Abstract
The development of an HIV vaccine will require a more precise understanding of the immunological and virological underpinnings of HIV infection. Magnetofection, the process of magnetizing HIV by coupling it to ferrous nanoparticles and coordinating infection using a magnetic field, synchronizes the viral replication cycle at attachment while recapitulating the events of natural infection. Although spinoculation also concentrates virus onto target cells to increase infection, it does not synchronize infection. The synchronization of HIV infection in vitro facilitates the study of events in the viral replication cycle and the antiviral immune response on timelines previously impossible. Furthermore, by infecting a high percentage of cells in a short time frame, magnetofection increases the throughput of in vitro assays. Once a virus stock is generated, magnetofection of target cells is rapid, requiring only 1-2 hours. Here we present a detailed protocol for this assay and review its applications to studying the immune response to HIV.
INTRODUCTION
Despite over 25 years of research since the identification of HIV as the causative agent of AIDS, the correlates of protection necessary for an effective prophylactic HIV vaccine remain elusive. Indeed, the failure of the Merck STEP trial underscores the need to understand the events of the viral replication cycle and antiviral immune response in greater detail. To this end, the development of new techniques is a necessary step to facilitate discoveries and insights into how best to overcome HIV’s subversion of the immune system. One such technique is magnetofection, which results in a synchronized infection in vitro.
Magnetofection is a generic term applied to the method of using magnetic force to bring vectors, such as nucleic acids, into close proximity to target cells. Indeed, the most widely used applications of this method are DNA transfection and siRNA gene silencing 1 2. Recently, however, several groups have begun to apply the magnetofection technique to HIV research 3-7 8-10 11 12-14 15. In this application of magnetofection, HIV or SIV virons are first rendered magnetically active via electrostatic associations of the viral Envelope protein with cationic ferrous nanoparticles. Next, a magnetic field is used to direct the magnetized virions to the surface of the target cell, thus overcoming the rate-limiting step of infection, which is the initial virus-cell interaction 11 14. A major benefit of this technique is that infecting virions attach to target cells within minutes of the addition of a magnetic field, thus synchronizing the infection at the attachment step of the viral replication cycle.
The synchronization of HIV infection in vitro allows investigators to study events in, and the cellular response to, the viral replication cycle on extremely small and precise timelines. Indeed, this technique has recently been successfully used to investigate the kinetics of SIV peptide epitope presentation to CD8+ 4,5,7 and CD4+ T cells 6, define conformational state dynamics of HIV Env 12, and determine a new parameter to measure the efficiency of neutralizing antibodies 13. Furthermore, because a large percentage of cells are infected in a short amount of time, magnetofection can increase the throughput of in vitro T cell assays which require virally-infected cells 3 15 9,10.
Comparison with spinoculation
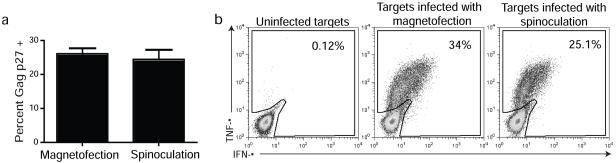
The HIV replication cycle initiates when the virion encounters and binds the CD4 molecule on a target cell with the viral Env glycoprotein gp120/41. Due to the colloidal nature of cell-free virus particles in suspension, the diffusion-dependent step of virus-cell interaction is extremely inefficient and is thus the rate-limiting step in the replication cycle. For example, in a traditional in vitro infection technique where target cells are incubated with virus for multiple hours in a small volume, only a fraction of viruses ever come into contact with cells and thus just a small percentage of cells become infected. Infection techniques that increase the efficiency of the initial attachment step by bringing virus particles into contact with target cells, therefore, result in higher levels of infection. Spinoculation, which uses centrifugal force 16, and magnetofection, which uses magnetic force 14, both increase infection efficiency by concentrating infectious virions onto target cells. Indeed, both methods result in similar levels of infected primary CD4+ T cells (Fig 1a). Furthermore, cells infected with either method behave in a similar fashion and efficiently process and present Gag peptide epitopes to Gag-specific CD8+ T cells (Fig 1b).
Figure 1. Magnetofection versus spinoculation.
