Abstract
Bacteria show remarkable adaptability in the face of antibiotic therapeutics. Resistance alleles in drug target-specific sites and general stress responses have been identified in individual endpoint isolates1–7. Less is known, however, about the population dynamics during the development of antibiotic-resistant strains. Here we follow a continuous culture of Escherichia coli facing increasing levels of antibiotic and show that the vast majority of isolates are less resistant than the population as a whole. We find that the few highly resistant mutants improve the survival of the population’s less resistant constituents, in part, by producing indole, a signaling molecule generated by actively growing, unstressed cells8. We show, through transcriptional profiling, that indole serves to turn on drug efflux pumps and oxidative stress protective mechanisms. The indole production comes at a fitness cost to the highly resistant isolates, and whole-genome sequencing reveals that this bacterial altruism is enabled by drug-resistance mutations unrelated to indole production. This work establishes a population-based resistance mechanism constituting a form of kin selection9 whereby a small number of resistant mutants can, at some cost to themselves, provide protection to other more vulnerable cells, enhancing the survival capacity of the overall population in stressful environments.
Antibiotic-resistant bacterial strains continually arise and their increasing prevalence poses significant clinical and societal challenges7,10. Functional analyses of resistant mutants and the study of the endogenous processes responsible for resistance by mutation have yielded valuable insights1–7,11,12. However, population dynamics and communal interactions that underlie the development of resistance through mutations are often overlooked. To study these neglected aspects, we tracked a bacterial population as it developed antibiotic resistance in a bioreactor.
Starting with an isogenic strain of wildtype Escherichia coli, we continuously challenged the population to progressively increasing concentrations of norfloxacin. To provide evolutionary pressure while maintaining a sizeable population, the concentration of antibiotic was chosen such that no more than 60% of growth was inhibited; we defined this concentration as the minimum inhibitory concentration (MIC). Every 24 hours, the daily population MIC was determined and the norfloxacin concentration adjusted as tolerated. From each daily sample, 12 individual isolates were randomly chosen and each of their MICs was determined.
