Abstract
The mechanism by which the apoptosome activates caspases during apoptosis has been controversial. Qi et al. (2010) now present a crystal structure of a funnel-shaped octameric apoptosome complex from the nematode Caenorhabditis elegans that challenges currently held assumptions about the human apoptosome structure.
During apoptosis, caspases cleave key intracellular target proteins to promote the death of the cell and disposal of the cell corpse. The first caspases activated during this process are the initiator caspases, which in turn cleave and activate the executioner caspases. The activation of initiator caspases is highly regulated, and it is thought to involve their dimerization, but the detailed mechanisms remain unclear. In mammals, activation of initiator caspase-9 requires the assembly of a heptameric wheel-like complex of Apaf-1 molecules known as the apoptosome. Once assembled the apoptosome recruits and activates initiator caspase-9 (Bao and Shi, 2007). However, here is where the controversies begin, in part because a high-resolution structure of the apoptosome has not been available, until now. In this issue, Qi et al. (2010) present an impressive feat—the first crystal structure of a complete apoptosome at 3.55 Å resolution.
The structure is an octamer of CED-4, the Caenorhabditis elegans ortholog of Apaf-1. CED-4 and Apaf-1 belong to a new class of signal transduction AAA+ proteins (Danot et al., 2009). They have an N-terminal caspase recruitment domain (CARD) and an NB-ARC region, which is composed of an α/β domain and a helical domain 1 (HD1), followed by a winged-helix domain (WHD) and a second helical domain (HD2) (Figure 1A). In healthy cells, CED-4 is sequestered by CED-9 (homologous to mammalian Bcl-2) into a (CED-4)2:CED-9 complex, in which the CED-4 molecules exist as an asymmetric dimer. In the presence of a death signal, an upstream protein EGL-1 (analogous to mammalian BH3-only proteins) interacts with CED-9 and releases the CED-4 dimer to become activated (Yan et al., 2005). The structural comparisons performed by Qi et al. show that the conformation of the asymmetric CED-4 dimer in the apoptosome is nearly identical to that found in the (CED-4)2:CED-9 complex, indicating that the CED-4 dimer sequestered by CED-9 does not undergo major conformational changes before assembling into an apoptosome. This hypothesis is supported by the fact that CED-4 binds ATP when it is sequestered by CED-9. This is unlike autoinhibited Apaf1, which is ADP bound and needs ADP/ATP exchange to undergo a conformational change before assembling into an apoptosome, and does not involve Bcl-2 proteins.
Figure 1. Structure of CED-4 and Apaf-1 Apoptosomes.
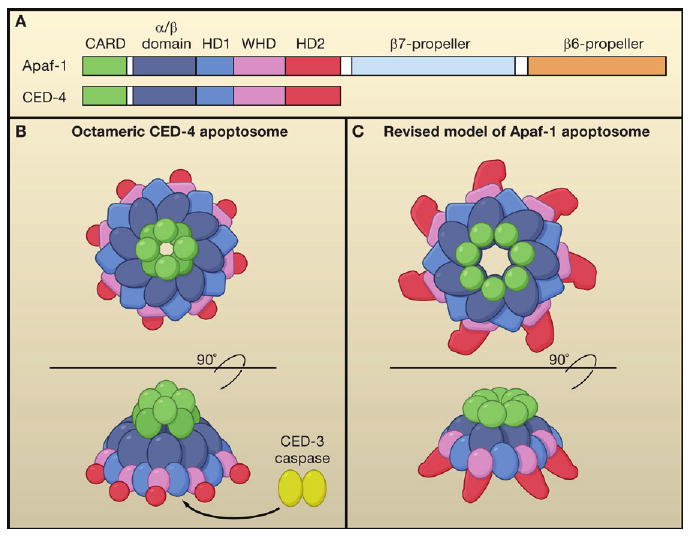
(A) Conserved domains between Caenorhabditis elegans CED-4 (571 amino acids) and human Apaf-1 (1237 amino acids).
(B) The C. elegans CED-4 octamer apoptosome as viewed from the top (upper) and side (lower).
(C) Human Apaf-1 heptamer apoptosome as viewed from the top (upper) and side (lower).
This structure contrasts with the tetramer model proposed earlier by this group (Yan et al., 2005), but the reason for this difference is now clear. A tetramer of asymmetric CARD dimers results in the protrusion of alternate N-terminal CARDs at the top of the apoptosome, explaining the unoccupied electron densities of the previous model (Figure 1B). Overall, the CED-4 apoptosome has an inverted funnel-like structure, rather than the flat structure proposed for Apaf-1.