(a) Duplicate samples of 1 × 106 activated CD4+ T cells were infected with equal amounts of sucrose-purified virus either by magnetofection or spinoculation. Infection efficiency was measured 24 hours post-infection by performing an intracellular Gag p27 stain. Cells were acquired on a FACSCalibur and the data analyzed using FlowJo software. Data are mean ± sd of duplicate samples from two independent experiments. (b) The activated CD4+ T cells shown in figure 1a were used as antigen presenting cells for Mamu-A*01-restricted Gag254-262 QI9-specific CD8+ T cells in an ICS assay 24 hours post-infection. Cells were acquired on a FACSCalibur and the data analyzed using FlowJo software.
Although similar levels of infection are achieved with both techniques, magnetofection has two major advantages over spinoculation. First, the spinoculation technique is time intensive as cells must be centrifuged with virus for 2 hours, followed by another incubation at 37 °C to allow viral fusion. In contrast, magnetofection is extremely rapid with incubation times as short as 1 minute sufficient for sedimentation of virions onto target cells 14. Secondly, because spinoculation requires a two-hour initial incubation time of virus with cells, the infection is not truly synchronized. While this is not necessary for many in vitro applications, this precludes the use of spinoculation for experiments examining the kinetics of events in the viral replication cycle in precise detail 5,12,13. Therefore, magnetofection is the only method currently available that results in a synchronized infection.
Limitations and applications of magnetofection
One major limitation of magnetofection is the residual presence of a small amount of magnetic nanoparticles following infection. Although we have encountered no problems in downstream applications of cells following magnetofection, the possibility exists that the presence of residual nanoparticles may interfere with very sensitive techniques. Furthermore, depending on the amount of virus used, magnetofection could result in super-physiological amounts of virus entering target cells. Therefore, it is highly recommended that a virus titration be performed to exclude this possibility and extend the results to physiological levels as described elsewhere 4.
The ability to synchronously direct the attachment of viral particles to target cells is a valuable tool in investigations of the viral replication cycle. For example, we have successfully used the technique to pinpoint the exact time required for CD8+ and CD4+ T cell epitopes to reach the cell surface and be presented by the MHC molecule to T cells 4-7. Furthermore, synchronous infections allow detailed investigations of the virus itself. Indeed, the technique has been used to study short-lived conformational states of the Envelope protein 12. These studies are important in understanding the mechanism of HIV entry inhibitors and neutralizing antibodies. Magnetofection, therefore, can be used to study the critical events in the virus replication cycle on a time frame not possible with other infection methods.
Experimental Design
Virus stocks and target cells
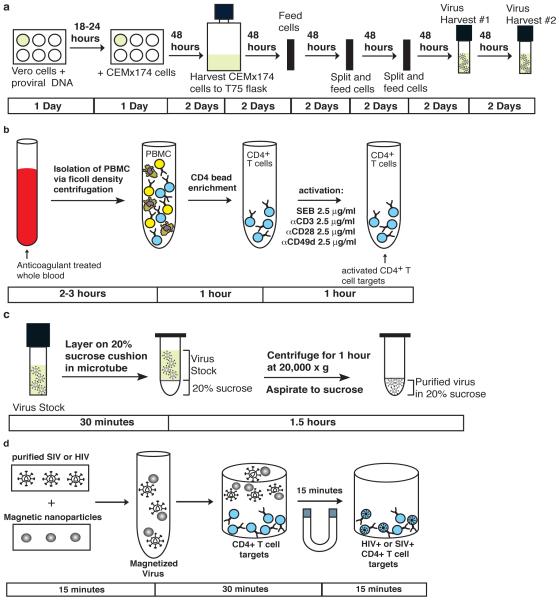
There are two main components of this method that must be prepared prior to beginning the assay. As shown in the experimental overview for magnetofection, they are the virus stock (Fig 2a) and the target cells (Fig 2b).
Figure 2. Schematic overview of the preparation and execution of magnetofection.
(a) Virus stocks are produced by transfecting vero cells and subsequently expanding the virus on CEMx174 cells. The virus-containing supernatant is then harvested, purified, and frozen in aliquots for use in figure 2c. Procedure steps 1 – 20. (b) Primary CD4+ T cells are obtained form whole blood via ficoll centrifugation and CD4 bead enrichment. These cells are then activated as shown. Procedure steps 21 – 30. (c) Virus stocks are thawed and then purified over a 20% sucrose cushion. Procedure steps 31 – 36. (d) The target cells generated in figure 2b are magnetofected with the purified virus from figure 2c. Procedure steps 37 – 47.
