Summary
Enterohemorrhagic E. coli (EHEC) colonizes the large intestine, causing attaching and effacing lesions (AE). Most of the genes involved in AE lesion formation are encoded within a chromosomal pathogenicity island termed the locus of enterocyte effacement (LEE). The LysR-type transcriptional factor QseA regulates the LEE by binding to the regulatory region of ler. We performed transcriptome analyses comparing WT EHEC and the qseA mutant to elucidate QseA’s role in gene regulation. During both growth phases, several genes carried in O-islands were activated by QseA, whereas genes involved in cell metabolism were repressed. During late-logarithmic growth, QseA activated expression of the LEE genes as well as non-LEE encoded effector proteins. We also performed electrophoretic mobility shift assays, competition experiments, and DNAseI footprints. The results demonstrated that QseA directly binds both the ler proximal and distal promoters, its own promoter, as well as promoters of genes encoded in EHEC-specific O-islands. Additionally, we mapped the transcriptional start site of qseA, leading to the identification of two promoter sequences. Taken together, these results indicate that QseA acts as a global regulator in EHEC, coordinating expression of virulence genes.
Introduction
Enterohemorrhagic Escherichia coli O157:H7 (EHEC) is a normal resident of cattle, but in humans EHEC causes severe disease characterized by bloody diarrhea and hemolytic uremic syndrome (Griffin et al., 1988). Infection commonly follows the ingestion of contaminated food or water (Carter et al., 1987, Honish, 1986, Riley et al., 1983, Ryan et al., 1986); however, infection may occur by direct contact with animals (Heuvelink et al., 2002) or via person-to-person transmission (Carter et al., 1987, Spika et al., 1986). EHEC survives passage through the acidic stomach and then enters the large intestine, where EHEC causes acute infection. There, EHEC senses the human hormones epinephrine and norepinephrine, as well as the bacterial hormone-like compound autoinducer 3 (AI-3) (Clarke et al., 2006, Sperandio et al., 2003). EHEC responds to these signals by activating expression of its flagella (Sperandio et al., 2002b) and of the locus of enterocyte effacement (LEE) (McDaniel et al., 1995). Upon reaching the epithelial cells, the LEE-encoded type three secretion system (TTSS) is expressed to translocate effector proteins directly into host epithelial cells (Jarvis et al., 1995). These effector proteins cause extensive cytoskeletal rearrangements and attaching and effacing (AE) lesion development (Campellone et al., 2004, Deng et al., 2004, Donnenberg & Kaper, 1991, Donnenberg et al., 1993, Garmendia et al., 2004, Jerse et al., 1990, Kenny et al., 1997, McKee et al., 1995, Mundy et al., 2004).
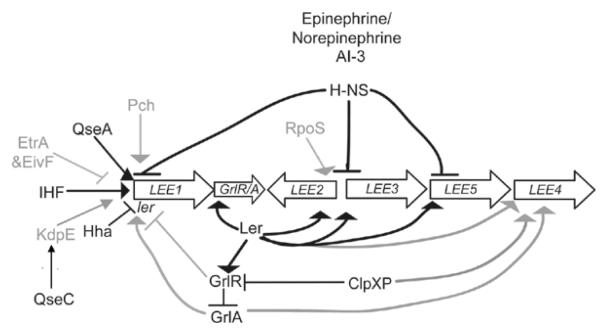
Most of the genes that encode for the TTSS and secreted proteins are located in the LEE (McDaniel et al., 1995). The LEE contains five operons (LEE1, LEE2, LEE3, LEE4, and LEE5) that are regulated directly or indirectly by more than 10 proteins (summarized in Fig. 1) (Barba et al., 2005, Bustamante et al., 2001, Deng et al., 2004, Friedberg et al., 1999, Haack et al., 2003, Hughes et al., 2009b, Iyoda & Watanabe, 2004, Iyoda & Watanabe, 2005, Mellies et al., 1999, Nakanishi et al., 2006, Sánchez-SanMartín et al., 2001, Sperandio et al., 2000, Sperandio et al., 1999, Umanski et al., 2002, Zhang et al., 2004). Regulation includes the global regulatory proteins integration host factor (IHF) that activates expression of ler (Friedberg et al., 1999) and H-NS that silences LEE transcription (Bustamante et al., 2001, Haack et al., 2003, Mellies et al., 1999, Sánchez-SanMartín et al., 2001, Sperandio et al., 2000, Umanski et al., 2002), as well as the environment-dependent regulator Hha that also acts to repress LEE transcription (Sharma and Zuerner, 2004). The sensor kinase QseC phosphorylates KdpE which subsequently activates expression of LEE1 (Hughes et al., 2009). The transcriptional regulators that are encoded by eivF and etrA in a second, nonfunctional TTSS are negative regulators of the LEE (Zhang et al., 2004). The ClpXP protease increases LEE transcription by inhibiting GrlR repression as well as by interacting with the stationary-phase sigma factor RpoS (Iyoda and Watanabe, 2005). RpoS positively regulates transcription of LEE3 (Iyoda & Watanabe, 2005, Sperandio et al., 1999), and the signaling molecule ppGpp can also increase transcription of the LEE (Nakanishi et al., 2006). The LEE-encoded regulator (Ler) that is encoded in LEE1 activates LEE2, LEE3 and LEE5 by binding to their promoters, displacing H-NS and allowing for transcription of these operons (Haack et al., 2003, Mellies et al., 1999, Russell et al., 2007, Sánchez-SanMartín et al., 2001, Sperandio et al., 2000). Also located in the LEE are grlR and grlA that encode for GrlR and GlrA, respectively (Deng et al., 2004). GrlR represses LEE transcription, whereas GrlA activates LEE transcription (Barba et al., 2005, Deng et al., 2004, Russell et al., 2007). The RNA chaperone protein Hfq negatively controls levels of GrlA and GrlR through post-transcriptional regulation, and is thus an indirect regulator of LEE expression (Hansen and Kaper, 2009; Shakhnovich et al., 2009). The pch genes that are homologous to perC in enteropathogenic E. coli encode for positive regulators of the LEE genes (Iyoda and Watanabe, 2004).
Figure 1.
Model of LEE regulation. Epinephrine/ norepinephrine and AI-3 are global regulators of virulence in EHEC. Black lines represent regulators whose direct interactions with the target promoter have been biochemically defined; grey lines represent interactions that occur indirectly or that have not been shown to bind biochemically to the target.
Additional regulation of the LEE also occurs via the LysR-type transcriptional regulator QseA. QseA was first identified as a regulator of the LEE genes as part of the cell-signaling cascade in the luxS system (Sperandio et al., 2002a). Initial studies demonstrated that an EHEC qseA mutant had a striking reduction in TTS activity, and secretion could be restored when qseA was complemented in trans (Sperandio et al., 2002a). Recent work has shown that QseA activates transcription of the LEE genes by directly binding to the ler promoter that encodes the master regulator of the LEE (Sharp and Sperandio, 2007). Moreover, QseA down-regulates its own expression (Sircili et al., 2004) as well as expression of qseE that encodes a sensor kinase involved in sensing epinephrine (Reading et al., 2009) and in regulation of AE lesion formation (Reading et al., 2007). In this study, our objective was to explore the extent of QseA gene regulation in EHEC as well as to characterize the molecular mechanism involved in QseA regulation.
Results
QseA plays an extensive role in EHEC gene regulation
To elucidate whether QseA’s role in regulating gene expression in EHEC was limited to ler and qseE, transcriptome analyses were undertaken, and these results were confirmed using real-time qPCR. We compared gene expression between wild-type (WT) EHEC strain 86-24 (Griffin et al., 1988) and the isogenic qseA mutant strain VS145 (Sperandio et al., 2002a) at both mid-exponential and late-exponential growth phases. The arrays contain approximately 10,000 probe sets for all 20,366 genes present in the genomes of the sequenced EHEC strains, EDL933 and Sakai, the uropathogenic E. coli strain CFT073, K-12 strain MG1655, and 700 probes covering intergenic regions that can encode non-annotated small ORFs, or small regulatory RNAs (sRNAs).
