Abstract
Productivity and predation are thought to be crucial drivers of bacterial diversity. We tested how the productivity–diversity of a natural bacterial community is modified by the presence of protist predators with different feeding preferences. In the absence of predators, there was a unimodal relationship between bacterial diversity and productivity. We found that three protist species (Bodo, Spumella and Cyclidium) had widely divergent effects on bacterial diversity across the productivity gradient. Bodo and Cyclidium had little effect on the shape of the productivity–diversity gradient, while Spumella flattened the relationship. We explain these results in terms of the feeding preferences of these predators.
Keywords: bacteria, productivity, biodiversity, protist, predation
1. Introduction
Productivity (here defined in terms of resource availability) is believed to be one of the principal drivers of bacterial community composition and diversity. While a large body of theory and data suggest that biodiversity is highest at intermediate levels of productivity, this relationship is neither universal for larger organisms (Mittelbach et al. 2001) nor for microbes (Smith 2007). Many mechanisms have been proposed to explain the variety of productivity–diversity relationships. We will not review this extensive literature, but one promising mechanism in bacterial communities is that predators modulate the response of the prey community to variation in productivity.
Predators modify the productivity–diversity relationship by altering the abundance and stability of competing prey species (Rosenzweig 1971; Chase et al. 2002). For example, simple models have shown that superior resource exploiters dominate unproductive environments, whereas resistance to predation is increasingly favoured as productivity increases (Leibold 1996), a hypothesis that has been confirmed using simplified microcosm communities containing a single strain of phage (predator) and bacteria (Bohannan & Lenski 2000).
Protists play a pivotal role in structuring natural bacterial communities (Pernthaler 2005). Bacterial communities undergo morphological and physiological changes when subjected to protist predators by forming predation-resistant filaments (Hahn et al. 2000; Jurgens & Sala 2000; Hahn & Hofle 2001) or by increasing cell size (Corno & Jurgens 2006), but there has been less experimental effort focused on the ecological effects of protist predators on bacterial community diversity. Studies have tended to either involve intraspecific morphological diversity (Meyer & Kassen 2007; Friman et al. 2008) simplified, constructed bacterial communities (Jiang & Morin 2005; Jiang & Krumins 2006; Meyer & Kassen 2007; Friman et al. 2008), or have been observational studies of natural environments (Hahn & Hofle 2001).
To bridge the gap between simple microcosm communities and the full complexity of a natural ecosystem, we advocate an experimental microcosm approach containing constructed protist communities together with natural bacterial communities. We tested the effects of three protist predators in microcosms that are laboratory analogues of water-filled tree-holes (Bell et al. 2005).
2. Material and methods
(a). Microcosms
Freshly fallen beech leaves were collected and stored at −20°C. Diverse bacterial communities were obtained by incubating 1 g of the beech leaves in 40 ml Chalkey's medium (CM: 10 g NaCl, 0.4 g KCl, 0.6 g CaCl2 per litre of water, autoclaved and adjusted to neutral pH) for 2 days at 20°C in closed microcosms. This mixture was vortexed and passed through a 1 µm filter to separate bacterial cells from larger organisms, and to obtain a bacterial community that was derived from the surface of the beech leaves. A beech leaf broth was obtained by autoclaving 50 g beech leaves in 500 ml CM at 120°C for 30 min. The resulting broth was passed through a 0.22 µm filter to remove debris and ensure sterility and serially diluted in CM to create the productivity gradient. The level of productivity is expressed as a proportion of the neat broth. Three 24-well plates containing 2 ml of broth were inoculated with 104 bacterial cells. The bacteria were incubated for 2 days (20°C) after which the appropriate microcosms were inoculated with 103 protist cells (Bodo sp., Spumella sp. or Cyclidium sp.) and incubated for 7 days. Each treatment combination was replicated three times.
(b). Bacteria
Bacteria and protists were fixed in 1 per cent paraformaldehyde and stained with the DNA specific stain SYBR Green I (Molecular Probes). Samples were analysed using a Becton Dickinson FACS Calibur flow cytometer using internal standards (2.49 µm beads). Protists were counted according to the methodology described by Zubkov et al. (2007). Bacterial diversity was assessed using terminal restriction fragment length polymorphism (tRFLP) where amplified bacterial DNA is cut using a site-specific endonuclease, and the sizes of the resulting restriction fragments are measured. Our tRFLP protocol is described by Thomson et al. (in press). Each restriction fragment was considered to be a taxonomic unit. The relative abundance of each taxon was calculated as the relative intensity of each tRFLP band within a sample.