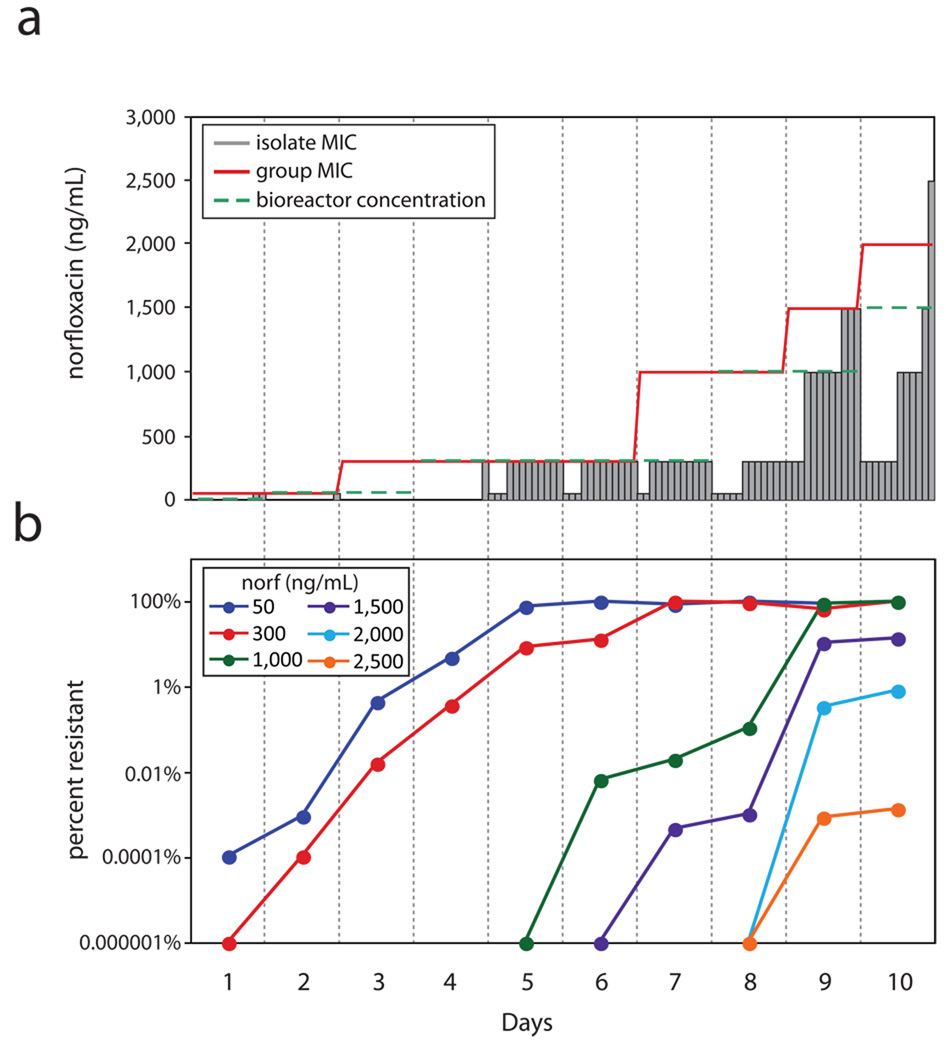
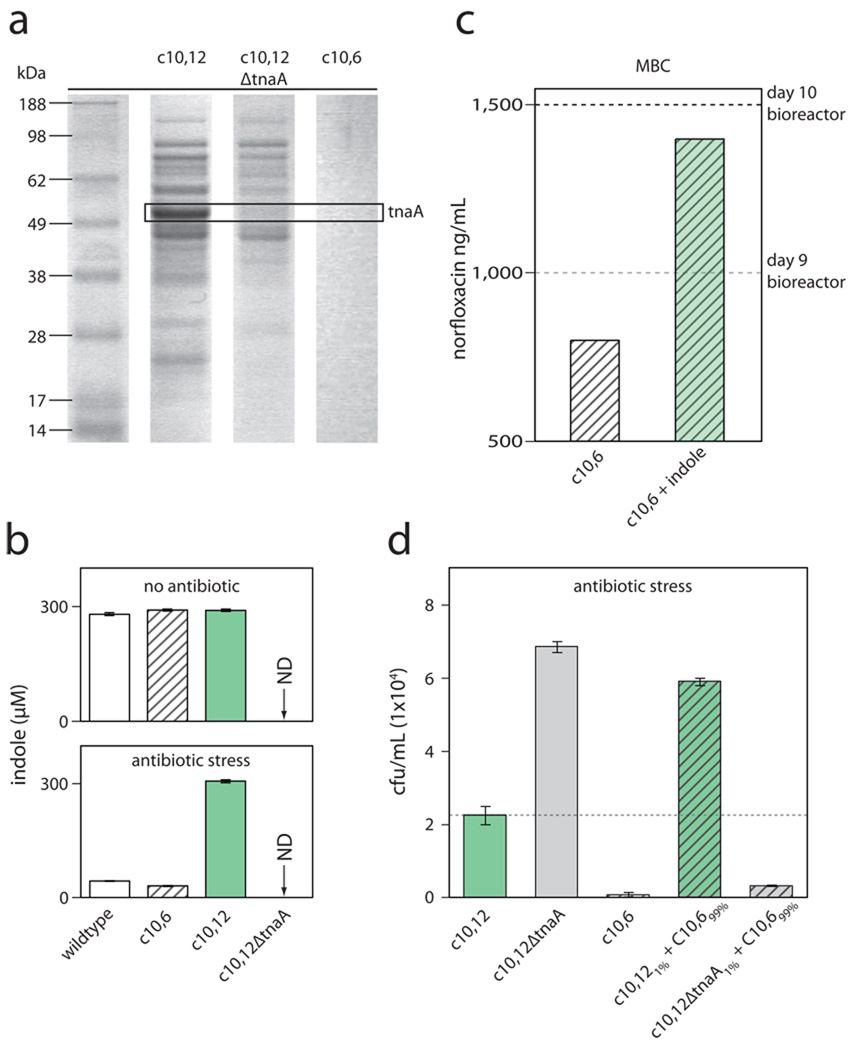
We found that the group’s MIC was not usually predictive of its constituencies’ MICs (Fig. 1a). The vast majority of individual isolates were actually less resistant isolates (LRIs); that is, isolates whose MICs were lower than the concentrations of norfloxacin in which they were found and, therefore, also lower than the group MIC. Intriguingly, we also isolated a mutant with an MIC higher than the bioreactor concentration, a highly resistant isolate (HRI). We suspected that our rare detection of HRIs was due to their low abundance in the population through most of the experiment. Indeed, when we plated daily populations under norfloxacin selection, we frequently detected low-abundance HRIs that emerged prior to increases in the group MIC (Fig. 1b). We were, however, surprised by the large number of LRIs in the population. We hypothesized that the few HRIs were generating a benefit for the numerous LRIs, thus allowing weaker isolates to endure more antibiotic stress than they could in isolation. Media conditioning by HRIs seemed a plausible mode of protection for LRIs. To test this hypothesis and to examine the conditioning that may be taking place, we focused our studies on the most resistant HRI that we detected: c10,12 – colony number 12 of those isolated on day 10. Supernatant from c10,12 following growth in the presence of norfloxacin was analyzed by protein gel electrophoresis. We detected a dominant protein band, along with several weaker protein bands, in the media. We next subjected the observed protein bands to mass spectrometry for identification (Supplement Table 1). The dominant protein band was identified as tnaA (Fig. 2a). We verified the identification of tnaA by creating the corresponding genetic mutant, c10,12ΔtnaA, and analyzing its supernatant for protein following growth under antibiotic stress. The dominant band was completely absent from the resulting gel. We also found this to be the case for c10,6, the least resistant isolate found on day 10. We next verified, by Sanger sequencing, that the promoter and coding regions of the tna operon of c10,12 had not undergone gain-of-function mutations. tnaA codes for the enzyme trypophanase, whose major enzymatic reaction is the breakdown of tryptophan into ammonia, pyruvate, and indole8.
