Abstract
Human leukemias harboring chromosomal translocations involving the mixed lineage leukemia (MLL, HRX, ALL-1) gene possess high-level expression, and frequent activating mutations of the receptor tyrosine kinase FLT3. We used a murine bone marrow transplant model to assess cooperation between MLL translocation and FLT3 activation. We demonstrate that MLL-AF9 expression induces acute myelogenous leukemia (AML) in approximately 70 days, whereas the combination of MLL-AF9 and FLT3-ITD does so in less than 30 days. Secondary transplantation of splenic cells from diseased mice established that leukemia stem cells are present at a very high frequency of approximately 1:100 in both diseases. Importantly, prospectively isolated granulocyte macrophage progenitors (GMPs) coinfected with MLL-AF9 and FLT3-ITD give rise to a similar AML, with shorter latency than from GMP transduced with MLL-AF9 alone. Cooperation between MLL-AF9 and FLT3-ITD was further verified by real-time assessment of leukemogenesis using noninvasive bioluminescence imaging. We used this model to demonstrate that MLL-AF9/FLT3-ITD-induced leukemias are sensitive to FLT3 inhibition in a 2–3 week in vivo assay. These data show that activated FLT3 cooperates with MLL-AF9 to accelerate onset of an AML from whole bone marrow as well as a committed hematopoietic progenitor, and provide a new genetically defined model system that should prove useful for rapid assessment of potential therapeutics in vivo.
Keywords: MLL, FLT3, imaging, murine models
Introduction
Recurrent chromosomal translocations are a hallmark of human acute leukemias. Their cloning and characterization has produced a greater understanding of leukemogenesis and in some cases new therapeutic approaches.1,2 Such translocations frequently produce chimeric fusion oncogenes and tend to involve genes that normally perform critical roles during hematopoietic development.3–5 This is particularly true for translocations targeting genes encoding DNA-binding proteins. Examples of such translocations are t(9;11), t(8;21) and t(15;17) encoding MLL-AF9, AML1-ETO and PML-RARα fusion proteins respectively. In all cases, the fusion protein presumably leads to aberrant gene expression programs resulting in disordered hematopoietic development, which represents a critical step in the leukemogenic pathway. Epidemiologic studies suggest that acquisition of the fusion oncoproteins is an initial step in leukemogenesis, and that subsequent mutations are needed for leukemia development.6 These studies combined with sophisticated murine models suggest a multistep pathogenesis of leukemia.
Translocations involving the mixed lineage leukemia (MLL, HRX, ALL-1) gene on chromosome 11q23 are found in approximately 10% of acute myelogenous leukemias (AML), acute lymphoblastic leukemias (ALL) and most cases of therapy-related AML (t-AML).7 The presence of an MLL translocation predicts a poor prognosis, particularly when identified in ALL blasts.8 More than 30 different MLL translocations have been cloned from acute leukemias, but all generate fusion genes that encode chimeric proteins possessing an N-terminal portion of MLL and the C-terminal portion of the fusion partner.1,7 The most common of the 11q23 translocations found in AML is the t(9;11)(p22;q23), whereas the most common translocation found in ALL is the t(4;11)(q21;q23).9,10
Accumulating data supports the notion that leukemias possess a hierarchy of cells, only a subset of which are capable of self-renewal. 11,12 These leukemia stem cells (LSCs) are presumably necessary for continued propagation of leukemia and therefore represent critical target cells for any successful therapy. Thus, genetic alterations that might affect LSC characteristics are of significant interest. Recent experiments have demonstrated MLL fusion proteins including MLL-AF9 are capable of imparting LSC-like properties on developing hematopoietic progenitors.13–15 However, MLL-translocations may not be sufficient to induce leukemia without cooperation from subsequent genetic events. This is supported by the latency of t-AML, where the MLL translocation is induced by chemotherapy treatment, but the disease does not develop until approximately 3–5 years after treatment. Also, an MLL fusion knock-in mouse model that expresses an MLL-AF9 fusion protein encoded by the t(9;11) develops AML but only after a 4–9 month latency.16,17 This is interpreted as a need for cooperating mutations in order for acute leukemia development to occur. Similarly, a conditional MLL-CBP knock-in mouse that recapitulates t-AML induced by the t(11;16) requires chemical or irradiation-induced mutagenesis for generation of a fatal myeloproliferative disease or leukemia.18 Furthermore, bone marrow transplant models progress to oligoclonal leukemias pointing to the need for cooperating mutations in MLL-induced leukemogenesis.19
Recent hypotheses as to the multistep nature of leukemogenesis suggest mutant signaling molecules are good candidates for cooperation with DNA-binding fusion oncogenes during leukemogenesis. 20 One mouse model indicates that this is indeed possible, as an activated receptor tyrosine kinase FLT3 induces an acute leukemia from an MLL-SEPT6-induced myeloproliferative disease.21 Activating FLT3 mutations are present in approximately 30% of cases of human AML including some AMLs harboring MLL-AF9 translocations.20,22,23 Recent studies suggest FLT3 mutations are present in human LSCs providing support for the importance of FLT3 signaling in AML.24 Furthermore, high-level expression of FLT3 is also associated with the presence of MLL translocations in both AML and ALL.23,25–27 We have previously identified frequent mutation of FLT3 in MLL-rearranged lymphoblastic leukemias, and demonstrated efficacy of a FLT3 inhibitor against MLL-rearranged lymphoblastic leukemias in a xenograft model system.28
Our previous findings that MLL-AF9 can transform a committed myeloid progenitor,14 along with the above mouse and human data, prompt assessment of cooperation between MLL-AF9 and FLT3 in a murine model system. We reasoned that development of such a model system would both determine if the oncogenes cooperate during leukemogenesis, and provide a model system for testing combination therapies against MLL fusion and FLT3-dependent disease. Using a bone marrow transplant assay, we show that MLL-AF9 and FLT3-ITD cooperate to produce a transplantable leukemia (AML) that is phenotypically similar to AML induced by MLL-AF9 alone, but with decreased latency. Even though the latency decreases from 70 to <30 days, the frequency of LSCs is approximately 1:100 in both MLL-AF9- and MLL-AF9/FLT3-ITD-dependent diseases. Importantly, the AML induced in the bone marrow transplant assays is similar to the disease that arises when mice are injected with granulocyte macrophage progenitors (GMPs) that have been transduced with either MLL-AF9 alone, or in tandem with FLT3-ITD. Finally, we have developed a luciferase transgenic mouse model from which we generated a luminescent MLL-AF9/FLT3-ITD-induced AML. Using this model we show that PKC412 is active against this disease in vivo in 2–3 weeks, indicating the in vivo dependence of this disease on FLT3-ITD. These data are consistent with previous findings that GMPs can be a cell of origin for MLL fusion expressing AML,13,14 and demonstrate that MLL-AF9 and FLT3-ITD can cooperate in the context of a committed progenitor cell. Furthermore we describe a model for rapid testing of new FLT3 inhibitors and combination therapies.