For now, the only available crystal structure of Apaf-1 is for the inhibited (ADP-bound) monomer, which lacks the WD-40 domain (Riedl et al., 2005). A heptameric Apaf-1 apoptosome structure was previously obtained by cryoelectron microscopy at 12.8 Å resolution. Docking the crystal structure of individual domains from the inhibited monomeric Apaf-1 into this EM structure of the Apaf-1 apoptosome reveals a flat wheel-like structure, with seven CARDs arranged as a ring in the center of the wheel (Yu et al., 2005). Qi et al. model the Apaf-1 apoptosome in a different way, providing a challenge to the previous structure. Although the overall structure of CED-4 differs from that of the inhibited monomeric Apaf-1, the structure of individual domains such as CARD, α/β, HD1, and WHD are very similar between CED-4 and Apaf-1. Therefore, the authors model the active Apaf-1 monomer by fitting the individual domains into their corresponding positions in the CED-4 apoptosome. Using this putative activated form of monomeric Apaf-1 to generate the Apaf-1 apoptosome structure, they derive a cone-shaped structure held together by an inner ring composed of the α/β domain (rather than the CARD) and a loose ring of CARDs positioned on top (Figure 1C). This position of the CARD of CED-4 is similar to a previous model of the Apaf-1 apoptosome derived from the structure of the classic AAA+ ATPase (STAND) complex (Danot et al., 2009), in which the center of the wheel is composed of the ATPase (α/β domain—HD1) domain, with the CARDs forming a looser ring on top (Diemand and Lupas, 2006). To provide experimental support for this revised model of the Apaf-1 apoptosome, Qi et al. generate point mutants predicted to disrupt the interactions between the α/β domain interfaces. They successfully identify four mutants that fail to form the apoptosome, supporting the notion that the Apaf-1 apoptosome is similar to the CED-4 apoptosome.
How does the funnel-shaped apoptosome activate caspases? The authors incubate processed CED-3 (the ortholog of caspase-9) with CED-4 to generate the CED-4:CED-3 apoptosome complex. Surprisingly, the molar ratio of CED-4 to CED-3 in this complex is 4:1, meaning that there is a single active CED-3 protease dimer per apoptosome. Structure analysis by cryoelectron microscopy indicates that the CED-3 caspase dimer resides inside the cone-shaped hutch of the CED-4 apoptosome, visible from the open bottom-side of the funnel. Biochemical experiments indicate that the CED-4 apoptosome promotes dimerization of processed CED-3 and enhances its protease activity. This finding is consistent with the previously hypothesized model in which caspase dimerization is driven by proximity.
It remains unclear how the CED-3 zymogen is recruited and activated by the apoptosome. With the CED-4 CARD perched on top of the apoptosome, it is not obvious how the CARD of CED-4 and the CARD of CED-3 would make contact when the active CED-3 dimer is inside the hutch. One caveat of this study is that the structure represents fully processed CED-3 lacking its CARD, as the CARD renders CED-3 insoluble. Therefore, the role of the CED-3 CARD in caspase recruitment and activation is an intriguing topic for future study.
The structural and biochemical analyses of the (CED-4)8:(CED-3)2 complex also has implications for caspase-9 activation by the Apaf-1 apoptosome. It is generally assumed that the CARD in the Apaf-1 apoptosome binds the CARD domain of caspase-9 to cause proximity-driven dimerization of caspase-9 (Pop et al., 2006). For example, forced dimerization of caspase-9 by replacing its CARD with a leucine zipper dimerization motif is sufficient for caspase-9 autoactivation. However, the oligomeric Apaf-1 apoptosome even more effectively enhances caspase-9 activity for its downstream target, caspase-3 (Yin et al., 2006). The exact action of the apoptosome on the caspase to achieve this level of activity is unclear, in part because the stoichiometry of the Apaf-1:caspase-9 complex in the holoenzyme is still debated. One possibility is that the Apaf-1 apoptosome recruits seven molecules of caspase-9 through CARD-CARD interactions at a 1:1 ratio with Apaf-1. An alternate view comes from an elegant biochemical study reporting that only one to two molecules of caspase-9 are recruited to the Apaf-1 apoptosome (Malladi et al., 2009). This is consistent with the (CED-4)8:(CED-3)2 model now reported by Shi's group. However, unlike CED-3, which has its CARD removed when the zymogen is matured by proteolysis, the activated caspases-9 retains its CARD. This difference raises the question of how caspase-9 is activated by the Apaf-1 apoptosome. If the CARD of Apaf-1 recruits the CARD of caspase-9, as generally assumed, then the activated caspase-9 will be sitting on the top of the apoptosome (or between higher-order multi-apoptosome complexes). This study suggests that the hutch under the CED-4 cone-shaped apoptosome may constrain the CED-3 caspase in a unique activation step. Assuming that the Apaf-1 apoptosome is similar in structure to the CED-4 apoptosome, an Apaf-1 hutch theoretically could drive dimerization and activation of caspase-9. Although this remarkable new structure challenges the dogma, there are now many new important questions to pursue.
References
- Bao Q, Shi Y. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Structure. 2009;17:172–182. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Diemand AV, Lupas AN. J Struct Biol. 2006;156:230–243. doi: 10.1016/j.jsb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB. EMBO J. 2009;28:1916–1925. doi: 10.1038/emboj.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandino S, Salvesen GS. Mol Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. Cell. 2010 this issue. [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, Wu JW, Kokel D, Li H, Hao Q, et al. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, Jiang X, Wu H. Mol Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. Structure. 2005;13:1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]