Figure 1. Tracking a population of Escherichia coli developing antibiotic resistance.
(a)A clonal wildtype Escherichia coli MG1655 population was continuously cultured in a bioreactor for 10 days with increasing concentrations of the quinolone, norfloxacin. MIC is defined as the drug concentration inhibiting no more than 60% of unstressed cell growth. The initial bioreactor concentration was set as the MIC of wildtype cells. Every 24 hours thereafter, the population MIC was measured, red lines. Following increases in group MIC, the bioreactor concentration, dashed green lines, was adjusted at the next sampling interval accordingly. Twelve individual isolates were selected from plating daily populations on non-selective plates and their MICs determined, grey bars. MICs shown are representative of biological duplicates. (b) Daily population analysis profiles, representing the fraction of the population resistant to each drug level, were taken throughout the 10 days of continuous culture. Daily populations were serially diluted and spotted on plates with a range of norfloxacin concentrations. Percent resistance, circles colored according to norfloxacin concentration, was calculated as the number of colonies at specific norfloxacin concentrations relative to the total number of cells (plated on non-restrictive plates). Results shown are representative of biological duplicates.
Figure 2. Indole production by isolates and the protective effect of extracellular indole.
(a) Proteins were detected in the supernatant of c10,12 when grown clonally under the bioreactor concentration of norfloxacin (1500ng/mL). These protein bands were subjected to mass spectrometry for protein identification. The top hit for the dominant protein band matched over 75% of residues for tnaA, which encodes the enzyme tryptophanase. The major enzymatic activity of tryptophanase yields indole. This dominant band was absent from the supernatant of c10,12ΔtnaA. No proteins were found in the supernatant of c10,6. (b) HPLC quantification of extracellular indole production by isolates with varying norfloxacin resistance: wildtype, white bars; c10,6, striped bars; c10,12, green bars; and c10,12ΔtnaA, not detected. With the exception of c10,12ΔtnaA, all isolates produce approximately 300µM indole in the absence of antibiotic stress. Under norfloxacin stress (1500ng/mL), c10,12 continued to produce up to 300µM of indole while wildtype and c10,6 produced <50µM of indole. No indole was detected for c10,12ΔtnaA. Results are means ± s.e.m (n≥3). (c) MBC, defined as the minimum concentration of norfloxacin that kills 99.9% of the cells in a culture, is shown for c10,6 with and without the addition of 300µM of indole. The bioreactor concentration for day 9 (1000ng/mL) and for day 10 (1500ng/mL) is also shown. (d) Total growth of mutants under norfloxacin stress (1500ng/mL) in isolation or in co-culture: c10,6, striped bars; c10,12, green bars; and c10,12ΔtnaA, grey bars. Each condition starts with the same total number of cells and co-cultures are mixed in a highly resistant isolate to less resistant isolate ratio of 1:100. Results shown are representative of biological replicates and expressed as means ± s.e.m.
Importantly, indole is a signaling molecule implicated in stress tolerance in Escherichia coli13–15. We hypothesized that indole produced by the highly resistant isolates was protecting its less resistant neighbors. To test this, we first quantified the extracellular indole produced by c10,12 (HRI), c10,6 (LRI), and wildtype using high-pressure liquid chromatography (HPLC). Under no antibiotic stress, these isolates were capable of producing up to approximately 300µM of indole (Fig. 2b). Under antibiotic stress, however, only c10,12 was capable of maintaining its indole production (Fig. 2b). We detected no indole from our tryptophanase mutant, c10,12ΔtnaA.
