
Heterologous expression of the catalytic subunits of the four K. stuttgartiensis ATPase gene clusters and subsequent antibody production
Parts of the catalytic beta (F-ATPases) and A (prokaryotic V-ATPase) subunits (Table S1) were cloned and expressed in E. coli. The purified protein fragments were used to raise antibodies in rabbit. All four antisera hybridized to the associated heterologous protein on dot blots (data not shown). The antisera were subsequently used to test for the presence of their target proteins in K. stuttgartiensis cell-free extract using SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis. The antiserum targeting F-ATPase-1 showed a band at the expected size (52 kDa) that was absent in both negative controls (incubation with pre-immune serum and incubation with only the secondary antibody) (Fig. 5). Incubation with the other antisera (anti-F-ATPase-2, -F-ATPase-3 and -V-ATPase-4) did not result in the detection of a protein of the expected size, nor of any other specific bands (data not shown). These results were consistent with the high levels of transcription and expression observed for F-ATPase-1 and the undetectable or at least very low levels of transcription and expression of the other membrane-bound ATPases under the applied growth conditions.
Fig. 5.

Immunoblot analysis of the antiserum directed at the catalytic beta subunit of the F-ATPase-1 gene cluster found in the K. stuttgartiensis genome. Lane 1: Marker (PageRuler™ Prestained Protein Ladder Plus, Fermentas); lane 2: incubation with anti-F-ATPase-1; lane 3: incubation with the pre-immune serum; lane 4: incubation with only the secondary antibody. Arrow: expected target size (52 kDa).
Anti-F-ATPase-1 was further tested for its specificity and affinity using immunofluorescence analysis of formaldehyde-fixed K. stuttgartiensis cells. This analysis showed that the antiserum specifically bound to whole K. stuttgartiensis cells (Fig. S2) while the pre-immune serum had only a very faint background at the same exposure time. This indicated that this antiserum was suitable for immunogold localization to identify the cellular location of this membrane-bound ATPase in the anammox cell.
Immunogold localization of F-ATPase-1 in K. stuttgartiensis
The antiserum targeting F-ATPase-1 was used in immunogold localization to determine its location in anammox cells prepared via the rehydration method (van Donselaar et al., 2007) and subsequent cryosectioning. All three membranes of the anammox cell, the outermost membrane (membrane 1), middle membrane (membrane 2) and innermost (anammoxosome) membrane (membrane 3), which appear as white trilamina in the cryosections, were significantly labelled (Figs 6 and 7 and Fig. S3) compared with the incubation with the pre-immune serum (the negative control). There was some background labelling in the anammoxosome and riboplasm but this was not significantly more in the incubation with the antiserum than in the incubation with the pre-immune serum. Since gold particles that could be attributed to more than one membrane were not taken into account, the labelling of the membranes is even underestimated. There was significantly more labelling on the outermost membrane and the innermost (anammoxosome) membrane compared with the middle membrane while there was no significant difference between the labelling of the outermost membrane and the innermost (anammoxosome) membrane. From these results, it can be concluded that the F-ATPase-1 is predominantly located on the innermost (anammoxosome) membrane (membrane 3) and the outermost membrane (membrane 1) and to a lesser extent on the middle membrane (membrane 2).
Fig. 6.
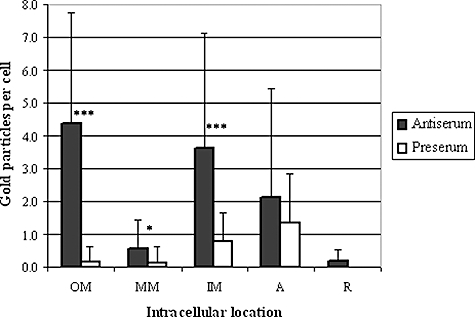
Immunogold labelling distribution (average gold particles per cell per location) in K. stuttgartiensis using the antiserum directed at F-ATPase-1 and its pre-immune serum. All three membranes of the anammox cell are significantly labelled compared with the incubation with the pre-immune serum (the negative control). OM, outermost membrane (membrane 1); MM, middle membrane (membrane 2); IM, innermost (anammoxosome) membrane (membrane 3); A, anammoxosome; R, riboplasm; ***, extremely statistically significant; *, statistically significant.
Fig. 7.
