SUMMARY
The p53 transcriptional network orchestrates alternative stress responses such as cell-cycle arrest and apoptosis. Here we investigate the mechanism of differential expression of p21, a key mediator of p53-dependent cell-cycle arrest. We demonstrate that the transcriptional activity of the p21 promoter varies greatly in response to distinct p53-activating stimuli. Chromatin immunoprecipitation analysis of the p21 locus indicates that histone acetyltransferases, general transcription factors, and Mediator subunits are assembled into alternative transcriptional complexes of different activity. Interestingly, core Mediator subunits MED1 and MED17 are recruited to the p21 locus regardless of the p53-activating stimuli utilized. In contrast, three subunits of the CDK module of Mediator (CDK8, MED12, and cyclin C) are exclusively recruited during conditions of strong p21 transcriptional activation. Furthermore, increased binding of CDK8 to p53 target genes correlates positively with transcriptional strength. RNAi experiments demonstrate that CDK8 functions as a coactivator within the p53 transcriptional program.
INTRODUCTION
The transcription factor p53 protects cells from malignant transformation, and the development of most tumors is associated with loss of p53 function. The p53 pathway is activated in response to potentially oncogenic signals, such as DNA damage, activation of oncogenes, and hypoxia. Upon activation, p53 manifests its tumor suppressor activity by inducing cell-cycle arrest, apoptosis, or senescence (Prives and Hall, 1999). How cells react to increased p53 activity varies greatly depending on the nature of the activation signal, cell type, and cellular context (Vousden and Lu, 2002). In the absence of cellular stress, p53 is repressed by HDM2, an E3 ubiquitin ligase that targets p53 for degradation and also masks its transactivation domain (Poyurovsky and Prives, 2006). Stress signals utilize distinct pathways to activate p53, which may involve p53 phosphorylation or activation of cellular repressors of HDM2, such as ARF (Ashcroft et al., 2000; Prives, 1998). Recently, small molecule competitive inhibitors of the p53-HDM2 interaction, termed Nutlins, have been developed. These compounds are capable of activating p53 without genotoxic stress by mimicking the three hydrophobic amino acids of p53 required for HDM2 binding (Vassilev et al., 2004). Interestingly, the cellular response to p53 activation by Nutlins is heterogeneous, with most cell types tested undergoing reversible cell-cycle arrest, whereas a few cell lines undergo apoptosis (Tovar et al., 2006).
The commitment to undergo cell-cycle arrest or apoptosis is clearly affected by the set of p53 target genes activated in different scenarios. Induction of cell-cycle arrest by p53 is mediated primarily by transcriptional activation of the cyclin-dependent kinase inhibitor p21/Cdkn1a (referred hereto as p21), whereas apoptosis is mediated by induced expression of numerous apoptotic genes functioning in the death-receptor pathway (e.g., Fas, DR5) and the intrinsic apoptotic pathway (e.g., Bax, Puma, Noxa) (reviewed in Vousden and Lu [2002]). The p53 transcriptional program is surprisingly flexible, and the existence of stimulus-specific p53-dependent gene expression profiles has been clearly demonstrated by DNA microarray analysis (Zhao et al., 2000). Differential expression of p53 target genes may be achieved through the action of stimulus- and gene-specific coregulators within the p53 transcriptional program.
Precise transcriptional control of thousands of RNA polymerase II (RNAP II)-dependent genes in a development-, tissue-, and signaling-specific manner is only possible through extensive combinatorial use of transcriptional regulators. This process requires communication between sequence-specific DNA binding proteins, often acting at distal enhancer elements, and the RNAP II machinery at the promoter region. Mediator is a large multisubunit complex conserved from yeast to humans that interacts directly with both DNA binding transcription factors and RNAP II and provides a regulatory link required for proper expression of most RNAP II genes. Interaction studies in yeast give us an overall view of Mediator in which a core complex of about 20 subunits is composed of three discrete modules termed head, middle, and tail. An additional four subunits (MED12, MED13, cyclin C, and CDK8) form a distinct, less strongly associated module, that contains the cyclin-dependent kinase CDK8 (SRB10 in yeast) (Guglielmi et al., 2004). Genetic studies lend credence to the modularity suggested by the structural model as deletions of subunits within a given module lead to similar changes in global gene expression (van de Peppel et al., 2005). The human Mediator family of transcriptional cofactors is comprised of a number of complexes that differ in subunit composition and activity (Taatjes et al., 2004). This diversity may reveal functional specialization of Mediator variants within cells, where different subunits or entire modules are used in a combinatorial manner to achieve regulatory specificity. These complexes can be loosely classified depending on the presence or absence of the CDK module. NAT, TRAP, ARC/DRIP, and SMCC are larger versions (L/MED) containing the CDK module. CRSP and PC2 are smaller Mediator preparations (S/MED) that lack the CDK module. Depending on the scenario, CDK8 can act as a negative or positive regulator of transcription (see the Discussion). A few examples illustrating this issue are the following: (1) as part of NAT, a specific variant of the L/MED complex, CDK8 can repress transcription via phosphorylation-dependent inactivation of TFIIH (Akoulitchev et al., 2000); (2) SRB10 can promote RNAP II transcription both in vitro and in vivo through a mechanism likely involving RNAP II phosphorylation (Liu et al., 2004); (3) the L/MED variant SMCC was shown to have a negative or positive effect on transcriptional activation depending on the assay conditions (Gu et al., 1999). Taken together, these studies suggest that CDK8 is a versatile regulator capable of playing diverse roles in transcriptional regulation.
Here we demonstrate that CDK8 functions as a stimulus-specific transcriptional coactivator within the p53 transcriptional program. In several cell types studied, strong transcriptional activation of the p21 locus is observed upon p53 activation by Nutlin 3, but not in response to DNA damage induced by ultraviolet light C (UVC). The core Mediator subunits MED1 and MED17 associate with the p21 locus in both scenarios. In contrast, CDK8, cyclin C, and MED12 are recruited to the p21 locus exclusively after Nutlin 3 treatment. CDK8 recruitment is also observed during p53-dependent activation of the Hdm2 locus, and the levels of CDK8 occupancy at p53 target genes correlates positively with gene activity. RNA interference and reporter assays indicate that CDK8 is indeed a positive regulator of p21 and Hdm2 transcription.