We next sought to determine the protective effect of extracellular indole on a less resistant isolate under antibiotic stress. We determined the minimum bactericidal concentration (MBC) of norfloxacin for c10,6, the least resistant isolate found on day 10, with and without addition of indole to the media. Added indole provided a stark survival benefit under drug stress (Fig. 2c). The MBC for c10,6 without indole is 800ng/mL, indicating that the day-9 bioreactor concentration (1000ng/mL) would have been sufficient to kill the isolate. Addition of indole boosts the MBC for c10,6 to 1400ng/mL, indicating that the effect of indole can account for much of the isolate’s ability to survive in the day-10 bioreactor concentration (1500ng/mL). It is possible that other protein products identified in the supernatant also contribute to the protective effect afforded by HRIs; this warrants further study. Taken together, our results suggested a plausible mechanism for altruistic communal interaction – a highly resistant isolate conditions the media, in part, through the production of indole, to benefit weaker and more vulnerable isolates.
To examine this putative protective communal interaction, we compared growth under antibiotic stress of three isolates – a less resistant isolate (c10,6), a highly resistant isolate (c10,12), and an indole-deficient mutant (c10,12ΔtnaA) – in isolation and in co-culture. Based on the results shown above, we reasoned that a population dominated by c10,6 (LRI) would be protected by a minority of c10,12 (HRI) and that this protective effect would be absent with an indole-deficient HRI, c10,12ΔtnaA. Of note, c10,12ΔtnaA outgrew c10,12 in isolation. Similarly, in a competitive fitness assay16 where the proportion of the two isolates was initially balanced, c10,12ΔtnaA outcompeted c10,12, leading to a relative fraction of 2.6:1 (c10,12ΔtnaA:c10,12), demonstrating that indole production by HRIs under antibiotic stress carries a fitness cost. For the co-culture experiments, we chose a 1:100 dilution of c10,12 (1%) to c10,6 (99%) to replicate the low abundance of highly resistant isolates. As shown in Fig. 2d, a mixture of c10,6 with c10,12 grew better than either isolate individually, indicating that the indole produced by c10,12 enhanced the survival capacity of c10,6 under antibiotic stress, boosting total growth beyond that of c10,12 alone. In contrast, the growth of a mixture of c10,6 with c10,12ΔtnaA achieved a level only slightly higher than c10,6 alone; this slight increase was due entirely to growth of c10,12ΔtnaA (see Supplement).
We next used genome-wide transcription profiling to explore the physiological role of indole. We measured total transcripts from an indole-deficient mutant with wildtype resistance, MG1655ΔtnaA, following exposure to the initial bioreactor concentration of norfloxacin (50ng/mL) supplemented with an intermediate concentration of indole (200uM). We generated an indole transcriptional signature by filtering for differentially expressed genes. Our analysis (see Supplement and Supplementary Table 2) showed that indole upregulated multi-drug efflux pumps such as mdtE (P = 0.026) and increased production of succinate through upregulation of astD14,15 (P = 0.025). Intriguingly, our analysis also revealed downregulation of oxyS (P = 0.004), a small RNA sensor for intracellular oxidative stress17. We also observed a decrease in iron-sulfur cluster repair and production through downregulation in iscU (P = 0.022), which has previously been shown to be central to a common mechanism of bactericidal antibiotic-mediated cell death18–20. Nitric oxide response systems were also activated through the downregulation of nsrR (P = 0.041) and the corresponding upregulation in hmp (P = 0.008), a central nitric oxide detoxification gene21. These findings indicate that indole induces two modes of antibiotic detoxification: physical export and activation of oxidative stress protective mechanisms.