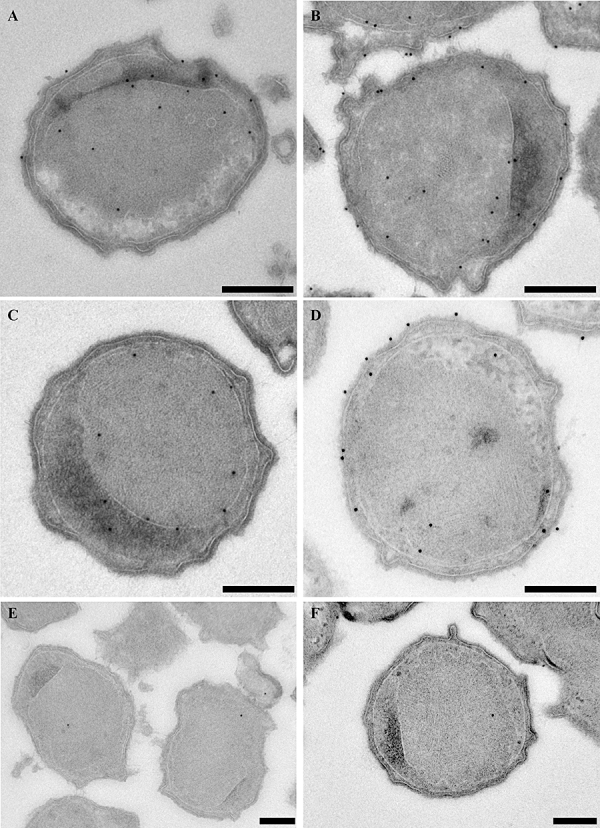
A–D. Immunogold localization of the antiserum directed at the catalytic beta subunit of the F-ATPase-1 gene cluster localizes this ATPase to all anammox membranes; the outermost membrane (membrane 1), middle membrane (membrane 2) and innermost (anammoxosome) membrane (membrane 3) in K. stuttgartiensis rehydrated cryosections. See Fig. S3 for annotation of specific gold labels. E and F. Negative control: incubation with the pre-immune serum instead of the antiserum. Scale bars: 250 nm.
Discussion
In this study, the intracellular location of membrane-bound ATPases in the anammox bacterium K. stuttgartiensis was addressed to investigate the hypothesis that the anammoxosome compartment and its membrane are used for the generation of energy and to identify the cytoplasmic membrane. Four putative ATPase gene clusters (F-ATPase-1, -2, -3, and prokaryotic V-ATPase-4) were identified in the K. stuttgartiensis genome. Transcriptomic, proteomic and immunoblot analyses with antisera directed at the catalytic subunits indicated that F-ATPase-1 was the most significant membrane-bound ATPase under the growth conditions investigated. Immunogold localization showed that this F-ATPase was predominantly present on the innermost (anammoxosome) membrane and outermost membrane of the anammox cell and to a lesser extent on the middle membrane.
Four ATPase gene clusters in the K. stuttgartiensis genome
The genome of K. stuttgartiensis encodes four putative ATPase gene clusters: one typical F-ATPase (F-ATPase-1), two atypical F-ATPases (F-ATPase-2 and -3) lacking the delta subunit and a prokaryotic V-ATPase (V-ATPase-4).
F-ATPase-1 resembled most the typical F-ATPase from E. coli but also the F-ATPase from Acetobacterium woodii (Müller et al., 2005). The K. stuttgartiensis transcriptome showed a high relative expression (ranging from 0.9–8.1) of all genes in the F-ATPase-1 gene cluster and in the proteome, peptides of six of the 11 genes were detected (Table 1). Further, the antiserum directed at the catalytic beta subunit specifically detected a protein of the expected size in immunoblot analysis (Fig. 5) and hybridized specifically to formaldehyde-fixed K. stuttgartiensis cells in immunofluorescence (Fig. S2). In conclusion, the transcriptome, proteome, immunoblot and immunofluorescence data indicated that the F-ATPase-1 gene cluster encodes the major functional membrane-bound ATPase in K. stuttgartiensis.
The gene cluster structure of F-ATPase-2 and F-ATPase-3 strongly resembled that of the atypical M. barkeri F-ATPase (Fig. 4) (Sumi et al., 1997). These gene clusters lacked a gene encoding the delta subunit that in F-ATPase connects the F1 and F0 static parts (Fig. 3). Without this subunit, the ATPase enzyme would probably be less stable. Therefore, if these gene clusters are expressed they would have to acquire the delta subunit from somewhere else, which would have to be the F-ATPase-1 gene cluster. Another possibility could be that the extended version of their b-subunit (255 aa and 246 aa compared with 169 aa in F-ATPase-1, Table 1) has functionally replaced the delta subunit. In M. barkeri it is doubtful whether this F-ATPase is functional because the amino acid sequences of the genes are so deviated from the typical F-ATPase and also because mRNA and protein products cannot be detected (Sumi et al., 1997). A functional F-ATPase in M. barkeri would be quite unusual anyway since Archaea (including M. barkeri) typically have prokaryotic V-ATPases and not F-ATPases. The transcriptome of K. stuttgartiensis showed a very low relative expression level (< 1.0) for both ATPase gene clusters and no peptides were detected in the proteome (Table 1). Antibodies generated against the catalytic subunits also could not detect their target proteins in K. stuttgartiensis cell-free extract under the cultivation conditions of this study. In conclusion, the predicted instability of both enzymes by the absence of the delta subunit, the very low expression level in the transcriptome and absence of peptides in the proteome and the inability of the antisera to detect their target proteins in cell-free extract make it very unlikely that these two gene clusters encode functional F-ATPases in K. stuttgartiensis. Perhaps these two gene clusters are cryptic, were acquired through lateral gene transfer and were duplicated in a subsequent rearrangement event.