RESULTS
Differential Expression of p21 during Stimulus-Specific Responses to p53 Activation
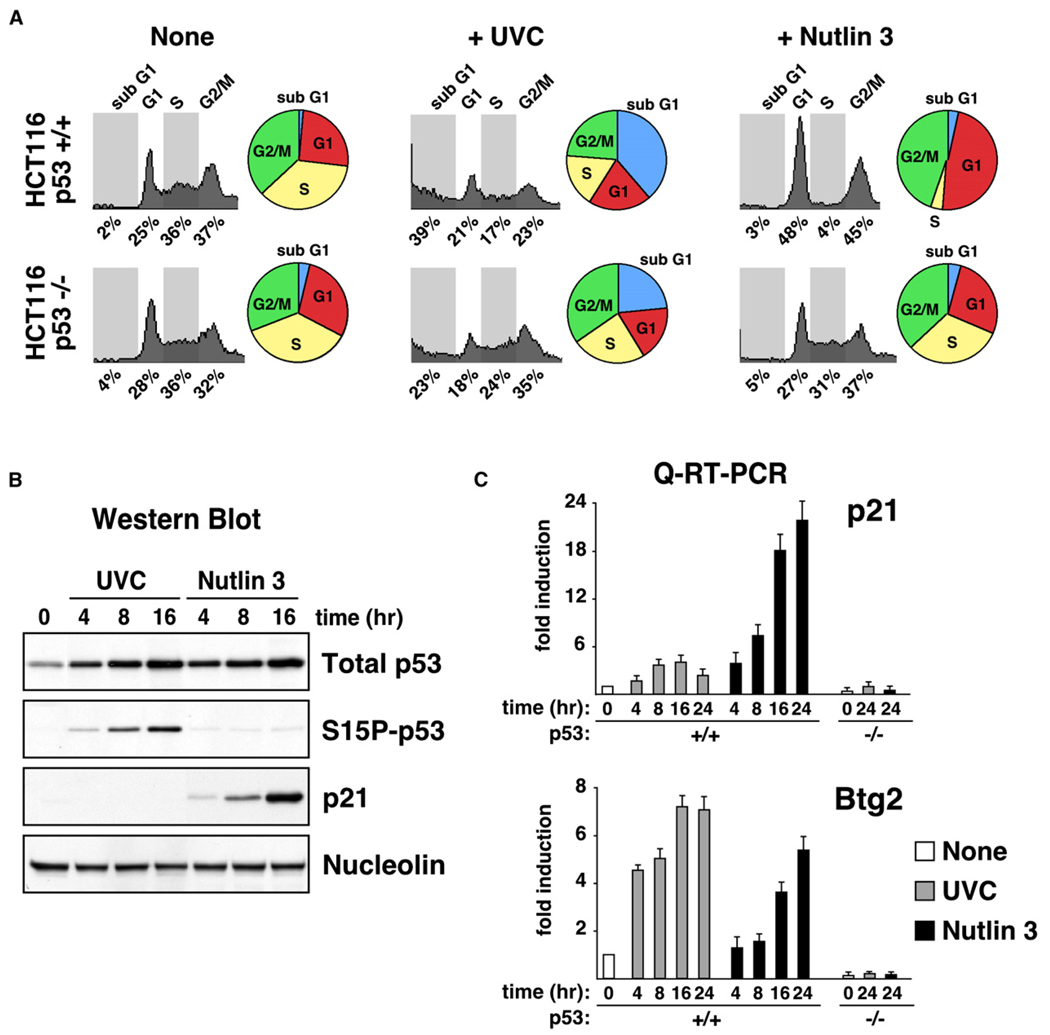
In order to study mechanisms of stimulus-specific transcriptional activation within the p53 network, we investigated the cellular response to several p53-activating agents in the colon cancer cell line HCT116+/+ (wild-type p53). In the first part of this work, we focus on UVC and Nutlin 3, two agents that act by different mechanisms and induce distinct cellular responses. FACS analysis demonstrates that UVC irradiation of HCT116+/+ cells leads to apoptosis, whereas Nutlin 3 treatment leads to cell-cycle arrest (Figure 1A). Actively growing cultures of untreated HCT116+/+ cells distribute evenly in G1, S, and G2/M phases of the cell-cycle. Twenty-four hours post-UVC irradiation, HCT116+/+ cells show a marked increase in the sub-G1 population, a hallmark of DNA degradation and cell death. The apoptotic fate of the culture was confirmed by Annexin V staining (data not shown). Apoptosis triggered by UVC is attenuated, but not absent, in the isogenic cell line HCT116−/− (p53 null), indicating that p53 plays a proapoptotic role in this scenario and that UVC also triggers p53-independent cell death. In contrast, HCT116+/+ cells treated with 10 µM Nutlin 3 undergo cell-cycle arrest, as evidenced by a depletion of the S phase population and an increase of the G1 and G2/M populations. Expectedly, HCT116−/− cells are unaffected by Nutlin 3 treatment.
Figure 1. Differential Expression of p21 during Stimulus-Specific Responses to p53 Activation.
(A) Cell-cycle profiles of HCT116 cells 24 hr posttreatment with UVC (25 J/m2) or Nutlin 3 (10 µM) were determined by FACS analysis of DNA content. Pie charts show the percentage of cells in each phase of the cell cycle.
(B) Western blot analysis of HCT116+/+ cells at the indicated times following treatment with UVC or Nutlin 3 showing accumulation of total p53, phospho-Ser15 p53 (S15P-p53), and p21.
(C) Real-time PCR analysis of p21 and Btg2 mRNA induction following UVC and Nutlin 3 treatment expressed as fold induction over untreated cells after normalization to 18S rRNA levels. Error bars represent the standard deviation.
The distinct responses to p53 activation by UVC and Nutlin cannot be attributed to differences in p53 stabilization and accumulation. Western blot analysis indicates that p53 accumulates with similar kinetics and to an equal extent after each treatment (Figure 1B). UVC irradiation leads to p53 stabilization through a DNA damage response pathway involving p53 phosphorylation (Prives, 1998). Accordingly, UVC irradiation produces a significant increase in phosphorylation of Ser15 in the p53 N terminus (Figure 1B). In contrast, Nutlin 3 is a nongenotoxic drug that activates p53 without detectable phosphorylation at multiple phosphoacceptor sites (Thompson et al., 2004). Accordingly, we did not detect Ser15 phosphorylation in Nutlin-treated HCT116+/+ cells (Figure 1B).
Analysis of p53 target gene expression revealed a significant difference in the levels of p21 expression that correlated with the observed cell fate choice. Western blot analysis demonstrates that the p21 protein accumulates in Nutlin 3-treated cells, but not in UVC-treated cells (Figure 1B). The differences in p21 protein accumulation can be attributed in part to a significantly stronger induction of p21 mRNA levels in Nutlin 3-treated cells (Figure 1C). However, UVC does produce some p21 mRNA accumulation that may indicate that other repressive mechanisms are at work, such as UVC-induced p21 protein degradation (Bendjennat et al., 2003). Expectedly, p21 mRNA accumulation is not observed in HCT116−/− cells after either treatment. The striking differences in p21 mRNA accumulation among UVC- and Nutlin-treated cells are not observed for the p53 target gene Btg2, suggesting the existence of gene-specific regulatory mechanisms. The distinct regulation of both genes cannot be attributed to their different overall fold induction in this cell line, as differential p21 expression is also observed in cell types displaying a high fold induction for Btg2 after both treatments (see later, Figure 4).
