Abstract
The fungal species Candida albicans and the bacterial species Staphylococcus aureus are responsible for a majority of hospital-acquired infections and often coinfect critically ill patients as complicating polymicrobial biofilms. To investigate biofilm structure during polymicrobial growth, dual-species biofilms were imaged with confocal scanning laser microscopy. Analyses revealed a unique biofilm architecture where S. aureus commonly associated with the hyphal elements of C. albicans. This physical interaction may provide staphylococci with an invasion strategy because candidal hyphae can penetrate through epithelial layers. To further understand the molecular mechanisms possibly responsible for previously demonstrated amplified virulence during coinfection, protein expression studies were undertaken. Differential in-gel electrophoresis identified a total of 27 proteins to be significantly differentially produced by these organisms during coculture biofilm growth. Among the upregulated staphylococcal proteins was l-lactate dehydrogenase 1, which confers resistance to host-derived oxidative stressors. Among the downregulated proteins was the global transcriptional repressor of virulence factors, CodY. These findings demonstrate that the hyphae-mediated enhanced pathogenesis of S. aureus may not only be due to physical interactions but can also be attributed to the differential regulation of specific virulence factors induced during polymicrobial growth. Further characterization of the intricate interaction between these pathogens at the molecular level is warranted, as it may aid in the design of novel therapeutic strategies aimed at combating fungal–bacterial polymicrobial infection.
Keywords: Candida albicans, Staphylococcus aureus, polymicrobial, biofilm, proteome
Introduction
In nature, most microorganisms are associated with surfaces in multispecies biofilm consortia. A biofilm can be defined as a community of microorganisms embedded in a self-derived polymeric matrix, attached to a surface. In a polymicrobial biofilm where multiple microbial species are closely associated, mutually beneficial interactions may develop. Polymicrobial biofilms are found in nearly every niche in the human body; the oral cavity and gastrointestinal and urogenital tracts exhibit tremendous microbial phylogenetic diversity (Aas et al., 2005; Manson et al., 2008;). Although recent decades have witnessed a surge in the area of biofilm research, relatively little is known about the behavior of communities of mixed microorganisms, particularly fungal–bacterial biofilms. Biofilm-embedded organisms demonstrate a uniquely altered gene expression, and studies have suggested that amplified pathogenic phenotypes may emerge during multispecies interactions (Mastropaolo et al., 2005; O'Connell et al., 2006;). One particular biofilm-mediated microbial association, of medical interest, is that which exists between the prokaryotic pathogen Staphylococcus aureus and the eukaryotic pathogen Candida albicans (for a review, see Shirtliff et al., 2009).
Methicillin-resistant S. aureus (MRSA) is a gram-positive coccoid bacterium that is responsible for a significant and increasing number of hospital- and community-acquired infections worldwide (Klevens et al., 2007). This species possesses a number of virulence factors including adhesins, immunoavoidance factors, toxins, coagulase, and a variety of antimicrobial resistance genes (Gordon & Lowy, 2008). The multiple virulence factors of MRSA, coupled with its inherent ability to resist antibiotic therapy via antibiotic resistance gene expression and biofilm formation, have made this pathogen a significant burden to the medical community (Goetghebeur et al., 2007).
Candida albicans, a fungal species commonly colonizing human mucosal surfaces, has long been adapted to the human host. However, under conditions of immune dysfunction, C. albicans strains cause recurrent mucosal infections and life-threatening disseminated infections (de Repentigny et al., 2004). Multiple antifungal-resistant forms of C. albicans are also being increasingly encountered in the hospital setting (Ramage et al., 2002). As a polymorphic species, C. albicans is capable of switching morphology between yeast, hyphal, and pseudohyphal forms, a transition central to its pathogenesis. Once in the hyphal form, host epithelial layers can be pierced, a crucial step in the initiation of candidiasis (Sudbery et al., 2004).
Currently, S. aureus and Candida spp. are ranked among the top three bloodstream pathogens causing severe morbidity and mortality in hospitalized patients. Not only are C. albicans and S. aureus responsible for a substantial number of infections independently, there is increasing evidence suggesting that they are commonly associated as coinfecting organisms (Abe et al., 2001; Baena-Monroy et al., 2005;). The clinical outcomes of polymicrobial sepsis compared with monomicrobial sepsis are grave, with significantly higher mortality rates (Pulimood et al., 2002). A study by Klotz et al. (2007) examining the incidence of candidal bloodstream infections in hospitals reported an S. aureus–Candida spp. co-culture rate of up to 20%.
Candida albicans and S. aureus have also been coisolated from various mucosal surfaces including vaginal and oral mucosa in a biofilm mode of growth. Although S. aureus was thought to be a transient member of the oral microbial communities, increasing evidence from several culturing surveys suggests that it is a common isolate from the oral cavity in healthy children and adults, especially in saliva, supragingival plaque, and on the tongue (Miyake et al., 1991; Smith et al., 2003; Ohara-Nemoto et al., 2008;). More seriously, these pathogens have been coassociated with a number of polymicrobial diseases including ventilator-associated pneumonia, cystic fibrosis, superinfection of burn wounds, urinary tract infections, and denture stomatitis (Ekwempu et al., 1981; Dahlen et al., 1982; Siegman-Igra et al., 1988; Smith et al., 2003; Valenza et al., 2008;). Some of the most compelling evidence for this particular bacterial–fungal interaction was demonstrated through a series of studies by Carlson and colleagues (Carlson, 1983; Carlson & Johnson, 1985;). The findings from these studies demonstrated a 6–70 000-fold decrease in the lethal dose 50% of S. aureus when coinoculated intraperitoneally with C. albicans in mice compared with single-species infections. Despite the significance of these observations, limited studies have examined the interactions of C. albicans and S. aureus during biofilm development, their most common infectious mode of growth.
In this study, we elucidated the nature and spatial relationship of the interactions between these two diverse pathogenic species using confocal scanning laser microscopy (CSLM) as they coexist and interact during polymicrobial biofilm growth. We have also characterized proteomic changes specific to polymicrobial culture of this cross-kingdom biofilm using two-dimensional differential in-gel electrophoresis (DIGE) and identified differentially regulated metabolic, stress, and virulence proteins via matrix-assisted laser desorption/ionization time-of-flight/time-of-flight tandem MS (MALDI-ToF/ToF MS) analysis.