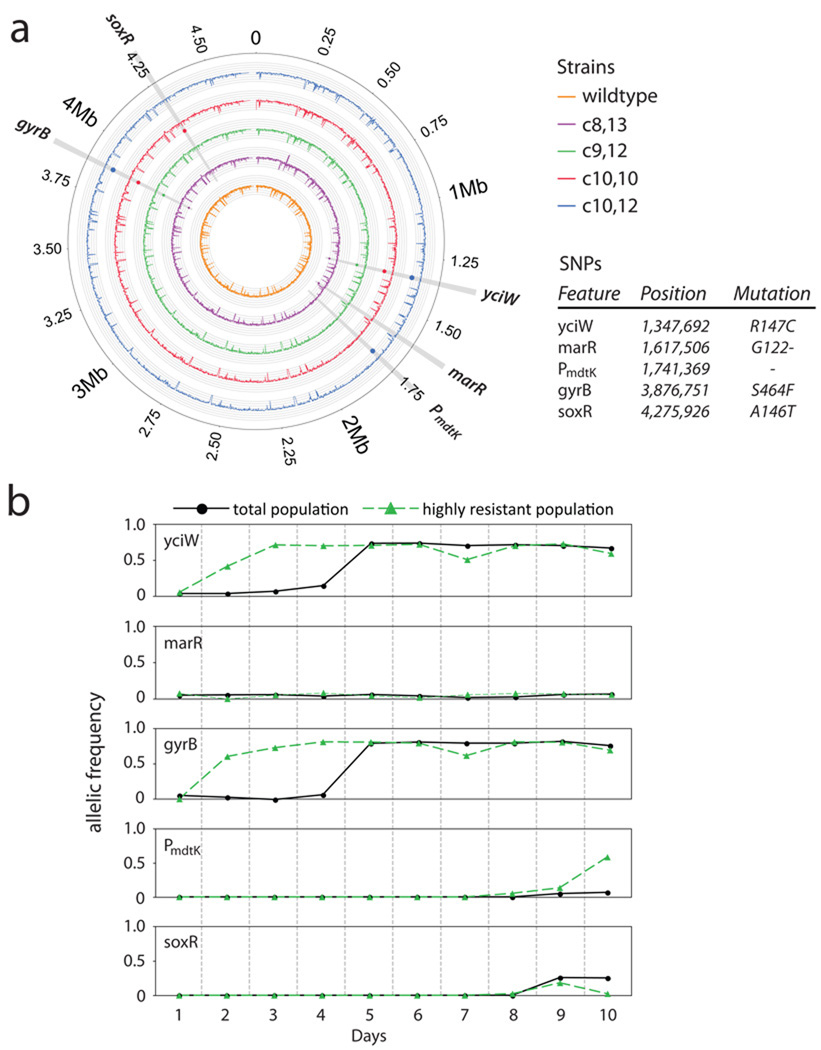
Finally, we employed whole-genome sequencing to explore the mutations in the highly resistant isolates and, more broadly, the mutations present in evolving populations. We selected three HRIs from days 8 through 10 along with one LRI from day 10; we also sequenced our initial wildtype for reference. We identified five single nucleotide polymorphisms (SNPs) not present in our initial wildtype (Fig. 3a). As expected, each resistant isolate carried a mutation in a subunit of a known norfloxacin target DNA gyrase, gyrB22. Interestingly, these mutants also carried a SNP in yciW, a widely conserved putative oxidoreductase. These two mutations were common to all evolved mutants, suggesting that the evolved isolates descended from the same ancestor. We also identified mutations in marR, a master regulator of drug efflux pumps, and its paralogue, soxR, whose regulon protects the cell from superoxide damage18,23. Lastly, we identified a mutation in the promoter for mdtK, an efflux pump with high norfloxacin specificity24. It is important to note that these mutations are unrelated to indole production, and instead likely serve to confer the level of drug resistance needed to maintain indole production under stress.
Figure 3. >Whole-genome sequencing of various mutants.
(a) Five total genomes were sequenced using the Solexa GA2: wildtype, three highly resistant isolates from days 8 through 10, and a less resistant isolate from day 10. Sequencing coverage for each isolate is plotted, according to color, on concentric tracks with the wildtype genome, orange, in the center. Intervals within each track represent 25x coverage per 1000 bases of the genome. Each SNP, represented by circles colored according to isolate, is marked at the appropriate genomic position on the genome(s) in which it was found. (b) Allelic frequency of each SNP over the course of the 10-day evolution experiment was estimated, using Sequenom’s iPLEX platform, in the total population, black circles, and in an enriched highly resistant population by norfloxacin selection, green triangles.
To track the emergence and fixation of each mutation in our evolution experiment, we genotyped, using mass spectrometry, our daily populations to estimate the allelic frequency of each SNP. To maximize detection of SNPs in rare abundance, we revived our daily populations in parallel: by non-restrictive growth and by norfloxacin selection with the respective daily bioreactor concentrations. This drug selection precedes the production of indole by the HRIs and therefore enriches for the low-abundance, highly resistant population. As shown in Fig. 3b, mutations in yciW and gyrB appeared simultaneously in the highly resistant population by the second day. The mutants carrying these two SNPs likely represented the earliest HRI that, through indole production, catalyzed the increase in group MIC. The truncated marR allele, found in the sequenced day-8 HRI, was so rare as to be undetectable by allelotyping, hinting at a diverse population of low-abundance HRIs. On day 9, mutations appear in soxR and mdtK. Though the soxR mutation is more abundant than the mdtK mutation in the total population, their abundances are roughly equivalent under drug selection, indicating that mutants harboring each are part of the HRI population. By day 10, however, despite the unchanged total abundance of each SNP, mdtK mutants begin to dominate the highly resistant subpopulation while soxR mutants are relegated to the less resistant subpopulation. This ebb and flow of resistant mutants suggests an environment supportive of a phenotypically diverse population.