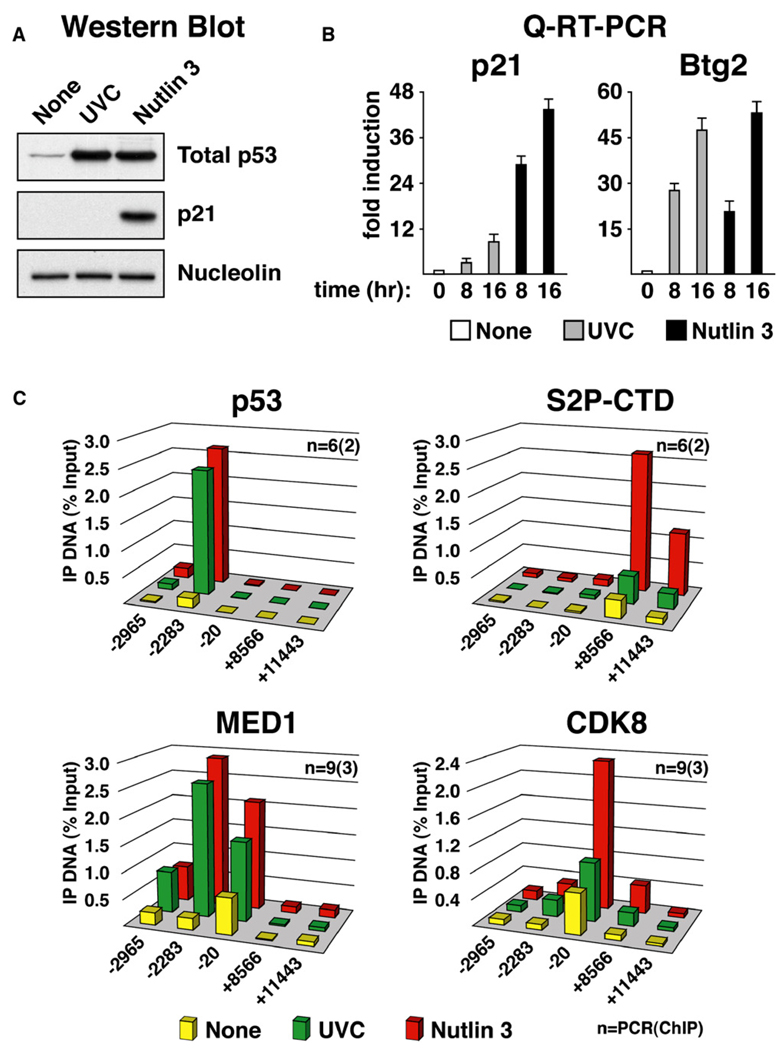
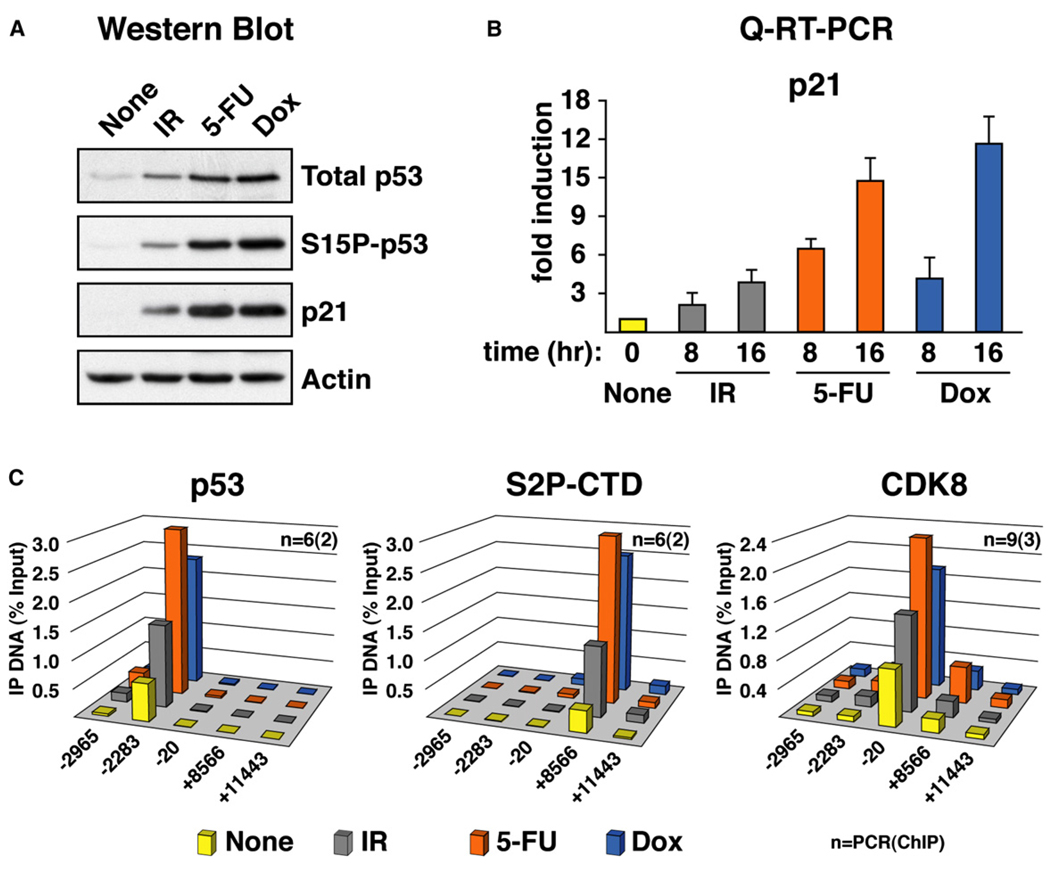
Figure 4. Differential p21 Expression and Stimulus-Specific Recruitment of CDK8 Are Conserved in SJSA-1 Cells.
(A) Western blot analysis of p53 and p21 induction in SJSA-1 cells 8 hr following treatment with UVC (50 J/m2) and Nutlin 3 (30 µM).
(B) Real-time PCR analysis of p21 and Btg2 mRNA induction at 8 and 16 hr following UVC and Nutlin 3 treatment expressed as fold induction over untreated cells and normalized to 18S ribosomal RNA levels. Error bars represent the standard deviation.
(C) ChIP analysis of the p21 locus was performed as in Figure 2 using antibodies against total p53 (p53), phosphorylated Ser-2 isoform of RNAP II (S2P-CTD), MED1, and CDK8. Numbers of PCR and ChIP replicates are indicated. Standard deviation is below 20%.
p53 Binding to Chromatin and Histone Acetylation Do Not Determine the Ultimate Activation Status of the p21 Gene
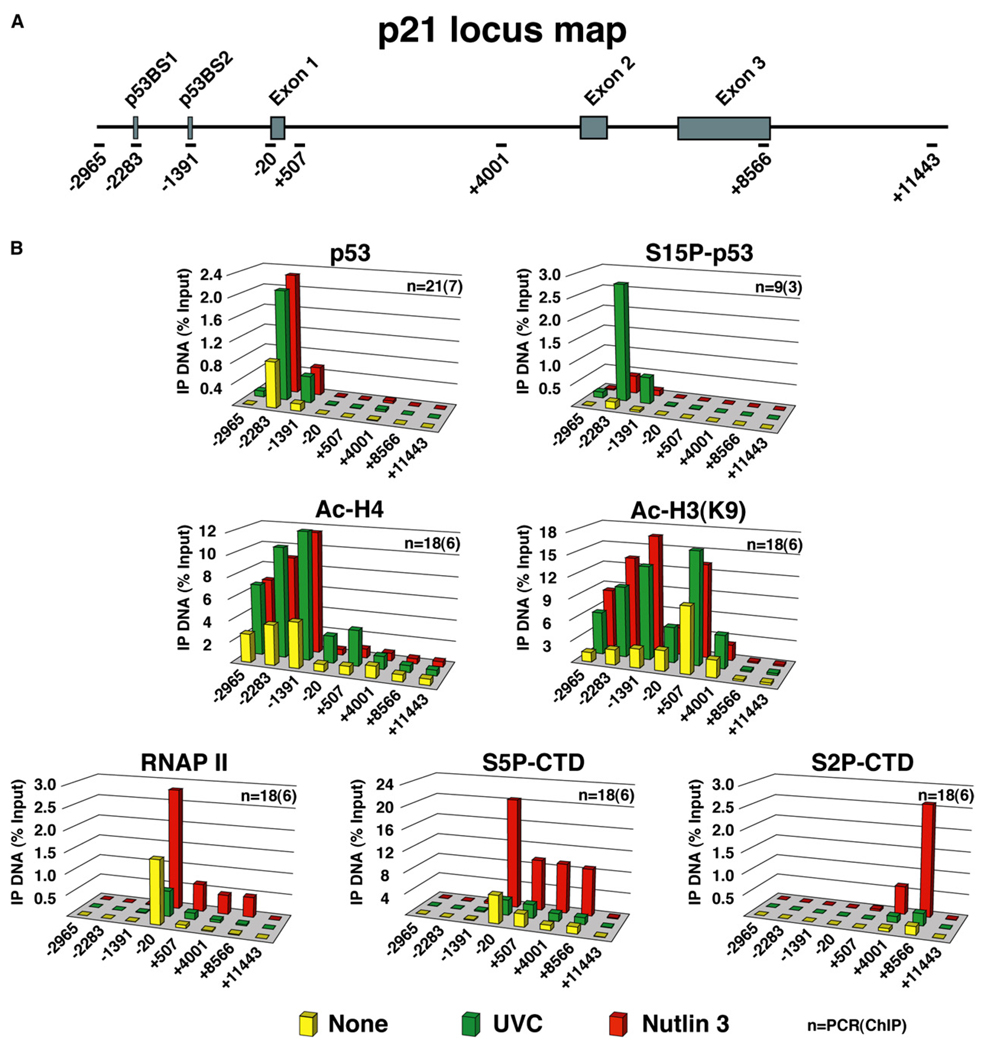
We hypothesized that stimulus-specific p21 mRNA accumulation could be attributed to differences in transcriptional control, which prompted us to perform chromatin immunoprecipitation (ChIP) analysis of the p21 locus in UVC- and Nutlin-treated cells (Figure 2). ChIP assays indicate that p53 binding to chromatin is increased to a similar extent in response to both treatments. Expectedly, only UVC treatment leads to an increase in chromatin-bound Ser15-phosphorylated p53. In vitro biochemical studies demonstrated that p53 can activate transcription of chromatin templates through the recruitment of histone modifying enzymes, including histone acetyltransferases (HATs) and arginine methyltransferases (An et al., 2004; Espinosa and Emerson, 2001). Interestingly, acetylation of histone H4 at four different lysines (Ac-H4) is observed mostly at the p21 upstream regulatory region and increases to similar levels after both treatments. Acetylation of lysine 9 of histone H3 (Ac-H3[K9]) is observed at both the upstream control region and the 5′ end of the intragenic region and is also increased to similar extent upon both p53-activating stimuli. Analysis of histone arginine methylation did not reveal the action of arginine methyltransferases at the p53 response elements in the p21 locus (data not shown). Overall, these results suggest that Ser15 phosphorylation is dispensable for p53-mediated recruitment of HATs and that inhibition of HDM2 binding is sufficient to increase histone acetylation at this locus. Increased histone acetylation upon UVC and Nutlin 3 treatment requires p53 activity, as it is not observed in the isogenic cell line HCT116−/− (data not shown) (Espinosa et al., 2003).