Materials and methods
Plasmids and virus production
The pMSCVneo plasmid was purchased from Clontech (Mountain View, CA, USA), and the pMSCV-IRES-GFP (pMIG) plasmid was obtained from the laboratory of Dr Stanley J Korsmeyer. The pMSCVneo MLL-AF9 fusion construct was obtained from Dr Jay Hess, and the pMIG-FLT3-ITD (N51)29 construct was generously provided by Dr Gary Gilliland. The pMSCV-IRES-EYFP-MLL-AF9 (pMIY-MLL-AF9) construct was generated by cloning MLL-AF9 into the EcoRI site of the pMIY vector, which was a generous gift from Dr Kate Vignali. For virus production, 10 μg of the above plasmid and 10 μg ψ-eco packaging vector were transfected into the 293 packaging cell line using Fugene (Roche, Indianapolis, IN, USA), and the resulting viral supernatants were harvested as previously described.30 The 293 cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal calf serum (FCS) (Invitrogen) and 100 units per ml penicillin/streptomycin (Invitrogen). Viral titer was determined by infection of BAF3 cells. All viruses used had a viral titer of approximately 5 × 105 infectious units per ml.
Mice and transplantation assays
Five days prior to bone marrow harvest, female C57BL/6 mice (Charles River Labs, Wilmington, MA, USA) were injected with 150 mgkg−1 5-fluorouracil (Sigma, St Louis, MO, USA). Whole bone marrow cells (4 × 106) were plated in 3ml ‘transplant media’ containing RPMI (Invitrogen), 10% FCS 6 ng ml−1 IL3 (Peprotech, Rocky Hill, NJ, USA), 10 ng ml−1 IL6 (Peprotech) and 100 ng ml−1 SCF (Peprotech) overnight. Plated cells were then infected with retrovirus plus 5 μg ml−1 polybrene, 7.5mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 6 ng ml−1 IL3, 10 ng ml−1 IL6, 100 ng ml−1 SCF and spun at 1800 r.p.m. for 90 min at 37 °C. Cells were reinfected after 24 h. After the second infection, cells were injected at a concentration of 106 cells in 300 μl phosphate-buffered saline (PBS) per recipient mouse. Recipient female C57BL/6 mice were irradiated (full body) with 6.0 Gy twice at a 3-h interval on the day of injection. Mice were killed when moribund. Blood, spleens and bone marrow were harvested and processed into single cell suspensions. Samples of lung, liver, kidney, bone and spleen were fixed in 10% neutral-buffered formalin for paraffin sectioning. For secondary transplantation, recipient female C57BL/6 mice were irradiated (full body) with 8.0 Gy once and the indicated number of leukemia cells was injected. For mice transplanted with sorted GMPs, the GMPs went through only a single round of infection, and then were sorted for GFP/YFP-positive cells on a Becton Dickenson FACSAria (BD, San Diego, CA, USA). The recipient mice received only one dose of radiation (6.0 Gy).
Luciferase mice and drug treatment assay
A plasmid with the human ubiquitin C promoter driving expression of firefly luciferase was created by subcloning the luciferase coding region from pGL3-basic (Promega, Madison, WI, USA) into pUB6/V5-His (Invitrogen). Prokaryotic sequences were removed by restriction digest and agarose gel purification. Transgenic mice were created by pronuclear injection of purified DNA into C57BL/6 eggs (Beth Israel Deaconess Medical Center Transgenic Core Facility, Boston, MA, USA). The presence of the luciferase gene was identified by PCR, and coisogenic mice were created by crossing of founders to C57BL/6 mice. A single transgenic line with high ubiquitous luminescence (UbC6-Luc) was subsequently maintained on a C57BL/6 background with homozygosity of the transgene.
In order to assess luciferase activity in hematopoietic stem cells (HSCs) derived from UbC6-Luc mice, recipient C57BL/6-TyrC mice (Jackson Labs, Bar Harbor, ME, USA) were irradiated with a split dose (500 and 450 cGy). A total of 1500 unfractionated bone marrow cells from a UbC6-Luc transgenic mouse, together with 1.2 × 105 nonluminescent C57BL/6 marrow cells (to ensure short-term survival), were transplanted into irradiated mice and recipient mice were imaged every 2–4 weeks.
For the in vivo bioluminescent leukemia assays, whole bone marrow was extracted from UbC6-Luc mice as mentioned above. Infection of the whole bone marrow and subsequent injection into Tyrosinase−/− C57BL/6 recipients was also carried out as above in wild-type C57BL/6 mice. Treatment of NOD-SCID mice (Jackson Labs) with PKC412 (Novartis, Basel, Switzerland) and luminescent imaging of the mice was performed as described previously.28
Immunophenotype Analysis
Peripheral blood was obtained immediately post-mortem. Blood smears were stained using the eosin and methylene blue Quickdip kit (Jorgensen Labs, Loveland, CO, USA). For leukemia immunophenotyping, cells isolated from spleen and bone marrow (as described above) were incubated with phycoerythrin-conjugated (PE) antibodies to CD3, Mac1 or c-kit (Caltag, Burlingame, CA, USA), and with allophycocyanin-conjugated (APC) antibodies to B220, Gr1 or Mac1 (Caltag), washed and analyzed.
DNA extraction and Southern blotting
Genomic DNA was extracted from spleens using the Gentra Systems kit (Gentra Systems, Minneapolis, MN, USA). For Southern analyses, 10 μg genomic DNA was digested with EcoRI (New England Biolabs, Ipswich, MA, USA) overnight at 37 °C and run on a 1% agarose gel. DNA was transferred overnight to a Genescreen Plus nylon membrane (Perkin Elmer, Boston, MA, USA). Membranes were probed with 106 CPM per ml of [α32P]dCTP (Perkin Elmer)-labeled enhanced green fluorescent protein (EGFP). The EGFP probe was derived from the 1 kb NcoI-HindIII fragment of the pMIG vector. Probes were radiolabeled using a random prime labeling kit (Prime-It II—Stratagene, La Jolla, CA, USA).
RNA Extraction and RT–PCR
Total RNA was isolated from spleens using Trizol (Invitrogen), and cDNA was generated using the Retroscript kit (Ambion, Austin, TX, USA). To amplify the MLL-AF9 breakpoint region the following primer pair was used: forward – 5′-AACCACC TCCGGTCAATAAGC-3′, reverse – 5′-TTCACGATCTGCTGCAG AATG-3′. To amplify the FLT3-ITD juxtamembrane domain, the following primer pair was used: forward – 5′-TGTCGAGCAGTA CTCTAAACATG-3′, reverse – 5′-CTTTCAGCATTTTGACGGC AACC-3′.
