The fundamental polymers of biology—proteins, DNA, and RNA—are products of repetitive condensation of simple amino acid or nucleotide building blocks and are comparatively easy to assemble. However, other biomolecules require additional reactions beyond condensation of building blocks. Examples are the fatty acids and the polyketide and nonribosomal peptide secondary metabolites. These molecules are produced by complex enzyme assembly lines that include multiple catalytic domains. Two new crystal structures—one reported recently (1), the other by Maier et al. on page 1315 of this issue (2)—enrich our understanding of how these mega-enzymes function as efficient factories to produce a remarkable range of metabolic products.
Maier et al. study the fatty acid synthase (FAS-I) responsible for de novo fatty acid synthesis in the cytosol of animal cells. FAS-I is homologous in sequence and architecture with the very large family of modular polyketide synthases (PKSs), which produce a wide variety of natural products with potential medicinal value. In 2006, the authors reported the structure of FAS-I from a 4.5 Å electron density map, in which most domains could be assigned but no details were visible (3). Their new structure provides sufficient detail to understand the fold and the connectivity of six of its eight domains; however, the flexibly tethered acyl carrier protein (ACP) and thioesterase (TE) domains remain invisible. The other structure, reported by Tanovic et al. (1), is of an intact module of a nonribosomal peptide synthetase (NRPS). Except for the peptidyl carrier protein (PCP), the primary NRPS domains are not related to those of the FAS and PKS systems, but the assembly-line approach is similar.
All data indicate that the ancestor of FAS-I was a set of monofunctional enzymes, presumably resembling the dissociated FAS (type II, FAS-II) that catalyzes fatty acid biosynthesis in modern bacteria and plant plastids (see the figure, panel A). Gene duplication, loss of function, and gene fusion gave rise to the 270-kD polypeptide that functions as the homodimeric FAS-I in mammals (see the figure, panel B). A different mega-enzyme fusion of monofunctional ancestors evolved in fungi (4). Of these two assembly-line architectures for fatty acid synthesis, the mammalian FAS-I proved the more adaptable, and it now exists not only for fatty acid biosynthesis but throughout the eubacterial and fungal world for synthesis of polyketides. Indeed, high-resolution structures of PKS components (5, 6) provide critical corroboration for the mammalian FAS structure.
Figure. Assembly-line proliferation.
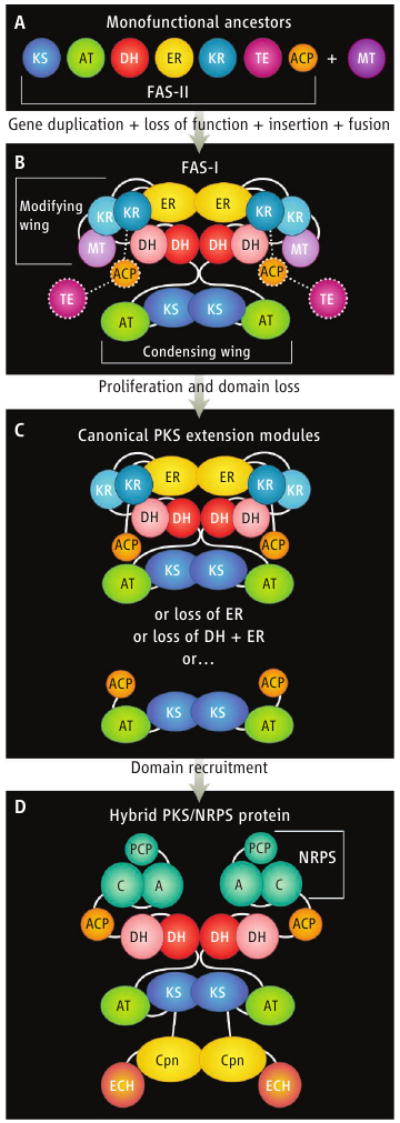
The dissociated FAS-II (A) evolved into the homodimeric FAS-I (B) [dotted lines outline disordered parts of the FAS-I structure (2); lighter shades indicate inactivated DH, KR, and MT domains]. Duplication of an ancestral FAS-I gene, followed by selective deletion, yielded the canonical extension modules of modern PKS pathways (C). An even richer diversity of polyketides arose by domain recruitment. For example, the CurF protein of the hybrid PKS/NRPS for curacin A (10) includes decarboxylase (ECH), cyclopropanase (Cpn), and NRPS domains (D).
Two key features of the FAS-I architecture explain its remarkable adaptability. First, the structure is segregated into two wings: a selection/condensing wing for addition of new building blocks, and a modifying wing for chemical processing of chain elongation intermediates (see the figure, panel B). The heart of the assembly line is the condensing wing, where an acyltransferase (AT) domain selects a building block and a ketosynthase (KS) domain adds it to the growing chain. The dimeric KS also contributes most of the dimer contacts in the complex.