Figure 2. Stimulus-Specific RNAP II Activation at the p21 Locus.
(A) Schematic of the p21 locus indicating the two p53 binding sites (high-affinity p53BS1 and low-affinity p53BS2) and overall gene structure. Amplicons used in real-time PCR quantification of ChIP-enriched DNA are named according to their relative distance (bp) to the transcription start site.
(B) ChIP analysis was carried out using protein extracts of untreated and UVC- or Nutlin-treated cells (8 hr) using antibodies against total p53 (p53), phospho-Ser15 p53 (S15P-p53), acetylated histone H4 (Ac-H4), acetylated histone H3 (Ac-H3[K9]), total RNAP II, and phosphorylated Ser5 and Ser2 forms of RNAP II (S5P-CTD and S2P-CTD). Numbers of PCR and ChIP replicates are indicated. Standard deviation is below 20%.
In untreateyd cells, significant amounts of RNAP II are observed at the p21 core promoter region, but not within the intragenic region, indicating that RNAP II is recruited yet paused before stimulation of p53 activity (Gomes et al., 2006). Remarkably, increased loading of RNAP II at the proximal promoter and enhanced occupancy within the intragenic region is only observed after Nutlin 3 treatment. Analysis of RNAP II phosphorylation confirms that sustained transcription occurs only after p53 activation by Nutlin 3. Ser5 phosphorylation occurs at the promoter clearance step and peaks at the 5′ region of the gene, whereas Ser2 phosphorylation accumulates during elongation and peaks at the 3′ end (Gomes et al., 2006). In contrast, UVC irradiation does not lead to sustained transcriptional activation of p21. After a short wave of transcription at early time points (data not shown) (Espinosa et al., 2003), levels of total and Ser5-phosphorylated RNAP II drop below basal levels by 8 hr postirradiation, indicating that the p21 promoter is effectively shut down at this point despite the presence of phosphorylated p53 and increased histone acetylation. Taken together, these results establish that regulatory events subsequent to p53 binding and histone acetylation define the ultimate activation status of the p21 gene.
CDK8 Is Recruited to the p21 Locus in a Stimulus-Specific Manner
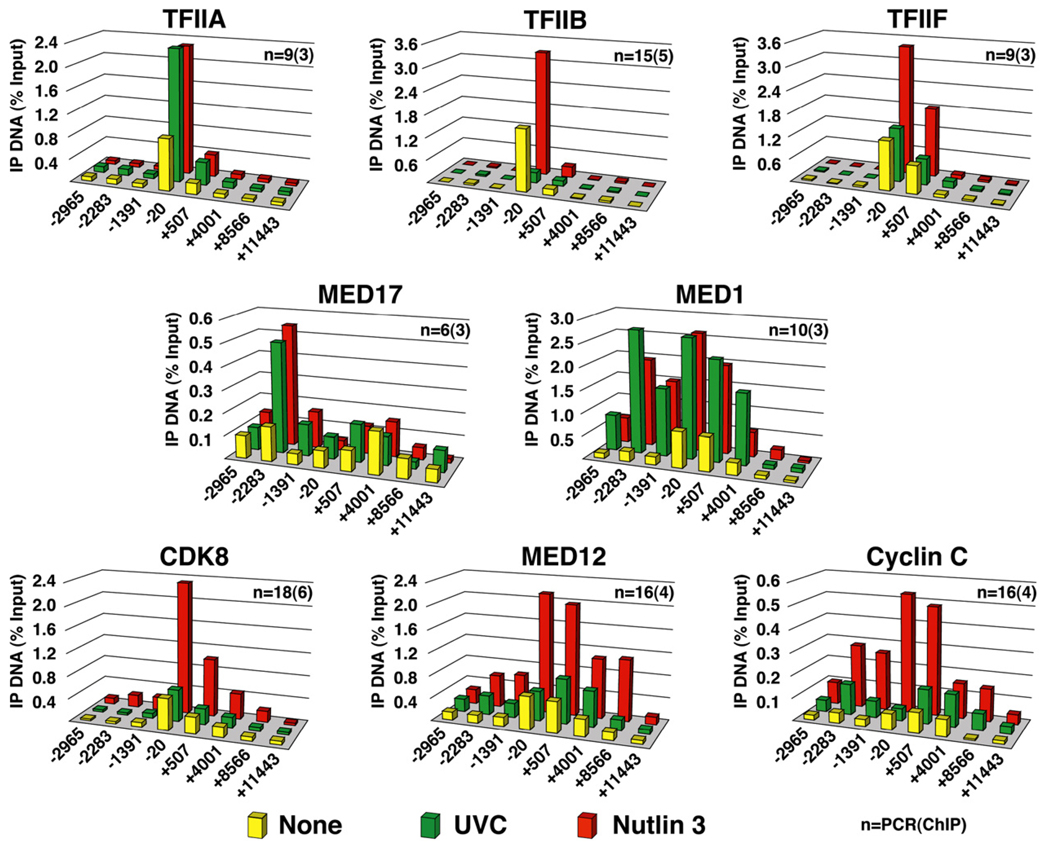
To gain further insight into the mechanism of stimulus-specific regulation of p21 transcription, we analyzed the recruitment of several subunits of general transcription factors (GTFs) and Mediator (Figure 3). The initiation factor TFIIA exhibits increased recruitment after both p53-activating stimuli, indicating that TFIIA responds to p53 activation in both scenarios but that its enhanced association does not define the ultimate transcriptional status of the p21 promoter. In contrast, binding of TFIIB oscillates with levels of total RNAP II, decreasing with UVC and increasing with Nutlin 3 treatment. TFIIF, another component of the preinitiation complex (PIC) shows increased association upon Nutlin 3 treatment and no changes in basal levels after UVC. These results reveal a striking flexibility in the association of components of the PIC to the p21 core promoter and allow us to identify differences between active and inactive conformations of the transcriptional machinery at this locus (see the Discussion). Next, we performed ChIP assays for different subunits of the Mediator complex, which identified other stimulus-specific regulatory events at the p21 locus. We chose to look at MED17 and MED1, two core Mediator subunits that are known to interact directly with p53 (Ito et al., 1999; Sato et al., 2004). Our ChIP analysis indicates that MED17, a core subunit of Mediator found in the “head module,” is recruited to the distal p53 response element after either stimulus. MED1, a subunit that is part of the “middle module” of the complex, is also recruited to the p21 locus regardless of the p53-activating stimuli utilized. MED1 shows a different occupancy profile than MED17, with significant levels of MED1 binding being observed at both the p53 binding sites and the p21 proximal promoter. In stark contrast, CDK8, a subunit of the CDK module, is recruited to p21 exclusively during conditions of sustained transcription upon Nutlin 3 treatment. ChIP analysis of MED12 and cyclin C suggest that the entire CDK module is recruited to the p21 locus in a stimulus-specific manner. Interestingly, CDK8 associates preferentially to the proximal promoter region of p21, a trend that is also observed, albeit to a lesser extent, for MED12 and cyclin C. Although our analysis identified several important differences in the composition of the transcriptional machinery acting on the p21 locus that could explain its stimulus-specific regulation, the rest of this work will focus on CDK8, an enigmatic transcriptional coregulator. Importantly, CDK8 recruitment to the p21 locus upon Nutlin 3 treatment is p53 dependent, as it is not observed in HCT116−/− cells (see Figure S1A in the Supplemental Data available with this article online). Furthermore, stimulus-specific recruitment of CDK8 cannot be attributed to different levels of cellular CDK8 in each scenario (Figure S1B).