The gene cluster structure of V-ATPase-4 resembled most that of B. burgdorferi prokaryotic V-ATPase (Fig. 4). These two gene clusters, and also the chlamydial prokaryotic V-ATPase gene clusters (Lolkema et al., 2003), do not contain homologues of subunits C and F (part of the central stalk), and G (part of the stator) (Fig. 3). It was not clear from the transcriptome and proteome whether gene cluster V-ATPase-4 could encode for a functional membrane-bound ATPase. Only the catalytic subunits (A and B) were detected, at a low level, in the transcriptome and only one peptide (subunit A) was detected in the proteome. Further, the antiserum targeting the catalytic subunit A could not detect its target protein in K. stuttgartiensis cell-free extract. In conclusion, either the V-ATPase-4 gene cluster is not expressed as a functional membrane-bound ATPase under the cultivation conditions of this study or it is expressed at such low levels that it is difficult to detect with the current methods.
The intracellular location of F-ATPase-1 in K. stuttgartiensis cells
All data indicated that the F-ATPase-1 gene cluster encoded a functional membrane-bound ATPase capable of ATP synthesis and that the other ATPase gene clusters were most likely not expressed in the cells used in this study. Therefore, only the antiserum targeting F-ATPase-1 was used in immunogold localization to determine its location in anammox cells prepared via the rehydration method and cryosectioning (van Donselaar et al., 2007). Note that, even if the atypical F-ATPase-2 and -3 could be expressed, cross-reactivity with the antiserum targeting the beta subunit of F-ATPase-1 was not to be expected since the antibody target of F-ATPase-1 showed only a 37% protein sequence identity with the beta subunits of the atypical F-ATPase-2 and -3. Anti-F-ATPase-1 located the F-ATPase to all three anammox membranes (Figs 6 and 7 and Fig. S3). The labelling was equally high on both the innermost (anammoxosome) membrane (membrane 3) and outermost membrane (membrane 1). Further, both of these membranes were significantly more densely labelled than the middle membrane (membrane 2) although the labelling on the middle membrane was still significantly higher than in the negative control. The immunogold labelling of the anammox membranes is even underestimated. First, gold particles that could be allocated to more than one membrane (i.e. within 25 nm distance from more than one membrane) were not taken into account. Second, labelling in the anammoxosome compartment could still be real innermost (anammoxosome) membrane labelling considering the high curvature of this anammoxosome membrane as visualized by electron tomography (van Niftrik et al., 2008b). These results indicate that the F-ATPase-1 is predominantly present on the innermost (anammoxosome) membrane and outermost membrane and to a lesser extent on the middle membrane and that thus all three membranes are energized and within the essential cell boundary. This is not consistent with the preliminary results using 31P NMR that indicated the presence of two pH peaks and thus most likely two energized membranes (van der Star et al., 2010). It is also inconsistent with the absence of genes encoding a putative nucleotide transporter in the genome of K. stuttgartiensis. Such a transporter would be necessary in a scenario with three energized membranes. Because most label was found on the anammoxosome and outer membranes we conclude that these membranes most likely have a proton-motive-force. Because the cytochromes are located inside the anammoxosome, F-ATPase-1 most likely produces ATP on this membrane. This ATP is then hydrolysed again by F-ATPase-1 on the outermost membrane. There are two explanations for the presence of F-ATPase in the middle membrane: it is the apo-complex en route to its final destination or it is functional, which we cannot explain at this moment.
In any case, the results so far indicate that the outermost membrane (membrane 1) is the cytoplasmic membrane as was initially proposed (Fig. 1, scenario 1) (Lindsay et al., 2001). To some extent, the ultrastructure and ATPase immunolabelling of the outermost membrane of K. stuttgartiensis resembles that of the Crenarchaeon Ignicoccus hospitalis (Küper et al., 2010). I. hospitalis also has a compartmentalized cell plan and an energized outermost membrane. However, in the I. hospitalis case, the outermost compartment has been defined as a periplasmic-like space based on its low electron density in transmission electron micrographs, which suggested the apparent absence of ribosomes and DNA. Consequently, when the outermost membrane was found to be energized, it was concluded that I. hospitalis has an energized ‘outer membrane’ (although this membrane is structurally not similar to the outer membrane of Gram-negative bacteria) and ATP synthesis in the ‘periplasm’. In a way, this resembles the K. stuttgartiensis case but in our case the compartments and membranes are defined differently. This raises the question what defines the cytoplasm, periplasm, cytoplasmic membrane and outer membrane?
The genetic potential found in the K. stuttgartiensis genome for producing a Gram-negative-like cell wall could be a cryptic result of lateral gene transfer or be a remainder of the evolutionary ancestor of anammox bacteria, which would then be a Gram-negative bacterium. However, another possibility is that there is indeed a Gram-negative-like cell wall in anammox bacteria but that this is lost during sample preparation for electron microscopy. Anammox bacteria certainly do not live in osmotically protected areas like the pathogenic cell wall-less mycoplasma species, so are in need of some form of structural integrity. Other planctomycetes possess proteinaceous cell walls and their cells show considerable structural integrity, even able in some cases to withstand treatment with 10% SDS at 100°C (König et al., 1984; Liesack et al., 1986), conditions under which Gram-negative cell walls and especially outer membranes would be expected to disintegrate. Perhaps in the anammox case, the ladderane lipids provide the structural integrity that most other bacteria derive from their cell wall. These lipids have been predominantly found in the anammoxosome membrane but also occur in one or both of the other two anammox cell membranes (Sinninghe Damstéet al., 2002). Alternatively, we may in fact lack some structural information and there may be yet another layer to the anammox cell. There is indeed some evidence of a regular protein surface layer (S-layer) lattice in K. stuttgartiensis from freeze-fracture replicas (Fuerst et al., 2006), but such an S-layer, if comparable to those protein or glycoprotein lattices of other bacterial and of archaeal cell walls, is unlikely to possess membrane properties. However, in Archaea, which often do not contain other cell wall components besides S-layers, S-layers are proposed to maintain cell shape and can be viewed as exoskeletons that contribute to mechanical and osmotic cell stabilization (Engelhardt, 2007). Perhaps in the anammox bacteria where, as in the Archaea, also no other cell wall components have been found, structural integrity is also derived from an S-layer lattice. Even though the complete loss of additional cell layers seems unlikely since many different fixation procedures have been applied to anammox bacteria (van Niftrik et al., 2008b), the presence of an outer membrane, exo- (S-layer) or endoskeleton (cytoskeleton) is currently under investigation by imaging anammox bacteria in their near-native state with cryoelectron microscopy.