Materials and methods
Strains and growth conditions
The MRSA hospital-acquired clinical isolate used in all the experiments was obtained from a patient with a biofilm-mediated infection at the University of Texas Medical Branch – Galveston and previously designated as strain M2 (Brady et al., 2006). The well-characterized C. albicans lab strain SC5314 was used for all the experiments (Gillum et al., 1984). In addition, S. aureus strain Seattle 1945 [containing a plasmid encoding for chloramphenicol resistance and green fluorescent protein (GFP) expression under control of the sarA promoter] and the constitutively GFP-expressing C. albicans strain CAF2-1 were also used (Morschhauser et al., 1998; Leid et al., 2002;). The following bacterial strains were also used: Staphylococcus epidermidis (clinical isolate), Pseudomonas aeruginosa (PA01), Streptococcus pyogenes (clinical isolate), Bacillus subtilis (ATCC #6633), and a laboratory strain of Escherichia coli (DH5-α).
For all studies, an aliquot of a glycerol stock of C. albicans strain SC5314 or GFP-expressing CAF2-1 was grown and maintained on Sabouraud dextrose agar (BBL, Cockeysville, MD). Cultures were grown overnight in yeast peptone dextrose (YPD) (BBL, Sparks, MD) in an orbital shaker (120 r.p.m.) at 37 °C under aerobic conditions. Yeast cells were harvested and washed twice in sterile phosphate-buffered saline (PBS). Starter cultures of clinical isolates of S. aureus (M2), GFP-expressing S. aureus (Seattle 1945), S. epidermidis (clinical isolate), P. aeruginosa (PA01), S. pyogenes (clinical isolate), B. subtilis (ATCC #6633), and a laboratory strain of E. coli (DH5-α) were grown in trypticase soy broth (TSB) (Remel, Lenexa, KS) and incubated overnight at 37 °C. Fresh log-phase bacterial starter cultures were grown by diluting the overnight culture 1 : 100 in fresh TSB for 3 h. Bacterial cultures were then washed twice in sterile PBS. Dual-species biofilms were grown in RPMI 1640 buffered with HEPES and supplemented with l-glutamine (Invitrogen, Grand Island, NY) and 5% heat-inactivated fetal bovine serum (RPMI–FBS) (Hyclone, Logan, UT) or YPD containing 5% FBS medium (YPD–FBS).
Biofilm growth
Staphylococcus aureus was grown as noted above and diluted to an OD600 nm of 0.1. Candida albicans overnight cultures were grown as described above and diluted to an OD540 nm of 1.0. Biofilms for protein nucleic acid (PNA)-FISH were grown for 24 h on glass coverslips in polystyrene 6-well plates (Corning, Lowell, MA) in 5 mL of RPMI–FBS. Dual-species biofilms were grown by inoculating wells with 50 μL of both species suspensions. PNA-FISH was performed as per the manufacturer's protocol (Advandx, Woburn, MA) with a Cy3-labeled C. albicans/fluorescein isothiocyanate (FITC)-labeled S. aureus PNA probe cocktail. Nonadherent cells were removed by washing with PBS before imaging. Fluorescence was captured with a Zeiss LSM 510 (Carl Zeiss, Thornwood, NY) confocal microscope using a × 20 objective and a FITC/Texas Red dual-band filter. In order to confirm the strain-independent interaction of S. aureus and C. albicans, dual-species biofilms of GFP-expressing strains were grown on glass coverslips in RPMI–FBS supplemented with 10 μg mL−1 chloramphenicol. Coverslips were processed for microscopy as described above. Finally, microbial protein samples for proteomic studies were prepared by growing mono- or dual-species biofilms in 6-well polystyrene plates as above in either 5 mL of RPMI–FBS (for experiments with hyphae) or YPD–FBS (for experiments with yeast cells) at 37 °C for 24 h.
Hyphal–bacterial attachment assay
Hyphae formation was induced by first growing C. albicans as described previously on glass coverslips in 6-well plates in 3 mL RPMI–FBS for 4 h. Nonadherent hyphae were removed by gently washing the coverslips in PBS, followed by the addition of 3 mL of fresh RPMI–FBS. Log-phase bacterial cell suspensions were washed in PBS, equalized to an OD600 nm of 0.1, and added to the C. albicans biofilms. Plates were placed on a rotary shaker to distribute the bacteria evenly and incubated for 1 h at 37 °C. Following incubation, nonadherent cells were removed by gently washing the coverslips in PBS and then examined using phase-contrast microscopy under a × 100 oil-immersion objective. The total number of bacterial cells per field and attached bacteria per hyphae were counted. Percent attachment was calculated by dividing the number of attached bacteria by the total number of bacteria. A total of 10 random fields per coverslip were analyzed.
Morphological specificity binding assay
Hyphal and blastospore biofilms were grown as described above in RPMI–FBS or YPD–FBS, respectively. Nonadherent cells were gently removed by washing in PBS. Log-phase staphylococcal cell suspensions were added to the C. albicans biofilms, shaken, and incubated for 1 h 37 °C. Following incubation, nonadherent cells were removed by gently washing the coverslips in PBS and then examined using phase-contrast microscopy under a × 100 oil-immersion objective. Attachment rates were calculated by counting the total number of yeast cells or hyphae per field as well as the number of attached S. aureus cells. These numbers were divided to calculate the average number of S. aureus attached per C. albicans cell. A total of 10 random fields per coverslip were analyzed.
Microbial viability assay
Polymicrobial biofilms were grown on glass coverslips as described previously using C. albicans SC5314 and S. aureus M2. Coverslips were removed from the incubator after 12, 24, and 40 h of growth. Biofilms were washed briefly in PBS, placed into sterile 6-well plates, and stained using the BacLight LIVE/DEAD viability kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The BacLight LIVE/DEAD system stains live cells green (Syto9), while dead cells appear red (propidium iodide). Coverslips were then mounted onto glass slides with Vectashield (Vector Laboratories, Burlingame, CA) and processed for CSLM. The spatial arrangement of the polymicrobial biofilm was determined by analysis of confocal z-axis image slices using the lsmix software package (Carl Zeiss).