The microarray data (available at GSE18118 (http://www.ncbi.nlm.nih.gov/geo/)) indicated that altered probe sets were both up-regulated as well as down-regulated indicating that QseA may act as an activator or repressor of its target genes (Table 1). Altered probe sets included genes derived from the E. coli K-12 strain MG1655, which contains a common E. coli backbone conserved among all E. coli pathovars (Rasko et al., 2008), as well as pathovar-specific genes (Table 2), suggesting a global role for QseA regulation.
Table 1.
Numbers of genes with altered expression in qseA mutant compared to WT.
| Increase | Decrease | No change | |
|---|---|---|---|
| Mid-log | 222 | 1874 | 7741 |
| Late-log | 55 | 605 | 8873 |
Table 2.
Comparison of K-12 and pathovar-specific genes with altered expression.
| Increase | Decrease | No change | ||
|---|---|---|---|---|
| MG1655- specific |
Mid-log | 60 | 3 | 3220 |
| Late-log | 12 | 15 | 3624 | |
| Pathovar-specific | Mid-log | 112 | 105 | 3409 |
| Late-log | 34 | 166 | 3907 |
Transcriptome comparisons between the qseA mutant and WT grown to mid-exponential phase revealed that QseA regulates genes involved in metabolism and virulence. We observed altered expression between the qseA mutant and WT for genes encoding for phosphonate metabolism and ethanolamine metabolism. Ethanolamine is abundant in the intestines (Lawhon et al., 2003; Sanatinia et al., 1995) and processed foods (Anderson, 1988; Collier et al., 1991) and may serve as a carbon, nitrogen, and energy sources during aerobic or anaerobic growth (Blackwell et al., 1976, Buchrieser et al., 2003, Kofoid et al., 1999).
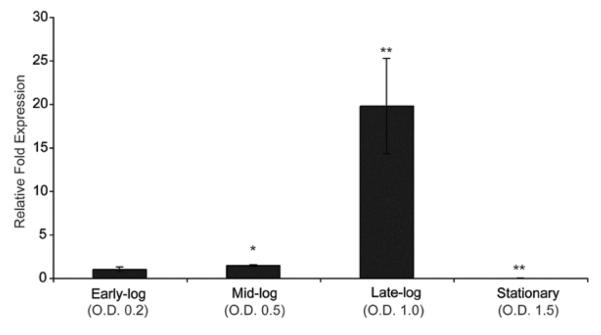
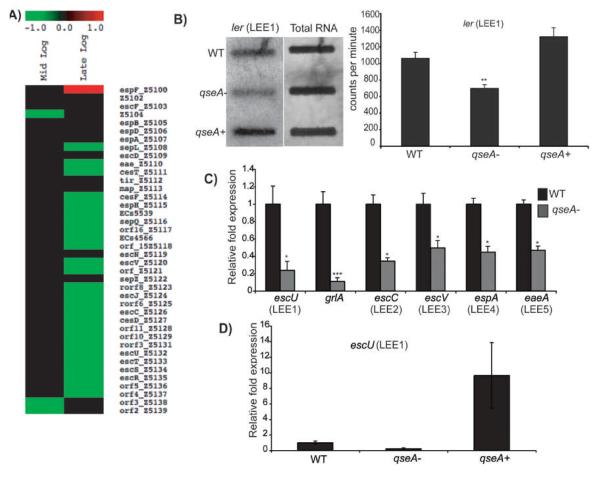
Microarray and qPCR data also revealed that qseA is most highly expressed at late-exponential growth (Fig. 2). Additionally, at late-exponential phase, the array data indicated that genes encoded in LEE1-5, including grlA, were significantly down-regulated (Fig. 3A) in the qseA mutant strain compared to WT. We confirmed that QseA is a positive regulator of the LEE by performing RNA slot blot analysis and by qPCR of ler and escU (both encoded within LEE1), grlA, and LEE1-5 (Fig. 3B, 3C, Table 3). Transcription of ler and escU could be restored to WT levels upon complementation with a functional copy of qseA (Fig. 3B and 3D). These data confirm that QseA promotes expression of the LEE by activating LEE1 and ler, thereby increasing expression of LEE2-5, as well as grlA.
Figure 2.
Transcriptional profile of qseA gene expression for WT EHEC grown to early-, mid-, late-exponential, and stationary growth phases as measured by qPCR and expressed as fold differences normalized to WT strain 86-24 at early-exponential growth phase. The error bars indicate the standard deviations of the ΔΔCT values (Anonymous, 1997). Significance is indicated as follows: one asterisk, P ≤ 0.05; two asterisks, P ≤ 0.005; and three asterisks, P ≤ 0.0005. The levels of rpoA transcript were used to normalize the CT values to account for variations in bacterial numbers.
Figure 3.
A) Heat map from microarray analysis representing LEE genes with altered expression in the qseA mutant compared to WT at mid-exponential and late-exponential growth phases; B) RNA slot blot hybridized with the ler probe. RNA was purified from WT, the qseA mutant, and the qseA complemented strains grown in DMEM to late-exponential phase of growth. The graph corresponds to direct quantification of the concentration of target DNA from three slot blots. Error bars indicate standard deviations; C) Transcriptional profile of LEE gene expression for WT EHEC and the isogenic qseA mutant as measured by qPCR and expressed as fold differences normalized to WT strain 86-24. The error bars indicate the standard deviations of the ΔΔCT values (Anonymous, 1997); D) qRT-PCR measuring gene expression of escU (LEE1) of the WT, qseA mutant, and qseA complemented strains The error bars indicate the standard deviations of the ΔΔCT values (Anonymous, 1997). For the qRT-PCR analyses, the levels of rpoA transcript were used to normalize the CT values to account for variations in bacterial numbers. In all graphs, significance is indicated as follows: one asterisk, P ≤ 0.05; two asterisks, P ≤ 0.005; and three asterisks, P ≤ 0.0005.
Table 3.
Relative change in expression in the qseA mutant strain VS 145 compared to WT strain 86-24.
| Gene | Fold change | P-value |
|---|---|---|
| escU (LEE1) | 4 | 0.0047 |
| grlA | 9 | 0.0005 |
| escC (LEE 2) | 3 | 0.0005 |
| escV (LEE 3) | 2 | 0.004 |
| espA (LEE 4) | 2 | 0.0005 |
| eaeA (LEE 5) | 2 | 0.0005 |
| Z5212 | 4 | 0.0021 |
| Z5213 | 5 | 0.0005 |
| Z5214 | 6 | 0.0027 |
| Z2967 | 83,000 | 0.0003 |
| Z2968 | 1794 | 0.0008 |
| Z2969 | 1224 | 0.0008 |
| Z2971 | 10,711 | 0.0001 |
| Z2972 | 1683 | 0.0001 |
| Z2973 | 65,566 | 0.0001 |
| Z2974 | 3577 | 0.0001 |
| Z2975 | 2592 | 0.0189 |
| Z2976 | 1310 | 0.0087 |
| Z2977 | 487 | 0.0059 |
| Z4181 | 3 | 0.0005 |
| Z4185 | 2592 | 0.0189 |
| Z4189 | 22 | 0.0001 |
| yqeI | 6 | 0.002 |
| ygeK | 36 | 0.0006 |
| Z0326 | 2 | 0.0009 |
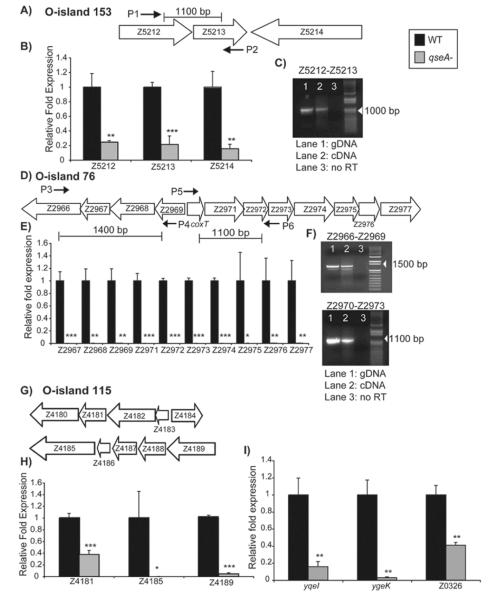
Moreover, the microarray and real-time qPCR analyses indicated that QseA regulated numerous genes located in O-islands, regions of the EHEC genome not found in K-12 (Hayashi et al., 2001, Perna et al., 2001), during late-logarithmic growth. Specifically, the genes encoded in O-island 153 (Fig. 4A) were down-regulated in qseA mutant compared to WT, and we confirmed these data by qPCR (Fig. 4B, Table 3). The genes in O-island 153 encode for the recently identified effector proteins EspX3′ (Z5212 and Z5213) and EspY5′ (Z5214) (Tobe et al., 2006). Reverse transcriptase PCR (RT-PCR) revealed that the Z5212 and Z5213 genes contained in O-island 153 are members of the same operon, and thus are expressed coordinately (Fig. 4C). Tobe et al. also reported that the prophage-like element Sp17 encodes secreted effector proteins (Tobe et al., 2006). Sp17 contains a cluster of genes from Ecs3512 to Ecs3508 that seem to comprise an operon (Abe et al., 2008). The microarray analyses revealed that these genes were down-regulated in the qseA mutant compared to WT.