Many lab-adapted and lab-isolated HIV and SIV virus variants are available as provirus-containing plasmids from reagent programs such as the NIH AIDS Research and Reference Reagent Program. Protocols to isolate and propagate clinical isolates of HIV are described elsewhere 17. The optimal choice of virus depends on the experimental parameter being investigated and should be chosen accordingly. For generating a virus stock starting from DNA, vero cells are first transfected with a plasmid containing the provirus of interest. CEMx174 cells are then overlaid on the virus-producing vero cells to initiate infection of the CEMx174 culture, which is subsequently expanded for twelve days. Finally, the virus-containing culture supernatant is purified of contaminating cells, aliquoted, and stored at −170 C in liquid nitrogen. Using this method, a large-scale, high titer virus stock of up to 1 L can be produced. Once aliquoted and stored in liquid nitrogen, these virus stocks are viable with no loss of infectivity for up to 5 years as described elsewhere 17. Because of the long-term stability of frozen virus stocks, this step can be performed at any time at least two weeks prior to performing magnetofection.
The choice of target cells is also dependent on the exact research parameter being investigated. Indeed, magnetofection has been used to infect primary CD4+ T cells 4, monocyte-derived macrophages 6, and immortalized CD4+ T cell clones 8. Because primary CD4+ T cells are the most broadly applicable and physiologically relevant cell type for in vitro assays, we will focus on the generation of these cells in this protocol (Fig 2 b). Infection of other cell types is performed exactly as described here. Furthermore, CD4+ T cell targets isolated and activated according to this protocol will propagate for up to a month in culture. Therefore, CD4+ T cell targets can be isolated and activated anywhere from the day before to one month prior to performing magnetofection. However, depending on whether activated or resting CD4+ T cells are desired as targets will ultimately determine when the cells are used post activation.
Once the virus stock (Fig 2a) and target cells (Fig2b) are prepared, magnetofection can be performed. First, the virus stock is purified by centrifugation over a 20% sucrose cushion in a tabletop microcentrifuge (Fig 2c). This results in a very pure and concentrated virus solution for infection. Next, the purified virus is incubated with the magnetic nanoparticles for 15 minutes and then brought into contact with the target cells via magnetic force (Fig 2d). As a negative control, target cells should be incubated with magnetic nanoparticles alone.
MATERIALS
Reagents
ViroMag beads (OZ Biosciences, cat. No. VM40100, VM40200, VM41000)
20% Sucrose solution (see reagent setup)
R10 media (see reagent setup)
R15-100 media (see reagent setup)
1x ACK red blood cell lysis media (see reagent setup)
RPMI 1640 media (Thermo Fisher, cat. No. SH30255.01)
Fetal bovine serum (FBS) (Thermo Fisher, cat. No. SH30070.03)
L-glutamine (100x) (Thermo Fisher cat. No. SH30034.01)
Antibiotic/antimycotic (100X) (Thermo Fisher, cat. No. SV3007901)
EDTA (0.5 M solution) (Gibco cat. No. 15575-020)
Ultracentrifuge-grade sucrose (Fisher cat. No. S5-500)
Trypsin (0.25%) (Thermo Fisher, cat. No. SH30042.01)
Recombinant human interleukin-2 (Proleukin) (Novartis)
CD4+ T cell microbead kit (Miltenyi Biotec)
-
Staphylococcal Enterotoxin B (SEB, 1 mg) (Sigma, cat. No. S4881)
CAUTION: SEB is a T cell superantigen and exposure via inhalation or ingestion can result in vomiting, pulmonary edema, and toxic shock syndrome. It is recommended to wear gloves and a surgical mask when handling SEB.