The localization of an F-ATPase to the anammoxosome membrane shows that this compartment is indeed used for the generation of energy analogous to the function of mitochondria in Eukaryotes. With its curved membrane to maximize the membrane area available for the catabolic processes to take place (van Niftrik et al., 2008b), the localization of the key anammox protein HAO (Lindsay et al., 2001) and cytochrome c proteins (van Niftrik et al., 2008a) to the anammoxosome compartment, and the presence of an ATP synthase on the anammoxosome membrane established here, it seems that anammox bacteria may indeed have evolved a bacterial organelle: a separate membrane-bounded compartment with a specific function inside the cell: energy metabolism.
Experimental procedures
Kuenenia stuttgartiensis cells
Samples containing an 80% enrichment culture of K. stuttgartiensis were taken from a 2 l sequencing batch reactor or a 15 l continuous reactor (modified from Strous et al., 1998).
Genome analysis
Kuenenia stuttgartiensis ORFs encoding putative membrane-bound ATPase subunits were blasted (blastp) against the NCBI protein database (http://www.ncbi.nlm.nih.gov/BLAST/). Putative homologues were subsequently aligned using the PIR pairwise alignment tool (http://pir.georgetown.edu/pirwww/search/pairwise.shtml).
Transcriptomic analysis
Kuenenia stuttgartiensis RNA was extracted using the Ribopure Bacteria Kit according to the manufacturer's instructions from Ambion (Foster City, CA, USA). First-strand cDNA was synthesized with random primers using the RevertAid™ H Minus First Strand cDNA Synthesis Kit, and the second strand was synthesized using DNA polymerase and manufacturer's instructions (Fermentas, Vilnius, Lithuania). The Solexa reads (3.5 million) were mapped using the CLC Workbench software version 3.7.1 (http://www.clcbio.com). The program reports expression as Reads Per Kilobase of exon model per Million mapped reads (RPKM).
Proteomic analysis
Kuenenia stuttgartiensis single-cell suspensions were harvested by centrifugation. The cell pellet was resuspended in one volume of 20 mM potassium phosphate buffer pH 8, and passed three times through a French pressure cell operated at 138 MPa. After centrifugation for 15 min at 1700 g at 4°C, a cell-free fraction was obtained as clarified supernatant. A sample of the cell-free extract was denatured by incubation with 60 mM Tris-HCl buffer pH 8 containing 5% β-mercaptoethanol, 2% sodium dodecyl sulphate (SDS) and 25% glycerol for 5 min at 100°C. SDS-PAGE was performed in 10% or 6% slab gels in 375 mM Tris hydrochlorideglycine buffer, pH 8.8, with approximately 50 µg of protein per lane. After separation of proteins and staining with colloidal Coomassie Brilliant Blue, the gel lane was cut into four slices, and each gel slice was destained with three cycles of washing with successively 50 mM ammonium bicarbonate and 50% acetonitrile (ACN). Sample analysis by LC-MS/MS, including protein reduction, alkylation and digestion, was performed as described previously (Ettwig et al., 2010).
Antibody production
For the antisera directed against the catalytic beta (F-ATPase-1, -2 and -3) or A subunits (V-ATPase-4), we expressed and purified parts of the K. stuttgartiensis kuste3795 (ATPase gene cluster 1), kuste4592 (ATPase gene cluster 2), kustc0572 (ATPase gene cluster 3) and kuste3866 (ATPase gene cluster 4) ORFs in E. coli as described previously (Harhangi et al., 2002), with the following changes. Primers were designed on the sequences (Table S1). All forward primers started on nucleotide position 1 of the ORF. For directional cloning, restriction sites were included in all primers. Further, stop codons were introduced in the reverse primers so as to express only an N-terminal His-tag. As an expression vector, pET30a-c(+) (Novagen, Darmstadt, Germany) was used, and as the host, Rosetta cells (Novagen, Darmstadt, Germany). The heterologous expressed protein fragments were purified using the nickel-nitrilotriacetic acid (Ni/NTA) protein purification system (Qiagen, Venlo, the Netherlands) with an 8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl buffer at different pH levels (6.3, 6.1, 5.9, 5.7, 5.5, 5.0 and 4.5) and imidazole concentrations (300, 250, 200, 150, 100, 50 and 20 mM). The identities of the expressed protein fragments were verified by MALDI-TOF MS peptide mass fingerprinting of a tryptic digest of the Ni-NTA purified protein (Harhangi et al., 2002). The expressed protein fragments were used to immunize rabbits in a 3-month immunization protocol (SEQLAB Sequencing Laboratories Göttingen GmbH, Göttingen, Germany). The antisera (anti-F-ATPase-1, -2, -3 and anti-V-ATPase-4) were then used as the primary antiserum (polyclonal, crude serum) in immunoblot analysis and (anti-F-ATPase-1) immunofluorescence and immunogold localization as described below.