3. Results
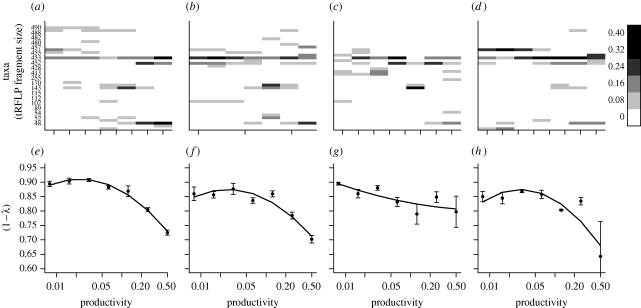
In the absence of predators, increasing resource concentration caused a progressive increase in the total number of bacterial cells (electronic supplementary material, figure S1; table 1). The increase did not correspond to an increase in bacterial diversity. Rather, a different suite of taxa was selected at high versus low productivity (figure 1a), producing the classic unimodal relationship.
Table 1.
Analysis of the experimental data. (Degrees of freedom (d.f.) and F-statistics associated with each of the experimental treatments (predators and productivity). Significant p-values are indicated with asterisks: *p < 0.05, **p < 0.005, ***p < 0.0005.)
| source of variance | d.f. | bacterial abundance | bacterial diversity |
|---|---|---|---|
| predators | 4 | 7.6*** | 4.5** |
| productivity | 1 | 772.5*** | 90.3*** |
| (productivity)2 | 1 | 310.9*** | 20.2*** |
| predators × productivity | 4 | 135.7*** | 1.4 |
| predators × (productivity)2 | 4 | 1.5 | 2.4* |
| residual | 90 |
Figure 1.
Composition and diversity of the microcosm bacterial communities. The top row of panels shows the mean relative abundance of each bacterial taxon across the productivity gradient for (a) no predators, (b) Bodo sp., (c) Spumella sp., and (d) Cyclidium. sp. Colour intensity indicates the relative abundance of each bacterial taxon. The tRFLP fragment size (number of base pairs) associated with each taxon is shown on the y-axis. (e–h) shows the bacterial biodiversity, measured as the complement of the Simpson index (1 − λ), across the productivity gradient for each predator treatment. Broadly similar results are obtained if the Shannon–Wiener index is used (electronic supplementary material, figure S2). Curves are fitted regression lines.
Bacterial abundance also increased across the productivity gradient when predators were present. Predators significantly dampened the increase at intermediate productivities but not at high or low productivities (electronic supplementary material, figure S1). The pattern was mirrored by higher predator abundance at intermediate productivities (electronic supplementary material, figure S1). Unlike bacterial abundance, predator abundance at high productivity was equal to their abundance at low productivity. The pattern of bacterial diversity was notably different for the different protists. The shape of the relationship was similar for Bodo and Cyclidium compared with the no-protist control (figure 1b,d). Spumella created bacterial communities that were markedly different because bacterial diversity was maintained even at high levels of productivity (quadratic term: F1,18 = 0.3, p = 0.62; figure 1c). The mode of the fitted relationship was within the range of productivity values used in the experiment for the no-protist control and for Bodo and Cyclidium (p < 0.05), but not for Spumella (p > 0.05) (Mitchell-Olds & Shaw 1987).
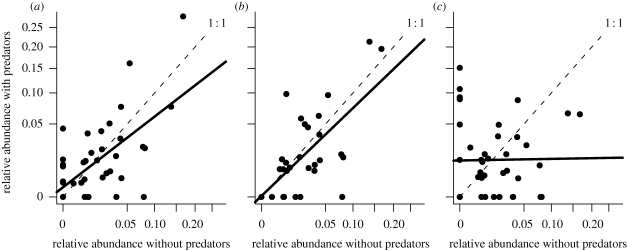
If we plot the relative bacterial abundance in the presence and absence of each predator (figure 2), the magnitude to which the slope of the regression deviates from the 1 : 1 line can be used as a measure of the degree to which the predator is a complete generalist (regression slope equals unity) or an extreme specialist that preferentially removes the most abundant prey species (regression slope is zero). Bodo grazed on all taxa (figure 2a), as demonstrated by a significantly positive relationship between bacterial abundance in the presence versus absence of Bodo (F1,38 = 23.8, p < 0.0005). The slope of the line was significantly different from unity (t38 = 2.3, p = 0.03) indicating a slight preference for more abundant bacterial taxa. A similar observation was made for microcosms containing Cyclidium (figure 2b, t38 = −1.20, p = 0.23). By contrast, Spumella removed the more dominant bacteria to a much greater extent than the rarer taxa, apparently allowing some previously rare bacteria to proliferate (figure 2c). The regression slope is consequently not different from zero (F1,38 = 0.01, p = 0.93).
Figure 2.
Relative abundance of each bacterial species in the presence of (a) Bodo sp., (b) Cyclidium sp., and (c) Spumella sp. as a function of their relative abundance in microcosms without predators. The axes are on an arcsine transformed square-root scale.