Proteomic analysis
Plates containing 24-h biofilms were gently shaken on a rotary shaker for 1 min and then the culture supernatants were discarded. To remove the biofilms from the wells, 1 mL of cell wash buffer (10 mM Tris, 5 mM Mg acetate, pH 8.0) supplemented with 3 mM phenylmethanesulfonyl fluoride was added and a cell culture tissue scraper was used to remove attached cells. Cells were then washed twice in cell wash buffer, resuspended in 1 mL lysis buffer (30 mM Tris, 4 M urea, 2 M thiourea, 1% CHAPS), and incubated on ice for 10 min. Cells were then mechanically disrupted in a FastPrep FP120 (ThermoSavant, Holbrook, NY) using 0.1 mm zirconia beads (Biospec Products, Bartlesville, OK) for 30 s, followed by a 2-min incubation on ice; the process was repeated for a total of 10 times. Suspensions were centrifuged for 10 min at 14 000 g and supernatants were removed and protein was quantified spectrophotometrically using the Advanced Protein Assay Reagent #2 (Cytoskeleton Inc., Denver, CO). Crude protein extracts were precipitated and purified with Perfect-Focus reagent as per the manufacturer's directions (G-Biosciences, Maryland Heights, MO) and stored at −70 °C until used.
Two-dimensional DIGE was performed according to the concepts of O'Farrell and Minden and outlined by Sauer and Camper (O'Farrell, 1975; Sauer & Camper, 2001; Minden, 2007;). Protein labeling was performed using the DIGE system (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. To achieve sufficient protein rehydration, 100 μg of each protein sample was resuspended in 150 μL of rehydration buffer (30 mM Tris, 7 M urea, 2 M thiourea, 2.5% CHAPS). Following rehydration, the pH was adjusted to 8.5 with dilute NaOH or HCl as needed. Candida albicans proteins were labeled with Cy2, S. aureus proteins were labeled with Cy3, and co-cultures were labeled with Cy5 at a ratio of 2 pmol CyDye μg−1 protein. Samples were incubated for 30 min on ice and kept protected from light. Following CyDye labeling, 15 μL of 10 mM lysine was added for 10 min to quench excess CyDye. Samples were combined and a final concentration of 35 mM DTT and 1.6% Pharmalyte 3-10 was added. Samples were applied to 24 cm, pH 3–10 (linear) Immobiline Dry-Strips (IPG) (GE Healthcare). Proteins were separated in the first dimension by their isoelectric point using a Multiphor II (Amersham) as per the manufacturer's directions. Before the second dimension, IPG strips were equilibrated and applied to 12% 26 cm × 20 cm sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Protein spots were resolved in the second dimension using a Höefer DALT Vertical System and fluorescence was captured using the Typhoon Imager 9400 (GE Healthcare). Following fluorescence scanning, gels were nondestructively silver stained for spot excision (Gharahdaghi et al., 1999). Protein spots that were upregulated in six out of six gels were selected for MALDI-ToF/ToF MS analysis as described previously (Brady et al., 2006).
Statistics
All studies were performed in triplicate at a minimum. In addition, all cell enumerations were performed on a minimum of 10 fields of view and at least 400 cells. A Student's t-test was used to compare microbial numbers, with a P<0.05 representing a statistical significance.
Results
Hyphal–bacterial attachment assay
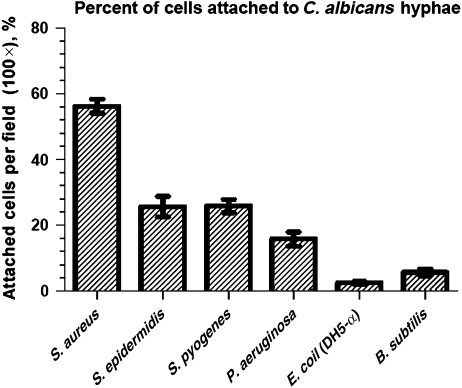
In order to assess the potential for hyphal–bacterial interactions, we tested a panel of various bacterial species displaying a wide variety of phenotypes including cell morphology, motility, ecological niche, and Gram stain identity for hyphal interaction. Candida albicans biofilms were grown overnight on glass coverslips, washed, and various bacterial strains added for 1 h. Hyphal binding was measured via phase-contrast microscopy as the number of attached bacterial cells to C. albicans hyphae divided by the total number of bacterial cells per microscopic field and reported as a percentage (Fig. 1). Percent counts demonstrated that S. aureus had the highest hyphal association (56%), followed by S. pyogenes and S. epidermidis (25%). Pseudomonas aeruginosa, a gram-negative motile rod and known hyphae binder, had a hyphal association of (17%), while E. coli, also a gram-negative rod, and B. subtilis, a gram-positive bacillus, demonstrated the lowest hyphal binding (5.7% and 2.5%, respectively).
Fig. 1.
Bacterial attachment assay. Candida albicans biofilms were grown for 3 h in RPMI to induce hyphae formation and incubated for 1 h with the following bacteria: Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus pyogenes, Pseudomonas aeruginosa, Bacillus subtilis, Escherichia coli (DH5-α). Nonadherent cells were removed by washing and the remaining cells were counted by phase-contrast microscopy. Percent hyphal attachment was assessed by counting the number of bacteria associated with the hyphae divided by the number of total bacteria per field. Ten fields were chosen at random and averaged; the experiment was repeated in triplicate. Error bars represent SD.
PNA-FISH
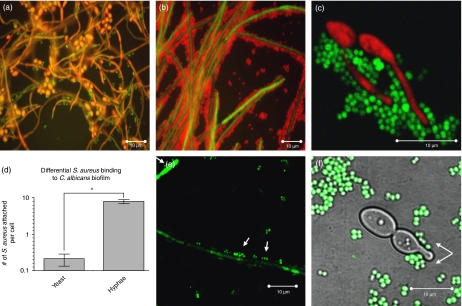
Because of the strong hyphal binding exhibited, fluorescence microscopy using species-specific PNA-FISH probes was used to visualize the physical interaction between C. albicans and S. aureus in an in vitro dual-species biofilm. Images revealed extensive adherence of S. aureus to C. albicans, with a preferential association to the invasive hyphal elements of C. albicans (Fig. 2a and c). In areas of dense hyphal biofilm growth, S. aureus could be seen completely covering C. albicans (Fig. 2b). To show the specificity of S. aureus for binding the hyphal form of C. albicans, polymicrobial interactions were assessed using both hyphae and yeast biofilms. Quantitative counts demonstrated a 30-fold increase in S. aureus binding to hyphae as compared with C. albicans yeast cells. These observations were confirmed by similar experiments performed using different GFP-expressing strains of C. albicans and S. aureus where a similar adherence pattern was demonstrated, confirming that this interaction is strain independent (Fig. 2e and f).