Anti-CD3 antibody (unconjugated, clone SP34-2) (BD, cat. No. 551916)
Anti-CD4 antibody (APC conjugated) (Miltenyi Biotec, cat. No. 130-091-232)
Anti-CD28 antibody (unconjugated, clone L293) (BD, cat. No 348040)
Anti-CD49d antibody (unconjugated, clone 9F10) (BD, cat. No 555502)
Anti-SIV Gag p27 or anti-HIV Gag p24 antibody (according to pathogen) (NIH AIDS Research and Reference Reagent Program)
Sterile PBS (1x DPBS, Cellgro, Mediatech Inc., cat. No. F4135)
Ficoll-Paque Plus (GE Healthcare, cat. No. 17-1440-03)
0.4% Trypan blue (wt/vol) in saline solution (Cellgro, Mediatech Inc., cat. No. 25-900-CI)
Vero cells (ATCC, cat. No. CCL-81)
CEMx174 cells (ATCC, cat. No. CRL-1992)
Lipofectamine 2000 transfection reagent (Invitrogen, cat. No. 11668-019)
-
Plasmid encoding HIV/SIV provirus of interest (NIH AIDS Research and Reference Reagent Program)
CAUTION: These plasmids encode infectious viruses that cause immunodeficiencies. It is recommended that work with these plasmids be conducted in a biosafety level 2 plus facility following level 2 plus safety measures. Biosafety level 2 plus requires at minimum the use of a biosafety level 2 facility with containment equipment, training, planning, and practices of a biosafety level 3. Thus, the work described in this protocol should be performed under the guidelines of a biosafety level 2 plus laboratory. All waste should be considered infectious and thus autoclaved and liquid waste should be inactivated with bleach.
1.8 ml cryotube vial (Fisher scientific, cat. No. 12-566-97)
Equipment
Super Magnetic Plate (OZ Biosciences, cat. No. MF10000)
Class I biological safety cabinet
Eppendorf microcentrifuge (Ependorf cat. No. 5407 000.317)
15-ml conical tubes (Fisher Scientific, cat no. 14-959-70C)
6-well and 24-well sterile plates (Fisher Scientific, cat. No. 07-200-84)
Vented T25 and T75 cell culture flasks (Fisher Scientific, cat. No. 10-126-28)
Disposable sterile 0.22 μm filter flasks (Fisher Scientific, cat. No. SCGP-U05-RE)
-
Sarstedt 1.5 ml screw cap micro tubes (Fisher Scientific, cat. No. 02-681-320)
CRITICAL: Tubes other than screw cap tubes may leak and present a safety risk. Therefore, use only screw-cap tubes.
Transfer pipettes (Fisher Scientific, cat. No. 13-711-20)
LS Columns (Miltenyi Biotec, cat. No. 130-042-401)
QuadroMACS separator (Miltenyi Biotec, cat. No. 130-091-051)
Hemacytometers (Hycor Biomedical, cat. No. 87144)
Reagent Setup
20% Sucrose solution: 2 mM EDTA in 1x PBS and supplemented with 20% sucrose (wt/vol). For 250 mls, dissolve 50 g of sucrose in 249.9 mls of warm 1x PBS. Mix until sucrose crystals fully dissolve. Add 104 ul of 0.5 M EDTA and mix. Sterile filter using disposable vacuum filter flask and store at 4 °C. Can be stored at 4 °C for up to one year.
SEB: Resuspend lyophilized SEB to 500 ug/ml using 2 mls of 1x PBS and divide into 20 ul aliquots. Can be stored at −20 °C for up to one year.
IL-2: Resuspend lyophilized IL-2 to 500,000 IU/ml in sterile 1x PBS and freeze in 50 ul aliquots. Resuspended IL-2 can be stored at −20 °C for up to one year.
R10 media: To 500 ml RPMI, add 55 ml FCS, 5.5 ml L-glutamine, 5.5 ml antibiotic/antimycotic. Sterile filter using vacuum filter flask. Can be stored at 4 °C for up to three weeks.
R15 media: To 124 ml RPMI, add 22.5 ml FCS, 1.5 ml L-glutamine, 1.5 ml antibiotic/antimycotic. Sterile filter using vacuum filter flask. Can be stored at 4 °C for up to three weeks.
R15-100 media: Add 10 ul IL-2 to 50 mls R15 media. Can be stored at 4 °C for up to one week.
1x ACK: Prepare a 20x ACK stock by adding 82.9 g NH4Cl, 10 g KCO3, and 4.2 g NA2 EDTA to 500 ml ddH20. The 20x ACK stock is stabile for up to a year at room temperature. Prepare 1x ACK by adding 25 mls 20x ACK to 475 mls ddH20 and sterile filter. 1x ACK can be stored for up to one year at room temperature (20°C – 25° C).
PROCEDURE
Production of high titer virus for in vitro studies (14 days)
1| Seed 5 × 105 vero cells into one well of a 6 well plate in 3 mls of R10 media the day before beginning production of virus. Incubate overnight in humidified incubator at 37 °C.