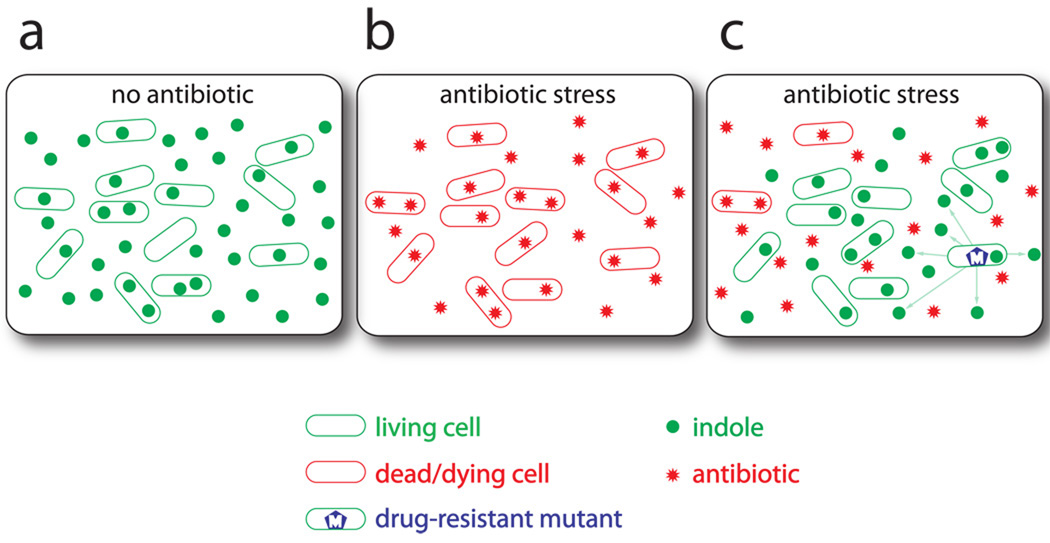
Our results establish a population-based antibiotic-resistance mechanism, illustrated schematically in Fig. 4, based on indole as a cell-signaling molecule. A population of E. coli in the absence of stress thrive and naturally exude the metabolic product indole (Fig. 4a). Under severe antibiotic stress, however, dead and dying cells no longer produce indole in significant quantities (Fig. 4b). An antibiotic-resistant mutant, once it emerges, endures a fitness cost to produce indole that protects the more vulnerable cells by inducing various antibiotic tolerance mechanisms (Fig. 4c).
Figure 4. A population-based antibiotic resistance mechanism.
A bacterial population is diagrammed. (a) In the absence of antibiotic stress, wild-type cells naturally produce indole. (b) Under antibiotic stress, wild-type cells stop producing indole and eventually die. (c) When a drug resistant mutant emerges, it is able to produce indole even under antibiotic stress. This indole allows the more vulnerable cells in the population to survive the antibiotic stress by inducing various antibiotic tolerance mechanisms, thereby boosting the survival capacity of the population.
Recognizing that indole would be protective under a variety of antibiotics, we hypothesized that this population-based resistance mechanism would also arise in response to other antibiotic treatments. To test this hypothesis, we repeated our bioreactor experiment, this time following a continuous culture of E. coli facing increasing concentrations of the aminoglycoside, gentamicin. As was the case for our earlier norfloxacin experiments, we found that individual isolates tended to be less resistant than the population as a whole and that a few highly resistant mutants appeared and preceded a change in the group MIC (Supplemental Fig. 1a,b). Upon further examination of isolates from day 7, we verified that a highly resistant isolate was capable of producing high levels of indole (>300uM) under antibiotic stress, while a less resistant isolate could not (Supplemental Fig. 1c). Importantly, co-culturing of these two isolates resulted in more robust growth than either of the isolates individually, indicating increased survival for the more vulnerable isolate (Supplemental Fig. 1d). These findings demonstrate that the population-based resistance mechanism illustrated in Fig. 4 is not drug specific.
This work shows that, under antibiotic stress, a few spontaneous drug resistant mutants will endure a fitness cost to produce and share among the population the metabolite indole, thus shielding the less resistant isolates from antibiotic insult. This altruism allows weaker constituents to survive and concurrently explore the space of beneficial mutations, a phenomenon akin to kin selection9. These few drug resistant mutants, by enhancing the survival capacity of the overall population in stressful environments, may also help to preserve the potential for the population to return to its genetic origins should the stress prove transient. Efforts to monitor and combat antibiotic resistance are complicated by these bet-hedging survival strategies and other forms of bacterial cooperation. Deeper exploration into the repertoire of strategic intracellular communication utilized by bacteria may prove critical for the rational design of effective clinical interventions to face a growing threat of resistant bacterial infections.