Fig. 2.
Biofilm architecture of Candida albicans and Staphylococcus aureus 24-h dual-species biofilm using PNA-FISH and GFP-expressing microorganisms. (a) Staphylococcus aureus (FITC-labeled probe, green) has a greater tropism for the hyphal form of C. albicans (TAMRA-labeled probe, red) compared with the yeast form. Field of view diameter is 150 μm. (b) An area of C. albicans (FITC-labeled probe, green) hyphal biofilm growth is completely covered by S. aureus (Cy3-labeled probe, red). (c) A × 63 zoom image showing staphylococci (FITC-labeled probe, green) binding to only the hyphal filaments of C. albicans (Cy3-conjugated probe, red). (d) Graph representing the average number of S. aureus cells attached per C. albicans cell during polymicrobial biofilm growth. Ten fields were chosen at random for counting and the experiment was repeated in triplicate. Error bars represent the SD. (e) Staphylococcus aureus (white arrows), expressing GFP under control of the sarA promoter, was found to be associated to GFP-expressing C. albicans hyphae. (f) Staphylococcus aureus (white arrows) demonstrating preferential binding to a C. albicans germ tube without binding to the yeast cell. Fluorescence was captured with a × 63 oil-immersion objective and FITC/DICIII, FITC/Texas Red filter sets. Asterisk (*) denotes a statistically significant difference at P<0.05.
Microbial viability assay
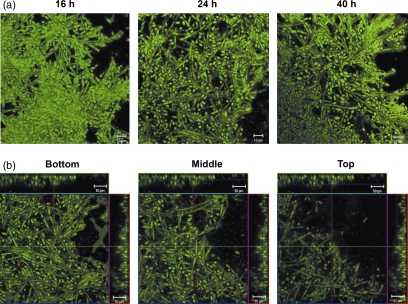
The BacLight LIVE/DEAD cell viability assay was used to determine whether fungal or bacterial cells were killed during polymicrobial biofilm growth and to assess the spatial arrangement of the biofilm. After 16, 24, and 40 h of growth, both cell types were viable as visualized by green fluorescent staining (Syto9) with an apparent lack of red fluorescence (propidium iodide) (Fig. 3a). In addition to staining for cell viability, the spatial arrangement of the dual-species biofilm was characterized by confocal z-stack imaging analysis. Bottom, middle, and top representative z-axis image slices from a 24-h polymicrobial biofilm show the presence of S. aureus attached to the hyphae of C. albicans throughout the entire biofilm architecture (Fig. 3b).
Fig. 3.
Viability and spatial arrangement in the dual-species biofilm. Candida albicans–Staphylococcus aureus biofilms were grown for various time points on glass coverslips, stained with BacLight LIVE/DEAD, and processed for CSLM. (a) At all the time points tested, both bacteria and fungi appear healthy as measured by the presence of green fluorescence (Syto9) and the absence of red (propidium iodide). (b) Representative confocal z-stack images of a typical 24-h dual-species biofilm demonstrating the presence of S. aureus attached to C. albicans hyphae throughout the bottom, middle, and top layers.
Proteomic analysis
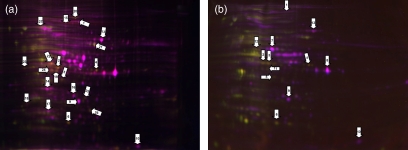
In order to identify other factors that may lead to increased virulence during coinfection, unfractionated, whole-cell proteins from 24 h in vitro biofilms were harvested, purified, and differentially lysine-labeled with NHS-ester CyDyes. Labeled proteins were then combined and subjected to isoelectric focusing and second dimension analysis. Representative gels from either mono- or dual-species biofilms composed of S. aureus and C. albicans yeast cells (Fig. 4a) or S. aureus and C. albicans hyphal cells (Fig. 4b) are shown. Spots were considered for MALDI-ToF/ToF MS identification if they were reproducible on six out of six gels. In this global proteomics screen, we identified 27 proteins that were upregulated in the co-culture biofilm. Among these were proteins important for growth and metabolism and others of hypothetical function. Most notable of interest were those proteins implicated in microbial stress and enhanced virulence of both species (Table 1).
Fig. 4.
Representative DIGE gel from mono- and dual-species biofilms. Whole-cell lysates, enriched in the cytoplasmic fraction, were obtained from 24-h biofilms. Proteins (100 μg) were differentially labeled with CyDye: Candida albicans labeled with Cy2 (blue), Staphylococcus aureus labeled with Cy3 (green), dual-species biofilm proteins labeled with Cy5 (red). Proteins were focused in the first dimension on pH 3–10 IEF strips and resolved in the second dimension on 12.5% polyacrylamide gels. (a) Representative gel from staphylococcal–yeast biofilms. (b) Representative gel from staphylococcal–hyphal biofilms.
Table 1.