2| Aliquot 250 μl serum-free medium (SFM) into two 15 ml conical tubes. Add 5 μg of virus-encoding DNA plasmid to SFM in one tube and vortex. To the other tube of SFM, add 10 μl of lipofectamine 2000 reagent and vortex. Incubate at room temperature for 5 minutes.
3| Add the DNA-containing SFM to the other tube and vortex to mix. Incubate at room temperature for 20 minutes.
4| Add the DNA/lipofectamine mixture to the vero cells drop wise, distributing the drops all over the surface of the well. Rock the plate gently to mix. CRITICAL STEP: Do not swirl to mix as this may concentrate the transfection reagent in the middle of the wells and lead to toxicity to the cells.
5| Incubate transected vero cells overnight in humidified incubator at 37 °C.
6| Centrifuge 2 × 106 CEMx174 cells for 8 minutes at 500g at room temperature to pellet. Decant supernatant and resuspend CEMx174 cells in 4 ml R10 media.
7| Remove culture supernatant from virus-producing vero cells and add the 4 ml of R10 media containing the CEMx174 cells.
8| Incubate cells for 24 hours in humidified incubator at 37 °C to initiate infection of CEMx174 cells.
9| Add 3 ml R10 media to feed cells. Incubate 24 hours in humidified incubator at 37 °C.
10| Collect the culture supernatant containing the CEMx174 cells using a 5 ml pipette. Tilt the plate and slowly wash the vero cells to dislodge any CEMx174 cells that might be stuck. CRITICAL STEP: Be careful not to dislodge vero cells.
11| Transfer CEMx174 cells to a T75 flask and add 14 ml R10 media. Total culture volume should now be 21 ml. Incubate 48 hours in humidified incubator at 37 °C.
12| Feed cells by adding 25 ml of R10 media, bringing total volume to 46 ml. Incubate 48 hours in humidified incubator at 37 °C.
13| Resuspend CEMx174 cells and remove and discard 23 ml culture. Add 23 ml fresh R10 media. Incubate 48 hours in humidified incubator at 37 °C.
14| Resuspend CEMx174 cells and remove 23 ml culture. If a larger volume of virus stock is desired, then transfer the freshly harvested 23 mls of CEMx174-containing media to a new T75 to expand the culture. Otherwise, simply discard the freshly harvested 23 mls of CEMx174-containing media. Add 23 ml fresh R10 media to bring T75 flask(s) to feed cells. Incubate 48 hours in humidified incubator at 37 °C.
15| To harvest the virus-containing supernatant, resuspend the cells and transfer culture to 50 ml conical tubes and centrifuge for 8 minutes at 500g at room temperature to pellet cells.
16| Pool cell-free supernatants and filter through 0.22 μm filter flask. Aliquot filtered supernatant into 1.8 ml cryotubes (aliquots of 1 ml) and store in liquid nitrogen freezer.
17| Resuspend CEMx174 cells in fresh R10 media to original culture volume, return to T75 flask, and incubate 48 hours in humidified incubator at 37 °C.
18| Transfer culture to 50 ml conical tubes and centrifuge for 8 minutes at 500g at room temperature to pellet cells.
19| Pool cell-free supernatants and filter through 0.22 μm filter flask. Aliquot filtered supernatant into 1.8 ml cryotubes (aliquots of 1 ml) and store in liquid nitrogen freezer. PAUSE POINT: Virus can be stored in liquid nitrogen for up to 10 years without significant loss of infectivity.
20| Test one aliquot from each harvest for vRNA copy number in QRT-PCR as described in detail elsewhere18.
? TROUBLESHOOTING.
Isolation and activation of CD4+ T cell targets (4 - 6 hours)
21| Slowly layer anticoagulant-treated blood on top of 5 ml of Ficoll-Paque in a 15 ml conical tube.
22| Centrifuge for 30 minutes at 1000g at room temperature with the centrifuge brake off.
23| Collect the band of cells that sits directly on top of the Ficoll layer with a transfer pipette and transfer to a new 15 ml conical tube. Fill conical to 15 mls with 1x PBS. Centrifuge for 8 minutes at 500g at room temperature.
24| Decant supernatant and resuspend cell pellet in 5 mls of 1x ACK buffer. Incubate at room temperature for 5 minutes to lyse residual red blood cells. Fill tube to 15 mls with 1x PBS to stop reaction. Centrifuge for 8 minutes at 500g at room temperature.