Myeloid progenitor sorting
Granulocyte macrophage progenitors were sorted as described previously.31 Briefly, bone marrow was isolated as described above and labeled for 30 min on ice with each of the following lineage-specific antibodies: anti-mouse CD3 (Cat# RM3400, Caltag), anti-mouse CD4 (Caltag), anti-mouse CD8a (Caltag), anti-mouse CD19 (Caltag), anti-mouse B220 (CD45R) (Caltag), anti-mouse Gr1 (Caltag), anti-mouse TER119 (Caltag) and anti-mouse CD127 (interleukin (IL)-7R) (Bioscience, San Diego, CA, USA), as well as anti-mouse Sca1 (Caltag). The cells are washed with PBS, then incubated with PE-Cy5-labeled anti-rat secondary antibody (Caltag) for 30 min on ice. Following a PBS wash, cells are resuspended in 200 μl PBS/25% rat IgG (Sigma) and incubate for 10 min on ice. The cells are then stained with FITC conjugated anti-CD34 (Becton Dickenson), PE conjugated FcγR II/III (Becton Dickenson), and APC-conjugated anti-c-Kit (Becton Dickenson) for 30 min on ice, then cells are washed and resuspended in 300μl PBS. Dead cells were labeled with 7-AAD (Invitrogen) for 15 min before the sorting. For analysis of leukemic progenitors, we replace the FITC-anti-CD34 antibody with a biotinylated anti-CD34 (Becton Dickenson), which is developed using a streptavidin-APC-Cy7 anti-rat antibody (Caltag). Progenitors, as well as cells expressing EGFP/EYFP, were sorted using a FACSAria multicolor cell sorter equipped with filters that separate EGFP and EYFP (BD).
Cell line derivation, maintenance and cell growth assays
Primary cell lines derived from whole bone marrow or spleen were maintained in ‘C10’ media containing RPMI, 10% FCS, 100 units per ml penicillin/streptomycin, 2mM L-glutamine (Invitrogen), 55 μM β-mercaptoethanol (Invitrogen), 0.1mM nonessential amino acids (Invitrogen), 1mM sodium pyruvate (Invitrogen) and 10mM HEPES (Invitrogen). Cell lines were initially grown in C10 media supplemented with 10 ng ml−1 IL3, 10 ng ml−1 IL6, 100 ng ml−1 SCF. For MLL-AF9+FLT3-ITD cell lines, all cytokines were withdrawn after a week in culture. For MLL-AF9 cell lines, only SCF and IL6 were withdrawn. For the cell growth assays, cell lines were plated in triplicate at a density of 106 cells per 1ml C10 media with or without 10 ng ml−1 IL3. Cells were subsequently counted manually every 2–3 days.
In vitro drug sensitivity assays
To assay for sensitivity to PKC412, the colorimetric Cell Proliferation Kit I – 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Roche) was utilized. Cells were plated at a density of 105 cells per 100 μl C10 media into 96-well microtiter plates (Corning, Corning, NY, USA) and then incubated for 48 h in the presence of specified concentration of PKC412 and/or 5 ngml−1 IL3. The MTT assay was then performed as per manufacturer protocol.
Immunoprecipitations and western blots
For immunoprecipitations, 2.5 × 106 cells were plated in the presence of 0, 1, 10 or 100 nM PKC412, with the 0 sample receiving an equivalent volume of dimethyl sulfoxide as a carrier control. Cells were harvested 4 h after PKC412 treatment and lysed in lysis buffer containing 20mM Tris-HCl pH 7.4, 150mM NaCl, 100mM NaF, 10% glycerol, 1% IGEPAL CA-630, 10mM EDTA, 1mM phenylmethylsulphonylfluoride, 100 units per ml leupeptin and 1mM Sodium Orthovanadate (all reagents from Sigma). Precleared lysates were then incubated with Protein A-sepharose beads in the presence of 1 μg α-hFLT3 antibody (R&D Systems, Minneapolis, MN, USA) for 4 h at 4 °C. Beads were then washed three times in lysis buffer. Proteins were eluted from the beads using sodium dodecyl sulfate sample buffer, and run on NuPAGE 10% Bis-Tris acrylamide gels (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen) for western analyses. Western blots were performed using the 4G10 mouse α-phosphotyrosine, goat α-hFLT3 (R&D Systems) or mouse α-pan-actin (Chemicon, Temecula, CA, USA).
Results
FLT3-ITD decreases the latency of MLL-AF9-induced leukemia
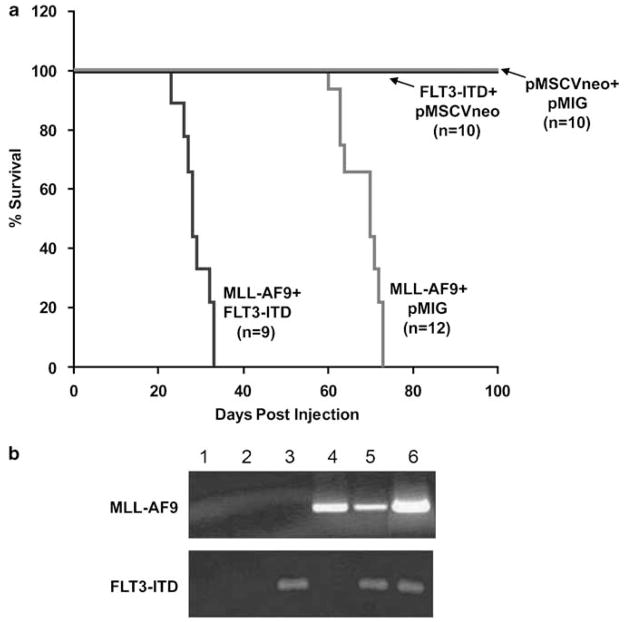
To assess for cooperation between a constitutively active FLT3 (FLT3-ITD) and MLL-AF9, we first utilized a bone marrow transplant assay.29 Bone marrow cells were harvested and transduced with various combinations of MSCV-MLL-AF9-pgkNeo (MLL-AF9), MSCV-FLT3ITD-IRES-EGFP (FLT3-ITD), MSCV-pgkNeo (MSCV-Neo) and MSCV-IRES-EGFP (MIG) retroviruses. Transduced bone marrow was then injected into lethally irradiated syngeneic recipient mice and monitored for disease development. Mice that received bone marrow cells transduced with MLL-AF9 and MIG retroviruses developed a lethal disease with a median latency of 68 days (Figure 1a and Table 1). The mice exhibited splenomegaly, elevated white blood cell counts, anemia and thrombocytopenia (Table 1). Mice that received cells transduced with both the MLL-AF9 and FLT3-ITD consistently developed a similar disease with a more rapid onset and a median latency of 28 days (Figure 1a and Table 1). Peripheral blood and organs from mice that received bone marrow transduced with both MLL-AF9 and FLT3-ITD revealed a disease phenotypically similar to the disease initiated by MLLAF9 alone (data not shown), which was consistent with an acute leukemia. Mice that received bone marrow cells transduced with FLT3-ITD plus MSCV-Neo or MSCV-Neo plus MIG did not develop obvious disease up to 150 days (Figure 1a). Total RNA was isolated from the enlarged spleens of moribund mice and reverse transcription (RT)–PCR was performed to assess the expression of the transduced genes in our system. The primer pairs used for the cDNA amplification were specific to the breakpoint region of the MLL-AF9 cDNA, and the juxtamembrane region of the human FLT3 cDNA encompassing the region containing the internal tandem duplication. As shown in Figure 1b, mRNA for MLL-AF9 and FLT3-ITD was expressed in appropriate spleen samples. Thus, expression of FLT3-ITD enhances the development of lethal MLL-AF9 leukemia.