Figure 4.
A) Schematic representation of O-island 153; B) Transcriptional profiles of Z5212, Z5213, and Z5214 gene expression for WT EHEC and the qseA mutant; C) RT-PCR analysis showing that Z5212 and Z5213 are co-transcribed. Primers flanking Z5212 and Z5213 show product when either the genomic DNA control (Lane 1) or cDNA is used (Lane 2), but show no product when no RT is added (Lane 3); D) Schematic representation showing the arrangement of O-island 76; E) Transcriptional profile of O-island 76 genes for WT EHEC and the qseA mutant; F) RT-PCR showing that Z2966 through Z2969 are co-transcribed and that Z2970 (coxT) through Z2973 are co-transcribed. Controls were set up as described in (C); G) Schematic representation showing the arrangement of O-island 115; H) Transcriptional profiles of Z4181, Z4185, and Z4189 for WT EHEC and the qseA mutant; I) Transcriptional profiles of yqeI, ygek, and Z0326 for WT EHEC and the qseA mutant. In figures B), E), H), and I), gene expression was measured by qPCR and expressed as fold differences normalized to WT strain 86-24. The error bars indicate the standard deviations of the ΔΔCT values (Anonymous, 1997). Significance is indicated as follows: one asterisk, P ≤ 0.05; two asterisks, P ≤ 0.005; and three asterisks, P ≤ 0.0005. The levels of rpoA transcript were used to normalize the CT values to account for variations in bacterial numbers.
Additionally, the microarray data indicated that QseA is involved in regulation of the genes located in the phage-encoded O-islands 76 (Fig. 4D) and 115 (Fig. 4G). The O-island 76 contains 12 genes that are down-regulated in the qseA mutant strain compared to WT (Fig. 4E, Table 3). RT-PCR confirmed that the first four genes in the operon (Z2966-Z2969) were co-transcribed and that the divergent genes (Z2970-Z2973) were also co-transcribed (Fig. 4F). The O-island 115 contains 10 genes, and we analyzed the effect of QseA on expression of three genes contained within this O-island. The real-time qPCR revealed that these genes are down-regulated in the qseA mutant compared to WT (Fig. 4H, Table 3). Many of these genes have not been functionally characterized; however, they are similar to genes that encode secreted effector proteins PrgK, PrgH, and SpaQ in Salmonella, YscS in Yersinia, and Spa9 in Shigella (Collazo & Galan, 1996, Klein et al., 2000, Li et al., 1995, Venkatesan et al., 1992). We also performed qPCR on yqeI, that encodes a predicted transcriptional regulator (Blattner et al., 1997), the gene ygeK that encodes for a response regulator protein (Yamamoto et al., 2005), as well as the prophage-encoded gene Z0326, and the data showed that these genes are down-regulated in the qseA mutant compared to WT (Fig. 4I, Table 3). Together, these analyses further support that QseA is an important regulator of virulence gene expression in EHEC.
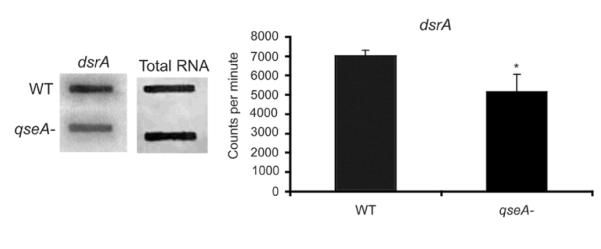
In addition to regulating genes that encode for proteins, the microarray data indicated that QseA regulates expression of small RNAs (sRNAs). More specifically, the sRNA downstream region sRNA (dsrA) was significantly decreased in the qseA mutant strain compared to WT (P ≤ 0.036) in RNA slot blot assays (Fig. 5). Moreover, the microarray data revealed that QseA regulates expression of a dicF and a dicF homolog- a sRNA involved in cell division (Bomchil et al., 2003; Faubladier et al., 1990)- as well as additional intergenic regions (i.e. sRNAs) that have not yet been characterized.
Figure 5.
RNA slot blot hybridized with the dsrA probe. RNA was purified from the WT and the qseA mutant strains grown in DMEM to late-exponential phase of growth. The graph corresponds to direct quantification of the concentration of target DNA from three slot blots. Error bars indicate standard deviations.
The qseA promoter contains two transcription start sites
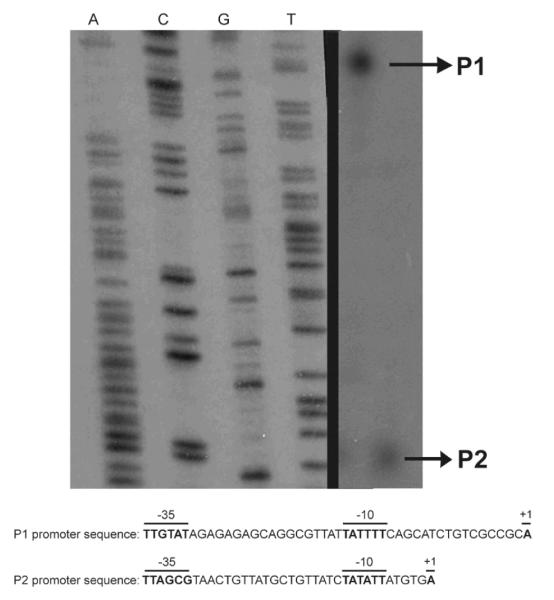
To characterize the mechanism of QseA regulation, we first performed primer extension analysis to determine the qseA transcriptional start site. A primer was designed approximately 100 base pairs downstream from the qseA translational start codon. Then, primer extension analysis was performed using cDNA synthesized from RNA that was purified from WT EHEC, transformed with pMK40, and grown to late-exponential phase in DMEM.
The primer extension results showed that qseA had two transcriptional start sites (TSSs) (Fig. 6A). We mapped the P2 (proximal) promoter TSS to 30 base pairs upstream of the qseA translation start site. For P2, the −10 sequence, TATATT, had one mismatch from the σ70 consensus TATAAT, and the −35 sequence, TTAGCG, had three mismatches from the consensus TTGACA (Fig. 6B). The −10 and −35 promoter sequences were separated by a 19 base pair spacer (Fig. 6B).
Figure 6.