25| Decant supernatant and resuspend cells in R10 media. Count cells using an automated cell counter or trypan blue and a hemacytometer. PAUSE POINT: PBMC in R10 media can be incubated in a vented T25 or T75 flask in humidified incubator at 37 °C for up to four days.
26| Isolate CD4+ T cells using the Miltenyi CD4+ T cell microbead enrichment kit according to the manufacturer’s instructions.
27| (Optional) Verify purity of CD4 enrichment by staining positively selected CD4+ T cells with 2.5 μl CD4-APC for 30 minutes.
28| Resuspend positively enriched CD4+ T cells in R15-100 to a concentration of 5 × 106/ml. Add 5 μl anti-CD3 Ab, 5 μl anti-CD28, 5 μl anti-CD49d, and 10 μl SEB per ml of enriched CD4+ T cells. Mix well and aliquot into a 24-well plate.
29| Incubate overnight (or 18-24 hours) in humidified incubator at 37 °C.
30| Harvest cells by pipetting up and down to resuspend cells and wash once with warm 1 x PBS. Resuspend cells in 5 mls R15-100 and culture in vented T25 culture flask in humidified incubator at 37 °C. PAUSE POINT: Target cells can be used for infections as soon as the following day or as late as four weeks later. For optimal infection, however, cells should be used five to ten days after activation and fed as media yellows.
? TROUBLESHOOTING
Purification of virus (1.5 – 2 hours)
31| Set microcentrifuge to cool to 4 °C.
32| Aliquot 100 μl 20% sucrose solution into Sarstedt microcentrifuge tube.
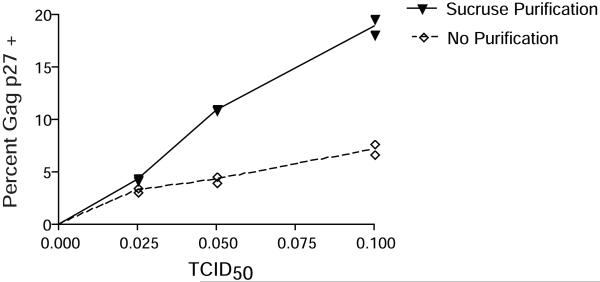
33| Carefully layer 1 ml of virus stock onto the sucrose. Up to 1.2 ml of virus stock can safely be layered onto sucrose solution. (Using 1 ml of virus stock is meant only to provide a starting point. Each virus stock should be titrated to determine the optimal level of virus per experimental needs. To this end, perform a titration of the amount of infecting virus used. For example, infect a fixed number of cells with 10-fold dilutions of sucrose-purified virus. Monitor infection efficiency via intracellular Gag p27 staining to determine virus needed for experimental purposes.) The effects of sucrose purification can be seen in Figure 3.
Figure 3. Sucrose purification is necessary to obtain high levels of infection in vitro.
Duplicate samples of 1 × 106 activated CD4+ T cells were magnetofected with increasing amounts of SIVmac239, as measured by TCID50, that were either unpurified (◇) or sucrose purified (▼). Infection efficiency was measured 24 hours post-infection by performing an intracellular Gag p27 stain. Cells were acquired on a FACSCalibur and the data analyzed using FlowJo software. Data points are duplicate samples and indicative of three independent experiments.
CRITICAL STEP: Mixing of the virus with sucrose will result in inefficient purification of the virus and thus lower levels of infection. Refer to figure 3 for the importance of the purification step.
34| Centrifuge in microcentrifuge for 1 hour at 20,000 x g at 4 °C.
35| Carefully aspirate all of the culture supernatant sitting on top of the sucrose solution. CRITICAL STEP: Residual culture supernatant will reduce infection efficiency.
36| Resuspend virus-containing sucrose solution with 100 μl cold R10 media. Pipette up and down to mix.
Magnetofection of activated CD4+ T cells (1 hour)
37| Vortex ViroMag beads and add 10 μl of ViroMag (VM) beads to virus-containing sucrose. Pipette up and down to mix. (While 10 ul of VM beads is a good starting point and is sufficient to bind up to 3 × 108 viral particles, the optimal ratio of virus to VM beads should be determined for each virus stock, amount of virus, and cell type. Perform infections using a fixed amount of cells and virus incubated with varying amounts (i.e. 1 μl to 20 μl) of VM beads to determine optimal ratio.) For mock infection, add VM beads to 100 μl cold R10.
38| Incubate at room temperature for 15 minutes.