Proteins upregulated in the dual-species biofilm
| Spot | MW (Da) | pI | Organism | Identity | Protein name | Peptide matches | Protein score confidence interval (%) | Accession number | Function |
|---|---|---|---|---|---|---|---|---|---|
| (A) Proteins upregulated in staphylococcal–yeast biofilms | |||||||||
| 1A | 84624.8 | 5.96 | C. albicans | Putative mitochondrial aconitate hydratase | Aco1p | 19 | 100 | 68479387 | Carbohydrate metabolism; tricarboxylic acid cycle |
| 14A | 21481.5 | 5.15 | C. albicans | Similar to heat shock protein 5 | Similar to Hsp5 | 9 | 100 | 68469633 | Cellular stress response; protein folding |
| 15A | 91795.2 | 6.35 | C. albicans | Heat shock protein 78 | Hsp78p | 16 | 100 | 31076745 | Cellular stress response; protein folding |
| 16A | 26893.9 | 5.74 | C. albicans | Triosephosphate isomerase | Tpi1p | 10 | 100 | 7270988 | Glycolysis; gluconeogenesis; fatty acid biosynthesis |
| 17A | 21960.3 | 4.98 | C. albicans | Thioredoxin peroxidase | Tsa1p | 7 | 99.99 | 68479826 | Cellular stress response; antioxidant |
| 18A | 49188.7 | 7.36 | C. albicans | Metal-binding activator 1 | Mac1p | 6 | 85.35 | 68471167 | Copper-binding transcriptional regulator; cellular stress response |
| 19A | 95340.7 | 5.68 | S. aureus | Alcohol dehydrogenase, iron containing | Adh | 15 | 100 | 57651152 | Carbon utilization; alcohol metabolism |
| 20A | 95397.8 | 5.73 | S. aureus | Putative aldehyde-alcohol dehydrogenase | AdhE | 16 | 100 | 49482391 | Carbon utilization; putative peroxide scavenger |
| 21A | 56138.6 | 6.02 | S. aureus | Probable malate:quinone oxidoreductase | Mqo1 | 21 | 100 | 82752186 | Carbohydrate metabolism; tricarboxylic acid cycle |
| 22A | 37820.9 | 5.14 | S. aureus | Ornithine carbamoyltransferase | ArgF | 12 | 100 | 49484831 | Amino acid biosynthesis |
| 23A | 35194.1 | 4.65 | S. aureus | Pyruvate dehydrogenase complex E1 component β | PdhB | 11 | 100 | 57651703 | Glycolysis; oxidoreductase |
| 24A | 35539.3 | 5.36 | S. aureus | Carbamate kinase | ArcC1 | 17 | 100 | 49484829 | l-Arginine degradation |
| 25A | 28737.4 | 5.87 | S. aureus | Transcriptional repressor CodY | CodY | 8 | 99.97 | 15924245 | Decreased hemolysin, biofilm, and quorum-sensing function |
| 26A | 23092.2 | 6.08 | S. aureus | Uracil phosphoribosyl transferase | Upp | 10 | 100 | 15925102 | Pyrimidine metabolism |
| 27A | 63331.2 | 5.2 | S. aureus | Pyruvate kinase | Pyk | 27 | 100 | 49483939 | Carbohydrate metabolism; glycolysis |
| (B) Proteins upregulated in staphylococcal–hyphal biofilms | |||||||||
| 1B | 93865.5 | 6.07 | C. albicans | Translation elongation factor 2 | Eft2p | 4 | 100 | 68481380 | Protein synthesis |
| 8B | 35924.7 | 6.61 | C. albicans | Glyceraldehyde 3 phosphate dehydrogenase | Thd1p | 15 | 100 | 68472227 | Carbohydrate metabolism; glycolysis |
| 11B | 33026.2 | 5.4 | S. aureus | Cysteine synthase | CysK | 4 | 100 | 82750220 | Cysteine biosynthesis |
| 3B | 29543.3 | 5 | S. aureus | l-Lactate dehydrogenase | Ldh1 | 9 | 100 | 87161566 | Growth during nitrosative stress |
| 12B | 40322.9 | 5.2 | S. aureus | Alanine dehydrogenase 1 | Ald1 | 15 | 100 | 21283057 | Cell wall synthesis; oxidation reduction |
| (C) Proteins upregulated in both biofilm conditions | |||||||||
| 9A,B | 21481.5 | 5.79 | C. albicans | Similar to phosphoglycerate mutase | Gpm1p | 16 | 100 | 68469783 | Carbohydrate metabolism; glycolysis |
| 10A,B | 17677.9 | 7.74 | C. albicans | Cyclophilin type peptidyl-prolyl cis–trans isomerase | Cyp1p | 4 | 100 | 68469052 | Protein folding; cellular stress response |
| 13A,B | 55751.8 | 6.54 | C. albicans | Pyruvate kinase | Pyk1p | 16 | 100 | 68482226 | Carbohydrate metabolism; glycolysis |
| 2A,B | 36423.8 | 5.34 | S. aureus | Alcohol dehydrogenase | Adh | 12 | 100 | 21282297 | Carbon utilization; alcohol metabolism |
| 5A,B | 29434.3 | 5.34 | S. aureus | 30s ribosomal protein S2 | RpsB | 6 | 100 | 57651825 | Protein synthesis; stress response |
| 6A,B | 18520.5 | 5.6 | S. aureus | Similar to universal stress protein family | Similar to UspA1 | 7 | 95.7 | 15924700 | Cellular stress response |
| 4A,B | 37381.6 | 6.08 | S. aureus | Threonine dehydratase | IlvA | 16 | 100 | 147733998 | Amino acid metabolism |
Discussion
Previous studies have identified that C. albicans–S. aureus intraperitoneal coinfections resulted in enhanced virulence and lethality in a mouse model, but a detailed description of the polymicrobial interactions between these pathogens has remained undefined (Carlson, 1983; Carlson & Johnson, 1985;). To this end, this study was designed to examine the physical interactions and the differential protein expression occurring during C. albicans–S. aureus polymicrobial biofilm growth.
Because C. albicans bacterial binding has been reported previously, the relative C. albicans hyphal-binding affinity of other bacteria was evaluated and compared with that of S. aureus (Fig. 1). Comparative adherence assays demonstrated that all bacterial species tested, including the more closely related species of S. pyogenes and S. epidermidis, associated with the hyphae of C. albicans significantly less than S. aureus. Differences in hyphal binding between various bacteria may be due to differences in surface protein expression or as yet unidentified microbial adhesins. Our assay also demonstrated significantly lower hyphal binding of the well-described C. albicans-interacting bacterial species P. aeruginosa compared with S. aureus. Even with this comparatively lower binding affinity, Hogan et al. (2004) have demonstrated that P. aeruginosa is capable of killing the hyphae of C. albicans, through a process involving the homoserine lactone quorum-sensing molecule, 3-oxo-C12. These observations indicate that, unlike the seemingly mutualistic C. albicans–S. aureus relationship demonstrated by our studies, the interaction between C. albicans and P. aeruginosa seems to be antagonistic.