A) The promoter of qseA was mapped using primer extension. The promoter region of qseA contains two promoters, a distal promoter P1 and a proximal promoter P2 responsive to QseA. The proximal (P2) and the distal (P1) promoters, sequences and conserved −10 and −35 regions are shown; B) The +1, −10, and −35 promoter sites are indicated by bold typeface for the P1 and P2 promoters, respectively.
The second promoter identified from primer extension was the P1 (distal) promoter located -125 upstream of the translational start site. In the P1 promoter, the distance between the −10 and −35 sequences was 18 base pairs (Fig. 6B). Four out of six base pairs matched the consensus sequence at the −10 promoter sequence, and three out of six matched with the consensus sequence at the −35 site (Fig. 6B).
QseA directly interacts with both the P1 and P2 ler promoters to activate transcription of ler
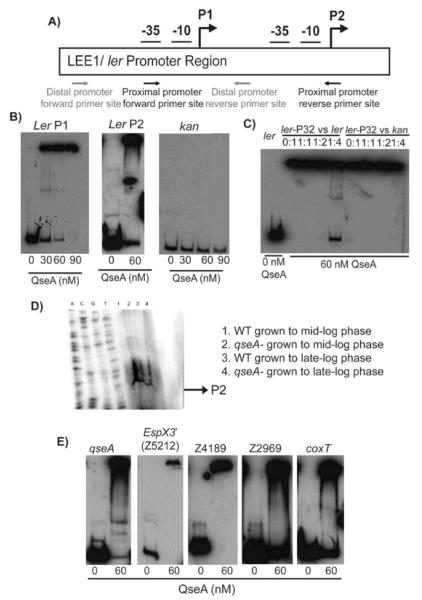
LysR-type transcriptional regulators may activate or repress transcription of its target genes by either direct or indirect action (Schell, 1993). Because microarray and real-time qPCR analyses cannot determine whether QseA regulation is direct or indirect, we performed electrophoretic mobility shift assays (EMSAs) with the promoter regions of several characterized and putative virulence genes identified in the transcriptome analyses. The EHEC LEE1 operon contains two promoters (Fig. 7A), the P1 distal promoter that is found in EPEC and Citrobacter rodentium and the P2 proximal promoter that is unique to EHEC (Deng et al., 2004, Mellies et al., 1999, Sharp & Sperandio, 2007, Sperandio et al., 2002a). Previously, it was shown that QseA binds to the P1 promoter to activate ler expression (Sharp and Sperandio, 2007), and our current work is consistent with those data (Fig. 7B and 8B). Additionally, our study revealed that QseA bound to the P2 promoter (Fig. 7B and 8C). To confirm specificity of binding, competition assays were performed (Fig. 7C). We showed that upon addition of unlabelled probe, QseA binding was competed out by the cold ler P2 promoter probe at a ratio of 1:4 (cold probe: labeled probe, volume/volume). The addition of unlabeled kan probe, as a negative control reaction, shows no competition, indicating that QseA specifically binds its target promoters.
Figure 7.
A) Schematic representation of the LEE1/ ler promoter region, indicating the P1 and P2 promoter regions; B) EMSA of the P1 promoter (−273 to −69 bp), the P2 promoter (−205 to −6), and the kan negative control promoter regions; C) Competition assays with QseA. The assay was performed using 60 nM of QseA in increasing amount of unlabeled ler promoter (−273 to −69 bp) probe. A competition assay was also performed using the kan promoter as a negative control; D) Primer extension of the ler P2 promoter. RNA was harvested from WT EHEC and the qseA mutant strains grown to mid- and late-exponential growth phases. The qseA-dependent promoter is indicated by the arrow; E) EMSAs of QseA with target promoters, including the −175 bp to +125 bp region of the qseA promoter, the −300 bp to +1 bp region (based on the translation start site) of the Z5212 promoter of O-153, the −300 bp to +1 bp region (based on the translation start site) of Z4189 of O-island 76, and the divergently transcribed Z2969 and coxT promoters from O-island 115 (−300 to +1 bp region based on the translation start site).
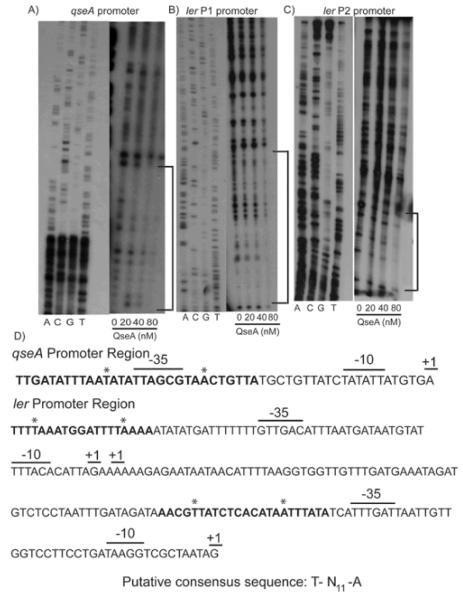
Figure 8.
DNase I footprints. A) DNase I footprint was performed using the qseA promoter (−127 to +125 bp based on the proximal promoter start site) and increasing amounts of QseA; B) DNase I footprint was performed using the ler distal promoter (−382 to −89bp based on the proximal promoter); C) DNase I footprint was performed using the proximal promoter (-205 to −6 bp) and increasing amounts of QseA; D) DNA sequences of the qseA and ler promoters regions including the +1 transcriptional start sites and the −10 and −35 recognition sequences. Bold lettering indicates the QseA binding sites, and asterisks indicate the putative consensus sequences T-N11-A.
Previous work indicated that QseA activated ler through the P1 promoter (Sperandio et al., 2002a); however, QseA interaction with the P2 promoter was not examined. Thus, we further characterized QseA regulation of ler by performing primer extension analysis of the P2 promoter (Fig. 7D). RNA was isolated from WT EHEC and the qseA mutant strains grown to both mid-logarithmic and late-logarithmic growth phases. In this analysis, a weaker band mapped to the P2 transcriptional start site in the qseA mutant RNA isolated from late-exponential growth phase compared to the qseA mutant RNA isolated from mid-exponential growth phase as well as RNA isolated WT EHEC at mid- or late-exponential growth.
QseA regulates transcription of qseA and secreted effectors through direct interaction
LysR regulators generally bind to and repress their own expression (Schell, 1993). To test if QseA binds to its own promoter, we designed primers from −300 to +1 base pairs upstream of the qseA translational start site. Accordingly, QseA binds to its own promoter within 300 nucleotides of the qseA ATG translational start site (Fig. 7E).
QseA was able to bind and shift radiolabeled DNA probes comprising the Z5212 (O-island 153), Z4189 (O-island 115), coxT and Z2969 (O-island 76) promoter regions (Fig. 7E). Thus, these results suggest that QseA activates transcription of Z5212 (O-island 153), Z4189 (O-island 115), coxT and Z2969 (O-island 76) by directly binding to the first gene in the respective operon to increase expression of the downstream genes.
QseA binds upstream of the −35 promoter element when activating ler and binds directly to the qseA −35 promoter element when repressing its own transcription
In order to confirm the results of the EMSA analyses and potentially define a QseA-recognition consensus sequence, we undertook DNase I footprinting experiments of both ler promoters as well as the qseA promoter region. The footprinting analyses indicated that QseA directly bound its −35 P2 promoter element (Fig. 8A, 8D). QseA bound nucleotides -23 to -52 bp with respect to its P2 transcription start site. QseA bound both ler promoters immediately upstream of their respective −35 promoter elements (Figs. 8B, 8C, and 8D). More specifically, QseA protected the promoter region of the ler P2 promoter located at −44 to −66 bp with respect to the P2 transcription start site. In the P1 promoter region, QseA protected the nucleotides located at −203 to −186 bp with respect to the P2 transcription start site.
The generally accepted LTTR (LysR-type transcriptional regulator) box consists of the sequence T-N11-A (Deghmane & Taha, 2003, Goethals et al., 1992, Maddocks & Oyston, 2008, Parsek et al., 1994b, Schell, 1993). We identified this sequence in both the ler promoters as well as in the qseA promoter sequences (Fig. 8D), and current studies using site-directed mutagenesis are being undertaken in order to confirm that these nucleotides are essential for QseA recognition of its target genes.