39| While the virus is incubating, count target CD4+ T cells using an automated counter or trypan blue exclusion. Place between 1 × 106 and 2 × 106 in a well of a 24 well plate. Although volume does not affect infection efficiency, allow enough volume for addition of virus/VM bead mixture.
40| Add virus/VM bead mixture to target CD4+ T cells and pipette to mix. Centrifuge for 2 minutes at 500g at room temperature to pellet cells.
41| Carefully remove plate and place on ViroMag magnetic plate.
42| Incubate for 15 minutes in humidified incubator at 37 °C.
43| Harvest infected CD4+ T cells by resuspending cells in warm 1x PBS. Transfer to fresh microcenterfuge tube. Wash out the well again with 1 x PBS to ensure all cells are harvested. Centrifuge in microcenterfuge for 5 minutes at 500g at room temperature to pellet cells.
44| (Optional) To remove surface-bound virions that failed to enter target cells, aspirate supernatant and resuspend cells in 1 ml trypsin solution. Incubate for 2 minutes in humidified incubator at 37 °C.
45| Aspirate supernatant and resuspend cells in 1 ml warm 1 x PBS to wash. Centrifuge for 5 minutes at 500g in microcenterfuge at room temperature to pellet cells.
46| Repeat wash in step 45.
47| Resuspend cells in 1 ml R15-100 and count using an automated counter or trypan blue exclusion. Plate cells in 24 well plate. Infected cells may be used immediately or incubated in humidified incubator at 37 °C until needed.
? TROUBLESHOOTING
TIMING
Steps 1 – 20, Producing high titer virus stocks: 14 days
Steps 21 – 30, Isolation and activation of CD4+ T cells: 4-6 hours
Steps 31 – 36, Purification of virus: 1.5 – 2 hours
Steps 37 – 47, Magnetofection of activated CD4+ T cells: 1 hour
ANTICIPATED RESULTS
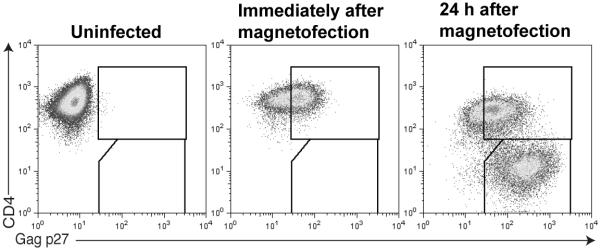
The process described here will result in a synchronous infection of a large percentage of target cells in vitro for downstream applications. For instance, these cells can be used as antigen presenting cells to study epitope presentation to T cells or be examined directly for events in the viral replication cycle such as Nef-mediated MHC-I down-regulation. Importantly, because the infection is synchronized at the step of attachment of the virion to the cell, the infected cells are ready for use directly following the end of this protocol. Immediately following magnetofection, the infected population can be visualized, for example, by staining for intracellular HIV/SIV Gag p27CA. As shown in figure 4, the infected cells are initially located within the CD4+, Gag p27+ gate due to the presence of Gag proteins from the infecting virion in the cell cytoplasm. By 24 hours post-infection, a distinct population of productively infected cells is detectable in the CD4-, Gag p27+ gate. The cells can be cultured up to an additional 48 hours per assay needs and, importantly, the percentage of infected cells will increase as each synchronized wave of progeny virions is produced and released.
Figure 4. Typical intracellular Gag p27 staining patterns following magnetofection.
CD4+ T cell targets were stained using CD4-APC and Gag p27-FITC before, directly following, or 24 hours following magnetofection. Cells were acquired on a FACSCalibur and analyzed using FlowJo software.
Table 1. TROUBLESHOOTING.