Figure 1.
Cotransduction of FLT3-ITD decreases the latency of MLL-AF9-induced leukemia. (a) A bone marrow transplant assay was performed using bone marrow cells transduced with pMSCVneo-MLL-AF9, pMIG-FLT3-ITD, both retroviruses or empty construct controls, followed by injection of transduced cells into lethally irradiated syngeneic C57/BL6 recipients. A Kaplan–Meyer curve assessing survival of recipient mice is shown. (b) RT–PCR was performed on total RNA isolated from splenic cells of diseased or control mice. The MLL-AF9 primers crossed the fusion point and the FLT3 primers were specific for human FLT3. The lane order is as follows: (1) negative control, (2) pMSCVneo+pMIG, (3) pMSCVneo+pMIG-FLT3-ITD, (4) pMSCVneo-MLL-AF9+pMIG, (5) pMSCVneo-MLL-AF9+pMIG-FLT3-ITD and (6) positive plasmid control.
Table 1.
Analysis of leukemic mice
| Mutation(s) | Latency (days) | WBC (× 103 per μl) | HCT (%) | PLT (× 103 per μl) | Spleen weight (mg) |
|---|---|---|---|---|---|
| Control | N/A | 11.8±7.6 | 48.7±1.8 | 1051±262.9 | 120±27.4 |
| FLT3-ITD | N/A | 10.1±3.5 | 47.6±2.7 | 847±475.3 | 110±14.1 |
| MLL-AF9 | 68±5 | 293.4±17 | 28.8±8.1 | 170.5±130 | 760±122.1 |
| MLL-AF9+FLT3-ITD | 28±3 | 75.9±49.3 | 30.8±16.7 | 102.2±64.2 | 527±142 |
Abbreviations: HCT, hematocrit; N/A, not applicable; PLT, platelet; WBC, white blood cell.
Control: n = 5 for spleen, n = 4 for WBC; FLT3-ITD: n = 2 for spleen, n = 3 for WBC; for all others n≥5.
MLL-AF9 and MLL-AF9/FLT3-ITD mice develop similar myeloid leukemias with expanded GMP-like compartments
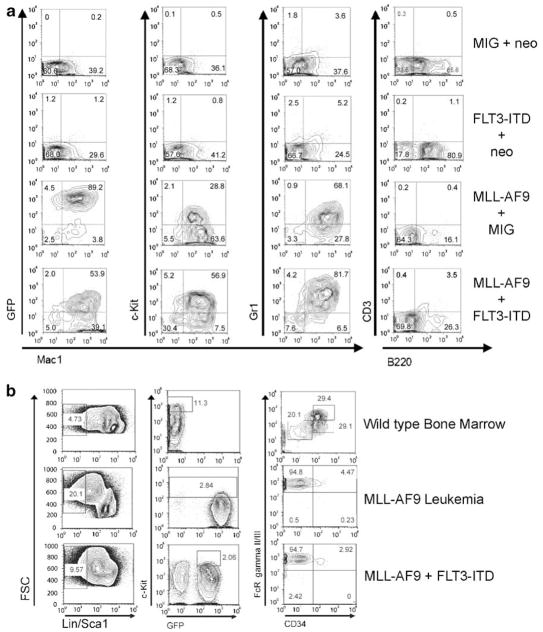
Immunophenotypic analysis of bone marrow and spleen cells (Figure 2a) from the diseased mice demonstrated an abundant population of cells positive for the myeloid lineage markers Mac-1 (CD11b) and Gr-1 (Ly-6G) from both MLL-AF9 mice and from MLL-AF9/FLT3-ITD mice. Additionally, a population of cells with variable positivity for the hematopoietic progenitor marker c-kit (CD117) was found in each case (Figure 2a). Cells from MLL-AF9 and MLL-AF9/FLT3-ITD mice were negative for the B cell marker B220 and the T cell marker CD3 (Figure 2a), as well as for the stem cell marker Sca1 (not shown). Furthermore, the majority of spleen cells from MLL-AF9/FLT3-ITD mice were EGFP positive, indicating they arose from transduced cells. The coexpression of Mac-1, Gr-1 and c-Kit, combined with the lack of expression of lymphoid markers is consistent with AML and similar to the phenotype described in previous murine models of MLL-AF9 fusion-induced AML.17 Thus, histologic and immunophenotypic analysis show that MLL-AF9 and MLL-AF9 combined with FLT3-ITD induce a similar leukemia in this system.
Figure 2.
Immunophenotyping demonstrate similar acute myeloid leukemias derived from MLL-AF9 and MLL-AF9+FLT3-ITD. (a) Flow cytometry was performed on spleen cells recovered from leukemic or control mice. Expression of the hematopoietic progenitor marker c-kit, the B cell marker B220, the T cell marker CD3 and the myeloid cell markers Mac1 and Gr1 were assessed. EGFP expression was also monitored. Coexpression of Mac-1 and Gr-1 along with variable c-kit expression is consistent with AML. (b) Myeloid progenitor analyses performed on bone marrow from leukemic mice indicates the prevalence of a Lin− Sca− c-kit+ FcγR+ CD34lo population in both MLL-AF9 and MLL-AF9/FLT3-ITD mice. Left panels show lineage marker/Sca1 expression. Center panels show Kit and green fluorescent protein (GFP) expression. Right panels show FcγR II/III and CD34 expression.
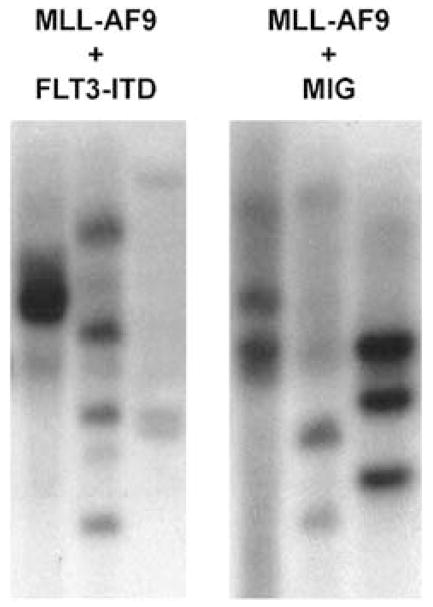
Recent experiments have shown that leukemias derived from MLL fusion proteins14,18 demonstrate expansion of a GMP-like population at the expense of other myeloid progenitors. In order to verify that this phenomenon occurs when a second oncogene is introduced along with MLL-AF9, we performed flow cytometry on bone marrow from normal and leukemic mice. Cells negative for lineage markers and for Sca1 (Lin− Sca−) cells (Figure 2b, left panels) were analyzed for c-kit and GFP expression. The c-kit+ GFP− population from normal bone marrow, and the c-kit+ GFP+ populations from leukemic marrow (Figure 2b, center panels) were further analyzed for FcγR and CD34 expression (Figure 2b, right panels). The FcγR+ CD34+ population shown in normal bone marrow represents GMPs, the FcγRlo CD34+ population demonstrates common myeloid progenitors (CMPs) and the FcγRlo CD34lo population megakaryocyte erythroid progenitors (MEPs). In leukemic bone marrow from both mice harboring MLL-AF9 alone, or in combination with FLT3-ITD, a GMP-like FcRγ+ CD34lo population represents the majority of cells with the CMP and MEP populations no longer present. Southern blot confirmed oligoclonality of both diseases (Figure 3). Thus, addition of FLT3-ITD does not alter the leukemic progenitor population produced by an MLL fusion protein.