Discussion
QseA belongs to the family of LysR-type transcriptional regulators. LysR-type transcriptional regulators are the most abundant class of bacterial DNA-binding proteins (Schell, 1993). Early studies suggested that LysR-type transcriptional regulators were involved only in activation expression of a divergently transcribed gene and auto-repressing their own transcription (Lindquist et al., 1989, Maddocks & Oyston, 2008, Parsek et al., 1994a, Schell, 1993). However, characterization of additional LysR-type regulators suggested that these proteins were global regulators, activating or repressing single or operonic genes that may be divergently transcribed or located throughout the chromosome (Deghmane et al., 2002, Deghmane et al., 2000, Hernández-Lucas et al., 2008, Heroven & Dersch, 2006, Maddocks & Oyston, 2008). Our study revealed that QseA regulates genes involved in metabolism, regulation, cell-to-cell signaling, and virulence (summarized in Fig. 10), and thus supports the paradigm that LysR-type proteins act as global regulators. QseA regulation of both K-12 and pathogen-specific genes may allow for integration and maintenance of horizontally-acquired virulence genes without disrupting central metabolic processes, as has been suggested for Pch (Abe et al., 2008).
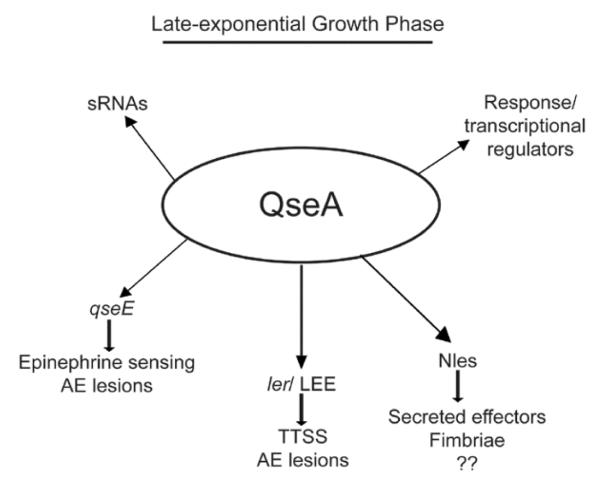
Figure 10.
Summary of QseA gene regulation in EHEC at late-exponential growth phase. Lines with arrows indicate that QseA promotes expression, and lines with bars indicate that QseA down-regulates expression of the respective genes encoding for the phenotypes listed.
Previous studies demonstrated that QseA regulates expression of ler, the master regulator of the LEE (Sharp & Sperandio, 2007, Sperandio et al., 2002a). This study has revealed that QseA regulation of ler affects expression of all the genes in the LEE pathogenicity island, including grlA, another positive regulator of ler (Deng et al., 2004) (Figs. 3A-D). We further characterized the interaction between QseA and ler using EMSA, primer extension, and footprinting analyses. Our results confirmed QseA interaction with the P1 promoter (Sharp & Sperandio, 2007, Sperandio et al., 2002a) (Figs. 7B, 8B). We also showed that QseA interacts directly with the P2 promoter (Figs. 7B, 7D, & 8C). LysR-type transcriptional regulators promote gene expression by typically binding to target sequences near the −35 RNA polymerase binding site (Schell, 1993). QseA is a positive activator of ler and binds to both the P1 and P2 promoter sites immediately upstream of their respective −35 promoter sequences (Fig. 8B, 8C, 9A). QseA activation of ler at both the P2 and P1 promoter sites occurs mainly at late-exponential growth phase (Fig. 7D and (Sperandio et al., 2002a)). In EHEC, the LEE genes are most highly expressed at late-exponential phase (Walters and Sperandio, 2006), thus QseA plays important role in coordinating the timing of LEE gene expression.
Figure 9.
Model of QseA transcriptional regulation in EHEC. A) When QseA act as a positive regulator of gene expression, QseA binds immediately upstream of the σ70 −35 recognition sequence, as occurs on both the ler P2 and P1 promoters; B) QseA represses its its own transcription by binding directly to the σ70 −35 promoter sequence, thereby occluding the RNA polymerase from binding to and transcribing the DNA.
EHEC encodes characterized and putative virulence factors outside the LEE, including fimbriae that may mediate attachment and effectors (Nles) that may enhance virulence (Coburn et al., 2007, Coombes et al., 2008, Deng et al., 2004, Garmendia et al., 2004, Hacker & Carniel, 2001, Hacker & Kaper, 1999, Kaper et al., 1999, Karmali et al., 2003, Shen et al., 2005, Tobe et al., 2006). Our study revealed that QseA is involved in promoting expression of genes encoding some Nles (Fig. 4A-I). QseA activates expression of Nles encoded for in O-islands 153, 76, and 115 by direct interaction with their promoter sequences (Fig. 7E). The O-island 153 encodes secreted proteins (Tobe et al., 2006), and the O-island 115 encodes putative virulence factors similar to the type III secretion system and secreted proteins of other enteric pathogens (Perna et al., 2001). The precise roles that these genes play in EHEC pathogenesis have not yet been elucidated.
Moreover, our study indicates that QseA regulates expression of sRNAs. sRNAs are increasingly being recognized as important in post-transcriptional gene regulation. QseA significantly down-regulated expression of the sRNA dsrA during mid-logarithmic growth and up-regulates this gene during late exponential growth (Fig. 5). In non-pathogenic E. coli, DsrA was initially identified as a regulator of capsule synthesis, and subsequent studies revealed that DsrA binds with Hfq to regulate the translation of two global regulatory proteins, RpoS and H-NS (Sledjeski and Gottesman, 1995). In EHEC, H-NS acts to silence transcription of the LEE, thus if QseA promotes transcription of dsrA, H-NS repression of the LEE might be over-ridden. Indeed, a recent study showed that when overexpressed, DsrA promoted transcription of ler (and thus the LEE) (Laaberki et al., 2006).
Finally, we characterized the molecular mechanism of QseA autoregulation. LysR-type transcriptional regulators generally negatively regulate their own expression (Schell, 1993). A previous study showed that QseA represses its own transcription, and in the current, we undertook EMSA analyses and confirmed that QseA binds to its promoter region to auto-repress transcription (Fig. 7E). Using, primer extension analyses, we determined that the qseA promoter sequence contains two promoter sites (Fig. 6). We determined that QseA binds to the P2 promoter site directly overlapping the −35 promoter sequence (Fig.8A, 8D). It is likely that QseA represses its own transcription by prohibiting RNA polymerase binding to the −35 promoter element (Fig. 9B).
LysR-type transcriptional regulators are the most common type of regulator found within the genomes of bacteria (Schell, 1993), thus better understanding of their contribution to virulence gene expression could lead to insights into disease progression, therapeutics, and vaccine development (Maddocks and Oyston, 2008). This study has elucidated the extensive role that QseA plays in EHEC gene expression and contributes to the understanding of LysR-type transcriptional regulators’ importance in bacterial pathogenesis.
Experimental procedures
Strains and growth media
All bacterial strains used in the study are listed in Table 4. Strains were grown at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) or in LB (Invitrogen). Antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; streptomycin, 50 μg/ml; and μg/ml 50 kanamycin.