SDS-PAGE and immunoblot analysis
Kuenenia stuttgartiensis proteins (cell-free extract prepared using French press) were separated (50 µg of protein per lane) on a 10% SDS-PAGE gel and transferred to a cellulose-nitrate membrane (Schleicher & Schuell GmbH, Dassel, Germany) with the semi-dry transfer cell blotting system (Bio-Rad, Veenendaal, the Netherlands). Blotting was performed at 50 mA for 3 h with a transfer buffer that consisted of 25 mM Tris, 192 mM glycine and 20% methanol. After blotting, the blot was dried and stored at 4°C until further use.
Blots stored at 4°C were washed in MilliQ water for 30 min and incubated in blocking buffer; 1% bovine serum albumin (BSA) in Tris-buffered saline (TBS; 10 mM Tris-HCl, 0.9% NaCl, pH 7.4) for 1 h. The blot was then incubated for 1 h in either blocking buffer or rabbit pre-immune serum diluted 500-fold in blocking buffer as the negative controls or primary antiserum diluted 500-fold in blocking buffer. The blot was washed three times for 10 min in TBS containing 0.05% Tween20 and incubated for 1 h in monoclonal mouse anti-rabbit IgG alkaline phosphatase conjugate (Sigma, Zwijndrecht, the Netherlands) diluted 150.000-fold in blocking buffer. The blot was washed two times for 10 min in TBS containing 0.05% Tween20 and three times for 10 min in TBS. The blot was incubated with the BCIP/NBT Liquid Substrate System (Sigma, Zwijndrecht, the Netherlands) for 8 min, rinsed in excess amounts of MilliQ water and dried. All blots were scanned with the same settings.
Immunofluorescence analysis
Kuenenia stuttgartiensis cells were washed with phosphate-buffered saline (PBS; 0.1 M phosphate, 0.137 M NaCl, 2.7 mM KCl, pH 7.4) and fixed in 3% formaldehyde in PBS for 120 min while rotating at 4°C. The cells were washed again with PBS, resuspended in 1:1 ethanol : PBS and stored at −20°C until further use.
Formaldehyde-fixed cells were transferred to 0.075% gelatine, 0.01% chromium coated, six-well diagnostic microscope slides (Menzel GmbH & Co. KG, Braunschweig, Germany) and allowed to air dry. Next, the cells were permeabilized by a 10 min incubation with 0.5% Triton X-100 in TBS and washed three times with TBS. A-specific binding sites were blocked by a 30 min incubation with blocking buffer (1% BSA in TBS) in a moist incubation chamber at room temperature after which the blocking buffer was removed and the cells were allowed to air dry. Cells were incubated at room temperature for 1 h with primary antiserum diluted 500- or 1000-fold in blocking buffer in a moist incubation chamber. Each specific well was washed twice with 0.1% BSA in TBS after which the entire slide was washed three times for 10 min in 100 ml of 0.1% BSA in TBS by shaking rigorously. Cells were allowed to air dry and were then incubated for 1 h with the secondary antibody, Cy-3-labelled sheep anti-rabbit IgG (Sigma, Zwijndrecht, the Netherlands), diluted 100-fold in blocking buffer at room temperature in a moist incubation chamber in the dark. Each specific well was washed twice with TBS after which the entire slide was washed three times for 10 min in the dark in 100 ml of TBS by shaking rigorously. The cells were allowed to air dry, Vectashield mounting medium with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, USA) was added to each well, the coverslip was sealed with nail polish and the slides were stored in the dark at 4°C until further investigation.
Several control treatments were used. All control treatments were performed on K. stuttgartiensis cells unless stated otherwise, substituting primary antiserum and/or secondary antibody respectively. Four negative controls were performed: blocking buffer combined with blocking buffer, blocking buffer combined with secondary antibody, rabbit pre-immune serum combined with secondary antibody and rabbit anti-Nitrosomonas sp. combined with secondary antibody. Two positive controls were performed: rabbit anti-anammox hydrazine/hydroxylamine oxidoreductase (Schalk et al., 2000) combined with secondary antibody and rabbit anti-Nitrosomonas sp. combined with secondary antibody on Nitrosomonas sp. cells.
Cells were investigated at 1.000× magnification with a Zeiss Axioplan 2 imaging epifluorescence microscope (Carl Zeiss B.V., Sliedrecht, the Netherlands).