Methods Summary
All experiments were performed with Escherichia coli MG1655 (ATCC 700926)-derived strains. tnaA deletion mutants were created by P1 transduction and derived from an E. coli single-gene knockout library25. In the bioreactor, E. coli were continuously cultured with a dilution rate of 0.48h−1. MICs and MBCs were determined by standard methods. Proteins were separated by denaturing polyacrylamide gel electrophoresis (SDS-PAGE) and identified by mass spectrometry26. Extracellular indole was quantified via high-pressure liquid chromatography (HPLC)27. The competition assay was performed in continuous culture and relative fractions were derived from Malthusian parameters16. Transcriptional profiling was performed as previously described18 and normalized with FARMS28. Whole genome sequencing was performed using custom adapters on a Solexa GA229. Allelic frequency was estimated through MALDI-TOF mass spectrometry using Sequenom’s iPLEX chemistry30.
Methods
Bacterial strains, media, and chemicals
All experiments were performed with Escherichia coli MG1655 (ATCC 700926)-derived strains. tnaA deletion mutants were created by P1 transduction and derived from an E. coli single-gene knockout library25. Bacterial cultures were grown at 37°C in Luria-Bertani (LB) media obtained from Fisher Scientific. Norfloxacin and gentamicin were purchased from Sigma. Kanamycin was purchased from Fisher Scientific. Indole was purchased from Acros Organics.
Bioreactor experiments
An overnight E. coli culture, grown from a single colony, was diluted 1:10,000 in the bioreactor and grown, dilution rate of 0.48h−1, to OD600 ~ 0.6. Media was then supplemented with norfloxacin and protected from heat and light. Every 24 hours, MICs were determined for the group and individual isolates, respectively. Daily populations, or overnight cultures of individual isolates, were diluted 1:10,000 into 100µL volume LB with a range of antibiotic concentrations in 96-well plates. Following overnight growth, OD600 was measured using a SPECTRAFluor Plus (Tecan) spectrophotometer and the MIC was determined as the concentration that inhibited no more than 60% of growth.
Protein identification and indole quantification
Supernatant from mutants grown overnight in LB with norfloxacin was purified with a 0.2µm nylon filter (Costar). Protein concentration of each sample was adjusted following quantification of total protein with a BCA protein assay (Pierce) and concentrated by acetone precipitation when needed. Proteins in each sample were separated by denaturing polyacrylamide gel electrophoresis (SDS-PAGE) with MES buffer on 10% Bis-Tris gels (Invitrogen) and stained with GelCode Blue Stain Reagent (Pierce). Whole gels were submitted to the Boston University Proteomics Core Facility for protein identification by mass spectrometry26. Extracellular indole was quantified via high-pressure liquid chromatography (HPLC), as previously described27. 50µL of filtered supernatant was injected into a Waters Xterra MS C18 column on an HP Agilent 1090 Series II. A standard curve, created by measuring signals of known indole concentrations in LB, was used.
MBC determination
A 1:1,000 dilution of an overnight culture of c10,6 was grown in LB and supplemented with 300µM indole or the equivalent concentration of methanol, the solvent for indole. OD600 was measured as described above following overnight incubation in a range of norfloxacin. Replicate cultures with OD600 ≤ 0.1 were subcultured on non-restrictive LB agar plates. The MBC was determined as the minimum norfloxacin concentration that killed at least 99.9% of an initial c10,6 inoculum.
Competition assay
Overnight cultures of c10,12 and c10,12ΔtnaA were combined at 1:1 and continuously cultured as described above. Media was supplemented with norfloxacin (1500ng/mL) and protected from heat and light. Samples were harvested after approximately five volume changes to avoid prolonged exposure to mutagenic effects of norfloxacin, and plated differentially on non-restrictive LB-agar plates and LB-agar plates supplemented with kanamycin (25ug/mL). Relative fractions of each isolate were derived from Malthusian parameters16 calculated from median cell counts.
Co-culture
Each isolate was grown overnight to an OD600 density > 1.5. Mixed cultures consist of a highly resistant isolate diluted 1:30,000 and a less resistant isolate diluted 1:300. In each clonal culture, an isolate was diluted 1:30,000+1:300 for a total number of cells equivalent to the mixed cultures. Cultures were incubated overnight in LB supplemented with norfloxacin (1500ng/mL). Cell counts were obtained by subculturing on non-restrictive LB-agar plates.