Table 4.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Genotype/description | Reference |
|---|---|---|
| Strain | ||
| 86-24 | Wild-type EHEC strain (serotype O157:H7) | (Griffin et al., 1988) |
| BL-21(DE3) | F– ompT hsdSB(rB–, mB–) gal dcm (DE3) | Invitrogen |
| VS145 | 86-24 qseA mutant | (Sperandio et al., 2002a) |
| VS151 | VS145 with plasmid pVS150 (qseA complement) | (Sperandio et al., 2002a) |
| MK11 | EHEC strain 86-24 with plasmid pMK40 | This study |
| MK14 | EHEC strain 86-24 with plasmid pVS23 | This study |
| MK15 | VS145 with plasmid pVS23 | This study |
| Plasmid | ||
| pET28 | Cloning/ expression vector | Novagen |
| pMK08 | EHEC 86-24 qseA in pET28 | This study |
| pVS23 | Regulatory region of LEE1 in pBluescript | (Sperandio et al., 2002a) |
| pRS551 | lacZ reporter gene fusion vector | (Simons et al., 1987) |
| pVS150 | EHEC 86-24 qseA in pACYC177 | (Sperandio et al., 2002a) |
| pMK40 | QseA promoter sequence in pRS551 | This study |
Recombinant DNA techniques
Standard methods were used to perform plasmid purification, PCR, ligation, restriction digests, transformations, and gel electrophoresis. All oligonucleotide primers are listed in Table 5.
Table 5.
Primers used in this study.
| Primers for qPCR and RNA slot blots |
|
|---|---|
| escU_RT_F1 | TCCACTTTGTATCTCGGAATGAAG |
| escU_RT_R1 | CAAGGATACTGATGGTAACCCTGAA |
| escC_RT_F1 | GCGTAAACTGGTCCGGTACGT |
| escC_RT_R1 | TGCGGGTAGAGCTTTAAAGGCAAT |
| escV_RT_F1 | TCGCCCCGTCCATTGA |
| escV_RT_R1 | CGCTCCCGAGTGCAAAA |
| grlA_RT_F1 | CCGGTTGTTCCAGGACTTTC |
| grlA_RT_R1 | TAAGCGCCTTGAGATTTTCATTT |
| espA_RT_F1 | TCAGAATCGCAGCCTGAAAA |
| espA_RT_R1 | CGAAGGATGAGGTGGTTAAGCT |
| eae_RT_F1 | GCTGGCCCTTGGTTTGATCA |
| eae_RT_R1 | GCGGAGATGACTTCAGCACTT |
| espX3′ (Z5212)_RT_F1 |
AACCACGCAGTTCCCCATAA |
| espX3′ (Z5212)_RT_R1 |
GTTAGACAATTTAGAAAAACGATTGAGATG |
| espX3′ (Z5213)_RT_F1 |
GGGACAAATTTTAGCGGTTCTACA |
| espX3′ (Z5213)_RT_R1 |
CGTCCACTTTGTTGGTGTCTTAAT |
| espY5′ (Z5214)_RT_F1 |
CGTCGTACTAAAGCGCCATTT |
| espY5′ (Z5214)_RT_R1 |
ACTGAGGACAAAGTTAAGAGATTTGAGA |
| ygeK_RT_F1 | TTTTCCATACGCATCCTTTCG |
| ygeK_RT_R1 | TTTGTAGCAAAGATGCCGTATATTG |
| yqe1_RT_F1 | GGCCTGAGCACCGAAAATTA |
| yqe1_RT_R1 | CTGGCTGGCGTTGTCAGA |
| Z0326_RT_F1 | GCAGGCTGTTCTGACACCAA |
| Z0326_RT_R1 | CCAAATGATGGCATATTCCTTTTT |
| Z2967_RT_F1 | CAAGACACTGTAATTGAACCTCATCA |
| Z2967_RT_R1 | CCGTCGCAATGCCTTTTAAA |
| Z2968_RT_F1 | CGGATTAATTCTGAAGCACGAA |
| Z2968_RT_R1 | TCGGCATCAAGCCACATAGA |
| Z2969_RT_F1 | CCTTTTCACTGATCGCACCTTT |
| Z2969_RT_R1 | TCGAGCAGCTTCGTTAGATGTG |
| Z2971_RT_F1 | GCCCACGCAATCCGTTT |
| Z2971_RT_R1 | GATTGTCGCCGCAGTTCAA |
| Z2972_RT_F1 | TTGATTTTCGCGGTGAACTG |
| Z2972_RT_R1 | TTTTTTCGGATGGAACAATTCC |
| Z2973_RT_F1 | GTGCAGCCCGTGACTCATT |
| Z2973_RT_R1 | TCAGCGTAAAAAGCAACGTCAT |
| Z2974_RT_F1 | CAGGGCAGCCCAGAAAGATA |
| Z2974_RT_R1 | GTGGCTATTGCGATGGTCAGT |
| Z2975_RT_F1 | CCTGGCGAACCGGAAAC |
| Z2975_RT_R1 | AACGTTATCGATGCGTTGCA |
| Z2976_RT_F1 | CGTCGTCACATCCAGACTGAA |
| Z2976_RT_R1 | TGCGCGACGCGGAT |
| Z2977_RT_F1 | CCTCTGCAATCGTCGATGTG |
| Z2977_RT_R1 | TCGGCATGGCGATGTGT |
| Z4181_RT_F1 | GAGCGGGTCAACGTCTCAA |
| Z4181_RT_R1 | GAGGTTTCATTGGTGGGAACA |
| Z4185_RT_F1 | TGGCAATCATCAAACGGCTAT |
| Z4185_RT_R1 | GAGAGCGCCATCAGGAAATTC |
| Z4189_RT_F1 | ACCAACTCCGGTTCAGAATATTTT |
| Z4189_RT_R1 | CGTCGGTTGTTAATTTTGTTGAGA |
| qseA _RT_F1 | CGCAAGGGAGATTTGTGACTAAT |
| qseA_RT_R1 | GGCACCCGCCGTTAGC |
| rpoA_RT_F1 | GCGCTCATCTTCTTCCGAAT |
| rpoA_RT_R1 | CGCGGTCGTGGTTATCTG |
| ler_dotblot_F1 | GAGATTATTTATTATGAATA |
| ler_dotblot_R1 | TCTTCTTCAGTGTCCTTCAC |
| dsrA_F2 | GCACAATAAAAAAATCCCGA |
| dsrA_R2 | ATTGCGGATAAGGTGATGAAC |
| Primers for RT-PCR | |
| Z2966-69_F1 | TTAATTGCCCATCAGGAAGC |
| Z2966-69_R1 | TTCCTGATGCTGCGTATTTG |
| Z2970-73_F1 | TCAATGTGAATGCCCCCTAT |
| Z2970-73_ R1 | ATGCGATATTCCCCAGCTTA |
| Z5212-Z5213_F1 | GGCACACCACCAAACCACGC |
| Z5212-Z5213_R1 | GCTCTTGCCTTACACTATA |
| Primers for EMSAs/ footprinting |
|
| QseA_pETF1 | CATATGGAACGACTAAAACGCATGTCGGTG |
| QseA_pET28R1 | AAGCTTTTACTTCTCTTTCCCGCGCCCGTG |
| LerProximal_F1 Ler promoter -173 F |
CGGGATCCCGATGATTTTCTTCTATATCATTG |
| LerProximal_R1 Ler promoter -42 R |
CGGAATTCCGCGACCTTATCAGGAAGGACC |
| LerDistal_F2 | CGGAATTCCTGGGGATTCACTCGCTTGC |
| LerDistal_R1 | TCTAATGTGTAAAATACAT |
| QseA_EMSA_F1 | TAGATACCTGTTGGCACAAGTACCCGGCACAC |
| QseA_EMSA_R1 | CATTATTCACTCTGACTTAAAAGTGATTTAGA |
| EspX3′ (Z5212)_EMSA_F1 |
TCCTCCGGCGTGTAATATTG |
| EspX3′ (Z5212)_EMSA_R1 |
TATCCATAAATCACCTTGAT |
| Z4189_EMSA_F1 | GCATCATTACAAACATTGAC |
| Z4189_EMSA_R1 | TGAAACAGATGACTTTGATA |
| Z2969_EMSA_F1 | GTCGCTCATGGCAACTTGAC |
| Z2969_EMSA_R1 | AGCTGTTAGCCAAAGCGTCT |
| coxT_EMSA_F1 | CAATCTTTCCCTTTCCTCAC |
| coxT_EMSA_R1 | TGATGGTTATACCTGACATC |
| kan_EMSA_F1 | CCGGAATTGCCAGCTGGGGCG |
| kan_EMSA_R1 | TCTTGTTCAATCATGCGAAACGATCC |
| amp_EMSA_F1 | GGAATTCGAAAGGGCCTCGTGATACGC |
| amp_EMSA_R1 | CGGGATCCGGTGAGCAAAAACAGGAAGG |
| Primers for primer extension |
|
| qseA_PE_F1 | CACGATAACGGGAAACAGACTCATGTTGACCT |
| qseA_PE_R1 | TGACACCGTCTGACTGATGGACGA |
| K983 | GGACTTGTTGTATGTG |
Plasmid pMK08 was constructed by amplifying the qseA gene from the EHEC strain 86-24 using AccuTaq polymerase (Sigma) using primers QseA_pETF1 and QseA_pET28R1 and cloning the resulting PCR product into the NdeI/HindIII cloning site of vector pET28.