Sample preparation for immunogold localization: cryofixation, freeze-substitution and cryosectioning (rehydration method) (van Donselaar et al., 2007)
Small aggregates of K. stuttgartiensis cells were cryofixed by high-pressure freezing and freeze-substituted in acetone containing 0.5% glutaraldehyde and 1% H2O as described previously (van Niftrik et al., 2008b). After freeze-substitution, fixation was continued for 60 min on ice. Samples were rehydrated in a graded acetone series on ice: 95%, 90%, 80% and 70% acetone in water containing 0.5% glutaraldehyde, then 50 and 30% acetone in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9) containing 0.5% glutaraldehyde, and finally 0.5% glutaraldehyde in PHEM buffer. Samples were rinsed in PHEM buffer and embedded in 12% gelatin in PHEM buffer. The gelatin-embedded cells were cut into small cubes (1–2 mm3) under the stereo microscope, infiltrated overnight at 4°C with 2.3 M sucrose in PHEM buffer and frozen in liquid nitrogen.
Samples were cryosectioned using a cryoultramicrotome UC6/FC6 (Leica Microsystems, Vienna, Austria). Ultrathin cryosections (55 nm) were picked up with a drop of 1% methyl cellulose and 1.15 M sucrose in PHEM buffer and transferred to carbon-formvar-coated grids (copper, 100 mesh, hexagonal) for immunogold localization.
Immunogold localization
Grids containing ultrathin cryosections of K. stuttgartiensis cells were washed for 30 min at 37°C with PBS, incubated for 10 min at room temperature on drops of PBS containing 20 mM glycine and blocked for 15 min on drops of PBS containing 1% BSA or 2% skim milk powder (Frema Reform, Germany). After blocking, the grids were incubated for 60 min with the primary antiserum diluted 50-fold in PBS containing 1% BSA or 2% skim milk powder and washed for 12 min on drops of PBS containing 0.1% BSA or 0.2% skim milk powder. Grids were incubated for 20 min with protein A coupled to 10 nm gold (PAG-10, CMC/UMC, Utrecht, the Netherlands), diluted 80-fold in PBS containing 1% BSA or 2% skim milk powder and washed for 14 min on drops of PBS. The cryosections on grids were fixed for 5 min with PBS containing 1% glutaraldehyde and washed for 10 min on drops of distilled water. Cryosections were post-stained for 5 min with 2% uranyl acetate in 0.15 M oxalic acid pH 7.4 and washed quickly on two drops of distilled waster and then on two drops of 1.8% methyl cellulose containing 0.4% aqueous uranyl acetate on ice. Finally, cryosections were embedded for 5 min in 1.8% methyl cellulose containing 0.4% aqueous uranyl acetate on ice after which they were air dried.
Several control treatments were performed. As a positive control, grids were incubated with rabbit anti-anammox hydrazine/hydroxylamine oxidoreductase (Schalk et al., 2000) as the primary antiserum. Negative controls were: incubation with pre-immune serum instead of primary antiserum, incubation with affinity-isolated rabbit anti-influenza haemagglutinin (anti-HA, H6908, Sigma, Zwijndrecht, the Netherlands) as a primary antiserum, and incubation with blocking buffer instead of primary antiserum. Further, PAG-10 was used instead of a secondary antibody (such as sheep anti-rabbit IgG) because PAG-10, unlike other secondary antibodies, gave no background in the negative control with only PAG-10. Also, we used 10 nm gold because there was sufficient labelling and it was not necessary to use smaller gold to prevent steric hindrance.
Grids containing ultrathin cryosections of K. stuttgartiensis cells were investigated at 80 kV in a transmission electron microscope (Tecnai12, FEI Company, Eindhoven, the Netherlands). Images were recorded using a CCD camera (MegaView II, AnalySis).
Statistical analysis of immunogold localization
The immunogold labelling of the different anammox membranes and compartments using the antiserum directed at F-ATPase-1 was analysed statistically. Gold particles were allocated to either a membrane [outermost membrane (membrane 1), middle membrane (membrane 2) or innermost (anammoxosome) membrane (membrane 3)], when in 25 nm distance (the approximate length of the antibody-PAG-10 complex) of this membrane or to a compartment (riboplasm or anammoxosome) when in more than 25 nm distance from a membrane. Labels that were within 25 nm distance of more than one membrane (and could thus be allocated to more than one membrane) were not taken into account. Further, the paryphoplasm as a compartment was not taken into account because the distance between the outermost membrane (membrane 1) and middle membrane (membrane 2) was usually less than 50 nm, so gold particles could always be allocated to either the outermost or middle membrane of anammox bacteria. In total, 50 cells were analysed in both the incubation with the antiserum and in the incubation with the pre-immune serum. Labelling was analysed statistically using the t-test (GraphPad: http://www.graphpad.com/quickcalcs/) for normal distributed data with equal variances, the Welch's t-test for normal distributed data with unequal variances (GraphPad) or the Mann–Whitney U-test (VassarStats: http://faculty.vassar.edu/lowry/VassarStats.html) for non-parametric data. A P-value between 0.05 and 0.01 was considered significant (*), a P-value between 0.01 and 0.0001 very significant (**) and a P-value of less than 0.0001 extremely significant (***).
Relevant accession numbers
NCBI (GenBank) (http://www.ncbi.nlm.nih.gov/)
Kuenenia stuttgartiensis: kuste3787 (CAJ74550) to kuste3796 (CAJ74559), kuste4592 (CAJ75354) to kuste4600 (CAJ75362), kustc0572 (CAJ71317) to kustc0579 (CAJ71324), kuste3864 (CAJ74627) to kuste3871 (CAJ74634). Escherichia coli K-12 MG1655: b3737 (AAC76760). Borrelia burgdorferi B31: bb0090 (aac66487). Prositehttp://www.expasy.ch/prosite/): ps00605.