Figure 3. Stimulus-Specific Recruitment of the CDK Module of Mediator to the p21 Locus.
ChIP analysis was carried out as in Figure 2 using antibodies against TFIIA-γ (TFIIA), TFIIB, RAP74 (TFIIF), MED17, MED1, CDK8, MED12, and cyclin C. Numbers of PCR and ChIP replicates are indicated. Standard deviation is below 20%.
Differential Recruitment of CDK8 Is Conserved in Cancer Cell Types of Different Origin
As a first step toward a better characterization of the role of CDK8 in p53-dependent transcriptional regulation, we decided to include other cell lines in our analysis. Differential accumulation of p21 protein following UVC and Nutlin 3 treatment was also observed in the osteosarcoma cell line SJSA-1. Similarly to what was observed for HCT116+/+ cells, p21 protein accumulates only after p53 activation by Nutlin 3 treatment, but not in response to UVC (Figure 4A). Unequal p21 protein expression could be explained partially by the reduced accumulation of p21 mRNA in UVC-treated as compared to Nutlin-treated cells (Figure 4B). Protein degradation mechanisms triggered by UVC may also be contributing to differential p21 protein expression in this cell type (Bendjennat et al., 2003). Once again, differential mRNA induction is not observed for the p53 target gene Btg2, which is strongly induced to similar levels after both stimuli in SJSA-1 cells. ChIP analysis indicates that, despite significant increases in the levels of chromatin-bound p53 after both stimuli, only Nutlin 3 treatment leads to significant increases in elongating RNAP II (S2P-CTD) in the p21 intragenic region (Figure 4C). Reproducing results observed in HCT116+/+ cells, CDK8 levels at the p21 core promoter region increase significantly only under conditions of sustained activation by Nutlin 3. Similar results were obtained when analyzing the colon carcinoma-derived cell line RKO (Figure S2). Despite certain quantitative differences in mRNA fold induction and ChIP signals among cell types, these results indicate that stimulus-specific regulation of p21 transcription and concurrent differential recruitment of CDK8 are conserved.
CDK8 Recruitment to the p21 Promoter Is Observed during Activation by Ionizing Radiation, 5-Fluorouracil, and Doxorubicin
Nutlin 3, while a very useful compound for studies of the p53 pathway, is not a natural stress stimulus. Thus, we decided to analyze p21 transcriptional regulation after p53 activation by forms of DNA damage other than UVC, such as ionizing radiation (IR). Additionally, we also employed the chemotherapeutic agents 5-fluorouracil (5-FU) and doxorubicin, two potent inducers of p53 activity. Treatment with any of these agents leads to p53 stabilization, phosphorylation of Ser15 and p21 protein accumulation in HCT116+/+ cells, albeit to different extents (Figure 5A). The different potency of these three p53-activating agents becomes more evident when analyzing p21 mRNA induction by Q-RT-PCR, with IR producing a weaker response than either 5-FU or doxorubicin (Figure 5B). ChIP analysis indicates that all three agents lead to an increase in chromatin-bound p53 and elongating RNAP II at the p21 locus (Figure 5C). CDK8 was also recruited to the p21 promoter after all three treatments, with a pattern that correlates with the levels of Ser2-phospho-RNAP II and mRNA accumulation at this time point (8 hr posttreatment). These results indicate that CDK8 recruitment is not exclusive to p21 transcriptional activation by Nutlin 3 treatment but that it also occurs in response to other p53-activating stimuli leading to a sustained increase of p21 transcription.
Figure 5. Recruitment of CDK8 to the p21 Locus during Activation by Distinct Stimuli.
(A) Western blot analysis of HCT116 p53+/+ cells 16 hr following treatment with IR (10 Gy), 5-fluorouracil (5-FU, 375 µM), and doxorubicin (Dox, 0.5 µM) showing accumulation of total p53 (p53), phospho-Ser15 p53 (S15P-p53), and p21.
(B) Real-time PCR analysis of p21 mRNA induction at the given times following IR, 5-FU, and doxorubicin treatment. Data is expressed as fold induction over untreated cells and normalized to 18S ribosomal RNA levels. Error bars represent the standard deviation.
(C) ChIP analysis of the p21 locus was performed as in Figure 2 using antibodies against total p53 (p53), phosphorylated Ser-2 isoform of RNAP II (S2P-CTD), and CDK8. Numbers of PCR and ChIP replicates are indicated. Standard deviation is below 20%.
CDK8 Is Recruited to the Intragenic Region of the p21 Locus during Activation
The low-density tiling shown in Figure 3 suggests that CDK8 is present inside the active p21 gene (compare amplicons +4001 and +8566 to amplicons −2965 and +11443). To confirm this, we performed high-density tilings of the p21 locus (Figure 6A). As compared to a control region 3 kb downstream of the 3′ end of the gene (arrow, +11443), statistically significant amounts of CDK8 are detected at every location analyzed within the p21 intragenic region post-Nutlin 3 treatment. The CDK8 occupancy profile, which resembles that of RNAP II, suggests that CDK8 may associate with the transcription elongation complex on the p21 locus. CDK8 signals in the body of the gene cannot be attributed to an artifact produced by the resolution of the ChIP assay, as shown by the occupancy profile of NELF-A, a subunit of the negative elongation factor that is detected only at the proximal promoter region.