Plasmid pMK40 was constructed by amplifying −200 bp to +100 bp in relation to the qseA translational start site using primers QseA_PE_F1 and QseA_PE_R1. The resulting product was cloned into the BamHI/EcoRI cloning site of pRS551. EHEC strain 86-24 was then transformed with pMK40 to create strain MK11.
RNA extraction
Cultures of strains 86-24 and VS145 were grown aerobically in LB medium at 37°C overnight, and then were diluted 1:100 in DMEM and grown at 37°C. RNA from three biological replicate cultures of each strain/condition was extracted at the early-exponential (OD600 of 0.2), mid-exponential growth phase (OD600 of 0.5), late exponential growth phase (OD600 of 1.0), or stationary growth phase (OD600 of 1.5) using the RiboPure Bacteria RNA isolation kit (Ambion).
Reverse-transcriptase PCR
The SuperScript™ First-Strand Syntheis System for RT-PCR was used to synthesize cDNA. The resulting cDNA was utilized for regular PCR with gene-specific primers that originated in Z2966 and Z2969, Z2970 and Z2973, Z5212 and Z5213, and Z5212 and Z5214 (listed in Table 5). A positive control with genomic DNA and a negative control of RNA without reverse-transcriptase-added were also used.
Microarray preparation and analyses
Affymetrix 2.0 E.coli gene arrays were used to compare gene expression in strain 86-24 to that in strain VS145 (qseA mutant) at both mid- and late-exponential growth phases grown aerobically in DMEM at 37°C. The GeneChip E. coli genome 2.0 array includes approximately 10,000 probe sets for all 20,366 genes present in the following four strains of E. coli: K-12 lab strain MG1655, uropathogenic strain CFT073, O157:H7 enterohemorrhagic strain EDL933, and O157:H7 enterohemorrhagic strain Sakai (http://www.affymetrix.com/products/arrays/specifi/ecoli2.affx). The RNA processing, labeling, hybridization, and slide-scanning procedures were performed as described in the Affymetrix Gene Expression Technical Manual (http://www.affymetrix.com/support/technical/manual/expressionmanual.affx).
The array data analyses were performed as described previously (Kendall et al., 2007). The output from scanning a single replicate of the Affymetrix GeneChip E. coli Genome 2.0 array for each of the biological conditions was obtained using GCOS v 1.4 according to the manufacturer’s instructions. Data were normalized using Robust Multiarray analysis (Irizarry et al., 2003) at the RMAExpress website (http:??rmaexpress.bmbolstad.com/) the resulting data were compared to determine features whose expression was increased or decreased in response to the qseA deletion. Custom analysis scripts were written in Perl to complete multiple array analyses. The results of the array analyses were further confirmed using qPCR as described below. We note that the isolate used in these studies has not been sequenced and thus is not fully contained on the array and that differences in genome content are evident. Expression data can be accessed using accession number GSE18118 at the NCBI GEO database.
Real-time qPCR
RNA was extracted as described above from three biological replicates each of strain 86-24 and strain VS145. The primers used in the real-time qPCR assays were designed using Primer Express v1.5 (Applied Biosystems) (Table 5). Amplification efficiency and template specificity of each of the primer pairs were validated and reaction mixtures were prepared as described previously (Walters and Sperandio, 2006). Real-time reverse transcription-PCR was performed in a one-step reaction using an ABI 7500 sequence detection system (Applied Biosystems).
Data were collected using the ABI Sequence Detection 1.2 software (Applied Biosystems). All data were normalized to levels of rpoA and analyzed using the comparative critical threshold (CT) method (Anonymous, 1997). The expression levels of the target genes under the various conditions were compared using the relative quantification method (Anonymous, 1997). Real-time data are expressed as the changes in expression levels compared to the WT levels. Statistical significance was determined by Student’s t test, and a P value of ≤0.05 was considered significant.
RNA slot blot
RNA was isolated as described above from strains MK14 and MK15 grown at 37°C to mid- (O.D.600, 0.5) and late-exponential (O.D.600, 1.0) phases in DMEM. RNA slot blotting was performed with 2 μg of total RNA in triplicate. The RNA was dissolved in 10 μl RNase-free water and then denatured 30 μl RNA denaturation solution (660 μl formamide, 210 μl of 37% (w/v) formaldehyde, and 130 μl of 10 x MOPS (morpholinepropanesulfonic acid, pH 6.8) at 65°C for 5 minutes. Then, the RNA was applied to a nylon membrane under vacuum by using a Bio-Rad dot blot apparatus. The well were then washed 2 times with 1 ml 10x SSC (1X SSC is 0.15 M NaCl and 0.015 M sodium citrate). The membranes were cross-linked, hybridized with either the ler or dsrA probe by using UltraHyb from Ambion at 42°C, then washed first with 1X SSC, 0.1% SDS (sodium dodecyl sulfate) and then three times with 0.5X SSC, 0.1% SDS. Bound probes were detected using the Amersham PhosphorImager. Liquid scintillation counting was used to quantify the concentration of target DNA.
DNA probes were generated with ler_dotblot_F1 and ler_dotblot_R1 or dsrA_F2 and dsrA_R2 primer sets and radio-labeled using the Ready-To-Go DNA Labeling Beads (GE Healthcare).
Purification of QseA in native conditions
In order to purify the His-tagged QseA protein, the E. coli strain BL-21 (DE3) (Invitrogen) containing pMK08 was grown at 37°C in LB to an OD600 of 0.5, at which point IPTG was added to a final concentration of 0.4 mM and allowed to induce for 3 h. Purification was then performed using nickel columns (Qiagen).
Electrophoretic mobility shift assays (EMSAs)
To determine the direct binding of QseA to target promoters, EMSAs were performed using the purified QseA-His and PCR amplified DNA probes (Table 5). DNA probes were then end-labelled with [γ-32P]-ATP (Perkin-Elmer) using T4 polynucleotide kinase (NEB) using standard procedures (Sambrook et al., 1989). End-labeled fragments were run on a 6% polyacrylamide gel, excised, and purified using the Qiagen PCR purification kit.
EMSAs were performed by adding increasing amounts of purified QseA protein (0-15 μg) to end-labeled probe (10 ng) in binding buffer (500 μg ml−1 BSA (NEB), 50 ng poly-dIdC, 60 mM Hepes pH 7.5, 5 mM EDTA, 3 mM dithiothreitol (DTT), 300 mM KCl, and 25 mM MgCl2) (Clarke and Sperandio, 2005) and incubated for 20 minutes at room temperature. A 5% ficol solution was added to the reactions immediately before loading the samples on the gel. The reactions were electrophoresed for approximately 8 h at 150 V on a 6% polyacrylamide gel, dried, and exposed to KODAK X-OMAT film.