4. Discussion
The results of our productivity–diversity experiments are largely consistent with the experimental and observational results for larger organisms (Mittelbach et al. 2001), as well as more simplified microcosm communities (Kassen et al. 2000; Hall & Colegrave 2007; Benmayor et al. 2008). Even though bacterial abundance increased across the productivity gradient, bacterial diversity increased slightly and then decreased with increasing productivity. That bacterial assemblages at high and low productivity are largely independent suggests a fitness trade-off between high and low productivity environments. At intermediate productivities, diversity is maximized because these assemblages start to cross over.
We have also demonstrated that predators of bacteria can have a wide variety of effects on bacterial diversity and community composition (figure 1). Such a result is unsurprising because previous studies have shown differential effects of protist predators (Simek et al. 1997), but the result does emphasize the need to think critically about how different kinds of predator affect bacterial communities. Some of the principal ideas of how predators influence competing prey species revolve around whether particular prey taxa are removed preferentially. Assessing the preferences of protists feeding on diverse bacterial communities containing hundreds or thousands of taxa is problematic, especially as many of the bacteria cannot be isolated and cultured. Feeding trials with individual bacterial strains are therefore not possible in practice, so we determined which bacterial taxa were removed compared with a predator-free control.
The consequence of a uniform per capita attack rate is that the addition of the predator will not alter the relative abundance of the prey species in the short term because each species is equally affected by the predator. The data therefore suggest that Bodo and Cyclidium are acting as generalist predators, at least in our experimental communities (figure 2a,b). By contrast, Spumella removed the more dominant bacterial species to a much greater extent than the rarer taxa (figure 2c), apparently allowing some previously rare bacterial species to proliferate. Generalist predators would decrease the abundance of every prey species resulting in overall lower diversity but a similar pattern of diversity across a productivity gradient is unaltered. Specialist predators simply allow predator-resistant taxa to proliferate resulting in a relatively flat relationship. This suggests a trade-off between predator resistance and competitive ability.
The experiment is similar to a recent study that constructed microcosm communities with three trophic levels containing bacteria, bacteriovores and either a specialist or a generalist predator (Jiang & Morin 2005). In apparent contrast to the current work, generalists reduced the variance in diversity across productivity levels, but the specialist had no effect. It is unclear how far these results can be generalized because the specialist predator had no apparent effect on community diversity, structure or density, whereas the generalist also consumed the prey's resource (bacteria). However, contrasting the two studies further emphasizes the importance of biological details in determining the impact of predators on community diversity.
The observations that predators had little effect on bacterial abundance at high productivity, and that predator abundance declined at high productivity despite higher bacterial abundance (electronic supplementary material, figure S1), strongly imply that the bacterial communities were able to withstand predation pressure. The results lend support to the hypothesis that the cost of resistance is lower at high productivity, resulting in bacterial communities that are able to resist protist predation. Such a conclusion is consistent with observations of the development of grazing-resistant forms across productivity gradients (Corno & Jürgens 2008). A recent chemostat experiment also investigated the effect of nutrient and predator limitation (Corno & Jürgens 2008). Although their experiment documented different patterns of predator and prey abundance across the productivity gradient (both increased linearly), they also concluded that the observed patterns of composition and diversity hinged on a trade-off between predator resistance and growth rate.
Overall, our findings are consistent with relatively simple underlying mechanisms: that there exist life-history trade-offs in natural bacterial communities, which allows predator-resistant prey to increase with resources availability. That relatively few assumptions are required suggest that these findings are likely to be relevant to a wide range of ecological communities and not only for bacterial communities. Moreover, we have shown that the interactive effects of key ecological variables can be studied in a meaningful way in natural microbial communities, containing largely ‘unculturable’ bacteria.
Acknowledgements
This work was supported by the NERC NE/F000286/1 and the Royal Society. We are grateful to M. Finke for piloting the methods used here.