Figure 3.
Diseases derived from MLL-AF9 with or without FLT3-ITD are oligoclonal. Genomic DNA was extracted from the spleen cells of leukemic mice. Southern blots were probed with the EGFP sequence to determine the clonality of the transplanted diseases. Shown are Southern blots of digested genomic DNA from six separate leukemic mice (three transduced with MLL-AF9/MIG and three transduced with MLL-AF9/FLT3-ITD).
Similar percentages of LSC are found in MLL-AF9 and MLL-AF9/FLT3-ITD leukemias
The presence and frequency of LSCs is generally assessed by serial transplantation. Thus, we determined if the malignancies induced by MLL-AF9 alone or theMLL-AF9/FLT3-ITD combination could be transferred to secondary recipient mice. Serial dilutions of 104, 103, 102 and 10 spleen cells harvested from diseased mice were injected into sublethally irradiated C57BL/6 recipient mice. The cells from both MLL-AF9 and MLL-AF9/FLT3-ITD leukemias induced disease with comparable disease latencies when equal numbers of cells were injected (Supplementary Figure 1a and b). Of interest, most secondary recipient mice injected with only 100 spleen cells developed leukemia. These data confirm findings that MLL fusion proteins give rise to a large LSC population14,15 and that these LSCs are present with a frequency of approximately 1:100 in both the MLL-AF9- and MLL-AF9/FLT3-ITD-induced diseases. Morphologic and immunophenotypic analysis showed that the transplanted leukemias were identical to the disease seen in the primary animals (data not shown).
FLT3-ITD cooperates with MLL-AF9 to induce leukemias derived from GMPs
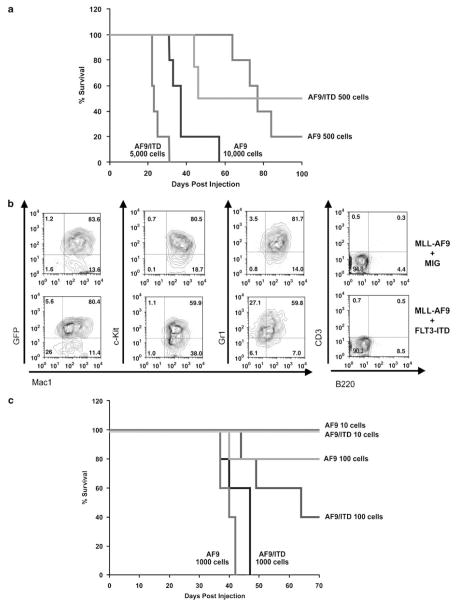
Recent studies have shown that MLL fusion proteins can induce leukemia from hematopoietic stem cells as well as more committed myeloid progenitors.13,14 Furthermore, MLL-AF9-induced AMLs give rise to a GMP-like population that is enriched for LSCs (one in six cells).14 Thus we wondered if MLL-AF9 and FLT3 might cooperate to induce leukemias from committed progenitors. We made use of an MSCV-IRES-EYFP-MLL-AF9 construct, so we could sort MLL-AF9 (YFP) and FLT3-ITD (GFP) double-positive cells. In this fashion, we could determine the number of MLL-AF9+FLT3-ITD-expressing cells injected into mice. The addition of FLT3-ITD to MLL-AF9-expressing GMPs decreased the latency of the MLL-AF9-mediated disease (Figure 4a). This disease could be initiated with as few as 500 cells. Importantly, 500 MLL-AF9-expressing GMPs also induced leukemia, indicating that the decreased latency time observed in MLL-AF9/FLT3-ITD mice is due to the more rapid generation of an aggressive disease rather than a difference in the number of injected cells. The MLL-AF9 expressing GMPs generate the same disease as from whole bone marrow as indicated by flow cytometry (Figure 4b). Next, we performed secondary transplants of spleen cells taken from mice with leukemias initiated in GMP. Similar to the myeloid leukemia we observed initiated from whole bone marrow, leukemic cells from both MLL-AF9 and MLL-AF9/FLT3-ITD mice induced diseases with comparable latencies for equivalent numbers of cells injected (Figure 4c). This confirms that MLL-AF9 and FLT3-ITD can cooperate in the context of a myeloid progenitor cell.
Figure 4.
FLT3-ITD also decreases the latency of leukemia induced by transduction of GMPs with MLL-AF9. (a) A transplant assay was performed using GMPs transduced with pMSCVneo-MLL-AF9 and pMIG empty vector or pMSCVneo-MLL-AF9 and pMIG-FLT3-ITD, followed by injection of the indicated number of transduced cells into lethally irradiated syngeneic C57/BL6 recipients. A Kaplan–Meyer curve assessing survival of recipient mice is shown. As seen in the bone marrow transplant assay in Figure 1, coexpression of FLT3-ITD and MLL-AF9 in GMPs leads to a disease with a more rapid onset than MLL-AF9 alone. (b) Immunophenotyping was performed as in Figure 2a. (c) Survival curves are shown for syngeneic secondary recipient mice that received the indicated number of leukemia cells.
The FLT3 inhibitor PKC412 is active against cell lines from MLL-AF9/FLT3-ITD-induced leukemias
In order to further investigate the role of FLT3 signaling in leukemias possessing both MLL-AF9 and FLT3-ITD, we generated multiple cell lines from leukemic mice. Spleen cells from either MLL-AF9 or MLL-AF9/FLT3-ITD-induced primary leukemias were plated in liquid culture in the presence of IL-3. Both MLL-AF9 and MLL-AF9/FLT3-ITD cells expanded and could be continuously propagated in the presence of IL-3 (Supplementary Figure 2a). However, only cell lines generated from leukemias coexpressing MLL-AF9 and FLT3-ITD were capable of growth in the absence of IL-3 (Supplementary Figure 2a). Immunophenotypic analysis demonstrated coexpression of Mac-1, Gr-1, c-kit and lack of expression of the lymphoid markers B220 and CD3 (data not shown). Western blot analysis for FLT3 demonstrated high-level FLT3 expression in the MLL-AF9/FLT3-ITD-derived leukemias but not the MLL-AF9-derived leukemias (Supplementary Figure 2b). Thus expression of FLT3-ITD induces cytokine independence in MLL-AF9-dependent myeloid leukemia cell lines.