Because of the significantly increased rates of staphylococcal–fungal association identified in the hyphal–bacterial attachment screen, polymicrobial growth was visualized by fluorescence microscopy in order to determine the architecture of the co-culture biofilm. Imaging analysis revealed S. aureus adhering to the invasive hyphal filaments of C. albicans, but not the round yeast cells (Fig. 2). Confocal z-stack imaging showed S. aureus to be distributed along the hyphal filaments throughout the entire biofilm architecture (Fig. 3b). These findings differ from the recent findings by Harriott & Noverr (2009) investigating increased drug resistance in polymicrobial biofilms in which S. aureus was noted to be attached to hyphal elements mostly in the uppermost layers of the biofilm. Differences in biofilm growth substratum and medium may partially account for these discrepancies. Preference for binding the hyphae of C. albicans has been reported in a number of other species, including S. pyogenes, Acinetobacter baumannii, and P. aeruginosa (Cunningham, 2000; Hogan & Kolter, 2002; Peleg et al., 2008; Bamford et al., 2009;). Many of these previously identified C. albicans–bacteria interactions result in fungal and/or bacterial killing during co-culture; however, the C. albicans–S. aureus interaction described in this study appears to be nonlethal for either organism as measured by the LIVE/DEAD cell viability assay (Fig. 3a). The lack of an antagonistic relationship during polymicrobial growth may have important implications for the enhancement of virulence during coinfection and may partially explain the relatively high rate of co-culture for these organisms. Combined, these important findings highlight the diversity of the interactions that take place between these human pathogens.
In light of the observed extensive association between S. aureus and C. albicans hyphae, we hypothesized that protein expression may be modulated in the dual-species environment, which could have important implications during coinfection. Hyphal binding may result in altered virulence factor production, augmenting immunoavoidance and/or damage to the host as has been seen in other species (Richard et al., 2002; Sibley et al., 2008;). In order to further characterize the molecular interactions between C. albicans and S. aureus and to identify the factors that may be responsible for their infectious synergism, a global proteomics approach was utilized; the upregulated proteins identified are listed in Table 1. Among the 27 differentially regulated proteins, some were upregulated either uniquely in the staphylococcal–yeast or staphylococcal–hyphae biofilms or in both co-culture conditions compared with mono-species cultures. These proteins were mainly involved in growth, metabolism, or response to stress including proteins that are inducible upon heat, oxidative, nutrient, and antibacterial stress.
Several stress-related proteins, known to be induced upon heat, oxidative, and antibacterial stress, were found to be consistently upregulated by S. aureus, indicating the presence of a stress response by S. aureus to both C. albicans yeast and hyphal forms (Table 1) (Kvint et al., 2003). Similarly, Cyp1p, a cis–trans isomerase involved in protein folding and upregulated during oxidative and nutritional stress, was upregulated in C. albicans (Dartigalongue & Raina, 1998; Andreeva et al., 1999; Wen et al., 2005;). The upregulation of the uspA-like protein, Cyp1p, and RpsB, a ribosomal protein, is consistent with the findings of upregulated proteins in vivo during Mycobacterium avium infection and emphasizes that these proteins may be important in resisting heat shock and stress inside the host (Hughes et al., 2007).
Many growth and metabolic proteins in both C. albicans and S. aureus were upregulated in the mixed biofilm. Contrary to our expectations, however, the majority of the upregulated proteins were present in the staphylococcal–yeast biofilm. Interestingly, C. albicans yeast cells demonstrated the upregulation of a significant number of proteins involved in cell stress, including the heat shock proteins, which are highly inducible upon cell stresses including heat, hypoxia, UV exposure, starvation, toxin exposure, and dehydration (Table 1) (Matthews & Burnie, 1992). It is possible that staphylococcal binding to C. albicans blastospores within the polymicrobial biofilm may have been evolutionarily selected against under seemingly ‘stressful’ conditions.
In C. albicans, Mac1p is a transcription factor that facilitates the uptake of copper. Copper is an important cofactor for a wide variety of cellular enzymes that carry out essential biological processes such as respiration (Marvin et al., 2003). Furthermore, copper is believed to play a detrimental role in protection against oxidative stress, which provides an additional explanation for the observed upregulation of Mac1p. This is corroborated by the observed aforementioned concomitant upregulation of various stress response proteins by C. albicans yeast cells. Combined, these findings clearly indicate that the presence of S. aureus induces a stress response by C. albicans.
Among the proteins of note found to be upregulated in C. albicans yeast cells was Tsa1p, a thioredoxin peroxidase important for detoxification after peroxide stress (Urban et al., 2005), and aconitate hydratase, which is highly susceptible to oxidation under stressed conditions (Tang et al., 2002; Matasova & Popova, 2008;). Few proteins were found to be upregulated in the staphylococcal–hyphae biofilm in either organism. In C. albicans, the expression of Tef2p, a GTP-binding translational elongation factor important for protein synthesis, was increased (Capa et al., 1998). In S. aureus, there was increased expression of alanine dehydrogenase, shown to be involved in the metabolism of alanine and suggested to have a role in bacterial cell wall synthesis (Andersen et al., 1992). In addition, cysteine synthase involved in the biosynthesis of cysteine was also found to be upregulated in S. aureus. Staphylococcus aureus mutants deficient in cysteine synthase are more susceptible to oxidative stress, acid, and phosphate-limiting conditions due to the role of cysteine in stress response and survival mechanisms (Lithgow et al., 2004).
Staphylococcal gene products that have been previously shown to play an important role in virulence and pathogenesis were also shown to be differentially regulated under coculture conditions compared with mono-species cultures. In the staphylococcal–yeast biofilm, CodY, a transcriptional repressor of a variety of S. aureus virulence factors exhibited increased expression (Levdikov et al., 2006). This protein has was shown to repress PIA-dependent biofilm formation, the production of hemolysins alpha and delta, and proteins involved in the global regulator of virulence, the agr-dependent quorum-sensing system (Frees et al., 2005; Majerczyk et al., 2008;). However, CodY was downregulated under the staphylococcal–hyphal biofilm growth conditions. Therefore, decreased CodY expression may enable enhanced toxin-mediated virulence and increased biofilm formation in S. aureus.