DNAse I footprinting
Dnase I footprints were performed as described previously (Sperandio et al., 2000). Briefly primers LerDistal R2, Ler −42R (Sharp and Sperandio, 2007), and QseA R1 (Table 5) were end labeled by standard procedures using [γ-32P]-ATP (Perkin Elmer). The resulting labeled primers were used in a PCR reaction with unlabeled primers Ler F4, Ler 173F (Sharp and Sperandio, 2007), or QseA F3, respectively. The resulting single-end-labeled PCR products were utilized in binding reactions (described in the EMSAs) with purified QseA for 20 minutes at 25°C. At this time, a 1:100 dilution of DNase I (Invitrogen) and the manufacturer-supplied buffer were added and allowed to digest at room temperature for 4 min. The digestion was stopped by adding 100 μl stop solution (200 mM NaCl, 2 mM EDTA, and 1% SDS). All protein was then extracted via phenol-chloroform, and the DNA was precipitated using 3 M potassium acetate, 100% ethanol, and 1 μl of glycogen. The DNase reactions were run in a 6% polyacrylamide gel next to a sequencing reaction (Epicentre). Amplified DNA from strain 86-24 was used to generate the sequencing ladder using primers LerProximal_F1 and LerProximal_R1, LerDistal_F2 and LerDistal_R1, and QseA_EMSA_F1 and QseA_EMSA_R1.
Primer extension
Primer extension analysis was performed as described previously (Mellies et al., 1999). Briefly, primer QseA_PE_R1 (Table 5), located 100 bp upstream of the ATG translational start site was end-labeled using [γ-32P]-ATP. A total of 40 μg of RNA, isolated was isolated as described above from strain MK11 and was used with the Primer Extension System—AMV Reverse Transcriptase kit (Promega). The resultant cDNA was precipitated, run on a 6% polyacrylamide-urea gel and visualized by autoradiography. A sequencing ladder (Epicentre) was run adjacent to the primer extension reaction. The sequencing ladder was generated from EHEC 86-24 genomic DNA amplified with primers QseA_PE_F1 and QseA_PE_R1.
Primer extension of the ler P2 promoter was done as described above using RNA isolated from strains MK14 and MK15 grown to mid- and late-exponential phases. The sequencing ladder was generated from EHEC 86-24 genomic DNA amplified with primers ler distal F2 and K983.
Acknowledgments
This work was supported by National Institutes of Health Grant AI053067, the Ellison Medical Foundation, the Burroughs Wellcome Fund, and a NIH Ruth L. Kirschstein Fellowship F32AI80115 to M.M.K. Its contents are solely the responsibility of the authors and do not represent the official views of the NIH NIAID.
References
- Abe A, Miyahara A, Oshima T, Tashiro K, Ogura Y, Kuhara S, Ogasawara N, Hayashi T, Tobe T. Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli. DNA Res. 2008;15:25–38. doi: 10.1093/dnares/dsm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. Biogenic amines in lactic acid-fermented vegetables. Lebensm_Wiss U Technol. 1988;21:68–69. [Google Scholar]
- Anonymous . Applied Biosystems Prism 7700 Sequence Detection System: user bulletin #2. Perkin-Elmer Corp.; Norwalk, Conn.: 1997. [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bomchil N, Watnick P, Kolter R. Identification and characterization of a Vibrio cholerae gene, mbA, involved in maintenance of biofilm architecture. J. Bacteriol. 2003;185:1384–1390. doi: 10.1128/JB.185.4.1384-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MB, Sperandio V. Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC) Molecular Microbiology. 2005;58:441–455. doi: 10.1111/j.1365-2958.2005.04819.x. [DOI] [PubMed] [Google Scholar]
- Collier PD, Cromiue DDO, Davies AP. Mechanisms of formation of chloropropanols present in protein hydrolysates. J. Am. Oil Chem. Soc. 1991;68:785–790. [Google Scholar]
- Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O’Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. Dissecting virulence: Systematic and functional analyses of a pathogenicity island. PNAS. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubladier M, Cam K, Bouché JP. Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing. J. Mol. Biol. 1990;212:461–471. doi: 10.1016/0022-2836(90)90325-G. [DOI] [PubMed] [Google Scholar]
- Friedberg D, Umanski T, Fang Y, Rosenshine I. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli 0157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- Hansen A-M, Kaper JB. Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 2009;73:446–465. doi: 10.1111/j.1365-2958.2009.06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuvelink AE, vanHeerwaaden C, Zwartkruis-Nahuis JT, vanOosterom R, Edink K, vanDuynhoven YT, deBoer E. Excherichia coli O157 infection associated with a petting zoo. Epidemiol. Infect. 2002;129:295–302. doi: 10.1017/s095026880200732x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DT, Clarke MB, Yamamoto K, Rasko D, A., Sperandio V. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC) PLoS Pathog. 2009;5:e10000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyoda S, Watanabe H. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157 : H7 to HEp-2 cells. Microbiology. 2004;150:2357–2571. doi: 10.1099/mic.0.27100-0. [DOI] [PubMed] [Google Scholar]
- Iyoda S, Watanabe H. ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J. Bacteriol. 2005;187:4086–4094. doi: 10.1128/JB.187.12.4086-4094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. PNAS. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall MM, Rasko DA, Sperandio V. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 2007;75:4875–4884. doi: 10.1128/IAI.00550-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laaberki MH, Janabi N, Oswald E, Repoila F. Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157:H7: interplay between Ler, GrlA, HNS and RpoS. Int J Med Microbiol. 2006;296:197–210. doi: 10.1016/j.ijmm.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M. Global regulation by CrsA in Salmonella typhimrium. Mol. Microbiol. 2003;48:1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x. al…, e. [DOI] [PubMed] [Google Scholar]
- Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. PNAS. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol. Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol. 2006;61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- Perna NT, Plunkett G.r., Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Rasko D,A, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 2008;190 doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading NC, Torres AG, Kendall MM, Hughes DT, Yamamoto K, Sperandio V. A novel two-component signaling system that activates transcription of an enterohemorrhagic E. coli (EHEC) effector involved in remodeling of host actin. J. Bacteriol. 2007;189:2468–2476. doi: 10.1128/JB.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading NC, Rasko DA, Torres AG, Sperandio V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci U S A. 2009;106:5889–5894. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanatinia H, Kofoid EC, Morrison TB, Parkinson JS. The smaller of two overlapping cheA gene products is not essential for chemotaxis in Escherichia coli. J Bacteriol. 1995;177:2713–2720. doi: 10.1128/jb.177.10.2713-2720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA. Molecular biology of the LysR family of transcriptional regulators. Ann. Rev. of Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- Shakhnovich EA, Davis BM, Waldor MK. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol Microbiol. 2009;74:347–363. doi: 10.1111/j.1365-2958.2009.06856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Zuerner RL. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2004;186:7290–7301. doi: 10.1128/JB.186.21.7290-7301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp FC, Sperandio V. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 2007;75:2432–2440. doi: 10.1128/IAI.02003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Sircili MP, Walters M, Trabulsi LR, Sperandio V. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 2004;72:2329–2337. doi: 10.1128/IAI.72.4.2329-2337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. PNAS. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Mellies JL, Delahay RM, Frankel G, Crawford JA, Nguyen W, Kaper JB. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Molecular Microbiology. 2000;38:781–793. doi: 10.1046/j.1365-2958.2000.02168.x. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Li CC, Kaper JB. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 2002a;70:3085–3093. doi: 10.1128/IAI.70.6.3085-3093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 2002b;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, Younis R, Mattthews S, Marches O, Frankel G, Hayashi T, Pallen MJ. An extensive repertoire of type III secretion effectors in Escherichia coli 0157 and the role of lambdoid phages in their dissemination. PNAS. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M, Sperandio V. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 2006;74:544–5455. doi: 10.1128/IAI.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chaudhuri RR, Constantinidou C, Hobman JL, Patel MD, Jones AC, Sarti D, Roe AJ, Vlisidou I, Shaw RK, Falciani F, Stevens MP, Gally DL, Knutton S, Frankel G, Penn CW, Pallen MJ. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 2004;72:7282–7293. doi: 10.1128/IAI.72.12.7282-7293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]