Transcriptional profiling
An indole-deficient mutant with wildtype resistance to norfloxacin, MG1655ΔtnaA, was seeded and continuously cultured as described above. Total transcripts were prepared and hybridized as previously described18 from samples harvested in serial representing three distinct conditions. The first sample was collected after a 30-minute exposure to norfloxacin (50ng/mL). The bioreactor media was then replaced, by dilution, with nonrestrictive LB media. 200µM of indole was added, followed by norfloxacin 30 minutes later. The second sample was collected after 30 minutes of simultaneous exposure to both indole and norfloxacin. The media was again purified as described. The third sample was collected after a re-exposure to norfloxacin. Resulting CEL files were normalized by FARMS and informative probes selected for analysis28.
Whole-genome sequencing and allelic frequency estimation
Genomic DNA (gDNA) was extracted from each sample using Qiagen’s genomic-tip kit. For whole-genome sequencing, gDNA of each isolate was sonicated (Covaris) to 200bp. Illumina single-end sequencing libraries for each isolate were prepared, using custom adapters29 and NEBNext DNA Sample Master Mix Set 1 (New England Biolabs). Libraries were submitted to the Boston University Sequencing Core. For allelic frequency estimation, 100bp surrounding each identified mutation was PCR-amplified from the total population gDNA and the enriched highly resistant population gDNA, respectively. Amplicons were submitted to Partners Healthcare Center for Personalized Medicine for MALDI-TOF mass spectrometry using Sequenom’s iPLEX chemistry30.
Supplementary Material
Acknowledgements
We thank Michael Chaparian and Qasim Beg for help with bioreactor instrumentation, We also thank Alan Herbert and Christine Ordija for use of the Covaris sonicator for preparing sequencing libraries. This work was supported by the National Institute of Health through the NIH Director’s Pioneer Award Program, grant number DP1OD003644, the National Science Foundation through RTG grant number DMS-0602204, and the Howard Hughes Medical Institute.
Footnotes
Author Contributions
All authors designed the study. HHL and MNM performed and analyzed the experiments with input from CRC and JJC. All authors prepared and commented on the manuscript.
Author Information
The microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE22833. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Albert TJ, et al. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat Methods. 2005;2(12):951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
- 2.Smith BT, Walker GC. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics. 1998;148(4):1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjedov I, et al. Stress-induced mutagenesis in bacteria. Science. 2003;300(5624):1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 4.Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- 5.Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50(6):2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37(3):311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45(5):1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gooder H, Happold FC. The tryptophanase-tryptophan reaction; the nature of the enzyme-coenzyme-substrate complex in. Biochem J. 1954;57:369–374. [PMC free article] [PubMed] [Google Scholar]
- 9.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4(8):597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 10.Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect. 2007;13(1):5–18. doi: 10.1111/j.1469-0691.2006.01492.x. [DOI] [PubMed] [Google Scholar]
- 11.Livermore DM. Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis. 2003;36(Suppl 1):S11–S23. doi: 10.1086/344654. [DOI] [PubMed] [Google Scholar]
- 12.Smith PA, Romesberg FE. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat Chem Biol. 2007;3(9):549–556. doi: 10.1038/nchembio.2007.27. [DOI] [PubMed] [Google Scholar]
- 13.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol. 2005;55(4):1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi A, Hirakawa H, Hirata T, Nishino K, Yamaguchi A. Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J Bacteriol. 2006;188(16):5693–5703. doi: 10.1128/JB.00217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, Lee J. Intercellular signal indole in microbial communities. FEMS Microbiol Rev. 2009 doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 16.Travisano M, Lenski RE. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics. 1996;143(1):15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90(1):43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 18.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130(5):797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 20.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135(4):679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tucker NP, Le Brun NE, Dixon R, Hutchings MI. There's NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 2010 doi: 10.1016/j.tim.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Mouneimne H, Robert J, Jarlier V, Cambau E. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43(1):62–66. doi: 10.1128/aac.43.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller PF, Gambino LF, Sulavik MC, Gracheck SJ. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 1994;38(8):1773–1779. doi: 10.1128/aac.38.8.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long F, Rouquette-Loughlin C, Shafer WM, Yu EW. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli. Antimicrob Agents Chemother. 2008;52(9):3052–3060. doi: 10.1128/AAC.00475-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiling K, et al. Comparison of proteomic and transcriptomic profiles in the bronchial airway epithelium of current and never smokers. PLoS One. 2009;4(4):e5043. doi: 10.1371/journal.pone.0005043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talloen W, et al. I/NI-calls for the exclusion of non-informative genes: a highly effective filtering tool for microarray data. Bioinformatics. 2007;23(21):2897–2902. doi: 10.1093/bioinformatics/btm478. [DOI] [PubMed] [Google Scholar]
- 29.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7281):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38(12):1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.