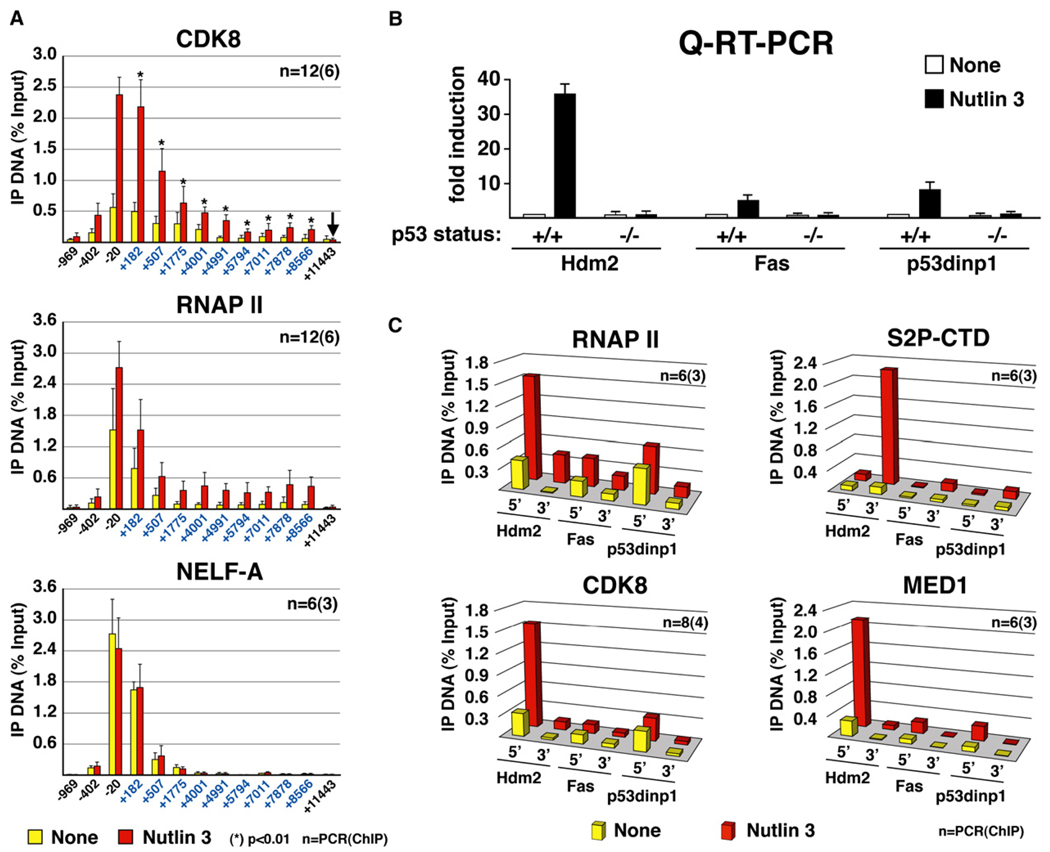
Figure 6. CDK8 Recruitment Positively Correlates with Transcriptional Activation of p53 Target Genes.
(A) High-density ChIP tiling of CDK8, RNAP II, and NELF-A at the p21 locus before and after Nutlin 3 treatment (8 hr) of HCT116+/+ cells. Significance of higher CDK8 occupancy at each intragenic amplicon (blue labels) relative to the control region +11443 (arrow) is indicated by asterisks (p values).
(B) Real-time PCR analysis of Hdm2, Fas, and p53dinp1 mRNA induction by Nutlin 3 treatment (16 hr) in HCT116+/+ and HCT116−/− cells expressed as fold induction over untreated cells and normalized to 18S ribosomal RNA levels. Error bars represent the standard deviation.
(C) ChIP analysis of RNAP II, phospho-Ser2 RNAP II (S2P-CTD), CDK8, and MED1 at the core promoter (5′) and the last exon (3′) of the Hdm2, Fas, and p53dinp1 genes before and after Nutlin treatment (8 hr). Numbers of PCR and ChIP replicates are indicated. Standard deviation is below 20%.
CDK8 Recruitment Correlates with Transcriptional Activity among p53 Target Genes
The simultaneous occurrence of active RNAP II and increased CDK8 recruitment to the p21 gene contrasts sharply with the observations made during the studies of other metazoan genes in which CDK8 presence correlates with transcriptional repression (Mo et al., 2004; Pavri et al., 2005). To generate further insight into this matter, we analyzed other p53 target genes. We observed that Nutlin 3 treatment produces very dissimilar levels of activation among p53 target genes. Q-RT-PCR analysis indicates that expression of Hdm2 is greatly induced in Nutlin 3-treated cells (30- to 40-fold), whereas activation of Fas and p53dinp1 is more modest (Figure 6B). ChIP analysis indicates that differences in mRNA accumulation among p53 target genes correlate tightly with RNAP II behavior at these loci (Figure 6C). Before Nutlin treatment, all three genes exhibit low levels of RNAP II at their proximal promoter region (5′ amplicons). Upon p53 activation by Nutlin, total RNAP II levels increase at all three loci, but significant quantitative differences are observed, with the Hdm2 promoter displaying signals 3- to 5-fold higher than Fas and p53dinp1. Furthermore, levels of elongating RNAP II are 10- to 20-fold higher at the 3′ end of the Hdm2 gene than at the corresponding region of the Fas and p53dinp1 loci (S2P-CTD, 3′ amplicons). Strikingly, Nutlin 3 treatment leads to a significant increase in CDK8 levels only at the Hdm2 proximal promoter. Furthermore, MED1 recruitment is also significantly higher at the active Hdm2 promoter, suggesting that CDK8 may be functioning in the context of MED1-containing Mediator at this locus. In summary, increased recruitment of CDK8 correlates positively with gene activity within the p53 transcriptional program.
CDK8 Is a Positive Regulator of p21 and Hdm2 Transcription
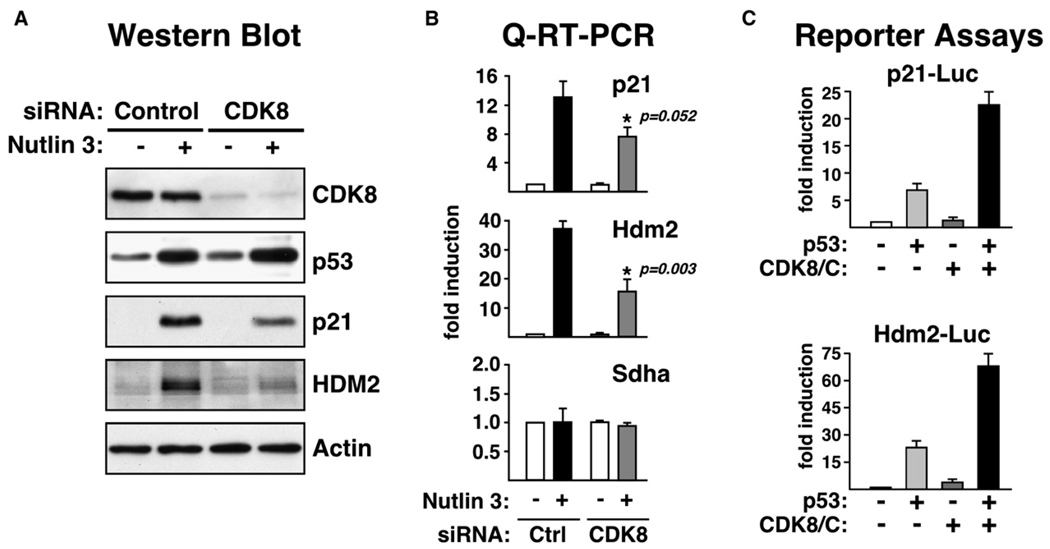
Recruitment of CDK8 to transcriptionally active p21 and Hdm2 discourages the notion that CDK8 may be a transcriptional repressor of these genes and suggests that CDK8 may function as a positive coregulator at these loci. To test this hypothesis, we performed RNAi knockdown and reporter assays. Specific siRNA duplexes were used to reduce endogenous CDK8 levels in HCT116+/+ cells. A 70%–80% reduction in CDK8 protein levels does not change basal or induced levels of p53 (Figure 7A). However, accumulation of p21 and HDM2 proteins upon Nutlin treatment is reduced by approximately 50% and 60%, respectively, in cells with decreased CDK8 levels. Q-RT-PCR confirms the decreased expression of p21 and Hdm2 mRNAs. In an alternative approach, we used luciferase reporter assays to measure the effect of CDK8 overexpression on p21 and Hdm2 promoter activity (Figure 7C). Saos-2 cells (p53 null) were transfected with the corresponding reporter constructs, as well as p53 and CDK8/cyclin C expression vectors. Expectedly, expression of p53 leads to increased activity of both reporters. Interestingly, coexpression of p53 and CDK8/cyclin C further enhanced reporter activity about 3- to 4-fold. In these assays, exclusion of cyclin C expression vector produced a modest decrease of CDK8 coactivation (data not shown). In conclusion, these results suggest that CDK8 is a positive regulator of these p53 target genes.
Figure 7. CDK8 Acts as a Positive Regulator of p53-Dependent p21 and Hdm2 Expression.
(A) Western blot analysis of untreated and Nutlin 3-treated (16 hr) HCT116+/+ cells following knockdown of CDK8.