Acknowledgments
We would like to thank Katinka van de Pas-Schoonen and Dr Boran Kartal for the K. stuttgartiensis enrichment cultures, Dr Bruno Humbel for operation of the high-pressure freezer and Professor Arie Verkleij and Dr Willie Geerts for stimulating discussions. L.v.N. is supported by the Netherlands Organisation for Scientific Research (NWO, VENI grant). Planctomycete research in the laboratories of J.A.F. and R.I.W. was supported by the Australian Research Council.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2005;437:349–355. doi: 10.1038/nature04159. [DOI] [PubMed] [Google Scholar]
- Boyer PD. The binding change mechanism for ATP synthase: some probabilities and possibilities. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- Boyer PD. The ATP synthase – a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- Brandes JA, Devol AH, Deutsch C. New developments in the marine nitrogen cycle. Chem Rev. 2007;107:577–589. doi: 10.1021/cr050377t. [DOI] [PubMed] [Google Scholar]
- Cirpus IEY, de Been M, Op den Camp HJM, Strous M, Le Paslier D, Kuenen JG, Jetten MSM. A new soluble 10 kDa monoheme cytochrome c-552 from the anammox bacterium Candidatus‘Kuenenia stuttgartiensis’. FEMS Microbiol Lett. 2005;252:273–278. doi: 10.1016/j.femsle.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Devol AH. Nitrogen cycle: solution to a marine mystery. Nature. 2003;422:575–576. doi: 10.1038/422575a. [DOI] [PubMed] [Google Scholar]
- van Donselaar E, Posthuma G, Zeuschner D, Humbel BM, Slot JW. Immunogold labeling of cryosections from high-pressure frozen cells. Traffic. 2007;8:471–485. doi: 10.1111/j.1600-0854.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- Drory O, Nelson N. The emerging structure of vacuolar ATPases. Physiology. 2006;21:317–325. doi: 10.1152/physiol.00017.2006. [DOI] [PubMed] [Google Scholar]
- Engelhardt H. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J Struct Biol. 2007;160:115–124. doi: 10.1016/j.jsb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- Fuerst JA. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology. 1995;141:1493–1506. doi: 10.1099/13500872-141-7-1493. [DOI] [PubMed] [Google Scholar]
- Fuerst JA, Webb RI, van Niftrik L, Jetten MSM, Strous M. Anammoxosomes of anaerobic ammonium-oxidizing planctomycetes. In: Shiveley JM, editor. Microbiology Monographs Volume 2: Complex Intracellular Structures in Prokaryotes. Berlin and Heidelberg: Springer-Verlag; 2006. pp. 259–283. [Google Scholar]
- van de Graaf AA, de Bruijn P, Robertson LA, Jetten MSM, Kuenen JJ. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology. 1996;142:2187–2196. [Google Scholar]
- Grüber G, Marshansky V. New insights into structure–function relationships between archeal ATP synthase (A1A0) and vacuolar type ATPase (V1V0) Bioessays. 2008;30:1096–1109. doi: 10.1002/bies.20827. [DOI] [PubMed] [Google Scholar]
- Grüber G, Wieczorek H, Harvey WR, Müller V. Structure–function relationships of A-, F- and V-ATPases. J Exp Biol. 2001;204:2597–2605. doi: 10.1242/jeb.204.15.2597. [DOI] [PubMed] [Google Scholar]
- Harhangi HR, Steenbakkers PJM, Akhmanova A, Jetten MSM, van der Drift C, Camp HJM. A highly expressed family 1 β-glucosidase with transglycosylation capacity from the anaerobic fungus Piromyces sp. E2. Biochim Biophys Acta. 2002;1574:293–303. doi: 10.1016/s0167-4781(01)00380-3. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase – a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler. 1993;374:166. [Google Scholar]
- Huston WM, Harhangi HR, Leech AP, Butler CS, Jetten MSM, Op den Camp HJM, Moir JWB. Expression and characterisation of a major c-type cytochrome encoded by gene kustc0563 from Kuenenia stuttgartiensis as a recombinant protein in Escherichia coli. Protein Expr Purif. 2007;51:28–33. doi: 10.1016/j.pep.2006.06.026. [DOI] [PubMed] [Google Scholar]
- König E, Schlesner H, Hirsch P. Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol. 1984;138:200–205. [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Küper U, Meyer C, Müller V, Rachel R, Huber H. Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic Archaeon Ignicoccus hospitalis. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.0911711107. doi: 10.1073/pnas.0911711107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Barker Jørgensen B, Kuenen JG, et al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature. 2003;422:608–611. doi: 10.1038/nature01472. [DOI] [PubMed] [Google Scholar]
- Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, et al. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci USA. 2005;102:6478–6483. doi: 10.1073/pnas.0502088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WCY, Rubinstein JL. Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound V0 motor. Proc Natl Acad Sci USA. 2010;107:1367–1372. doi: 10.1073/pnas.0911085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W, König H, Schlesner H, Hirsch P. Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group. Arch Microbiol. 1986;145:361–366. [Google Scholar]