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 20 | Low virus yields or non-infectious virus |
Unhealthy CEMx174 culture |
Ensure CEMx174 cells are healthy at start of protocol. Healthy CEMx174 cells form cell clumps visible to the naked eye. Produce new stock using healthy CEMx174 cells. If problem persists, obtain a new CEMx174 passage. |
| Slow/fast in vitro viral growth kinetics |
Monitor CEMx174 cell culture by microscopy every day for formation of syncytia. Harvest virus when >50 of cell clumps have syncytia. |
||
| 30 | CD4+ T cells not growing well |
Cells not activated properly |
Thoroughly mix cell culture following addition of stimulation cocktail. |
| 47 | Target cells poorly infected |
Improper ratio of virus and VM beads |
Titrate virus and VM bead ratio to obtain optimal ratio. |
| VM beads not vortexed prior to use/no VM beads added |
Vortex VM beads prior to use and ensure addition to virus. Cells should appear slightly brown under microscope following magnetofection. |
||
| Virus stock poorly infectious. |
Determine infectivity of virus by incubating with activated CD4+ T cells and culturing for 7 days. |
||
| Cells not at bottom of well during magnetofection. |
Be extremely careful when removing plate from centrifuge prior to placement on magnet. Ensure cells at bottom of well following 15 minutes incubation on magnet by observing through microscope. If cells not at bottom, centrifuge again. |
Acknowledgments
This work was supported by NIH grants RO1 AI076114 and R01 AI049120 to D.I.W. We thank Matthew Buechler and Laura Newman for technical assistance and Amanda Espinosa for administrative support.
Footnotes
The authors declare they have no competing financial interests.
REFERENCES
- 1.Mykhaylyk O, Antequera YS, Vlaskou D, Plank C. Generation of magnetic nonviral gene transfer agents and magnetofection in vitro. Nat Protoc. 2007;2:2391–2411. doi: 10.1038/nprot.2007.352. [DOI] [PubMed] [Google Scholar]
- 2.Buerli T, et al. Efficient transfection of DNA or shRNA vectors into neurons using magnetofection. Nat Protoc. 2007;2:3090–3101. doi: 10.1038/nprot.2007.445. [DOI] [PubMed] [Google Scholar]
- 3.Migueles SA, et al. Lytic Granule Loading of CD8(+) T Cells Is Required for HIV-Infected Cell Elimination Associated with Immune Control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sacha JB, et al. Gag-Specific CD8+ T Lymphocytes Recognize Infected Cells before AIDS-Virus Integration and Viral Protein Expression. J Immunol. 2007;178:2746–2754. doi: 10.4049/jimmunol.178.5.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacha JB, et al. Pol-specific CD8+ T cells recognize simian immunodeficiency virus-infected cells prior to Nef-mediated major histocompatibility complex class I downregulation. J Virol. 2007;81:11703–11712. doi: 10.1128/JVI.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacha JB, et al. Gag- and Nef-specific CD4+ T cells recognize and inhibit SIV replication in infected macrophages early after infection. Proc Natl Acad Sci U S A. 2009;106:9791–9796. doi: 10.1073/pnas.0813106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sacha JB, et al. Differential antigen presentation kinetics of CD8+ T-cell epitopes derived from the same viral protein. J Virol. 2008;82:9293–9298. doi: 10.1128/JVI.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minang JT, et al. Efficient inhibition of SIV replication in rhesus CD4+ T-cell clones by autologous immortalized SIV-specific CD8+ T-cell clones. Virology. 2008;372:430–441. doi: 10.1016/j.virol.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 9.Minang JT, et al. The Mamu B 17-restricted SIV Nef IW9 to TW9 mutation abrogates correct epitope processing and presentation without loss of replicative fitness. Virology. 2008;375:307–314. doi: 10.1016/j.virol.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minang JT, et al. Nef-mediated MHC class I down-regulation unmasks clonal differences in virus suppression by SIV-specific CD8(+) T cells independent of IFN-gamma and CD107a responses. Virology. 2009;391:130–139. doi: 10.1016/j.virol.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas JA, Ott DE, Gorelick RJ. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J Virol. 2007;81:4367–4370. doi: 10.1128/JVI.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haim H, et al. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haim H, Steiner I, Panet A. Time frames for neutralization during the human immunodeficiency virus type 1 entry phase, as monitored in synchronously infected cell cultures. J Virol. 2007;81:3525–3534. doi: 10.1128/JVI.02293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haim H, Steiner I, Panet A. Synchronized infection of cell cultures by magnetically controlled virus. J Virol. 2005;79:622–625. doi: 10.1128/JVI.79.1.622-625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maness NJ, et al. AIDS virus specific CD8+ T lymphocytes against an immunodominant cryptic epitope select for viral escape. J Exp Med. 2007;204:2505–2512. doi: 10.1084/jem.20071261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van ’t Wout AB, Schuitemaker H, Kootstra NA. Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat Protoc. 2008;3:363–370. doi: 10.1038/nprot.2008.3. [DOI] [PubMed] [Google Scholar]
- 18.Loffredo JT, et al. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. J Virol. 2007;81:2624–2634. doi: 10.1128/JVI.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]