The second key architectural feature is an open and flexible design that is ideal for insertion or deletion of catalytic domains, especially in the modifying wing. Each two-carbon addition (via malonate) to a fatty acid chain is followed by three reactions—keto reduction (KR), dehydration (DH), and enoyl reduction (ER)—carried out in the modifying wing of the FAS-I. A major source of chemical diversity in polyketides arises from deletion or inactivation of one or more of these modifying domains (see the figure, panel C), providing the chemical variation that is lacking in fatty acids.
In FAS-I and most fungal PKSs, the assembly line is used for iterative synthesis: Each enzyme domain performs the same reaction at each extension step on the growing substrate. In contrast, in most bacterial PKSs, polyketide synthesis is sequential: Each extension step is carried out by an individual FAS-I–like “module,” offering the possibility to vary the building block identity and modification chemistry at each step. This scheme greatly expands genetic and protein complexity. Several modules (up to 20 or more) are required to build a complex polyketide, and specific interactions of sequential modules must be faithfully maintained by fusion or by docking domains (7, 8).
A big surprise of the new FAS structure is a vestigial methyltransferase (MT) domain at the periphery of the dimer, following the DH in the polypeptide sequence. Thus, the megaenzyme ancestor of FAS-I appears to have had a methylation reaction as part of its fatty acid biosynthetic cycle. Was there a prokaryotic methyl branched-chain fatty acid, unknown to us today? The MT domain lost its function in FAS-I, was deleted from most PKS systems, but exists in some PKSs as an active methyltransferase. And herein lies a conundrum; the ubiquity of PKS pathways in bacteria and elsewhere strongly argues that the original FAS-I evolved in a prokaryote. However, other than Mycobacterium tuberculosis and related species that generate unusual fatty acids, we know of no modern prokaryote that uses a FAS-I for normal membrane lipid fatty acid biosynthesis (9).
In many PKS modules, the open FAS-I architecture has been augmented with a variety of other catalytic domains, such as S-acetyltransfer, halogenase, cyclopropanase, decarboxylase, and even entire NRPS modules (see the figure, panel D) (10, 11). The new structure of the terminal module of the surfactin NRPS (1) shows how it, too, is highly adaptable. Like FAS-I, the NRPS has a solid platform for condensation, including an adenylation (A) domain to select the amino acid building block and a condensation (C) domain to form a peptide link to the growing chain (1). The monomeric C-A didomain (analogous to KS-AT in the FAS-I condensing wing) is fused to a PCP and a terminal TE domain. As in the FAS-I structure, the PCP is flexibly linked to the synthetase by tethers long enough for it to deliver substrate to the active sites of all catalytic domains. Unlike the FAS-I structure, the PCP and TE domains are well ordered in the NRPS module.
The three assembly line types use homologous domains (ACP or PCP) to carry the growing fatty acid, polyketide, or peptide via a pantetheine-linked thioester. The common thioester chemistry and the adaptable architecture have resulted in the proliferation of hybrid PKS-NRPS and even PKS-FAS-I pathways found in phylogenetically diverse bacteria (9, 12). The rich diversity of PKS, NRPS, and hybrid systems demonstrates that nature has not employed a Henry Ford–like assembly line, from which the customer could have any color car so long as it was black. Rather, we see a modular assembly line that is easily copied, modified, and adapted to new function; this is the secret to its success.
Footnotes
Fatty acid synthases and related megaenzymes are highly adaptable to new functions as a result of their modular architecture.
Contributor Information
Janet L. Smith, Email: janetsmith@umich.edu.
David H. Sherman, Email: davidhs@umich.edu.
References
- 1.Tanovic A, Samel SA, Essen LO, Marahiel MA. Science. 2008;321:659. doi: 10.1126/science.1159850. published online 26 June 2008. [DOI] [PubMed] [Google Scholar]
- 2.Maier T, Leibundgut M, Ban N. Science. 2008;321:1315. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 3.Maier T, Jenni S, Ban N. Science. 2006;311:1258. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 4.Jenni S, Leibundgut M, Maier T, Ban N. Science. 2006;311:1263. doi: 10.1126/science.1123251. [DOI] [PubMed] [Google Scholar]
- 5.Keatinge-Clay AT, Stroud RM. Structure. 2006;14:737. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. Proc Natl Acad Sci U S A. 2006;103:11124. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thattai M, Burak Y, Shraiman BI. PLoS Comput Biol. 2007;3:1827. doi: 10.1371/journal.pcbi.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nougayrède JP, et al. Science. 2006;313:848. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 9.Gokhale RS, Saxena P, Chopra T, Mohanty D. Nat Prod Rep. 2007;24:267. doi: 10.1039/b616817p. [DOI] [PubMed] [Google Scholar]
- 10.Chang Z, et al. J Nat Prod. 2004;67:1356. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 11.Gu L, et al. Science. 2007;318:970. doi: 10.1126/science.1148790. [DOI] [PubMed] [Google Scholar]
- 12.Fischbach MA, Walsh CT, Clardy J. Proc Natl Acad Sci U S A. 2008;105:4601. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]