- Lindsay MR, Webb RI, Fuerst JA. Effects of fixative and buffer on morphology and ultrastructure of a freshwater planctomycete, Gemmata obscuriglobus. J Microbiol Methods. 1995;21:45–54. [Google Scholar]
- Lindsay MR, Webb RI, Fuerst JA. Pirellulosomes: a new type of membrane-bounded cell compartment in planctomycete bacteria of the genus Pirellula. Microbiology. 1997;143:739–748. doi: 10.1099/00221287-143-3-739. [DOI] [PubMed] [Google Scholar]
- Lindsay MR, Webb RI, Strous M, Jetten MSM, Butler MK, Forde RJ, Fuerst JA. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch Microbiol. 2001;175:413–429. doi: 10.1007/s002030100280. [DOI] [PubMed] [Google Scholar]
- Lolkema JS, Chaban Y, Boekema EJ. Subunit composition, structure, and distribution of bacterial V-type ATPases. J Bioenerg Biomembr. 2003;35:323–335. doi: 10.1023/a:1025776831494. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Müller V, Ruppert C, Lemker T. Structure and function of the A1A0-ATPases from methanogenic Archaea. J Bioenerg Biomembr. 1999;31:15–27. doi: 10.1023/a:1005451311009. [DOI] [PubMed] [Google Scholar]
- Müller V, Lemker T, Lingl A, Weidner C, Coskun Ü, Grüber G. Bioenergetics of Archaea: ATP synthesis under harsh environmental conditions. J Mol Microbiol Biotechnol. 2005;10:167–180. doi: 10.1159/000091563. [DOI] [PubMed] [Google Scholar]
- van Niftrik L, Geerts WJC, van Donselaar EG, Humbel BM, Webb RI, Fuerst JA, et al. Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage and localization of cytochrome c proteins. J Bacteriol. 2008a;190:708–717. doi: 10.1128/JB.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niftrik L, Geerts WJC, van Donselaar EG, Humbel BM, Yakushevska A, Verkleij AJ, et al. Combined structural and chemical analysis of the anammoxosome: a membrane-bounded intracytoplasmic compartment in anammox bacteria. J Struct Biol. 2008b;161:401–410. doi: 10.1016/j.jsb.2007.05.005. [DOI] [PubMed] [Google Scholar]
- van Niftrik L, Geerts WJC, van Donselaar EG, Humbel BM, Webb RI, Harhangi HR, et al. Cell division ring, a new cell division protein and vertical inheritance of a bacterial organelle in anammox planctomycetes. Mol Microbiol. 2009;73:1009–1019. doi: 10.1111/j.1365-2958.2009.06841.x. [DOI] [PubMed] [Google Scholar]
- Rastogi VK, Girvin ME. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature. 1999;402:263–268. doi: 10.1038/46224. [DOI] [PubMed] [Google Scholar]
- Schalk J, de Vries S, Kuenen JG, Jetten MSM. Involvement of a novel hydroxylamine oxidoreductase in anaerobic ammonium oxidation. Biochemistry. 2000;39:5405–5412. doi: 10.1021/bi992721k. [DOI] [PubMed] [Google Scholar]
- Sinninghe Damsté JS, Strous M, Rijpstra WIC, Hopmans EC, Geenevasen JAJ, van Duin ACT, et al. Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature. 2002;419:708–712. doi: 10.1038/nature01128. [DOI] [PubMed] [Google Scholar]
- Sinninghe Damsté JS, Rijpstra WIC, Geenevasen JAJ, Strous M, Jetten MSM. Structural identification of ladderane and other membrane lipids of planctomycetes capable of anaerobic ammonium oxidation (anammox) FEBS J. 2005;272:4270–4283. doi: 10.1111/j.1742-4658.2005.04842.x. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Wehmeyer U, Liesack W. 16S ribosomal RNA- and cell wall analysis of Gemmata obscuriglobus, a new member of the order Planctomycetales. FEMS Microbiol Lett. 1986;37:289–292. [Google Scholar]
- van der Star WRL, Dijkema C, de Waard P, Picioreanu C, Strous M, van Loosdrecht MCM. An intracellular pH gradient in the anammox bacterium Kuenenia stuttgartiensis as evaluated by 31P NMR. Appl Microbiol Biotechnol. 2010;86:311–317. doi: 10.1007/s00253-009-2309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE. The rotary mechanism of ATP synthase. Curr Opin Struct Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- Strous M, Heijnen JJ, Kuenen JG, Jetten MSM. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl Microbiol Biotechnol. 1998;50:589–596. [Google Scholar]
- Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- Sumi M, Yohda M, Koga Y, Yoshida M. F0F1-ATPase genes from an archaebacterium, Methanosarcina barkeri. Biochem Biophys Res Commun. 1997;241:427–433. doi: 10.1006/bbrc.1997.7809. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Imamura H. Rotation, structure, and classification of prokaryotic V-ATPase. J Bioenerg Biomembr. 2005;37:405–410. doi: 10.1007/s10863-005-9480-1. [DOI] [PubMed] [Google Scholar]
- Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffé M. Direct visualization of the outer membrane of Mycobacteria and Corynebacteria in their native state. J Bacteriol. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