(B) Real-time PCR analysis of p21 and Hdm2 mRNA induction in cells transfected with control of CDK8-specific siRNA. Data is expressed as fold induction over untreated cells and normalized to 18S ribosomal RNA levels. Error bars represent the standard deviation.
(C) p21 and Hdm2 reporter assays in Saos-2 cells transfected with p53, CDK8, and cyclin C expression vectors as noted. Normalized values are shown as fold induction relative to empty vector alone. Error bars represent the standard deviation.
DISCUSSION
The p53 transcriptional response is intriguingly complex. Distinct sets of p53 target genes are activated in response to different p53-activating stimuli (Zhao et al., 2000). A major challenge in the p53 field is to elucidate the regulatory mechanisms that drive specific p53-dependent cellular responses. In this report, we demonstrate that CDK8, a subunit of the Mediator complex often associated with transcriptional repression, functions as a stimulus-specific positive coregulator within the p53 transcriptional program.
The present study was motivated by our desire to investigate the mechanisms of stimulus-specific p53 transcriptional responses in arresting versus apoptotic cells. To this end, we employed two mechanistically distinct p53 activating agents, Nutlin 3 and UVC. This led to the observation that sustained transcriptional activation of p21 occurs in arresting, Nutlin-treated cells, but not in apoptotic, UVC-treated cells. ChIP assays indicate that p53 binding to chromatin and p53-mediated histone acetylation is not sufficient for sustained transcriptional activation of p21, suggesting that downstream events define the ultimate transcriptional status of this gene. A recent report from the Prives lab arrived at similar conclusions while studying p53 action in response to DNA replication block by hydroxyurea (HU) (Mattia et al., 2007). In HU-treated cells, p53 is stabilized, binds to the p21 locus, and mediates histone acetylation. However, p21 transcriptional activation is drastically impaired in HU-treated cells as compared to cells in which p53 is activated by the DNA-damaging agent daunorubicin. Mattia et al. demonstrated that RNAP II is effectively recruited to the p21 promoter in response to HU treatment but that it fails to elongate into the intragenic region, revealing the importance of postinitiation mechanisms in regulation of p21 transcription. In our paradigm, p21 transcriptional activation is impaired after UVC treatment, albeit by a different mechanism than observed in HU-treated cells. After UVC irradiation, we observe a net loss of preloaded RNAP II on the p21 core promoter. Time course analyses reveal that UVC does produce an early transient wave of p21 transcription but that activation is not sustained over time, suggesting that RNAP II fails to reinitiate transcription in this scenario (Espinosa et al., 2003).
What defines the ultimate activation status of the p21 promoter? Our results reveal stimulus-specific recruitment of several transcriptional regulators to the p21 locus, including components of the PIC and Mediator complex. Our ChIP analysis allows us to distinguish two different conformations of the transcriptional apparatus acting on the p21 locus. A transcriptionally inactive conformation is observed post-UVC treatment. This assembly displays increased levels of p53, histone acetylation, TFIIA, MED17, and MED1, along with decreased levels of TFIIB and RNAP II. A transcriptionally active conformation is observed after Nutlin 3 treatment, which is distinguished from the inactive assembly by the increased levels of TFIIB, TFIIF, CDK8, MED12, cyclin C, and RNAP II. Given the known positive role of TFIIB in transcription initiation, it is possible that a decrease in TFIIB association explains the lack of reinitiation by RNAP II in UVC-treated cells, but a direct cause-effect relationship cannot be established at this point. TFIIF is a multifunctional GTF playing positive roles in transcription initiation, promoter escape, and elongation (Yan et al., 1999), thus raising the possibility that its increased recruitment is required for sustained RNAP II activation in Nutlin-treated cells. Recruitment of CDK8 and its associated proteins MED12 and cyclin C to an activated p21 locus could not have been predicted from our collective knowledge of this protein kinase, because, depending on the scenario, CDK8 has been described as a negative or positive transcriptional regulator.
Salient evidence for CDK8 as a repressor could be itemized as follows. (1) The presence of CDK8 in Mediator preparations correlates with reduced coactivation potential in vitro (Taatjes et al., 2002). Furthermore, NAT, an L/MED variant can repress transcription by CDK8-mediated inactivation of TFIIH (Akoulitchev et al., 2000). (2) A molecular switch from L/MED to S/MED has been observed during activation of RARα and C/EBPβ target genes in vivo (Mo et al., 2004; Pavri et al., 2005). (3) Microarray analysis of Mediator subunit mutants in S. cerevisiae revealed a mostly repressive role for the CDK module (van de Peppel et al., 2005). (4) SRB10, the yeast CDK8 homolog, phosphorylates and inactivates the yeast transcriptional activators STE12, GCN4, and MSN2 (Chi et al., 2001; Nelson et al., 2003). However, a uniform view of CDK8 function is prevented by several reports demonstrating that CDK8 also plays positive roles in transcription. These findings can be summarized as follows. (1) Some L/MED preparations display coactivator function for the thyroid hormone receptor (TR) and chimeric activators in in vitro transcription assays (Fondell et al., 1999; Furumoto et al., 2007). (2) Genome-wide occupancy analysis shows no correlation between CDK8 binding and gene activity in S. pombe. In fact, the CDK module is commonly bound to highly transcribed genes (Zhu et al., 2006). (3) SRB10 can promote RNAP II transcription in S. cerevisiae. Inhibition of SRB10 leads to decreased RNAP II activity in vitro and diminished occupancy at the ADH1 and PMA1 genes in vivo (Liu et al., 2004). (4) SRB10 is a coactivator of the yeast transcription factors GAL4 and SIP4 (Hirst et al., 1999; Vincent et al., 2001). (5) Knockdown of CDK8 levels attenuates activation by the chimeric transcription factor Gal4-VP16 in human cells (Furumoto et al., 2007). In summary, it is evident that CDK8 is a flexible regulator capable of playing diverse roles in transcriptional regulation and that the ultimate outcome of CDK8 action varies depending on the context. Our results indicate that CDK8 functions as a positive regulator of transcription within the p53 transcriptional program.
CDK8, MED12, and cyclin C are recruited to p21 during activation. The fold increase in CDK8 recruitment equals or surpasses that of many other known activation events at this locus, such as increases in total p53, histone acetylation, and RNAP II recruitment. CDK8 levels correlate tightly with the varying amounts of elongating RNAP II observed during activation of the p21 locus after IR, 5-FU, and doxorubicin. CDK8 is also recruited to the Hdm2 locus during Nutlin 3-induced activation. Furthermore, quantitative analysis indicates that the transcriptional strength of different p53 target genes after Nutlin 3 treatment correlates positively with increased RNAP II activity and CDK8 recruitment. Manipulation of cellular levels of CDK8 suggests that it is indeed a positive regulator in this context.
Our detailed ChIP analysis reveals that different Mediator subunits have distinct behaviors in terms of both response to p53-activating stimuli and spatial localization within the p21 locus. Proteomic analysis of the human Mediator reveals that MED17, a subunit of the “head module,” is present in virtually every Mediator variant isolated so far (Sato et al., 2004). Furthermore, ablation of this subunit abolishes transcription of nearly all protein coding genes in yeast (Holstege et al., 1998). MED17 interacts directly with several activators, including p53 itself (Ito et al., 1999; Taatjes et al., 2004). Accordingly, MED17 is recruited to the distal, high-affinity p53 binding site in the p21 locus regardless of the p53-activating stimuli utilized. The presence of MED1 at the p21 locus is perhaps more informative. Work from the Roeder lab revealed that MED1 only exists in a Mediator subpopulation representing less than 20% of the total Mediator pool and that MED1-containing Mediator associates tightly with RNAP II to near stoichiometeric levels (Zhang et al., 2005). This “holoenzyme complex” supports both basal and activated transcription in vitro without the need for additional RNAP II. Importantly, this Mediator variant carries the CDK module, suggesting that CDK8 is not always a transcriptional repressor in the context of MED1-containing L/MED complexes. MED1 also interacts directly with p53 (Ito et al., 1999). In our ChIP analysis, we observe significant MED1 association to both the distal p53 binding sites and the proximal promoter. It is generally accepted that one function of Mediator is to serve as a “molecular bridge” between DNA binding transcription factors and RNAP II. The ChIP occupancy profile of MED1 at the p21 locus supports this idea. Interestingly, CDK8 is recruited preferentially to the proximal promoter and to a lesser extent to the intragenic region of p21. This pattern is also observed for MED12 and cyclin C, albeit to a different extent. This result favors the notion that the CDK module has a flexible interaction with the rest of Mediator. This is further supported by the fact that, unlike MED17 and MED1, association of CDK module subunits is increased only during conditions of sustained p21 transcription. In summary, this study provides new in vivo evidence testifying for the modular and heterogeneous nature of the human Mediator complex.