References
- Bell T., Ager D., Song J.-I., Newman J. A., Thompson I. P., Lilley A. K., Van der Gast C. J.2005Larger islands house more bacterial taxa. Science 308, 1884 (doi:10.1126/science.1111318) [DOI] [PubMed] [Google Scholar]
- Benmayor R., Buckling A., Bonsall M. B., Brockhurst M., Hodgson D. J.2008The interactive effects of parasites, disturbance and productivity on experimental adaptive radiations. Evolution 62, 467–477 (doi:10.1111/j.1558-5646.2007.00268.x) [DOI] [PubMed] [Google Scholar]
- Bohannan B. J. M., Lenski R. E.2000Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3, 362–377 (doi:10.1046/j.1461-0248.2000.00161.x) [Google Scholar]
- Chase J. M., Abrams P. A., Grover J. P., Diehl S., Chesson P., Holt R. D., Richards S. A., Nisbet R. M., Case T. J.2002The interaction between predation and competition: a review and synthesis. Ecol. Lett. 5, 302–315 (doi:10.1046/j.1461-0248.2002.00315.x) [Google Scholar]
- Corno G., Jurgens K.2006Direct and indirect effects of protist predation on population size structure of a bacterial strain with high phenotypic plasticity. Appl. Env. Microbiol. 72, 78–86 (doi:10.1128/AEM.72.1.78-86.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corno G., Jürgens K.2008Structural and functional patterns of bacterial communities in response to protist predation along an experimental productivity gradient. Env. Microbiol. 10, 2857–2871 (doi:10.1111/j.1462-2920.2008.01713.x) [DOI] [PubMed] [Google Scholar]
- Friman V. P., Hiltunen T., Laakso J., Kaitala V.2008Availability of prey resources drives evolution of predator–prey interaction. Proc. R. Soc. B 275, 1625–1633 (doi:10.1098/rspb.2008.0174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., Hofle M. G.2001Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35, 113–121 (doi:10.1111/j.1574-6941.2001.tb00794.x) [DOI] [PubMed] [Google Scholar]
- Hahn M. W., Moore E. R. B., Hofle M. G.2000Role of microcolony formation in the protistan grazing defense of the aquatic bacterium Pseudomonas sp. MWH1. Microb. Ecol. 39, 175–185 [DOI] [PubMed] [Google Scholar]
- Hall A. R., Colegrave N.2007How does resource supply affect evolutionary diversification? Proc. R. Soc. B 274, 73 (doi:10.1098/rspb.2006.3703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Krumins J. A.2006Consumer vs environmental productivity control of bacterial diversity and bacteria-mediated organic matter decomposition. Oikos 114, 441–450 (doi:10.1111/j.2006.0030-1299.14961.x) [Google Scholar]
- Jiang L., Morin P. J.2005Predator diet breadth influences the relative importance of bottom-up and top-down control of prey biomass and diversity. Am. Nat. 165, 350–363 (doi:10.1086/428300) [DOI] [PubMed] [Google Scholar]
- Jurgens K., Sala M. M.2000Predation-mediated shifts in size distribution of microbial biomass and activity during detritus decomposition. Oikos 91, 29–40 (doi:10.1034/j.1600-0706.2000.910103.x) [Google Scholar]
- Kassen R., Buckling A., Bell G., Rainey P. B.2000Diversity peaks at intermediate productivity in a laboratory microcosm. Nature 406, 508–512 (doi:10.1038/35020060) [DOI] [PubMed] [Google Scholar]
- Leibold M. A.1996A graphical model of keystone predators in food webs: trophic regulation of abundance, incidence, and diversity patterns in communities. Am. Nat. 147, 784–812 [Google Scholar]
- Meyer J. R., Kassen R.2007The effects of competition and predation on diversification in a model adaptive radiation. Nature 446, 432–435 (doi:10.1038/nature05599) [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T., Shaw R. G.1987Regression analysis of natural selection: statistical inference and biological interpretation. Evolution 41, 1149–1161 (doi:10.2307/2409084) [DOI] [PubMed] [Google Scholar]
- Mittelbach G. G., Steiner C. F., Scheiner S. M., Gross K. L., Reynolds H. L., Waide R. B., Willig M. R., Dodson S. I., Gough L.2001What is the observed relationship between species richness and productivity? Ecology 82, 2381–2396 (doi:10.1890/0012-9658(2001)082[2381:WITORB]2.0.CO;2) [Google Scholar]
- Pernthaler J.2005Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3, 537–546 (doi:10.1038/nrmicro1180) [DOI] [PubMed] [Google Scholar]
- Rosenzweig M. L.1971Paradox of enrichment: destabilization of exploitation ecosystems in ecological time. Science 171, 385–387 (doi:10.1126/science.171.3969.385) [DOI] [PubMed] [Google Scholar]
- Simek K., Vrba J., Pernthaler J., Posch T., Hartman P., Nedoma J., Psenner R.1997Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Env. Microbiol. 63, 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V. H.2007Microbial diversity–productivity relationships in aquatic ecosystems. FEMS Microbiol. Ecol. 62, 181–186 (doi:10.1111/j.1574-6941.2007.00381.x) [DOI] [PubMed] [Google Scholar]
- Thomson B. C., Ostle N., McNamara N., Bailey M. J., Whiteley A. S., Griffiths R. I.In press Vegetation affects the relative abundances of dominant soil bacterial taxa and soil respiration rates in an upland grassland soil. Microb. Ecol. (doi:10.1007/s00248-009-9575-z) [DOI] [PubMed] [Google Scholar]
- Zubkov M. V., Burkill P. H., Topping J. N.2007Flow cytometric enumeration of DNA-stained oceanic planktonic protists. J. Plankton Res. 29, 79–86 (doi:10.1093/plankt/fbl059) [Google Scholar]