The FLT3 inhibitor PKC412 inhibits FLT3 autophosphorylation and induces growth arrest and apoptosis in FLT3-dependent leukemias.32 Thus, we assessed if the FLT3 inhibitor PKC412 inhibits phosphorylation of FLT3-ITD at a concentration that coincided with a decrease in cell number in our cell lines. Western analyses of immunoprecipitated FLT3-ITD from samples treated with increasing concentrations of PKC412 showed nearly complete inhibition of phosphorylation at a concentration of 10 nM (Supplementary Figure 2d). Next, we determined if PKC412 was more potent in its ability to inhibited growth of cell lines derived from leukemias expressing MLL-AF9 and FLT3-ITD as compared to those expressing MLL-AF9 alone. PKC412 demonstrated remarkable potency against MLL-AF9/FLT3-ITD cell lines proliferating in the absence of IL3 with an IC50 of approximately 10 nM (Supplementary Figure 2c). In contrast, MTT assays demonstrated that both MLL-AF9/FLT3-ITD and MLL-AF9 cell lines growing in the presence of IL3 were minimally affected up to a dose of 100 nM (Supplementary Figure 2c). These data confirm the dependency of cells expressing the FLT3-ITD on continued FLT3 signaling.
Development of an in vivo imaging system for rapid assessment of leukemogenesis and new therapeutic agents
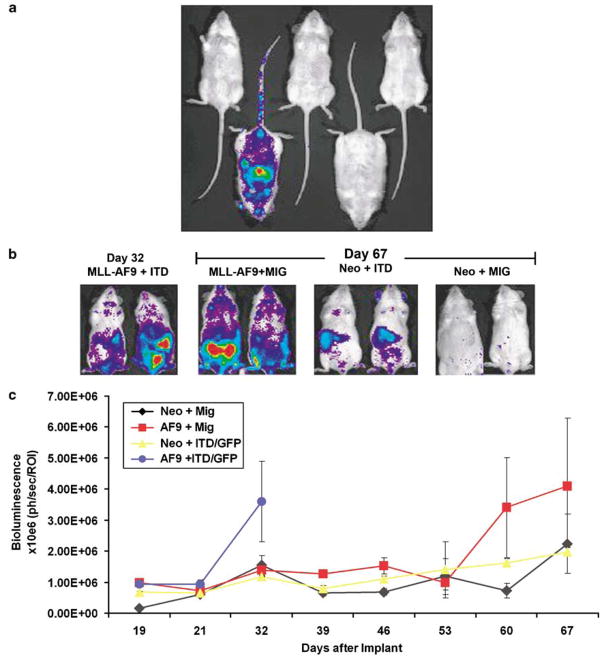
While the development of disease was very uniform in this genetically defined model system (Figure 1a), we wished to further refine the system so that disease burden, and the effect of therapeutic interventions, could be directly quantified, rather than relying on animal death as an end point. We established a transgenic mouse strain, UbC6-Luc, in which firefly luciferase was ubiquitously expressed from the human ubiquitin C promoter. Although the human ubiquitin C promoter has been shown to be broadly expressed in solid organs in transgenic animals,33 we wished to specifically assess luminescence in hematopoietic cells. As such, we transplanted HSCs at limiting dilution from UbC6-Luc transgenic mice into albino coisogenic recipients (C57BL/6-TyrC), which provides improved imaging sensitivity by comparison to pigmented recipients. In a cohort of mice transplanted with a total of 1500 bone marrow cells, stable luminescence was readily apparent in one recipient mouse 12 weeks after transplantation (Figure 5a). Since the frequency of HSCs is 1:10 000–1:100 000 within the bone marrow, the luminescence in this mouse is likely the progeny emanating from a single transplanted HSC. These results demonstrate adequate luciferase expression in UbC6-Luc hematopoietic cells for noninvasive bioluminescence imaging.
Figure 5.
A bone marrow transplant model using luciferase transgenic mice can be used to follow disease progression and quantitate drug efficacy in vivo. (a) Five C57BL/6-TyrC mice were injected with 1500 bone marrow cells from the UbC6-Luc transgenic mice and imaged every 2–4 weeks. Imaging at 12 weeks shows stable engraftment and luminescence from one mouse as would be expected based on the frequency of hematopoietic stem cells in murine bone marrow. (b) Donor bone marrow was taken from UbC6-Luc transgenic mice and was transduced with the indicated viruses followed by injection into lethally irradiated C57BL/6-TyrC mice. Leukemia development was assessed by serial imaging. MLL-AF9/FLT3-ITD mice showed increased luminescent signal at 32 days, whereas MLL-AF9 mice demonstrated increased luminescence at 67 days. (c) Quantitation of luciferase expression (photonic flux) is shown for all cohorts of mice. The increase in luminescent signal coincides with the development of AML.
For noninvasive visualization of leukemogenesis, bone marrow from UbC6-Luc mice was harvested and infected as above with combinations of MLL-AF9, FLT3-ITD, MSCV-Neo and MIG retroviruses. Transduced bone marrow cells were injected into C57BL/6-TyrC recipients, and the mice were monitored for disease by bioluminescence imaging. Rapid expansion of transduced cells could be detected in MLL-AF9 + FLT3-ITD recipients within 32 days (Figures 5b and c), with lethal disease in all animals before 39 days. The MLL-AF9 recipients showed a similar increase in luminescence but only after 60 days (Figures 5b and c). Thus, the latency of the diseases in this set of experiments is consistent with those from our original bone marrow transplant experiments (Figure 1a). These data not only provide independent verification of cooperative disease induction by MLL-AF9 and FLT3-ITD, but also provide us with large numbers of luminescent leukemia cells that can rapidly induce disease upon transplantation into secondary recipients. This genetically defined syngeneic imaging model can be used to generate cohorts of mice to rapidly test new therapeutics directed against either FLT3 or MLL-AF9.
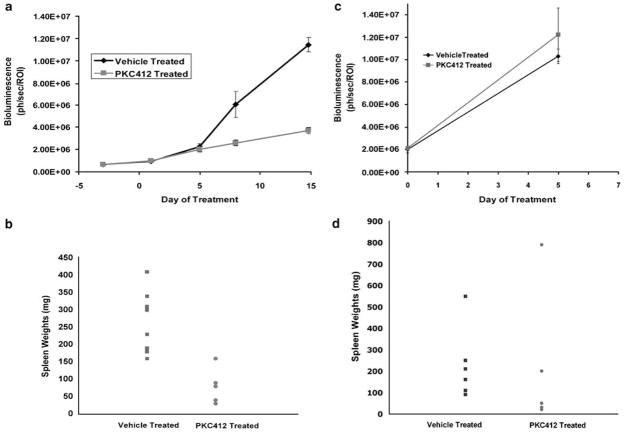
As a test for this system, and to validate FLT3-dependence of the MLL-AF9/FLT3-ITD-induced disease in vivo, we assessed the efficacy of PKC412 against mice transplanted with leukemia. One million spleen cells from the luciferase positive MLL-AF9/FLT3-ITD leukemic mice (Figure 5b and c) were injected into sublethally irradiated syngeneic secondary recipients and tumor burden was noninvasively quantified by bioluminescence imaging. After inoculation with leukemia cells, engraftment and expansion of leukemia was followed by imaging, and homogeneous cohorts of mice with progressive leukemia (that is established disease) were divided into treatment groups 10 days after transplant. Mice were treated with either 150mgkg−1 PKC412 or vehicle control once daily and tumor burden was assessed by bioluminescence imaging every 2–5 days. Treatment with PKC412 resulted in significant reduction in tumor growth after 7 days of treatment (Figure 6a). Consistent with this finding, the PKC412-treated mice had spleen weights that were significantly decreased as compared to the vehicle-treated mice after 14 days of treatment (Figure 6b). To assess the specificity of the FLT3 inhibitor in vivo, one million spleen cells from the luciferase positive MLL-AF9 (without FLT3-ITD) leukemic mice (Figures 5a and b) were injected into syngeneic recipients, and the experiment was carried out as in Figure 6a. In contrast to the previous experiment, PKC412-treated mice had no noticeable change in luminescence compared to the vehicle-treated mice (Figure 6c). Likewise, spleen weights of MLL-AF9 mice treated with vehicle or PKC412 were not significantly different (Figure 6d). These data show not only that the leukemia cells expressing FLT3-ITD remain dependent upon activated FLT3 for proliferation/survival and exhibit a greater sensitivity to FLT3 inhibitors, but also demonstrate the utility of this model system for rapid real-time assessment of therapeutics within a 2–3 week experimental window.