Our current knowledge about the mechanism of p53-dependent transcriptional control of p21 may provide some clues as to why CDK8 functions as a positive coregulator in this context. First, p21 transcription is clearly controlled at steps after RNAP II recruitment. Significant amounts of RNAP II are present at the p21 promoter in exponentially growing cells expressing very low levels of p21 mRNA. The p21 promoter harbors more preloaded RNAP II than any other p53 target gene promoter that we have analyzed so far (Espinosa et al., 2003) (data not shown). Preloaded RNAP II is hypophosphorylated in Ser5, as indicated by a higher fold induction of the S5P-CTD signal compared to total RNAP II, suggesting that it may be paused at a step prior to promoter clearance (Gomes et al., 2006). Interestingly, conversion of paused RNAP II into an elongation competent form coincides with a significant increase in the low basal levels of Mediator subunits at the p21 locus. Is Mediator acting at postrecruitment steps on the p21 promoter? Such a role for Mediator would require further investigation but is not unprecedented, as studies on MED23−/− cells concluded that Mediator acts at both recruitment and postrecruitment steps in Elk-1-dependent transcriptional activation (Wang et al., 2005). Another peculiarity of p21 transcriptional control is that this gene is effectively transcribed even in the absence of active CDK9 and RNAP II CTD Ser2 phosphorylation (Gomes et al., 2006). p21 belongs to a select group of p53 target genes that are activated in response to treatment with the CDK9 inhibitor DRB, revealing the existence of alternative mechanisms of transcription elongation control at these genes. Do these observations relate to the fact that CDK8 is observed within the p21 intragenic region? Genome-wide occupancy analysis in S. pombe detected binding of subunits of the CDK module to numerous intragenic regions, further supporting the idea that CDK8 may play a role at postinitiation steps (Zhu et al., 2006). Future in vivo and in vitro mechanistic studies will be necessary to determine which steps of the transcription process are affected by CDK8 action and how this may explain its context-dependent roles in transcriptional repression and activation.
EXPERIMENTAL PROCEDURES
Cell Culture
HCT116, RKO, SJSA-1, and Saos-2 cells were maintained in McCoy’s 5A, MEM, RPMI, and D-MEM media (Gibco-Invitrogen), respectively, supplemented with 10% fetal bovine serum and antibiotic/antimycotic mix. For FACS, western blot, Q-RT-PCR, and ChIP, cell cultures at 50%–70% confluency were stimulated with UVC (25–50 J/m2, Stratalinker 2400, Stratagene), Nutlin 3 (10–30 µM, a generous gift of Dr. Lyubomir Vassilev, Hoffman LaRoche), IR (10 Gy, Torrex 120 D irradiator, EG&G Astrophysics Research), 5-FU (375 µM, Sigma), or doxorubicin (0.5 µM, Sigma).
FACS and Western Blots
For cell-cycle distribution profiles, cells were harvested by trypsinization, washed twice with cold PBS, resuspended in PBS/2 mM EDTA, and fixed overnight in 70% ethanol. Cells were washed, treated with RNase A, stained with 50 µg/ml propidium iodide, and analyzed using a FACScan instrument (Becton Dickinson). For western blots, 10 µg of total protein extract were loaded onto 10% SDS-PAGE and transferred to PVDF membranes. Antibody information can be found in Table S1.
ChIPs and Q-RT-PCR
These assays were performed as described in Gomes et al. (2006). For ChIP assays, cells were fixed with 1% formaldehyde and harvested for whole-cell lysate preparation. Protein lysate (1 mg) was used per ChIP with indicated antibodies (Table S1). ChIP-enriched DNA was analyzed by Q-PCR as described in Gomes et al. (2006). For RT-PCR, total RNA was isolated using the RNeasy Mini Kit (Qiagen), and cDNA was generated with the SuperScriptase II (Invitrogen). See Tables S2–S4 for primer sequences.
siRNA Knockdown
Cells were transfected with control siGlo siRNA (Dharmacon) or CDK8-specific siRNA (Invitrogen, target sequence: 5′-AUAUAAUAGUGACUUCACCAUUCCC-3′), using Lipofectamine 2000 (Invitrogen). Thirty-two hours posttransfection, cells were treated with 10 µM Nutlin 3 for 16 hr before harvesting for western blot and RT-PCR analysis.
Luciferase Reporter Assays
Saos-2 cells were transfected using Lipofectamine 2000 (Invitrogen) with the following plasmids as described in the text: 500 ng WWP-luc (p21-Luc), 500 ng pGL2-HDM2 (Hdm2-Luc), 100 ng pCMV-lacZ, 100 ng pCB6-p53, 1 µg pCMV-CDK8, and 1 µg pCMV-cyclin C. Twenty-four hours posttransfection, cells were harvested and luciferase activity was determined using the Dual-Light System (ABI).
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Espinosa lab for support and advice, Glen Bjerke for technical assistance, Dr. Lubo Vassilev for Nutlin 3, Dr. Vogelstein for cells, and Dr. Royal Robins for the RNWF. This work was supported by grants from the National Institutes of Health (NIH) (CA117907), the March of Dimes (5-FY05-1217), DOD-CDMRP (CM05054), the Leukemia and Lymphoma Society, and SPORE in Lung Cancer. J.M.H. was supported by the Undergraduate Research Opportunity Program (UROP-CU-Boulder).
Footnotes
Supplemental Data
Supplemental Data include four tables and two figures and can be found with this article online at http://www.molecule.org/cgi/content/full/27/1/121/DC1/.
REFERENCES
- Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, Sarasin A, Fotedar A, Fotedar R. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114:599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Fondell JD, Guermah M, Malik S, Roeder RG. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc. Natl. Acad. Sci. USA. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto T, Tanaka A, Ito M, Malik S, Hirose Y, Hanaoka F, Ohkuma Y. A kinase subunit of the human mediator complex, CDK8, positively regulates transcriptional activation. Genes Cells. 2007;12:119–132. doi: 10.1111/j.1365-2443.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattia M, Gottifredi V, McKinney K, Prives C. p53-dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol. Cell. Biol. 2007;27:1309–1320. doi: 10.1128/MCB.01520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBP beta. Mol. Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Poyurovsky MV, Prives C. Unleashing the power of p53: lessons from mice and men. Genes Dev. 2006;20:125–131. doi: 10.1101/gad.1397506. [DOI] [PubMed] [Google Scholar]
- Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- Prives C, Hall PA. The p53 pathway. J. Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Naar AM, Andel F, 3rd, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, Xu Q, Wahl GM, Heimbrook DC, Vassilev LT. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J. Biol. Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl. Acad. Sci. USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TT, van Leenen D, Holstege FC. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vincent O, Kuchin S, Hong SP, Townley R, Vyas VK, Carlson M. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:5790–5796. doi: 10.1128/MCB.21.17.5790-5796.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Yan Q, Moreland RJ, Conaway JW, Conaway RC. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J. Biol. Chem. 1999;274:35668–35675. doi: 10.1074/jbc.274.50.35668. [DOI] [PubMed] [Google Scholar]
- Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol. Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.