The virulence-associated l-lactate dehydrogenase 1 (Ldh1), an enzyme involved in the generation of l-lactate during fermentation, was upregulated in the staphylococcal–hyphal biofilm, but not in the staphylococcal–yeast biofilm. Recently, biochemical studies by Richardson and colleagues demonstrated that S. aureus Ldh1 is uniquely inducible under nitrosative stress conditions, enabling S. aureus to persist in the host in the presence of host-derived nitric oxide. Furthermore, an S. aureus ldh1 mutant exhibited attenuated virulence compared with wild-type S. aureus in a mouse model of systemic infection (Richardson et al., 2008). Closely related staphylococcal species, S. epidermidis and Staphylococcus saprophyticus, lack Ldh1 and therefore cannot survive under conditions of nitric oxide stress as encountered in host macrophages and neutrophils.
The increased expression of CodY and downregulation of Ldh1 lead us to hypothesize that S. aureus may downregulate its virulence while coexisting with C. albicans yeast cells at a mucosal surface such as at vaginal, gastrointestinal, or oral tracts as a strategy to remain in a commensal state at these sites, thereby evading detection and clearance by the host immune system. Conversely, candidal germination appears to induce S. aureus virulence and biofilm formation capability through the downregulation of CodY expression. The simultaneous increase in Ldh1 expression could potentially combat nitric oxide produced by the host in response to C. albicans hyphal invasion (Oliveira et al., 2007). While these proteomics studies are not a comprehensive analysis of the entire proteome, they do demonstrate the plasticity of global protein expression unique to polymicrobial growth. Further experiments to address these polymicrobial-enhanced immunoavoidance and virulence mechanisms, as well as the possible differential expression of cell wall proteins and secreted factors, are warranted and currently underway in our laboratories.
In conclusion, this study characterizes a unique microbial association within the context of a polymicrobial biofilm, in which S. aureus binds the hyphal elements of C. albicans. In addition, it establishes the presence of a robust and dynamic interaction between two diverse and significant human pathogens by demonstrating the upregulation of several putative virulence factors specific to polymicrobial growth. The findings generated from this investigation will contribute to our understanding of the complex and clinically significant interactions that take place between microbial species as they coexist in the host and during infectious processes. Therefore, continued epidemiologic and laboratory research is needed to better characterize and understand these pathogens in the context of complicated polymicrobial infections, allowing for improved diagnostic and therapeutic strategies in the future.
Acknowledgments
We graciously thank AdvanDx for the generous donation of various PNA-FISH probes. We also thank Joachim Morschauser at the University of Würzburg, Germany, for providing the GFP-expressing C. albicans strain. We also thank Anthony Haag at the Mass Spectrometry Core of the Biomolecular Resource Facility at the University of Texas Medical Branch for conducting MALDI-ToF/ToF MS and database analyses. These studies were funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health grant R01 AI69568 and the National Institute of Dental and Craniofacial Research, National Institutes of Health grants 1R01DE20939 and 5T32 DE007309.
Statement
Re-use of this article is permitted in accordance with the Terms and Conditions set out at http://www3.interscience.wiley.com/authorresources/onlineopen.html
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe S, Ishihara K, Okuda K. Prevalence of potential respiratory pathogens in the mouths of elderly patients and effects of professional oral care. Arch Gerontol Geriat. 2001;32:45–55. doi: 10.1016/s0167-4943(00)00091-1. [DOI] [PubMed] [Google Scholar]
- Andersen AB, Andersen P, Ljungqvist L. Structure and function of a 40,000-molecular-weight protein antigen of Mycobacterium tuberculosis. Infect Immun. 1992;60:2317–2323. doi: 10.1128/iai.60.6.2317-2323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva L, Heads R, Green CJ. Cyclophilins and their possible role in the stress response. Int J Exp Pathol. 1999;80:305–315. doi: 10.1046/j.1365-2613.1999.00128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Monroy T, Moreno-Maldonado V, Franco-Martinez F, Aldape-Barrios B, Quindos G, Sanchez-Vargas LO. Candida albicans, Staphylococcus aureus and Streptococcus mutans colonization in patients wearing dental prosthesis. Med Oral Patol Oral Cir Bucal. 2005;10(suppl 1):E27–E39. [PubMed] [Google Scholar]
- Bamford CV, d'Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady RA, Leid JG, Camper AK, Costerton JW, Shirtliff ME. Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect Immun. 2006;74:3415–3426. doi: 10.1128/IAI.00392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capa L, Mendoza A, Lavandera JL, Gomez de las Heras F, Garcia-Bustos JF. Translation elongation factor 2 is part of the target for a new family of antifungals. Antimicrob Agents Ch. 1998;42:2694–2699. doi: 10.1128/aac.42.10.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E. Effect of strain of Staphylococcus aureus on synergism with Candida albicans resulting in mouse mortality and morbidity. Infect Immun. 1983;42:285–292. doi: 10.1128/iai.42.1.285-292.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E, Johnson G. Protection by Candida albicans of Staphylococcus aureus in the establishment of dual infection in mice. Infect Immun. 1985;50:655–659. doi: 10.1128/iai.50.3.655-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlen G, Linde A, Moller AJ, Ohman A. A retrospective study of microbiologic samples from oral mucosal lesions. Oral Surg Oral Med Oral Pathol. 1982;53:250–255. doi: 10.1016/0030-4220(82)90299-7. [DOI] [PubMed] [Google Scholar]