Figure 6.
Quantitative analysis of PKC412 efficacy against MLL-AF9/FLT3-ITD-dependent AML in vivo. (a) Spleen cells from leukemic MLL-AF9/FLT3-ITD mice (from Figure 5b) were injected into sublethally irradiated NOD/SCID mice. After leukemic cell engraftment, mice were treated with either the FLT3 inhibitor PKC412 or vehicle and imaged every 2–5 days. Disease burden increases rapidly in mice treated with vehicle alone, while PKC412 mice show much less luminescence (P= 0.011 at day 7, P= 0.00001 at day 14). (b) Spleens from treated and untreated mice were harvested and weighed. PKC412-treated mice exhibited decreased spleen weights as compared to the vehicle-treated mice (P= 0.0001), an independent measure of leukemia response to PKC412. (c) Spleen cells from leukemic MLL-AF9 mice (from Figure 5b) were injected into sublethally irradiated NOD/SCID mice as in panel a. After leukemic cell engraftment, mice were treated with either the FLT3 inhibitor PKC412 or vehicle and imaged after 5 days. Disease burdens from both treated and untreated mice increased drastically (P= 0.6344 at day 5). Mice in both groups became moribund at or around the last time point. (d) Spleens from treated and untreated MLL-AF9 mice were harvested and weighed as in panel b. Spleen weights from both sets of mice were statistically indistinguishable (P= 0.5154).
Discussion
There is accumulating evidence that points to a multistep pathogenesis for leukemia development and progression. In such models of leukemogenesis, the initial genetic event often leads to the expression of chimeric fusion oncogenes encoded by recurrent chromosomal translocations, while subsequent mutations may activate specific signaling pathways.25,34,35 Given that leukemias bearing translocations involving the MLL gene on chromosome 11q23 demonstrate high level expression and frequent mutation of the receptor tyrosine kinase FLT3, we hypothesized that MLL fusion proteins and constitutively active FLT3 are cooperating genetic events in the multistep pathogenesis of MLL-rearranged leukemias. Furthermore, we reasoned that combination of bioluminescence technology and bone marrow transplant assays would provide a model system for rapid assessment of targeted therapeutics in vivo. Here we provide evidence for cooperation between the MLL fusion protein MLL-AF9 and the constitutively active FLT3-ITD in a murine model system. In bone marrow transplant assays, mice receiving MLL-AF9-transduced marrow develop leukemia with a disease latency of approximately 70 days, whereas coexpression of MLL-AF9 and FLT3-ITD produced a similar disease with a shortened latency of 28 days. Secondary transplant assays demonstrated that the diseases are both highly transplantable AMLs that possess LSCs with a frequency of approximately 1:100. Cell lines derived from leukemias expressing MLL-AF9 and FLT3-ITD were capable of cytokine-independent growth in contrast to cell lines derived from leukemias expressing MLL-AF9 alone which remain dependent upon IL-3 for growth and survival. Finally, we developed a luciferase transgenic mouse model that allows monitoring of leukemia progression in live animals. We used this system to demonstrate that MLL-AF9 leukemias expressing FLT3-ITD were dependent upon FLT3 signaling as the FLT3 inhibitor PKC412 showed efficacy and specificity in vitro and in vivo. These data demonstrate that MLL-AF9 and activating mutations of FLT3 cooperate during leukemogenesis, and provide evidence that this model can be used to test therapeutic approaches.
Previous murine models have demonstrated a cooperative role for FLT3-ITD in the development of acute promyelocytic leukemia (APML). In these studies transgenic mice expressing the PML-RARα fusion encoded by t(15;17)(q22;q11.2) progress to an APML-like disease with a long latency that decreases when FLT3-ITD is expressed in bone marrow cells.36 Data supporting cooperation of FLT3 activating mutations and MLL translocations come mostly from human leukemias where high-level FLT3 expression is correlated with the presence of MLL rearrangements in ALL and AML.23,25,27,37 Also, activating FLT3 mutations are found in both MLL-rearranged leukemias of myeloid and lymphoid origin even though FLT3 mutations are rare in most other types of ALLs.23,28,38,39 A recent study demonstrated a potential role for FLT3 in the progression from an MLL fusion-induced myeloproliferative disease to acute leukemia.21 In our studies leukemias generated by either MLL-AF9 alone or MLL-AF9/FLT3-ITD had an immunophenotype consistent with AML. Furthermore, either disease could be transferred to secondary recipients with as few as 100 cells. Thus, our model demonstrates that constitutively active FLT3 can cooperate with MLL-AF9, a fusion that induces a disease more consistent with de novo acute leukemia. Demonstration of cooperation between MLL-AF9 and FLT3 during the development of AML supports the notion that at least two different classes of mutations are necessary for leukemia development.20,40
Recent studies suggest leukemias are comprised of a mixture of cells, only a fraction of which possess self-renewal properties necessary to maintain the disease in vivo, and that eradication of these LSCs will be necessary for successful leukemia therapies.11 Thus the biological characteristics of LSCs are of significant interest. The assay most commonly used to detect the presence LSCs is initiation of leukemia in secondary recipient mice, as only cells with self-renewal properties are capable of reproducing the leukemia. When such assays are performed using human leukemias, approximately 104 AML cells are required to obtain leukemia in immunodeficient mice.12 Recent studies assessing the frequency of LSCs in murine models of MLL-AF9 leukemias have demonstrated LSC to be present with higher frequency whether the leukemias are initiated from whole bone marrow or committed progenitors.14,15 The data presented here are in keeping with these studies and further demonstrate that an MLL-AF9 and FLT3-ITD-induced leukemia possesses LSCs with a frequency of approximately 1:100 in bone marrow. Studies assessing the frequency of LSCs in a murine model of MOZ-TIF2 induced AML demonstrated a much lower frequency of 1:104.41 Thus, MLL-AF9 appears to be quite potent in its ability to produce LSCs and FLT3 appears to cooperate in this context. The fact that MLL-AF9 and FLT3-ITD cooperate to produce a similar frequency of LSCs in 28 days as MLL-AF9 alone gives in 70 days suggests that FLT3 is providing a proliferative/survival advantage to LSCs and is not merely inducing proliferation of more differentiated cells incapable of self-renewal. Furthermore, these studies demonstrate rapid transformation of a committed progenitor via two oncogenes, which supports the hypothesis that GMPs are one possible cell of origin for MLL-rearranged AML, and may give rise to LSC in MLL-AF9-induced human leukemia. The fact that the diseases from GMPs are transplantable with comparable numbers to that from whole bone marrow disease further supports the idea that GMPs are a cell of origin for MLL-AF9 leukemia.