- Dartigalongue C, Raina S. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Repentigny L, Lewandowski D, Jolicoeur P. Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin Microbiol Rev. 2004;17:729–759. doi: 10.1128/CMR.17.4.729-759.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwempu CC, Lawande RV, Egler LJ. Microbial flora of the lower genital tract of women in labour in Zaria, Nigeria. J Clin Pathol. 1981;34:82–83. doi: 10.1136/jcp.34.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frees D, Thomsen LE, Ingmer H. Staphylococcus aureus ClpYQ plays a minor role in stress survival. Arch Microbiol. 2005;183:286–291. doi: 10.1007/s00203-005-0773-x. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Goetghebeur M, Landry PA, Han D, Vicente C. Methicillin-resistant Staphylococcus aureus: a public health issue with economic consequences. Can J Infect Dis Med Microbiol. 2007;18:27–34. doi: 10.1155/2007/253947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46(suppl 5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Ch. 2009;53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Kolter R. Pseudomonas–Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- Hughes V, Smith S, Garcia-Sanchez A, Sales J, Stevenson K. Proteomic comparison of Mycobacterium avium subspecies paratuberculosis grown in vitro and isolated from clinical cases of ovine paratuberculosis. Microbiology. 2007;153:196–205. doi: 10.1099/mic.0.29129-0. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Klotz SA, Chasin BS, Powell B, Gaur NK, Lipke PN. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn Micr Infec Dis. 2007;59:401–406. doi: 10.1016/j.diagmicrobio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Kvint K, Nachin L, Diez A, Nystrom T. The bacterial universal stress protein: function and regulation. Curr Opin Microbiol. 2003;6:140–145. doi: 10.1016/s1369-5274(03)00025-0. [DOI] [PubMed] [Google Scholar]
- Leid JG, Shirtliff ME, Costerton JW, Stoodley AP. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002;70:6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J Biol Chem. 2006;281:11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]
- Lithgow JK, Hayhurst EJ, Cohen G, Aharonowitz Y, Foster SJ. Role of a cysteine synthase in Staphylococcus aureus. J Bacteriol. 2004;186:1579–1590. doi: 10.1128/JB.186.6.1579-1590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson JM, Rauch M, Gilmore MS. The commensal microbiology of the gastrointestinal tract. Adv Exp Med Biol. 2008;635:15–28. doi: 10.1007/978-0-387-09550-9_2. [DOI] [PubMed] [Google Scholar]
- Marvin ME, Williams PH, Cashmore AM. The Candida albicans CTR1 gene encodes a functional copper transporter. Microbiology. 2003;149:1461–1474. doi: 10.1099/mic.0.26172-0. [DOI] [PubMed] [Google Scholar]
- Mastropaolo MD, Evans NP, Byrnes MK, Stevens AM, Robertson JL, Melville SB. Synergy in polymicrobial infections in a mouse model of type 2 diabetes. Infect Immun. 2005;73:6055–6063. doi: 10.1128/IAI.73.9.6055-6063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasova LV, Popova TN. Aconitate hydratase of mammals under oxidative stress. Biochemistry. 2008;73:957–964. doi: 10.1134/S0006297908090010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews R, Burnie J. The role of hsp90 in fungal infection. Immunol Today. 1992;13:345–348. doi: 10.1016/0167-5699(92)90169-8. [DOI] [PubMed] [Google Scholar]
- Minden J. Comparative proteomics and difference gel electrophoresis. Biotechniques. 2007;43:739–743. doi: 10.2144/000112653. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Iwai T, Sugai M, Miura K, Suginaka H, Nagasaka N. Incidence and characterization of Staphylococcus aureus from the tongues of children. J Dent Res. 1991;70:1045–1047. doi: 10.1177/00220345910700070501. [DOI] [PubMed] [Google Scholar]
- Morschhauser J, Michel S, Hacker J. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol Gen Genet. 1998;257:412–420. doi: 10.1007/s004380050665. [DOI] [PubMed] [Google Scholar]
- O'Connell HA, Kottkamp GS, Eppelbaum JL, Stubblefield BA, Gilbert SE, Gilbert ES. Influences of biofilm structure and antibiotic resistance mechanisms on indirect pathogenicity in a model polymicrobial biofilm. Appl Environ Microb. 2006;72:5013–5019. doi: 10.1128/AEM.02474-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ohara-Nemoto Y, Haraga H, Kimura S, Nemoto TK. Occurrence of staphylococci in the oral cavities of healthy adults and nasal oral trafficking of the bacteria. J Med Microbiol. 2008;57:95–99. doi: 10.1099/jmm.0.47561-0. [DOI] [PubMed] [Google Scholar]
- Oliveira MA, Carvalho LP, Gomes Mde S, Bacellar O, Barros TF, Carvalho EM. Microbiological and immunological features of oral candidiasis. Microbiol Immunol. 2007;51:713–719. doi: 10.1111/j.1348-0421.2007.tb03960.x. [DOI] [PubMed] [Google Scholar]
- Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC, Jr, Mylonakis E. Prokaryote–eukaryote interactions identified by using Caenorhabditis elegans. P Natl Acad Sci USA. 2008;105:14585–14590. doi: 10.1073/pnas.0805048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulimood S, Ganesan L, Alangaden G, Chandrasekar P. Polymicrobial candidemia. Diagn Micr Infec Dis. 2002;44:353–357. doi: 10.1016/s0732-8893(02)00460-1. [DOI] [PubMed] [Google Scholar]
- Ramage G, Bachmann S, Patterson TF, Wickes BL, Lopez-Ribot JL. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemoth. 2002;49:973–980. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- Richard M, Ibata-Ombetta S, Dromer F, Bordon-Pallier F, Jouault T, Gaillardin C. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol Microbiol. 2002;44:841–853. doi: 10.1046/j.1365-2958.2002.02926.x. [DOI] [PubMed] [Google Scholar]
- Richardson AR, Libby SJ, Fang FC. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science. 2008;319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 2009;299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegman-Igra Y, Schwartz D, Konforti N. Polymicrobial bacteremia. Med Microbiol Immun. 1988;177:169–179. doi: 10.1007/BF00232896. [DOI] [PubMed] [Google Scholar]
- Smith AJ, Robertson D, Tang MK, Jackson MS, MacKenzie D, Bagg J. Staphylococcus aureus in the oral cavity: a three-year retrospective analysis of clinical laboratory data. Brit Dent J. 2003;195:701–703. doi: 10.1038/sj.bdj.4810832. [DOI] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Tang Y, Quail MA, Artymiuk PJ, Guest JR, Green J. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology. 2002;148:1027–1037. doi: 10.1099/00221287-148-4-1027. [DOI] [PubMed] [Google Scholar]
- Urban C, Xiong X, Sohn K, Schroppel K, Brunner H, Rupp S. The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans. Mol Microbiol. 2005;57:1318–1341. doi: 10.1111/j.1365-2958.2005.04771.x. [DOI] [PubMed] [Google Scholar]
- Valenza G, Tappe D, Turnwald D, Frosch M, Konig C, Hebestreit H, Abele-Horn M. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros. 2008;7:123–127. doi: 10.1016/j.jcf.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Wen ZT, Suntharaligham P, Cvitkovitch DG, Burne RA. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect Immun. 2005;73:219–225. doi: 10.1128/IAI.73.1.219-225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]