The development of an in vivo luminescent murine model of MLL-AF9- and FLT3-dependent AML provides a needed resource for the assessment of novel therapeutics. The ongoing genomic assessment of human malignancy combined with high-throughout approaches has provided a number of novel potential therapeutic approaches that will require assessment in preclinical models of malignancy. Furthermore, accumulating evidence suggests that successful targeted therapies will require the combination of multiple agents in order to prevent resistance.42 Thus models systems of cancer are desperately needed to rapidly test new therapeutic combinations. Human cancer cell lines represent the most accessible preclinical models, but most such lines have been propagated in culture for extended periods therefore likely acquiring mutations that select for growth in vitro. Injection of human cell lines into immunodeficient mice may provide a more relevant context for cancer cell survival, but the concern of acquired mutations remains, and a significant proportion of cell lines do not engraft in murine bone marrow. Testing of therapeutics on primary leukemia samples represents an attractive possibility, but the fact that primary human leukemia samples tend not to proliferate well in most culture systems confounds testing of therapeutics, such as kinase inhibitors, where proliferation of cells is likely to be required to obtain an accurate assessment of efficacy. Also, access to primary patient samples continues to be a challenge. Thus, we have developed a murine model system dependent upon two oncogenes, MLL-AF9 and FLT3, in which we can demonstrate efficacy of the FLT3 inhibitor PKC412 in as few as 2–3 weeks. This system takes advantage of a newly developed transgenic mouse that expresses luciferase under the control of the ubiquitin promoter to provide primary leukemia cells that can be visualized using bioluminescent imaging. This transgenic model should provide an important resource for further development of diverse models of cancer that can be used for rapid assessment of potential molecularly targeted therapies.
Supplementary Material
Acknowledgments
We thank D Gary Gilliland and members of the Korsmeyer laboratory for helpful discussions and Elise Schindler for administrative assistance. Also, we thank the late Stanley J Korsmeyer, a tremendous mentor and friend, for his guidance throughout this work. This work was supported by NIH grants CA92551, the Charles H Hood Foundation, Friends of the Dana Farber Cancer Institute and the Leukemia Lymphoma Society (SAA).
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 3.Shivdasani RA, Orkin SH. The transcriptional control of hematopoiesis. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 4.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 5.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 6.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 7.Dimartino JF, Cleary ML. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br J Haematol. 1999;106:614–626. doi: 10.1046/j.1365-2141.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen CS, Sorensen PH, Domer PH, Reaman GH, Korsmeyer SJ, Heerema NA, et al. Molecular rearrangements on chromosome 11q23 predominate in infant acute lymphoblastic leukemia and are associated with specific biologic variables and poor outcome. Blood. 1993;81:2386–2393. [PubMed] [Google Scholar]
- 9.Huret JL, Dessen P, Bernheim A. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia. 2001;15:987–989. doi: 10.1038/sj.leu.2402135. [DOI] [PubMed] [Google Scholar]
- 10.Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- 11.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 12.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 13.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 15.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 16.Corral J, Lavenir I, Impey H, Warren AJ, Forster A, Larson TA, et al. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 17.Dobson CL, Warren AJ, Pannell R, Forster A, Lavenir I, Corral J, et al. The mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 1999;18:3564–3574. doi: 10.1093/emboj/18.13.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Iwasaki H, Krivtsov A, Febbo PG, Thorner AR, Ernst P, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 21.Ono R, Nakajima H, Ozaki K, Kumagai H, Kawashima T, Taki T, et al. Dimerization of MLL fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis. J Clin Invest. 2005;115:919–929. doi: 10.1172/JCI22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- 23.Libura M, Asnafi V, Tu A, Delabesse E, Tigaud I, Cymbalista F, et al. FLT3 and MLL intragenic abnormalities in AML reflect a common category of genotoxic stress. Blood. 2003;102:2198–2204. doi: 10.1182/blood-2003-01-0162. [DOI] [PubMed] [Google Scholar]
- 24.Levis M, Murphy KM, Pham R, Kim KT, Stine A, Li L, et al. Internal tandem duplications of the FLT3 gene are present in leukemia stem cells. Blood. 2005;106:673–680. doi: 10.1182/blood-2004-05-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong SA, Golub TR, Korsmeyer SJ. MLL-rearranged leukemias: insights from gene expression profiling. Semin Hematol. 2003;40:268–273. doi: 10.1016/s0037-1963(03)00196-3. [DOI] [PubMed] [Google Scholar]
- 26.Tsutsumi S, Taketani T, Nishimura K, Ge X, Taki T, Sugita K, et al. Two distinct gene expression signatures in pediatric acute lymphoblastic leukemia with MLL rearrangements. Cancer Res. 2003;63:4882–4887. [PubMed] [Google Scholar]
- 27.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 29.Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 30.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 32.Weisberg E, Boulton C, Kelly LM, Manley P, Fabbro D, Meyer T, et al. Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412. Cancer Cell. 2002;1:433–443. doi: 10.1016/s1535-6108(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 33.Schorpp M, Jager R, Schellander K, Schenkel J, Wagner EF, Weiher H, et al. The human ubiquitin C promoter directs high ubiquitous expression of transgenes in mice. Nucleic Acids Res. 1996;24:1787–1788. doi: 10.1093/nar/24.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dash A, Gilliland DG. Molecular genetics of acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14:49–64. doi: 10.1053/beha.2000.0115. [DOI] [PubMed] [Google Scholar]
- 35.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 36.Kelly LM, Kutok JL, Williams IR, Boulton CL, Amaral SM, Curley DP, et al. PML/RARalpha and FLT3-ITD induce an APL-like disease in a mouse model. Proc Natl Acad Sci USA. 2002;99:8283–8288. doi: 10.1073/pnas.122233699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 38.Armstrong SA, Mabon ME, Silverman LB, Li A, Gribben JG, Fox EA, et al. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103:3544–3546. doi: 10.1182/blood-2003-07-2441. [DOI] [PubMed] [Google Scholar]
- 39.Taketani T, Taki T, Sugita K, Furuichi Y, Ishii E, Hanada R, et al. FLT3 mutations in the activation loop of tyrosine kinase domain are frequently found in infant ALL with MLL rearrangements and pediatric ALL with hyperdiploidy. Blood. 2004;103:1085–1088. doi: 10.1182/blood-2003-02-0418. [DOI] [PubMed] [Google Scholar]
- 40.Gilliland DG. Targeted therapies in myeloid leukemias. Ann Hematol. 2004;83 (Suppl 1):S75–S76. doi: 10.1007/s00277-004-0850-2. [DOI] [PubMed] [Google Scholar]
- 41.Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 42.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.